Scientists worldwide are racing to develop effective vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the COVID-19 pandemic. An important and perhaps underappreciated aspect of this endeavor is ensuring that the vaccines being developed confer immunity to all viral lineages in the global population. Toward this end, a seminal study published in PNAS (1) analyzes 27,977 SARS-CoV-2 sequences from 84 countries obtained throughout the course of the pandemic to track and characterize the evolution of the novel coronavirus since its origination. The principle conclusion reached by the authors of this work is that SARS-CoV-2 genetic diversity is remarkably low, almost entirely the product of genetic drift, and should not be expected to impede development of a broadly protective vaccine.
Although errors introduced during genome replication are a major source of genetic variation in all virus populations, limiting the fitness costs of accumulated errors is especially critical for coronaviruses, the RNA genomes of which are the largest known. For this reason, coronaviruses evolved nonstructural protein 14 (nsp14), which accompanies viral replicases during RNA synthesis and excises misincorporated ribonucleotides from nascent strands before they can be extended, thus preventing errors from becoming permanent. This error-correcting capacity was unknown among RNA viruses prior to its discovery in SARS-CoV-1 (2, 3), and it contributes to a replication error rate more than 10-fold lower than that of other RNA viruses (4, 5). This activity also likely contributes to the low genetic diversity of SARS-CoV-2, although to our knowledge nsp14 function in the novel coronavirus has yet to be investigated.
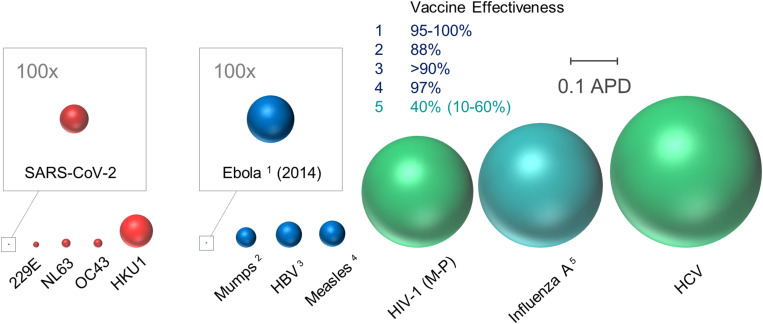
For many viruses, surface glycoproteins contain not only elements required for specific binding of cellular receptors, membrane fusion, and virus entry into the host cell but also epitopes recognized by neutralizing antibodies produced as part of an effective adaptive immune response. Hence, tracking genetic variation in the SARS-CoV-2 surface glycoprotein is of paramount importance for determining the likelihood of vaccine effectiveness or immune escape. To put this variation in perspective, Fig. 1 shows a graphical illustration of comparative genetic diversity among surface glycoproteins of select human pathogenic viruses, including SARS-CoV-2, correlated with the availability and effectiveness of respective preventive vaccines.
Fig. 1.
Comparative genetic diversity among coronaviruses and select viral pathogens. As indicated by the scale bar, sphere radius reflects average pairwise distances (APD) of viral surface glycoprotein gene sequences among different viruses. Diversities among coronaviruses (for which no vaccines have been developed to date) are indicated in red, and those of other viruses for which effective vaccines are available or unavailable are shown in blue and green, respectively. Since 2005, the average effectiveness of combination influenza seasonal vaccines (influenza A: H1N1, H2N3, influenza B) has been 40%. Accordingly, genetic diversity of influenza A is depicted by blue-green shading to reflect an intermediate level of vaccine effectiveness. Sequences were obtained from public databases and identical sequences were included only once. MEGA7 software was used to calculate APD among gene segments encoding proteins involved in attachment/entry: Spike or Spike-like human coronaviruses (SARS-CoV-2, 229E, NL63, OC43, and HKU1), spike glycoprotein (Ebola), HN (mumps), S (HBV), H (measles), Env (HIV-1), HA (influenza A), and E1 (HCV). More specifically, HIV-1 Group M subtypes A–D, F–H, J–K, CRF01_AE, and CRF02_AG; HBV serotypes A–H; HCV genotypes 1a–c, 2a–b, 4a, 5a, 6a, 6k, and 6m; and influenza A H1N1 pdm09, seasonal H1N1, H3N2, and H5N1 were included. Majority-rule consensus of unique sequences for HIV-1 (Group M, N, O, and P), HBV, HCV, and influenza A was performed in Seaview v4.7. Total numbers of sequences analyzed: SARS-CoV-2 (21,554), 229E (25), NL63 (52), OC43 (79), HKU1 (38), Ebola (578), mumps (341), HBV (10,271), measles (38), HIV-1 (5,603), influenza A (133), and HCV (439).
