Significance
Gastric cancer (GC), a deadly malignancy in the world, particularly in East Asia, remains a major clinical challenge given a lack of effective targeted treatments for advanced HER2-negative cases (80 to 88%). This work uncovers a PHF8/c-Jun-mediated axis that upregulates the expression of PKCα, triggering Src activation and PTEN inhibition. High PHF8-PKCα expression predicts worse prognosis in GC. Importantly, targeting the PKCα-Src-PTEN pathway by pharmacological inhibitors represents a new intervention strategy for HER2-negative gastric cancers.
Keywords: HER2-negative gastric cancer, histone lysine demethylase, PHF8, PKCα, PTEN
Abstract
Targeted treatments for advanced gastric cancer (GC) are needed, particularly for HER2-negative GC, which represents the majority of cases (80 to 88%). In this study, in silico analyses of the lysine histone demethylases (KDMs) involved in diverse biological processes and diseases revealed that PHD finger protein 8 (PHF8, KDM7B) was significantly associated with poor clinical outcome in HER2-negative GC. The depletion of PHF8 significantly reduced cancer progression in GC cells and in mouse xenografts. PHF8 regulated genes involved in cell migration/motility based on a microarray analysis. Of note, PHF8 interacted with c-Jun on the promoter of PRKCA which encodes PKCα. The depletion of PHF8 or PKCα greatly up-regulated PTEN expression, which could be rescued by ectopic expression of a PKCα expression vector or an active Src. These suggest that PTEN destabilization occurs mainly via the PKCα-Src axis. GC cells treated with midostaurin or bosutinib significantly suppressed migration in vitro and in zebrafish models. Immunohistochemical analyses of PHF8, PKCα, and PTEN showed a positive correlation between PHF8 and PKCα but negative correlations between PHF8 and PTEN and between PKCα and PTEN. Moreover, high PHF8-PKCα expression was significantly correlated with worse prognosis. Together, our results suggest that the PKCα-Src-PTEN pathway regulated by PHF8/c-Jun is a potential prognostic/therapeutic target in HER2-negative advanced GC.
Gastric cancer (GC) is the second leading cause of death among all malignancies worldwide (1). More than 50% of new GC cases occur in the World Health Organization Western Pacific region (1). Surgical resection remains the gold standard in GC therapy, particularly for early-stage GC (2). However, GC is usually asymptomatic at the early stage and reaches an advanced stage at diagnosis, particularly in regions lacking an implemented screening system (3). According to a metaanalysis by Wagner et al. (4), the prognosis of metastatic GC remains poor, with a median survival of 4.3 mo for patients who receive best supportive care and 11 mo for those who receive combination chemotherapy. The survival of patients with GC treated with chemotherapy for the last two decades has remained steady owing to a dearth of major breakthroughs in the development of new cytotoxic agents (5). A recent trend is to combine targeted therapy with chemotherapy. In a multicenter Trastuzumab for Gastric Cancer (ToGA) study, the median overall survival was 13.8 mo in an anti-HER2 targeted treatment (trastuzumab) group as compared with 11.1 mo in a chemotherapy alone group (6), suggesting that patients with HER2-positive GC (12 to 20%) may benefit from this approach (7). However, no effective targeted treatments are available for advanced HER2-negative cases.
Genome-based molecular characterization provides an avenue for patient stratification, prognosis, and the customization of treatment (3). The Cancer Genome Atlas data for 295 primary GC tissues without chemotherapy and radiotherapy reveal differential patterns of DNA methylation, somatic gene alterations, gene expression, and proteomic events (8). Key genetic alternations are primarily found in oncogenes/tumor suppressor genes, including TP53, KRAS, ARID1A, PIK3CA, ERBB3, PTEN, and HLA-B (8). A global gene-expression profiling analysis and targeted sequencing of 300 GC samples by the Asian Cancer Research Group corroborated common recurrent driver mutations, including mutations in TP53, ARID1A, PIK3CA, KRAS, PTEN, and ERBB3 (9). Apart from mutational patterns revealed from these frontier studies, epigenetic irregularities that contribute to progression and even chromosome remodeling and increased instability cannot be overlooked (10, 11). Among epigenetic regulators (12), histone lysine demethylases (KDMs) have drawn substantial attention, as they catalyze the removal of key methyl groups from histones, which can greatly impact gene expression and the chromatin spatial organization and even rewire tumorigenic programs with increased malignant capability (13). For instance, KDM5A (RBP2), which demethylates the H3K4me3 sites of target genes, including CDKIs, is overexpressed in GC and is involved in GC progression (14). KDM4B is also often amplified in GC; it promotes carcinogenesis in the presence of Helicobacter pylori and up-regulates the expression of IL-8, MMP1, and ITGAV via its H3K9me3/2 demethylating activity (15–17).
