Significance
Infections are known to perturb intestinal microbiome composition, but the underlying mechanisms and the implications of specific commensal species for antiviral immunity remain largely unknown. Here, we found that infection of mice with a fast-spreading and persistent, but not slow-spreading acute, viral isolate induced profound microbial dysbiosis, including blooming of Akkermansia muciniphila, that was largely caused by CD8 T cells and/or CD8 T cell-induced anorexia. Notably, administration of A. muciniphila into infected mice attenuated selected aspects of CD8 T cell responses, supporting its immunosuppressive potential in this context. Our findings bring about a role for CD8 T cells in promoting anorexia and microbial shifts after infection, including the enrichment of a fasting-associated commensal that can attenuate CD8 T cell responses.
Keywords: CD8 T cell, LCMV, microbiome, Akkermansia, anorexia
Abstract
Infections elicit immune adaptations to enable pathogen resistance and/or tolerance and are associated with compositional shifts of the intestinal microbiome. However, a comprehensive understanding of how infections with pathogens that exhibit distinct capability to spread and/or persist differentially change the microbiome, the underlying mechanisms, and the relative contribution of individual commensal species to immune cell adaptations is still lacking. Here, we discovered that mouse infection with a fast-spreading and persistent (but not a slow-spreading acute) isolate of lymphocytic choriomeningitis virus induced large-scale microbiome shifts characterized by increased Verrucomicrobia and reduced Firmicute/Bacteroidetes ratio. Remarkably, the most profound microbiome changes occurred transiently after infection with the fast-spreading persistent isolate, were uncoupled from sustained viral loads, and were instead largely caused by CD8 T cell responses and/or CD8 T cell-induced anorexia. Among the taxa enriched by infection with the fast-spreading virus, Akkermansia muciniphila, broadly regarded as a beneficial commensal, bloomed upon starvation and in a CD8 T cell-dependent manner. Strikingly, oral administration of A. muciniphila suppressed selected effector features of CD8 T cells in the context of both infections. Our findings define unique microbiome differences after chronic versus acute viral infections and identify CD8 T cell responses and downstream anorexia as driver mechanisms of microbial dysbiosis after infection with a fast-spreading virus. Our data also highlight potential context-dependent effects of probiotics and suggest a model in which changes in host behavior and downstream microbiome dysbiosis may constitute a previously unrecognized negative feedback loop that contributes to CD8 T cell adaptations after infections with fast-spreading and/or persistent pathogens.
Adaptability, the capacity to adjust or adapt to improve fitness in the face of environmental changes is a trait shared among all forms of life (1). Such adaptations occur at the single cell, organismal, and population levels and involve mechanisms with different time requirements to be implemented and exhibiting distinct degree of reversibility (1). Mammalian hosts and their immune systems often adapt to the presence of pathogens and engage different coping mechanisms depending on their virulence (2). In this regard, infections with fast-spreading and/or persistent pathogens elicit potent inflammatory responses that contribute to pathogen control, but can also undermine host survival, if unrestrained (2–6). Immune adaptations that attenuate the magnitude or amplitude of antipathogen responses are therefore necessary to allow pathogen clearance during acute infections, and/or partial pathogen control during chronic infections, while avoiding host death due to excessive immunopathology. Such adaptations encompass the innate and adaptive immune system, involve multiple layers of cell-intrinsic transcriptional, epigenetic, posttranscriptional, and metabolic regulation, and are triggered in response to environmental changes (e.g., abundant pathogen-associated molecular patterns and antigens, an inflammatory milieu, and altered nutrient and oxygen levels) (3, 7). Among the most studied adaptations that attenuate immune responses, CD8 T cell exhaustion, a unique cellular state characterized by diminished effector functions and expression of inhibitory receptors, is highly conserved not only among persistent infections in mice and humans, but also in tumor settings (7).
More recently, it has become evident that the character and strength of immune responses can be regulated by the microbiome (8). In particular for chronic pathogens, the intestinal microbiome has been shown to regulate host resistance to infections caused by persistent lymphocytic choriomeningitis virus (LCMV) (9), Plasmodium yoelii (10), and Mycobacterium tuberculosis (11, 12). It has also been shown that persistent pathogens such as HIV (13), hepatitis C virus (HCV) (14), and Hepatitis B virus (HBV) (15) in humans as well as simian immunodeficiency virus (SIV) (16) in macaques and Plasmodium species (17, 18) in mice induce significant changes in the intestinal microbiome composition or dysbiosis. On the other hand, acute infections caused by respiratory viruses (19–21) or enteropathogenic bacteria (22, 23) can also alter the composition of the gut microbiome. Notably, some of these studies have drawn a number of correlations between specific bacterial taxa and diverse disease parameters (13–15) that suggest important roles for the microbiome changes after infections. Studies addressing the mechanisms that dictate dysbiosis and/or the functional effects of specific taxa are, however, scarce.
In the present study, we use the well-established LCMV mouse model system to compare side-by-side microbiome changes induced at different times after infection with a fast-replicating persistent vs. a slow-replicating acute isolate. We found that the most profound microbiome alterations occurred after infection with the persistent, but not acute, LCMV isolate and were mostly transient (i.e., at day 8 but not at day 20 postinfection [p.i.]) despite continuously high viral titers at the latter time point. The microbiome changes unique to the infection with the fast-replicating persistent virus involved transiently increased Verrucomicrobia and reduced Firmicute/Bacteroidetes ratio at the phylum level as well as overrepresentation of Akkermansia, Bacteroides, and Sutterella among other taxa. We also detected reductions in Lactobacillus and Turicibacter, the latter of which was maintained at day 20 p.i. Intriguingly, CD8 T cells were responsible for modulating over half of the dysbiotic taxa after infection with the fast-replicating persistent LCMV isolate, thus revealing a key role for CD8 T cells in promoting intestinal dysbiosis. Coincidently, and only early after infection with the fast-replicating persistent isolate, CD8 T cells induced a profound anorexic behavior and a significant fraction of the taxa uniquely changed after this infection were commonly altered in other fasting conditions. Among these fasting-associated taxa, we validated (at the species level) that the commensal Akkermansia muciniphila bloomed after infection with the fast-replicating persistent LCMV, and its abundance was remarkably increased by lack of food consumption in the absence of infection. We further observed a significant decrease in the levels of A. muciniphila in infected mice in which anorexia was reverted via CD8 T cell depletion. These observations supported the conclusion that anorexia induction was one of the means via which CD8 T cells caused intestinal dysbiosis after infection with persistent, fast-replicating, LCMV. On the other hand, experimental enrichment of A. muciniphila after LCMV infection suppressed Granzyme B (GrzB) and T-BET expression, and H3K27 trimethylation in distal virus-specific CD8 T cells. Together, these data contrast the specific microbiome changes that occur after infection with two closely related viral isolates, uncoupling dysbiosis from sustained viral loads and instead relating it to both CD8 T cell responses and downstream anorexia occurring during the early phase of infection with a fast-replicating, persistent virus. In addition, our data support a potential role for a CD8 T cell-driven, fasting-associated commensal, A. muciniphila, in regulating CD8 T cells and attenuating selected aspects of their effector responses.
Results
Infection with a Fast-Spreading Persistent LCMV Isolate Induced Profound Intestinal Dysbiosis That Was Mostly Transient and Uncoupled from Sustained Viral Loads.