Although genetic diversity is only one of many determinants of vaccine efficacy, there is a clear inverse correlation between these two metrics among viral pathogens examined in our analysis. Presumably due to its relatively recent origins, genetic diversity in the SARS-CoV-2 surface glycoprotein, spike, encoded by the S gene, is exceedingly low, even in comparison to other human coronaviruses. Toward the opposite extreme, diversity among influenza A surface glycoproteins is 437-fold greater than that measured in SARS-CoV-2. The relative age of influenza A (dating at least back to the 16th century) is certainly a major factor in this disparity, as is reassortment of genome segments encoding influenza A surface antigens hemagglutinin (HA) and neuraminidase (NA) (6). Indeed, sudden emergence of influenza A virus variants containing HA–NA combinations not previously encountered by contemporaneous human populations caused the pandemics of 1918 (H1N1), 1957 (H2N2), 1968 (H3N2), and 2009 (H1N1pdm09). Although coronavirus genomes are not segmented like those of influenza viruses, they are nevertheless capable of high rates of recombination. Hence, future emergence of new virulent derivatives of SARS-CoV-2 paralleling those observed with influenza A is a possibility that will require global monitoring of both animal and human reservoirs.
As differences in biology and epidemiology among these human viral pathogens are considerable, so is the extent of sequence divergence in genes encoding their respective envelope glycoproteins. HIV-1, for example, has fueled the AIDS pandemic for more than 40 y, during which time genetic diversity was acquired through both recombination and propagation of replication errors (7). Similarly, widespread sustained prevalence contributed to genetic diversity in hepatitis B virus (HBV) (8) and hepatitis C virus (HCV) (9), both causative agents of ongoing chronic hepatitis pandemics. Since these viruses cause chronic infections, their evolution is also shaped by immune pressure to a degree not possible with SARS-CoV-2, given the typical short course of COVID-19. However, with respect to our analysis, it is perhaps most important to recognize that the genetic diversities of human coronaviruses (i.e., 229E, NL63, OC43, HKU1, and now SARS-CoV-2), some of which may have been circulating in the population for centuries, are less than or comparable to those measured for mumps, measles, hepatitis B, and Ebola viruses, against which vaccines have been developed that are at least 88% effective (https://www.cdc.gov/vaccines/).
The measured and well-supported conclusions of Dearlove et al. (1) markedly contrast with an early study of SARS-CoV-2 evolution that raised alarm at the emergence and spread of a “strain” more “aggressive” than the original (10). It was argued that the novel coronavirus population was divided into S and L “strains” distinguishable by two mutations at genome positions 8,782 (ORF1ab) and 28,144 (ORF8). In an addendum, the authors acknowledged that they provided no evidence supporting any epidemiological conclusion regarding the virulence or pathogenicity of SARS-CoV-2, and that their description of the “L type” as being more “aggressive” was inappropriate. That word was omitted from the subsequent print version of the article, each instance being replaced by a variation of “more frequently observed.” Unfortunately, online reports derived from this article were not as self-correcting or restrained, using phrases or titles such as “At least eight strains of the coronavirus are making their way around the globe, creating a trail of death and disease that scientists are tracking by their genetic footprints” (11), “the coronavirus is continuously mutating to overcome the immune system resistance of different populations” (12), and “Coronavirus: Are there two strains and is one more deadly?” (13) to describe and interpret the scientific findings presented in the aforementioned paper. It is hard to argue that these reports accurately portrayed the means, degree, and consequences of low-level accumulation of genetic diversity in SARS-CoV-2 to the public, and we hope such information is relayed more carefully and conscientiously in the future.
Despite the remarkable wealth of data currently available, careful temporally and geographically resolved analyses of genetic diversity in large SARS-CoV-2 datasets do not always produce consensus. One recent concern has been the basis for emergence of a mutation encoding a D614G amino acid substitution in the SARS-CoV-2 spike protein. First observed in Germany in late January 2020, this variant is now the dominant form among SARS-CoV-2 viruses worldwide. Korber et al. recently concluded that the ascendency of 614G was not a consequence of genetic drift but instead occurred because the mutation renders the virus more infectious (14). This conclusion was initially based on their observation that the proportion of sequences carrying the D614G mutation progressively increased in every region in Asia, Europe, Oceania, and North America that was well-sampled in the GISAID database (https://www.gisaid.org/). Moreover, subsequent analyses showed that pseudotyped virus containing the 614G mutation spread more rapidly in cell culture, probably due to a structural alteration that reduced shedding of the S1 spike protein subunit (14–16).
Dearlove et al. (1) acknowledge that emergence of the 614G mutation may constitute an exception to their overarching conclusion that SARS-CoV-2 genetic variation is overwhelmingly due to genetic drift. However, as a caveat to accepting this determination prematurely, they cite a parallel finding that A82V and other mutations in the ebolavirus surface glycoprotein were associated with increased infectivity. In this case, subsequent analysis in cell culture showed that the degree of increased infectivity varied with cell type (17) and no phenotypic differences were observed when mutant viruses were evaluated in animal models (18). Moreover, the authors argue that because the 614G variant has relatively rarely been sampled in China, and there is no evidence for convergent evolution independently producing the same or a similar mutation, the hypothesis that 614G emerged as a consequence of a genetic bottleneck during spread of the virus from Asia to Europe remains viable.