To identify KDM member(s) associated with poor clinical outcomes for HER2-negative patients with GC, we conducted in silico Oncomine and Kaplan–Meier plotter analyses. PHF8 (also known as KDM7B) was not only significantly overexpressed in tumors but was also associated with worse overall survival and earlier progression for HER2-negative patients. PHF8 contains a plant homeodomain (PHD) and a catalytic jumonji C domain; it acts as a transcriptional activator to remove epigenetic methyl marks (H3K9me2/1, H3K27me2, and H4K20me1) (18, 19). We demonstrate that PHF8 functions as a coactivator of c-Jun and directly regulates cell migration-related genes including PRKCA that encodes PKCα, which is required for SRC-mediated PTEN degradation. These findings were further substantiated in a zebrafish xenograft model. Together, our results suggest that the PKCα-Src-PTEN pathway regulated by the PHF8-c-Jun complex serves as a crucial factor for advancement to a more malignant phase and is a prognostic/therapeutic target in HER2-negative metastatic GC.
Results
Overexpression of PHF8 Is Associated with Worse Clinical Outcomes in HER2-Negative Gastric Cancer.
We first evaluated the expression of 21 KDM members using Oncomine GC datasets with patient number greater than 50 (www.oncomine.org/) (SI Appendix, Table S1) (20). SI Appendix, Figs. S1A and S2 revealed that KDM1B, KDM2A, and PHF8/KDM7B were significantly up-regulated in tumor specimens as compared to normal tissues (P < 0.05) (21–24). We next evaluated the clinical relevance of the up-regulated KDMs with respect to the endpoints of 5-y overall survival (OS) and first progression (FP) for HER2-negative patients with GC using data retrieved from the Kaplan–Meier plotter (KM plotter) (SI Appendix, Table S2) (25). Higher PHF8/KDM7B expression was significantly associated with worse OS and FP for HER2-negative cases (SI Appendix, Fig. S1 B and C). KDM1B and KDM2A, however, exhibited inconsistent results depending on the probe (SI Appendix, Fig. S1 B and C). These results together indicate that PHF8/KDM7B (hereafter termed PHF8) is a prognostic epigenetic regulator.
Effects of PHF8 in Metastatic GC Cells In Vitro and In Vivo.
PHF8 was noted to have an even significantly higher expression at the metastatic sites of GC in Oncomine analysis (SI Appendix, Fig. S2A) (21). To characterize the biological role of PHF8 in advanced GC, two HER2-negative, MKN28 and MKN45 lines resembling the chromosomally unstable tumors (CIN) subtype (26) were utilized. MKN45 was obtained from the liver metastasis of a patient with a poorly differentiated primary GC of diffuse histology and was characterized as microsatellite unstable tumors (MSI)-low and Epstein-Barr Virus (EBV)-negative. MKN28 was derived from a lymph node metastasis with an intestinal differentiation primary GC and had moderate copy number alterations and a moderate number of genes with single nucleotide variants.