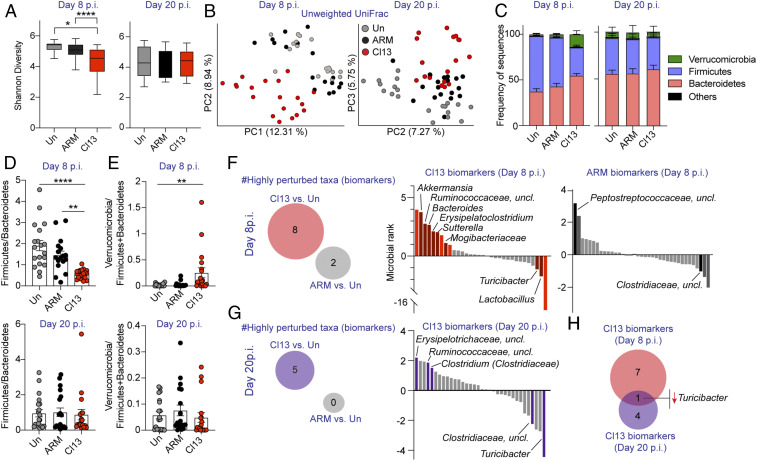
To investigate side-by-side alterations in intestinal microbiome composition after infection with a fast-replicating persistent vs. a slow-replicating acute virus, we performed 16S rRNA gene amplicon sequencing of caecal and colonic contents from mice infected with two closely related LCMV isolates: Armstrong53b (ARM), which spreads more slowly and is cleared in a week, and clone13 (Cl13), which is fast-spreading and capable of persisting in C57BL/6 mice (6, 24). Two independent repeats with uninfected, ARM- and Cl13-infected mice (10 mice/group) were performed at days 8 and 20 p.i. Sampling of caecal and colonic contents was chosen over stool pellets, as it allows detection of taxa abundant inside the large intestine, which may not necessarily be eliminated in stools (25, 26). Alpha-diversity determined by the Shannon diversity index, which combines richness and evenness (27), was selectively decreased upon Cl13 infection at day 8, but not at day 20 p.i. (Fig. 1A). We next identified specific differences in microbial composition by computing beta-diversity using the phylogenetic unweighted UniFrac metric or nonphylogenetic Jaccard and Bray–Curtis indexes followed by principal coordinates analysis (PCoA). Although ARM-infected and uninfected mice appeared to be significantly distant by permutational ANOVA (PERMANOVA) analysis with at least two of the aforementioned metrics on days 8 and 20 p.i. (SI Appendix, Fig. S1 A–C), there was no clear separation by any of the three main principal coordinates (PCs) at the two time points studied (Fig. 1B and SI Appendix, Fig. S1 D–F). On the other hand, the intestinal microbiome from day-8 Cl13-infected mice formed a prominent cluster by PC1 and/or 2 (Fig. 1 B, Left and SI Appendix, Fig. S1 E and F, Left Top) which was significantly distant from both uninfected and ARM-infected mice (SI Appendix, Fig. S1 A–C). No separation was observed along PC3 (SI Appendix, Fig. S1 D–F, Left Bottom). Remarkably, despite sustained viral loads in circulation and intestinal tissues (28, 29), at day 20 p.i. the microbiome of Cl13-infected mice segregated closer (Fig. 1B and SI Appendix, Fig. S1 D–F, Right), albeit still significantly different (SI Appendix, Fig. S1 A–C), to the samples from uninfected or ARM-infected mice. Note that in all cases the PCoA clustering was consistent in different cages (SI Appendix, Fig. S1G) and could not be explained by reproducible differences in dispersion (SI Appendix, Fig. S1 A–C, PERMDISP).
Fig. 1.
LCMV Cl13 infection induced intestinal dysbiosis that was mostly transient, uncoupled from viral loads, and characterized by reduced Firmicutes-to-Bacteroidetes ratio and increased Verrucomicrobia. See also SI Appendix, Fig. S1 and Datasets S1–S4. C57BL/6 mice were infected with LCMV ARM, Cl13, or left uninfected (Un) and 16S rRNA gene amplicon sequencing was performed on colonic and caecal contents from days 8 and 20 (p.i.). (A) Alpha-diversity by the Shannon diversity index at indicated time points. (B) Beta-diversity PCoA with unweighted Unifrac distance at indicated time points. PC1 was omitted for PCoA from the day 20 p.i. time point as it captured the basal differences of the cohorts used in the two experiments, rather than infection type. (C) Frequency of sequences at the phylum level at indicated time points. (D and E) Frequency of phylum Firmicutes divided by Bacteroidetes (D) or Verrucomicrobia divided by Firmicutes and Bacteroidetes (E) for each individual mouse at indicated time points. (F and G) Songbird MR analysis of genera in mice infected with ARM or Cl13 vs. Un on day 8 p.i. (F) or day 20 p.i. (G). Taxa with microbial ranks higher than 1 or lower than −1 in two independent repeats are considered highly perturbed (i.e., biomarkers). (Left) Total number of genera highly perturbed by Cl13 or ARM infections. (Right) The x axes correspond to individual taxa, taxa highly perturbed are indicated by text and colored in red (F) or violet (G) for Cl13 infection and black (F) for ARM infection. (H) Venn diagram overlapping biomarkers identified in F and G for day 8 (red) and day 20 (violet) after Cl13 infection. (A, C, and D) Graphs show averages plus min and max (A) or ± SEM (C–E). (A–E) Data are represented by pooling n = 8 to 10 mice/group from two independent experiments. (F–H) Data are representative of two independent experimental repeats. (A) Pairwise Kruskal–Wallis with false discovery rate (FDR) (Benjamini–Hochberg) correction. (D and E) Kruskal–Wallis with Dunn’s correction; *p-val < 0.05, **<0.01, ****<0.0001.
Importantly, when analyzing bacterial phylum composition at day 8 p.i., we detected profound reduction of the Firmicutes-to-Bacteroidetes ratio (Fig. 1 C and D), concomitant with increases in the relative abundance of phylum Verrucomicrobia over Firmicutes and Bacteroidetes in Cl13-infected vs. uninfected mice (Fig. 1 C and E) that were specific for Cl13-infected mice. In contrast, alterations in the Firmicutes-to-Bacteroidetes ratio or phylum-level changes in Verrucomicrobia were not detected upon ARM infection (Fig. 1 C–E) and were not maintained at day 20 after Cl13 infection (Fig. 1 C–E). We next performed multinomial regression (MR) analysis with Songbird (30), with cutoffs at rank values of 1 and −1, to identify the most perturbed taxa at days 8 and 20 after the different infections. We restricted our analysis to taxa that were reproducibly perturbed in both independent experimental repeats. With these rigorous criteria, we defined biomarkers for each infection type and time point as taxa that were highly and reproducibly perturbed vs. uninfected controls processed in parallel. We identified eight biomarkers at day 8 after Cl13 infection, and two different ones at the same time point after ARM infection (Fig. 1F and Datasets S1 and S2). Dysbiosis in day-8 Cl13-infected mice was characterized by reductions in the relative abundance of Lactobacillus and Turicibacter as well as enrichment of Bacteroides, Sutterella, Akkermansia, and Erysipelatoclostridium (i.e., Clostridium within the Erysipelotrichaceae family), among others (Fig. 1F and Dataset S1). In contrast, ARM infection promoted increases of unclassified genera within the Peptostreptococcaceae family, while decreasing genera within the Clostridiaceae family (Fig. 1F and Dataset S2). By day 20 p.i., a total of five taxa were selectively perturbed by Cl13 infection while no genus was highly affected in ARM-infected mice (Fig. 1G and Datasets S3 and S4). This late dysbiosis in day-20 Cl13-infected mice included decreased relative abundance of Turicibacter and unclassified genera within the Clostridiaceae family concomitant with overrepresentation of unclassified genera within the Erysipelotrichaceae and Ruminococcaceae families, and Clostridium (Clostridiaceae family) (Fig. 1G and Dataset S3). Intriguingly, only Turicibacter was persistently altered in the same direction during both the early and the chronic phase of Cl13 infection (Fig. 1H). It should be noted that it is possible that the highly stringent criteria that we have chosen to identify the most reliable infection biomarkers have left out taxa that might still be biologically significant, such as Roseburia, which was reduced at days 8 and 20 p.i. in one experiment (rank day 8 p.i. −2.66; rank day 20 p.i. −1.53) (Datasets S1 and S3) but could not be validated in the second repeat, likely due to very low basal amounts of this taxon in uninfected controls from different cohorts (e.g., 0.29% ± 0.25 in cohort 1; 0.07% ± 0.12 in cohort 2).