It is perhaps even more important to note that the question of whether the 614G mutation increases infectivity has no bearing on the expected efficacy of vaccines currently under development. Indeed, amino acid position 614 is not located within the receptor binding domain, the motif expected to house epitopes most frequently recognized by neutralizing antibodies, and cell culture studies confirm that viruses pseudotyped with 614D or 614G spike variants are neutralized with equal effectiveness (19, 20). Taken together, these results are consistent with the central conclusion of Dearlove et al. (1) that the current state of SARS-CoV-2 genetic diversity should not be expected to impede development of a broadly protective vaccine.
It could be argued that maintaining the ∼30-kb RNA genome of SARS-CoV-2 reduces its tolerance for genetic diversity, rendering the novel coronavirus perhaps more susceptible to control by widespread immunization than might be expected for other RNA viruses. However, it is equally valid to suggest that because SARS-CoV-2 has infected and spread within an immunologically naïve population it has yet to experience the sort of immune pressure that helped shape the evolution of the endemic viruses shown in Fig. 1, and its own capacity to evolve remains unknown. Accordingly, we must continue to be diligent in tracking genetic changes in the novel coronavirus, both to follow their spread and quickly identify antigenic shifts should they occur. Yet, it is equally important to recognize that what we have observed to this point is slow genetic drift characteristic of a virus with a highly stable genome and to keep these and future observations on SARS-CoV-2 genetic diversity in the appropriate perspective, especially when communicating them to the general public.
Acknowledgments
We gratefully acknowledge the authors and the originating and submitting laboratories for their shared sequence data. We thank Michael Bale, Wei Shao, and Valerie Boltz for collection and analyses of the publicly available sequence data and for useful discussions on viral genetic diversity. We thank John Coffin for useful discussions and suggestions.
Footnotes
The authors declare no competing interest.
See companion article, “A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants,” 10.1073/pnas.2008281117.
References
- 1.Dearlove B. et al., A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. U.S.A. 117, 23652–23662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denison M. R., Graham R. L., Donaldson E. F., Eckerle L. D., Baric R. S., Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 8, 270–279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minskaia E. et al., Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 5108–5113 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy S., Shackelton L. A., Holmes E. C., Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 9, 267–276 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Eckerle L. D. et al., Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6, e1000896 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H., Webster R. G., Webby R. J., Influenza virus: Dealing with a drifting and shifting pathogen. Viral Immunol. 31, 174–183 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Hemelaar J., The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 18, 182–192 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z. H. et al., Genetic variation of hepatitis B virus and its significance for pathogenesis. World J. Gastroenterol. 22, 126–144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timm J., Roggendorf M., Sequence diversity of hepatitis C virus: Implications for immune control and therapy. World J. Gastroenterol. 13, 4808–4817 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang X. et al., On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev., 10.1093/nsr/nwaa036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weise E., 8 strains of the coronavirus are circling the globe. Here’s what clues they’re giving scientists. USA Today, 27 March 2020. https://www.usatoday.com/story/news/nation/2020/03/27/scientists-track-coronavirus-strains-mutation/5080571002/. Accessed 1 September 2020.
- 12.Tingson N., Coronavirus Has THREE distinct strains, according to study; US suffering from original variation. Tech Times, 10 April 2020. https://www.techtimes.com/articles/248721/20200410/coronavirus-has-three-distinct-strains-according-to-study-us-suffering-from-original-variation.htm. Accessed 1 September 2020.
- 13.Hamzelou J., Coronavirus: Are there two strains and is one more deadly? NewScientist, 5 March 2020. https://www.newscientist.com/article/2236544-coronavirus-are-there-two-strains-and-is-one-more-deadly/. Accessed 1 September 2020. [Google Scholar]
- 14.Korber B. et al.; Sheffield COVID-19 Genomics Group , Tracking changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q. et al., The impact of mutations in SARS-CoV-2 Spike on viral infectivity and antigenicity. Cell, 10.1016/j.cell.2020.07.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L., et al. , The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv:10.1101/2020.06.12.148726 (12 June 2020).
- 17.Wong G. et al., Naturally occurring single mutations in ebola virus observably impact infectivity. J. Virol. 93, e01098-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marzi A., et al. , Recently identified mutations in the Ebola virus–Makona genome do not alter pathogenicity in animal models. Cell Rep. 23, 1806–1816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueda M. T. et al., Functional mutations in spike glycoprotein of Zaire ebolavirus associated with an increase in infection efficiency. Genes Cells 22, 148–159 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Wang M. K., Lim S. Y., Lee S. M., Cunningham J. M., Biochemical basis for increased activity of Ebola glycoprotein in the 2013-16 epidemic. Cell Host Microbe 21, 367–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]