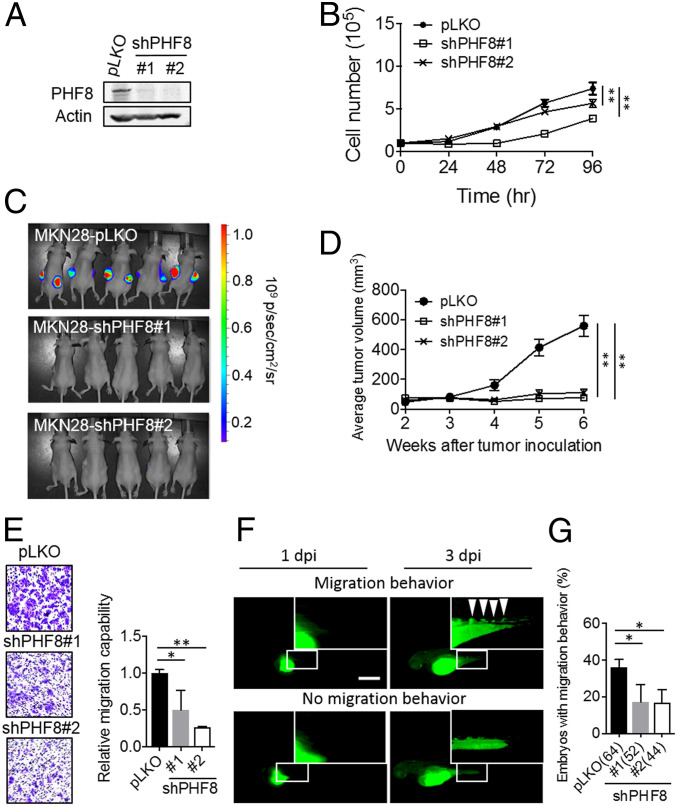
We first prepared control (pLKO) or PHF8-depleted (shPHF8#1 and shPHF8#2) for MKN28 (Fig. 1A) using a lentiviral approach. The depletion of PHF8 significantly reduced the levels of MKN28 cell proliferation (Fig. 1B) as compared with those of control cells. Importantly, tumor growth of MKN28 xenografts was significantly impaired for the PHF8-KD MKN28 cells (Fig. 1 C and D). The depletion of PHF8 also significantly suppressed cell migration (Fig. 1E) as compared with control cells. Similar results also found for MKN45 PHF8-depleted (shPHF8#1 and shPHF8#2) cells as compared with those of control cells (SI Appendix, Fig. S3 A–C).
Fig. 1.
PHF8 is crucial for MKN28 cell proliferation and migration in vitro and in vivo. (A) Analysis of PHF8 in MKN28 infected with lentivirus carrying control pLKO or shPHF8 constructs (#1 or #2), followed by puromycin selection. PHF8 expression was analyzed by Western blotting. Actin was the internal control. (B) The depletion of PHF8 exhibited a reduced degree of cell proliferation in MKN28. (C and D) Luciferase-expressing MKN28 cells (pLKO [n = 5], shPHF8#1 [n = 5], and shPHF8#2 [n = 5]) were grown on nude mice. Images of control and shPHF8 MKN28-luc xenografts were taken after 6 wk of injection (C). Two weeks after implantation, the tumor volumes were measured every week up to 6 wk (D). Data are presented as mean ± SD, with the SD derived from five mice. **P < 0.01 (one-way ANOVA test). (E) The depletion of PHF8 exhibited a reduced degree of cell migration in MKN28 using Transwell cell migration assay. (F and G) Zebrafish migration assays. MKN28 labeled with CFSE (green fluorescence dye) were ectopically injected into the yolk-sac parts of 2-d-old zebrafish embryos. Fluorescence microscopic analysis was conducted at 1 dpi and 3 dpi. Representative fluorescence images of a zebrafish embryo displaying cell dissemination (Upper) or no migration (Lower) at 3 dpi (F). (Scale bar, 100 µm.) The depletion of PHF8 exhibited a significantly reduced proportion of embryos with migration behavior in MKN28 (G). Total number of embryos (pLKO, shPHF8#1, or shPHF8#2) is shown in the bracket. Data were obtained from three independent studies. *P < 0.05 (two-tailed Student’s t test). In B, E, and G data are presented as the average of three replicates ± SD *P < 0.05, **P < 0.01 (two-tailed Student's t test).