Overall, these data are the first defining specific intestinal microbiome perturbations occurring after infection with two closely related viral isolates that exhibit contrasting capability to spread and persist, revealing that: 1) infection with a fast-spreading persistent virus caused large-scale microbial dysbiosis that did not occur upon infection with a closely related slow-spreading acute virus; 2) the most dramatic microbiome alterations after chronic infection were uncoupled from sustained pathogen loads and, instead, occurred transiently during the early phase of the infection; 3) transient microbiome alterations early after infection with the fast-spreading persistent virus were characterized by reduced Firmicutes-to-Bacteroidetes ratios (e.g., increased Bacteroides as well as reduced Lactobacillus and Turicibacter) and increased Verrucomicrobia (i.e., Akkermansia); and 4) reductions in Turicibacter were sustained during the chronic phase of infection.
CD8 T Cells Drove Great Part of the Intestinal Dysbiosis Identified after Infection with LCMV Cl13.
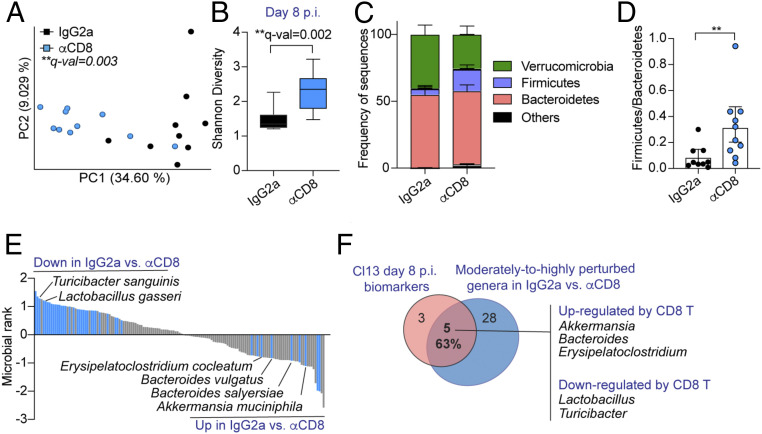
Given that microbial dysbiosis coincided with peak CD8 T cell responses at day 8 p.i. (28), we next investigated the role of CD8 T cells in driving the microbiome changes uniquely identified after LCMV Cl13 infection. For that, we performed two independent experiments shown in Fig. 2 and SI Appendix, Fig. S2, respectively, where we treated 10 Cl13-infected mice per group with anti-CD8 or isotype control antibodies, followed by shotgun metagenomics sequencing of caecal and colonic contents at day 8 p.i. As expected, treatment with anti-CD8 antibody significantly reduced the number of total and virus-specific CD8 T cells (SI Appendix, Fig. S2A). Remarkably, Cl13-infected mice treated with anti-CD8 depleting antibodies formed a distinct cluster via beta-diversity PCoA when compared to their isotype-treated counterparts (Fig. 2A and SI Appendix, Fig. S2B). This treatment also increased gut microbial alpha-diversity in one out of the two independent experiments (Fig. 2B and SI Appendix, Fig. S2C), which, as described above (Fig. 1A), was selectively reduced at day 8 after Cl13 but not ARM infection. We also detected overrepresentation of Firmicutes upon CD8 T cell depletion (Fig. 2C and SI Appendix, Fig. S2D), which explained an elevation of the Firmicutes-to-Bacteroidetes ratio (Fig. 2D and SI Appendix, Fig. S2E) toward the value described above for uninfected and ARM-infected mice (Fig. 1D). As shown in Fig. 2E and SI Appendix, Fig. S2F, blue bars, and Dataset S5, subsequent MR analysis with Songbird and with rank cutoffs of 0.5 and −0.5, to include moderately to highly perturbed taxa, indicated that treatment with anti-CD8 antibodies reverted the changes of commensal species belonging to three of the most increased (i.e., Bacteroides, Akkermansia, and Erysipelatoclostridium) and two of the most decreased (i.e., Lactobacillus and Turicibacter) genera described above as biomarkers for day-8 Cl13-infected mice (Fig. 1F). Overall, CD8 T cells modulated (in a moderate and/or high degree) the abundance of 63% of the biomarkers selectively altered upon Cl13 infection (Figs. 1F and 2F) including Turicibacter, which was underrepresented at both days 8 and 20 after Cl13 infection (Fig. 1H).
Fig. 2.
CD8 T cells drove great part of the intestinal dysbiosis identified after LCMV Cl13 infection. See also SI Appendix, Fig. S2 and Dataset S5. C57BL/6 mice were infected with LCMV Cl13, injected i.p. with isotype control (IgG2a) or CD8-depleting antibodies (αCD8) followed by shotgun metagenomics sequencing of colonic and caecal contents on day 8 p.i. (A) Beta-diversity PCoA with Jaccard distance. (B) Alpha-diversity by the Shannon diversity index. (C) Frequency of sequences at the phylum level. (D) Frequency of phylum Firmicutes over Bacteroidetes for each mouse. (E) Songbird MR analysis of species in IgG2a- vs. αCD8-treated Cl13-infected mice. Taxa with rank cutoff values of −0.5 and 0.5 that were perturbed in the same direction in two independent experimental repeats are highlighted in blue and indicated by name. (F) Overlap between Cl13-specific biomarkers at day 8 p.i. (as defined in Fig. 1F) and altered genera upon CD8 T cell depletion as defined in E. (B–D) Averages and min or max (B) or ± SEM (C and D). (A–F) Representative data from one independent experiment with n = 9 to 10 mice/group. (A) PERMANOVA with 999 permutations, (B) Kruskal–Wallis test, (D) Mann–Whitney test; *p or q-val < 0.05, **<0.01.
These data indicated that, despite their exhausted state (28, 31, 32), CD8 T cells are responsible for great part of the dysbiosis observed after infection with a fast-spreading persistent virus. Specifically, CD8 T cells were, at least partially, responsible for the transient reduction in Firmicutes-to-Bacteroidetes ratio (i.e., enriching Bacteroides and Erysipelatoclostridium while reducing Lactobacillus and Turicibacter), as well as for the overrepresentation of Akkermansia. These findings situate CD8 T cells as key drivers of the microbial changes that take place early after infection with a virus capable of spreading quickly and/or establishing persistence.
CD8-Driven Anorexia Is Associated with a Fasting-Related Microbiome Early after LCMV Cl13 Infection.
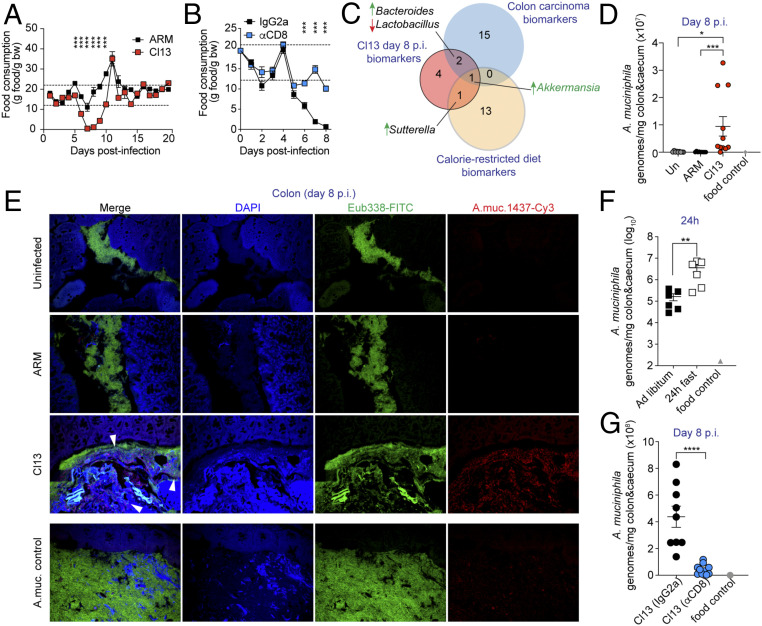
We next sought to investigate the mechanisms via which CD8 T cells promoted transient (and profound) dysbiosis after infection with the fast-spreading persistent LCMV Cl13 isolate. We considered that, among the day-8 Cl13 taxa biomarkers altered in a CD8 T cell-dependent manner, the phylum Verrucomicrobia (i.e., Akkermansia genus), has been previously reported to increase in anorexic human patients (33) as well as in calorie-restricted rodents, some fish, and snakes (34). We, therefore, investigated potential changes in eating behavior by comparing food consumption in ARM- vs. Cl13-infected mice. We observed that Cl13, but not ARM infection, induced profound anorexia spanning from day 6 through 9 p.i. (Fig. 3A), that was accompanied by 18% ± 3.5 weight loss at day 9 p.i. (SI Appendix, Fig. S3A). Importantly, continuous CD8 T cell depletion completely restored food consumption (Fig. 3B) and attenuated weight loss (SI Appendix, Fig. S3B), which provided causal evidence that CD8 T cells can drive anorexic behavior. Intriguingly, efficient CD4 T cell depletion (SI Appendix, Fig. S3C) or IFN-I receptor (IFNAR) blockade did not prevent the induction of anorexia, even though IFNAR blockade (but not CD4 T cell depletion) consistently ameliorated its magnitude (SI Appendix, Fig. S3 D and E) and attenuated weight loss (SI Appendix, Fig. S3 F and G).
Fig. 3.
CD8 T cell-driven anorexia is associated with a fasting-related microbiome, including natural blooming of A. muciniphila, early after LCMV Cl13 infection. See also SI Appendix, Figs. S3 and S4 and Dataset S6. C57BL/6 mice were infected with LCMV ARM, Cl13, or left uninfected (Un) (A, D, and E) or infected with LCMV Cl13 and injected i.p. with isotype control (IgG2a) or CD8-depleting antibodies (αCD8) (B and G). (A and B) Grams (g) of food consumed per gram of body weight (bw) over time in indicated groups. Dotted lines mark the healthy range of food consumption determined in uninfected mice as described in SI Appendix, Materials and Methods. (C) Taxa overlap of Cl13 d 8 p.i. biomarkers defined as in Fig. 1E (red), mouse colon carcinoma biomarkers defined by linear discriminant analysis effect size in ref. 80 of mice s.c. transplanted with colon cancer cells (blue), as reported in ref. 35 and caloric restriction biomarkers defined after MR analysis with Songbird of the stools of mice after 3 wk on a calorie-restricted diet, from a previous study (37) (yellow). (D, F, and G) Absolute A. muciniphila genomes in colonic and caecal contents were quantified by qPCR in uninfected mice or mice infected with LCMV ARM or Cl13 at day 8 p.i. (D), in uninfected mice fed ad libitum, or fasted for 24 h (F) and in Cl13-infected mice treated with CD8-depleting or isotype control antibodies (G). DNA from a food pellet was used as a negative control (food control). (E) FISH images with Eubacteria (Eub338, green) or A. muciniphila (A.muc.1437, red)-specific probes on colon sections from Un, ARM-, and Cl13-infected mice at day 8 p.i. One uninfected mouse was intraorally (i.o.) inoculated with ∼1.2 × 108 cfu of live A. muciniphila 24 h prior to necropsy and used as positive control (A.muc. control). (A, B, D, F, and G) averages ± SEM. Data are representative of one out of three (A and B), two (E and G), or pooled from two (F) or three (D) independent experiments with n = 3 to 10 mice/group. (A and B) Unpaired two-tailed Student’s t test per time point, (F and G) Mann–Whitney test, (D) one-way ANOVA with Tukey’s correction; *p-val < 0.05, **<0.01, ***<0.001, ****<0.0001.
To investigate whether such anorexic behavior may explain some of the transient changes in the intestinal microbiome of Cl13-infected mice, we next compared the Cl13-driven microbiome to recently proposed microbiome biomarkers of colon carcinoma-bearing mice, which suffer from anorexia (35, 36). We observed that three out of the eight Cl13-infection-specific biomarkers at day 8 p.i., as defined in Fig. 1F, overlapped with those from colon carcinoma, including reductions in Lactobacillus as well as enrichment in Bacteroides and Akkermansia (Fig. 3C red vs. blue circles). Of note, perturbation of these three taxa was reverted when anorexia was corrected, via CD8 T cell depletion, in Cl13-infected mice (Figs. 2 E and F and 3B). To elucidate which of these taxa biomarkers were common to anorexia, independent of inflammation, we applied Songbird MR analysis using rank cutoff values of 1 and −1 on publicly available 16S rRNA gene amplicon data from uninfected mice under a caloric restricted diet (37) and compared the identified fasting biomarkers (Dataset S6) with the Cl13-infection-driven microbiome. We observed that two, Sutterella and Akkermansia, of the eight microbial alterations detected during the early phase of Cl13 infection were commonly perturbed, in the same direction, during calorie restriction (Fig. 3C, red vs. yellow circles) and Akkermansia was overrepresented in the three juxtaposed conditions (Fig. 3C).
These data indicated that infection with a fast-spreading, and potentially persistent virus induces a profound, CD8 T cell-dependent anorexic behavior. Our results also demonstrated that a significant fraction of the taxa uniquely altered during the anorexic phase, including overrepresentation of Bacteroides and Akkermansia as well as reductions in Lactobacillus, were commonly perturbed in noninfectious fasting conditions and were no longer altered when anorexia was reverted, via CD8 T cell depletion during infection. Thus, these findings situated anorexia as one possible mechanism via which CD8 T cell responses may induce dysbiosis after viral infection.
A. muciniphila Blooms in a CD8 T Cell-Dependent Manner after LCMV Cl13 Infection and Is Enriched upon Lack of Food Consumption.
Because the Akkermansia genus emerged as a common fasting biomarker in the inflammatory and noninflammatory conditions that we compared in our analysis (Fig. 3C) we selected it for further validation and followup studies. Notably, within the genus Akkermansia, the commensal A. muciniphila is the only species that colonizes the human gut (38). We therefore evaluated whether A. muciniphila was uniquely increased in Cl13- vs. ARM-infected mice. Quantification of A. muciniphila genomes by qPCR in colon and caecal contents from Cl13-infected vs. both uninfected and ARM-infected mice demonstrated its selective enrichment upon Cl13 infection (Fig. 3D). We next evaluated the localization of A. muciniphila by performing fluorescence in situ hybridization (FISH) with an A. muciniphila-specific probe. This analysis further supported overrepresentations of A. muciniphila in Cl13-infected vs. ARM-infected and uninfected mice and mapped A. muciniphila to colon and caecal compartments (Fig. 3E and SI Appendix, Fig. S4).
Finally, we sought to investigate a causal link between the blooming of A. muciniphila and the CD8 T cell-induced anorexia after Cl13 infection. For that, we first quantified A. muciniphila genomes in colon and caecal contents from uninfected mice fed ad libitum or starved for 24 h. We observed that such a short interruption in food consumption alone, without viral infection, significantly increased the amount of A. muciniphila genomes by ∼20-fold (Fig. 3F). Moreover, reversion of the anorexic behavior via CD8 T cell depletion in Cl13-infected mice (Fig. 3B) was accompanied by a significant reduction in intestinal A. muciniphila genomes (Fig. 3G).
Together, these results demonstrated that an absolute increase of A. muciniphila is part of the profound dysbiosis that occurred downstream of CD8 T cell responses during the early phase of infection with a fast-spreading and persistent virus, and causally linked the lack of food consumption with A. muciniphila blooming in the intestinal compartment. Importantly, these findings further support a role for anorexia as one mechanism via which CD8 T cells may promote microbial dysbiosis, including enrichment of the Akkermansia genus, after viral infection.
Oral Administration of A. muciniphila Attenuated Antiviral CD8 T Cell Responses after LCMV Infection.
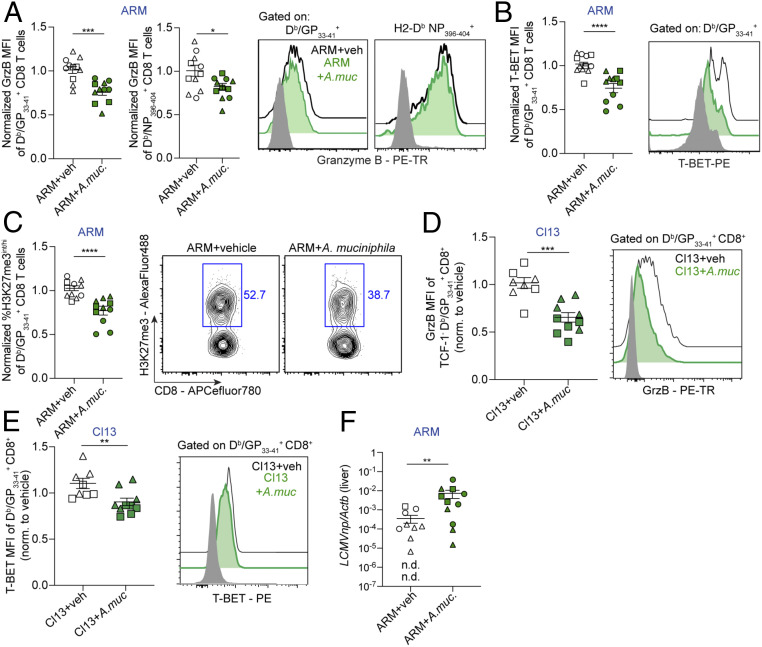
To investigate the potential bidirectional modulation of A. muciniphila and CD8 T cells, we experimentally induced A. muciniphila enrichment in ARM-infected mice, which neither become anorexic nor exhibit intestinal blooms of this commensal (Fig. 3 A, D, and E). We chose to inoculate ARM-infected mice with live A. muciniphila or vehicle control from days 3.5 to 7.5 p.i. to maintain comparable innate immune responses and CD8 T cell priming between the two groups. We next evaluated the impact on CD8 T cell responses within the small intestinal intraepithelial lymphocyte (IEL) compartment, which is in closest proximity to the intragastrically administered bacteria, and spleen, as a representative distal lymphoid organ. Analysis of LCMV-specific CD8 T cell responses from IEL demonstrated similar percentages and numbers (SI Appendix, Fig. S5A) as well as equivalent expression of the cytotoxic molecule GrzB (SI Appendix, Fig. S5B) and frequencies of killer cell lectin like receptor G1 (KLRG1) positive or CD103+ cells (SI Appendix, Fig. S5C), which mark cells with effector and tissue-resident phenotypes, respectively (39–41). We also observed equivalent accumulation of splenic LCMV-specific CD8 T cell populations (SI Appendix, Fig. S6A) as well as their cytokine production (SI Appendix, Fig. S6B) and proportions of short-lived, memory precursor, and early effector cells (SLEC, MPEC, and EEC) (41–43), in both groups (SI Appendix, Fig. S6C). Strikingly, however, we observed a significant reduction in GrzB levels among splenic CD8 T cells specific for the immunodominant LCMV epitope GP33–41 (Db/GP33–41+), and to a lesser extent among CD8 T cells specific for NP396–404, in ARM-infected mice inoculated with A. muciniphila vs. control mice (Fig. 4A). We also found reduced levels of the transcription factor (TF) T-BET among Db/GP33–41+ CD8 T cells (Fig. 4B), which is known to promote GrzB expression and CD8 T cell effector fate (41, 44). Given that trimethylation of lysine 27 residue on histone 3 (H3K27me3) has been shown to reinforce effector CD8 T cell differentiation (45, 46) we next evaluated the potential effect of A. muciniphila on CD8 T cell H3K27me3 marks. As shown in Fig. 4C, A. muciniphila-inoculated mice exhibited reduced H3K27me3 in virus-specific CD8 T cells compared to control mice. Importantly, A. muciniphila inoculation did not increase splenic Treg accumulation (SI Appendix, Fig. S6D). Of note, reduced GrzB and T-BET were also observed in splenic CD8 T cells after Cl13-infected mice were inoculated with A. muciniphila (Fig. 4 D and E), indicating that the effects of this commensal on CD8 T cell responses were conserved in the context of functional (acute infection) or exhausted (chronic infection) CD8 T cells. Finally, although A. muciniphila inoculation into ARM- or Cl13-infected mice did not alter weight loss (SI Appendix, Fig. S7 A and B), eating behavior (SI Appendix, Fig. S7 C and D), or the amount of infectious virus in liver, lung, and/or blood (SI Appendix, Fig. S7 E–I), it did cause a significant delay in the clearance of LCMV ARM genomes from the livers (Fig. 4F).
Fig. 4.
Oral administration of A. muciniphila into LCMV ARM- or Cl13-infected mice attenuated selected CD8 T cell effector features. See also SI Appendix, Figs. S5–S8. C57BL/6 mice were infected with LCMV ARM (A–C and F) or LCMV Cl13 (D and E) and inoculated i.o. with vehicle (veh) or ∼1.2 × 108 cfu of live A. muciniphila (A.muc.) daily from days 3.5 to 7.5 p.i. All analyses were done at day 8 p.i. (A–E) Mean fluorescence intensity (MFI) for Granzyme B (GrzB) (A and D) and T-BET (B and E) among gated LCMV tetramer Db/GP33–41+ CD8+ cells from the spleen. (C) Frequency of H3K27me3int/hi cells among Db/GP33–41 or Db/NP396–404 tetramer+ CD8+ in the spleen, as indicated. (A–E) Values are normalized to the average of the vehicle control group. (A–E) Representative FACS histograms are shown for the indicated groups; gray histogram was gated on CD8+ from Un mice. (F) Liver levels of LCMVnp normalized to Actb. (A–F) Pooled data from three (A–C and F) or two (D and E) experiments with n = 3 to 5 mice/group. Individual experiments are indicated with different shapes (i.e., circles, squares, triangles). (A–F) Averages ± SEM. (A–F) Unpaired two-tailed Student’s t test. Statistically significant differences were detected in three out of three (B and C), two out of three (A, for Db/GP33–41), two out of two (D) or one out of two (E) independent experiments. Even when not reaching statistical significance, individual experiments showed a consistent trend that, in all cases, reached significance after pooling data from independent repeats. For A (Db/NP396–404) and F, differences only reached statistical significance after pooling independent experimental repeats; *p-val < 0.05, **<0.01, ***<0.001, ****<0.0001.
Together, these data indicated that A. muciniphila inoculation attenuated splenic, but not small intestinal, CD8 T cell responses, specifically diminishing expression of GrzB and T-BET as well as reducing H3K27me3 marks. Such CD8 T cell suppression seemed to be accompanied with a delayed clearance of the acute infection in mice receiving A. muciniphila. Interpretation of these results should take into consideration that, consistent with a previous report that used a similar dose of this bacterium (47), qPCR in caecal and colonic compartments did not detect significant enrichment of A. muciniphila after its oral administration (SI Appendix, Fig. S8). Such low levels of A. muciniphila in the experimentally treated mice raises the possibility that the phenotypes we observed after inoculation of this commensal into infected mice may represent an underestimation of the effects caused by the profound A. muciniphila blooming that naturally arises after Cl13 infection.
Discussion
Host adaptations to fast spreading and/or persistent pathogen replication are often required to ensure host survival in various host species and include mechanisms such as the attenuation of both innate and adaptive arms of the immune system (2–4). Here, we found that infection with a fast-spreading persistent virus induced profound microbiome alterations, which were different from the ones induced by a closely related acute virus that spreads at a slower rate. Interestingly, most microbiome alterations induced by the fast-spreading persistent virus were restricted to the early phase of infection and did not associate with continuous viral replication into the chronic phase. Instead, such microbiome dysbiosis was driven by CD8 T cell responses and partially associated with CD8 T cell-induced anorexia. Remarkably, among the most enriched taxa detected early after infection with the fast-spreading persistent virus, we identified the fasting-associated commensal A. muciniphila, which bloomed in a CD8 T cell-dependent manner and was able to suppress selected aspects of distal CD8 T cell responses. Our results demonstrate that the endogenous intestinal microbiome of an infected host differentially changes after infection with closely related viruses that exhibit distinct capacity to spread and persist and suggest that infection-induced changes in eating behavior and microbiome could act as both regulators and targets of CD8 T cell responses.
It is remarkable that the intestinal microbiome was more profoundly and transiently altered during the early vs. late phase of persistent LCMV infection, despite comparable viral replication at both time periods. Furthermore, our data demonstrated that such early dysbiosis was caused, in great part, by CD8 T cell responses. Importantly, while the intestinal microbiome has been previously shown to modulate CD8 T cell responses (48–51) a role for CD8 T cells in modulating gut microbiome composition has never been reported. In this regard, the transitory Verrucomicrobia and Akkermansia overrepresentation that we observed on day 8 (but not day 20) p.i. and its connection to fasting across several species (34) helped us identify a profound anorexic behavior that was restricted to the early phase of LCMV Cl13 infection. This result is in agreement with an independent study that showed anorexia in the same LCMV model system (52). Here we validated these findings and expanded them by demonstrating that the aforementioned anorexia observed early after infection with the fast-spreading LCMV isolate is dependent on CD8 T cells. Given that anorexia has been observed at time points when strong CD8 T cell responses are detected after other chronic and acute infections (53–57), it is tempting to speculate that CD8 T cell-induced anorexia may represent a conserved response to tissue damage or excessive inflammation upon infection with pathogens that overwhelmingly replicate in vital organs and/or are still present at high titers when CD8 T cell responses peak.
Consistent with the aforementioned anorexic behavior in LCMV infection, we observed that a significant fraction of the microbiome changes were commonly detected in other inflammatory and/or noninflammatory fasting conditions. Among them, we demonstrated that the species A. muciniphila naturally bloomed in the cecum and colon during the early phase of infection with the LCMV Cl13 isolate, it was rapidly boosted in response to lack of food consumption, and was significantly reduced when anorexia was reverted by CD8 T cell depletion. Given that fiber-deprived diets promote blooms of A. muciniphila (58), it is possible that the lack of dietary fiber intake early after LCMV Cl13 infection may have provided a niche for the outgrowth of this bacterium. Regardless, our observations linked A. muciniphila blooming with CD8 T cell-driven anorexia and supported the conclusion that anorexia is one means via which CD8 T cells induce intestinal dysbiosis after chronic infection. It should be noted, however, that not all taxa modulated by CD8 T cells during LCMV Cl13 infection were commonly altered in the noninfectious fasting conditions that we analyzed here, suggesting the existence of alternative, anorexia-independent mechanisms employed by CD8 T cells to modulate gut microbial composition. On the other hand, the phylum Verrucomicrobia is also augmented after infection with influenza virus (59), which induces anorexia with a similar kinetic as LCMV Cl13 (60), suggesting that A. muciniphila and possibly other fasting-associated commensals may bloom, perhaps also in a CD8 T cell-dependent manner, during some acute infections.
While it was previously demonstrated that the intestinal microbiome as a whole promotes preinfection, tonic, expression of genes related to antiviral immunity in macrophages, and enhances resistance to a subsequent chronic infection (9), the potential impact of the changes in specific commensal species that are induced after a chronic infection has been initiated remained unknown. In this regard, we found that enrichment of A. muciniphila via oral administration into specific pathogen-free (SPF) LCMV-infected mice (that had unmanipulated microbial communities) suppressed selected effector features (i.e., GrzB, T-BET, and/or H3K27me3) in virus-specific CD8 T cells and appeared to delay viral control in the liver. These observations were particularly unexpected, given the beneficial effects that A. muciniphila has been ascribed during checkpoint blockade therapy (61). We speculate that the differential microbial communities into which A. muciniphila is introduced may account for some of these discrepancies, as individual bacterial species are heavily influenced by the complex microbial environments surrounding them (62). Alternatively, given that different CD8 T cell subsets are present at early vs. late time points after chronic infections (63, 64) and in tumors (65, 66), it is possible that the effect of A. muciniphila on CD8 T cell responses may vary depending on the time of administration. Despite that, the effects we observed in virus-specific CD8 T cells upon early inoculation of A. mucinipihila, were consistent in the context of LCMV infections with any of the two viral isolates studied, and were particularly impressive in the light of a previous report where modulation of CD8 T cell responses was only achieved after combined administration of 11 different taxa (50). Thus, our data support the possibility that the natural enrichment of A. muciniphila that we uncovered in the early phase after infection with a fast-spreading virus may contribute to the attenuation of the initial virus-specific CD8 T cell responses, most likely in conjunction, and possibly in a redundant fashion, with other fasting-associated commensals and with previously described T cell-inhibitory mechanisms (3, 4).
Interestingly, we observed that A. muciniphila attenuated distal CD8 T cells in the spleen but did not seem to have an effect on IEL CD8 T cells from the small intestine, despite their closer proximity to intestinal bacteria. Even though we cannot rule out effects in the large intestine, this is particularly unexpected since A. muciniphila has been recently shown to induce bacteria-specific immune responses in the small intestine (67). Given that A. muciniphila is known to strongly influence whole-body metabolism (47, 68, 69), it is possible that it suppresses CD8 T cell effector features by changing the nutrients available to fuel ensuing immune responses, which in turn can affect functional capacity and epigenetics in a cell-type- or tissue-specific manner (70). Indeed, nutrient requirements may vary among tissues, providing a possible explanation for the differential effects of A. muciniphila in spleen vs. small intestinal IEL CD8 T cells. Regardless of the mechanism underlying A. muciniphila effects in distal vs. local tissues, our findings describe a commensal regulating distal, virus-specific CD8 T cell responses, without causing similar phenotypes in the small intestine IEL, broadening our current understanding of microbiome-mediated immune system regulation.
In conclusion, by revealing CD8 T cell-dependent anorexia, CD8 T cell-mediated blooming of A. muciniphila and CD8 T cell attenuation by this commensal, our study suggests a potential way of CD8 T cell autoregulation that involves changes in host behavior and microbiome. In this context, we propose a model in which the blooming of A. muciniphila that we described here may be part of a whole-organism negative feedback loop initiated by CD8 T cell-driven changes in eating behavior, that can lead to microbiome alterations, which may, in turn, contribute to the attenuation of CD8 T cell responses. Suppression of CD8 T cell responses through previously described inhibitory mechanisms (3, 7) along with the contribution of the aforementioned negative feedback loop may explain the recovery from the CD8 T cell-dependent anorexic behavior alongside with the mostly transient nature of the CD8 T cell-dependent dysbiosis, as the host transitions from the acute into the chronic phase of a persistent infection. Last but not least, it is important to emphasize that there is great interest in A. muciniphila as a potential probiotic (71–73), that is reflected by recently finalized (68) and ongoing clinical trials (NCT02086110; NCT03547440; NCT03749291; NCT04038619; NCT04058392). Our work unveils additional effects of this potential new-generation probiotic in regard to its capacity to attenuate ensuing CD8 T cell responses. As non-FDA regulated probiotics are being consumed by 3.9 million individuals in the United States alone (74) increased understanding of context-dependent consequences of probiotic administration is needed to maximize their beneficial properties while preventing undesired side effects.
Materials and Methods
Mice, Infections, and Antibody Treatments.
All studies were performed with 6- to 10-wk-old female C57BL/6 mice bred at The Jackson Laboratory. For microbiome studies, purchased mice were randomly distributed in cages containing five mice each and housed in that way for 3 wk before the day of infection per guidelines of the Earth Microbiome Project (75). Mice were infected i.v. with 2 × 106 plaque forming units (PFU) of LCMV ARM or Cl13. Viral stocks were grown, identified, and quantified as reported previously in ref. 76. Viral loads were determined by standard plaque assay (76) of serum or tissue homogenates. In vivo CD8 and CD4 T cell depletion as well as IFNAR blockade studies were performed as specified in SI Appendix. The detailed procedure for food consumption measurements, for the establishment of a “healthy range” of daily food consumption and for acute starvation experiments can be found in SI Appendix. Mice were maintained in a closed breeding facility at the University of California San Diego (UCSD), where mice were housed in ventilated cages containing HEPA filters. Mouse handling conformed to the requirements of the National Institutes of Health and the Institutional Animal Care and Use Guidelines of UCSD.
Lymphocyte Isolation from Spleen and Small Intestinal IEL.
Single-cell suspensions from the spleen and the small intestinal IEL were obtained as detailed in SI Appendix.
Quantitative PCR.
To quantify A. muciniphila genomes, previously frozen colonic and caecal contents were thawed, weighed, and DNA was extracted with DNeasy PowerSoil Kit (Qiagen) by following the manufacturer’s protocol. qPCR from samples as well as from an A. muciniphila standard was performed as detailed in SI Appendix. For quantification of transcripts in liver, tissues were processed and the expression of LCMV nucleoprotein relative to Actb was quantified as detailed in SI Appendix.
Flow Cytometry.
For surface staining of splenocytes and IEL, cells were first stained with a fixable viability dye followed by staining with MHC-I tetramers as detailed in SI Appendix. For intracellular staining after ex vivo peptide restimulation, cells were fixed in 1% paraformaldehyde (PFA) and stained with antibodies in 1× perm/wash buffer (Thermo Scientific) for 30 min at 4 °C. Alternatively, for intranuclear staining, cells were fixed with the Foxp3 Transcription Factor Staining Buffer Set Kit (Thermo Scientific) per vendor’s recommendation and stained with antibodies in 1× perm/wash buffer for 1 h at room temperature. Antibodies used in this study can be found in SI Appendix. Cells were acquired using a Digital LSR II flow cytometer (Becton Dickinson) or a ZE5 Cell Analyzer (Bio-Rad). Flow cytometric data were analyzed with FlowJo software v9.9.6 and v10.
Ex Vivo T Cell Stimulation.
Splenocytes were cultured in the presence of Brefeldin A (1 μg/mL; Sigma) and 1 μg/mL of the MHC class-I-restricted LCMV NP396–404 or GP33–41 peptides (all >99% pure; Synpep) and stained with surface and intracellular antibodies, as detailed in SI Appendix.
Culture and Inoculation of A. muciniphila.
A. muciniphila (ATTC BAA-835) was purchased from ATCC. A detailed procedure on how to prepare glycerol and mucin stocks as well as anaerobic brain heart infusion (BHI) media supplemented with mucin (BHI+mucin) can be found in SI Appendix. To initiate A. muciniphila cultures for oral inoculation, glycerol stocks were brought into the anaerobic chamber on dry ice, scraped with a sterile tip that was then placed in 3 to 5 mL of BHI+mucin media in 15-mL stopper cap falcon tubes and cultured at 37 °C in the anaerobic chamber for 3 to 4 d. Control tips with no A. muciniphila and media alone were cultured in parallel as a control for contamination and to be inoculated as vehicle control, respectively. Initially, it was determined that after 3 to 4 d of A. muciniphila glycerol stock culture in these conditions, the optical density at 600 nm was 0.5 to 0.6, which after plating at serial dilutions corresponded to 5.5 to 6 × 108 colony forming units (cfu)/mL 200 µL of these cultures (i.e., ∼1.2 × 108 cfu) or vehicle control were inoculated per mouse at indicated time points. Note that very similar amounts of bacteria per dose have been used previously ref. 47. A. muciniphila cultures were routinely plated in BHI+mucin media and 1% agar to confirm the homogeneity of the culture and to rule out potential contamination.
Bacterial FISH.
Caecal and colonic tissue samples from respective experimental groups were harvested and fixed in Carnoy’s fixative (60% ethanol, 30% chloroform, 10% glacial acetic acid) and processed further as previously described in ref. 77. Tissue sections (5 μM) were deparaffinized followed by hybridization and staining, as specified in SI Appendix. Samples were viewed and imaged on an Olympus IX81 inverted microscope in combination with a FV1000 confocal laser scanning system.
Library Generation and Sequencing for 16S rRNA and Shotgun Metagenomics Studies.
A detailed description of the sample collection and the DNA extraction procedures for 16S rRNA V4 amplicon sequencing and shotgun metagenomics sequencing can be found in SI Appendix. Sequencing was done following standard protocols from the Earth Microbiome Project (78, 79). Extracted DNA was amplified by using barcoded primers. Each sample was PCR amplified in triplicate and V4 pair-end sequencing or whole-genome sequencing using a miniaturized version of the KAPA HyperPlus kit was performed using Illumina MiSeq or HiSeq.
16S rRNA Gene Amplicon Data Analysis.
Preprocessing and taxonomy assignation for our 16S rRNA raw data were done as detailed in SI Appendix. Raw 16S rRNA sequencing data from the stools of mice fed a calorie-restricted diet for 3 wk (37) was obtained from National Center for Biotechnology Information (NCBI) Short Read Archive (SRA: PRJNA480387), uploaded to Qiita, and processed as described in SI Appendix. We calculated alpha-diversity, beta-diversity, principal coordinates analysis, permutational ANOVA, and differential dispersion as detailed in SI Appendix. To identify differentially abundant genera among groups in our 16S rRNA sequencing data as well as in caloric-restriction data (37), we used Songbird multinomial regression analysis (30), as detailed in SI Appendix.
Shotgun Metagenomic Data Analysis.
Preprocessing of these data were done as detailed in SI Appendix. A table in .biom format with taxonomic predictions at the species level was then rarefied to a depth of 200,000 or 450,000. The detailed procedure to compute alpha-diversity, beta-diversity, principal coordinates analysis, and permutational ANOVA can be found in SI Appendix. To identify differentially abundant species between groups, we collapsed our rarefied .biom tables to the species level and used Songbird multinomial regression analysis, as specified in SI Appendix.
Statistical Analysis.
Unpaired two-tailed Student’s t test was used to compare two groups. If variances were not equal by F-test, data were tested using the nonparametric Mann–Whitney U test. Significant differences among more than two groups were determined based on one-way ANOVA with Tukey’s multiple comparison correction or, in the case of unequal variances, nonparametric Kruskal–Wallis test with Dunn’s multiple comparison correction. Statistical tests were performed using GraphPad Prism v8.
Supplementary Material
Acknowledgments
We thank members of the Zúñiga laboratory for discussions and critical reading of the manuscript. We thank Dr. Julia M. Gauglitz for initial multinomial regression analyses; Dr. Clarisse A. Marotz and Dr. Pedro Belda-Ferre for key assistance with anaerobic cultures; Dr. Gail Ackermann for data upload and metadata curation on Qiita; Dr. Tomasz Kosciolek for guidance with Qiita/QIIME; “Qiita help” personnel for assistance with Qiita; as well as personnel at the animal facility and the IGM Genomics center sequencing core at UCSD.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003656117/-/DCSupplemental.
Data Availability.
All 16S rRNA gene amplicon and shotgun metagenomics sequencing data are publicly available on the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home, accession number ERP123227) and on Qiita (https://qiita.ucsd.edu, study ID 11043).
References
- 1.Yona A. H., Frumkin I., Pilpel Y., A relay race on the evolutionary adaptation spectrum. Cell 163, 549–559 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R., Schneider D. S., Soares M. P., Disease tolerance as a defense strategy. Science 335, 936–941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuniga E. I., Macal M., Lewis G. M., Harker J. A., Innate and adaptive immune regulation during chronic viral infections. Annu. Rev. Virol. 2, 573–597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Virgin H. W., Wherry E. J., Ahmed R., Redefining chronic viral infection. Cell 138, 30–50 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Oldstone M. B. A. et al., Lymphocytic choriomeningitis virus Clone 13 infection causes either persistence or acute death dependent on IFN-1, cytotoxic T lymphocytes (CTLs), and host genetics. Proc. Natl. Acad. Sci. U.S.A. 115, E7814–E7823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q. et al., Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science 323, 1726–1729 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto M. et al., CD8 T cell exhaustion in chronic infection and cancer: Opportunities for interventions. Annu. Rev. Med. 69, 301–318 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Belkaid Y., Hand T. W., Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abt M. C. et al., Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villarino N. F. et al., Composition of the gut microbiota modulates the severity of malaria. Proc. Natl. Acad. Sci. U.S.A. 113, 2235–2240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan N. et al., Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front. Immunol. 7, 529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majlessi L. et al., Colonization with Helicobacter is concomitant with modified gut microbiota and drastic failure of the immune control of Mycobacterium tuberculosis. Mucosal Immunol. 10, 1178–1189 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Dillon S. M., Frank D. N., Wilson C. C., The gut microbiome and HIV-1 pathogenesis: A two-way street. AIDS 30, 2737–2751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue T. et al., Gut dysbiosis associated with hepatitis C virus infection. Clin. Infect. Dis. 67, 869–877 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Yang R., Xu Y., Dai Z., Lin X., Wang H., The immunologic role of gut microbiota in patients with chronic HBV infection. J. Immunol. Res. 2018, 2361963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vujkovic-Cvijin I. et al., Gut-resident Lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Rep. 13, 1589–1597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooney J. P. et al., Inflammation-associated alterations to the intestinal microbiota reduce colonization resistance against non-typhoidal Salmonella during concurrent malaria parasite infection. Sci. Rep. 5, 14603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniguchi T. et al., Plasmodium berghei ANKA causes intestinal malaria associated with dysbiosis. Sci. Rep. 5, 15699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J. et al., Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 211, 2397–2410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groves H. T. et al., Respiratory disease following viral lung infection alters the murine gut microbiota. Front. Immunol. 9, 182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deriu E. et al., Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog. 12, e1005572 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupp C. et al., Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Gillis C. C. et al., Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23, 54–64.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed R., Oldstone M. B., Organ-specific selection of viral variants during chronic infection. J. Exp. Med. 167, 1719–1724 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanca A. et al., Metaproteogenomics reveals taxonomic and functional changes between cecal and fecal microbiota in mouse. Front. Microbiol. 8, 391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu S. et al., Bacterial community mapping of the mouse gastrointestinal tract. PLoS One 8, e74957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon C. E., The mathematical theory of communication. 1963. MD Comput. 14, 306–317 (1997). [PubMed] [Google Scholar]
- 28.Wherry E. J., Blattman J. N., Murali-Krishna K., van der Most R., Ahmed R., Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77, 4911–4927 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beura L. K. et al., Lymphocytic choriomeningitis virus persistence promotes effector-like memory differentiation and enhances mucosal T cell distribution. J. Leukoc. Biol. 97, 217–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton J. T. et al., Establishing microbial composition measurement standards with reference frames. Nat. Commun. 10, 2719 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallimore A. et al., Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187, 1383–1393 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry E. J. et al., Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Mack I. et al., Weight gain in anorexia nervosa does not ameliorate the faecal microbiota, branched chain fatty acid profiles, and gastrointestinal complaints. Sci. Rep. 6, 26752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mack I. et al., Is the impact of starvation on the gut microbiota specific or unspecific to anorexia nervosa? A narrative review based on a systematic literature search. Curr. Neuropharmacol. 16, 1131–1149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bindels L. B. et al., Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. ISME J. 10, 1456–1470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bindels L. B. et al., Increased gut permeability in cancer cachexia: Mechanisms and clinical relevance. Oncotarget 9, 18224–18238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabbiano S. et al., Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell. Metab. 28, 907–921.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M., Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Masopust D., Vezys V., Wherry E. J., Barber D. L., Ahmed R., Cutting edge: Gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J. Immunol. 176, 2079–2083 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Sheridan B. S. et al., Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity 40, 747–757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi N. S. et al., Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plumlee C. R. et al., Early effector CD8 T cells display plasticity in populating the short-lived effector and memory-precursor pools following bacterial or viral infection. Sci. Rep. 5, 12264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaech S. M. et al., Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Cruz-Guilloty F. et al., Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206, 51–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray S. M., Amezquita R. A., Guan T., Kleinstein S. H., Kaech S. M., Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity 46, 596–608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kakaradov B. et al., Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Everard A. et al., Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachem A. et al., Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51, 285–297.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Pearson J. A. et al., Altered gut microbiota activate and expand insulin B15-23-reactive CD8+ T cells. Diabetes 68, 1002–1013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanoue T. et al., A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez-Perez G. et al., Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J. Immunol. 196, 3768–3779 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Baazim H. et al., CD8+ T cells induce cachexia during chronic viral infection. Nat. Immunol. 20, 701–710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatter J. A. et al., Toxoplasma gondii infection triggers chronic cachexia and sustained commensal dysbiosis in mice. PLoS One 13, e0204895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cumnock K. et al., Host energy source is important for disease tolerance to malaria. Curr. Biol. 28, 1635–1642.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Horne-Debets J. M. et al., PD-1 dependent exhaustion of CD8+ T cells drives chronic malaria. Cell Rep. 5, 1204–1213 (2013). [DOI] [PubMed] [Google Scholar]
- 56.John B. et al., Dynamic imaging of CD8(+) T cells and dendritic cells during infection with Toxoplasma gondii. PLoS Pathog. 5, e1000505 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruibal P. et al., Unique human immune signature of Ebola virus disease in Guinea. Nature 533, 100–104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desai M. S. et al., A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bartley J. M., Zhou X., Kuchel G. A., Weinstock G. M., Haynes L., Impact of age, caloric restriction, and influenza infection on mouse gut microbiome: An exploratory study of the role of age-related microbiome changes on influenza responses. Front. Immunol. 8, 1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang A. et al., Opposing effects of fasting metabolism on tissue tolerance in bacterial and viral inflammation. Cell 166, 1512–1525.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Routy B. et al., Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Morin M., Pierce E. C., Dutton R. J., Changes in the genetic requirements for microbial interactions with increasing community complexity. eLife 7, e37072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z. et al., TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51, 840–855.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hudson W. H. et al., Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51, 1043–1058.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beltra J. C. et al., Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zander R. et al., CD4. Immunity 51, 1028–1042.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ansaldo E. et al., Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Depommier C. et al., Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greer R. L. et al., Akkermansia muciniphila mediates negative effects of IFNγ on glucose metabolism. Nat. Commun. 7, 13329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krautkramer K. A. et al., Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J., Lin S., Vanhoutte P. M., Woo C. W., Xu A., Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in apoe-/- mice. Circulation 133, 2434–2446 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Wu W. et al., Protective effect of Akkermansia muciniphila against immune-mediated liver injury in a mouse model. Front. Microbiol. 8, 1804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blacher E. et al., Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572, 474–480 (2019). [DOI] [PubMed] [Google Scholar]
- 74.The Lancet Gastroenterology Hepatology , Probiotics: Elixir or empty promise? Lancet Gastroenterol. Hepatol. 4, 81 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Thompson L. R. et al.; Earth Microbiome Project Consortium , A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmed R., Salmi A., Butler L. D., Chiller J. M., Oldstone M. B., Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 160, 521–540 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swidsinski A. et al., Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm. Bowel Dis. 13, 963–970 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Caporaso J. G. et al., Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gilbert J. A., Jansson J. K., Knight R., The Earth microbiome project: Successes and aspirations. BMC Biol. 12, 69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segata N. et al., Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA gene amplicon and shotgun metagenomics sequencing data are publicly available on the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home, accession number ERP123227) and on Qiita (https://qiita.ucsd.edu, study ID 11043).