We next evaluated migration behavior using an embryonic zebrafish xenotransplantation assay, an efficient in vivo system that utilizes a small number of cells (100 to 200 cells) to accurately monitor cell migratory activity within a couple of days (27). Cells (pLKO vs. shPHF8) were labeled with carboxyfluorescein succinimidyl ester (CFSE), an amine-reactive green fluorescent dye, and injected into zebrafish embryos. Migration activity was monitored at 1 d-postinjection (1 dpi) and 3 dpi by fluorescence microscopy. Both control cells (MKN28, Fig. 1F and MKN45, SI Appendix, Fig. S3D) disseminated to the distal parts at 3 dpi (Upper panel) as compared with cells that remained in the embryo. The quantification of embryos harboring distal tumor foci revealed a significantly higher degree of metastatic activity for the pLKO group (36.1%) than for the shPFH8 group (shPHF8#1, 17.1%; shPHF8#2, 16.7%) (Fig. 1G). Likewise, there was a significantly lower degree of migration in the PHF8-knockdown MKN45 group than in the control group (pLKO, 31.3%; shPHF8#1, 8.9%; and shPHF8#2, 11.8%) (SI Appendix, Fig. S3E). These results together suggest that PHF8 is crucial for tumor growth and migration.
PHF8 Promotes GC Progression by Regulating PKCα and ICAM-1.
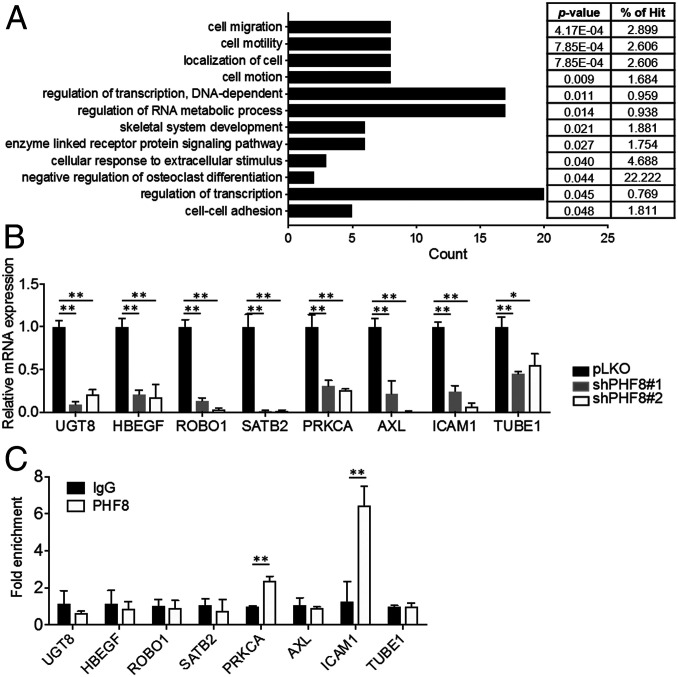
To characterize the molecular mechanisms by which PHF8 contributes to GC progression, we performed a comparative microarray analysis of pLKO and shPHF8 MKN28 cells (GSE117980). DAVID functional annotation indicated that genes that were down-regulated (less than or equal to twofold; n = 150) in shPHF8 cells were primarily involved in cell migration (n = 8, P = 0.00041) and cell motility (n = 8, P = 0.00079) (https://david.ncifcrf.gov/) (Fig. 2A). qRT-PCR confirmed that the eight genes associated with both cell migration and motility (UGT8, HBEGF, ROBO1, STATB2, PRKCA, AXL, ICAM1, and TUBE1) had significantly lower expression levels in two independent shPHF8 cell lines than in pLKO cells (Fig. 2B).
Fig. 2.
PHF8 regulates the expression of genes involved in cell migration and cell motility. (A) DAVID functional annotation analysis of the genes with twofold or less alterations in PHF8-knockdown MKN28 in a microarray analysis. (B) qRT-PCR analysis of down-regulated genes in the cell migration/cell motility/cell motion category. The data were normalized by GAPDH mRNA levels. (C) ChIP analysis of PHF8 occupancy on the promoter region of genes involved in cell migration category in MKN28 using IgG or anti-PHF8 antibodies. (B and C) Data are presented as the average of three replicates ± SD *P < 0.05, **P < 0.01 (two-tailed Student's t test).
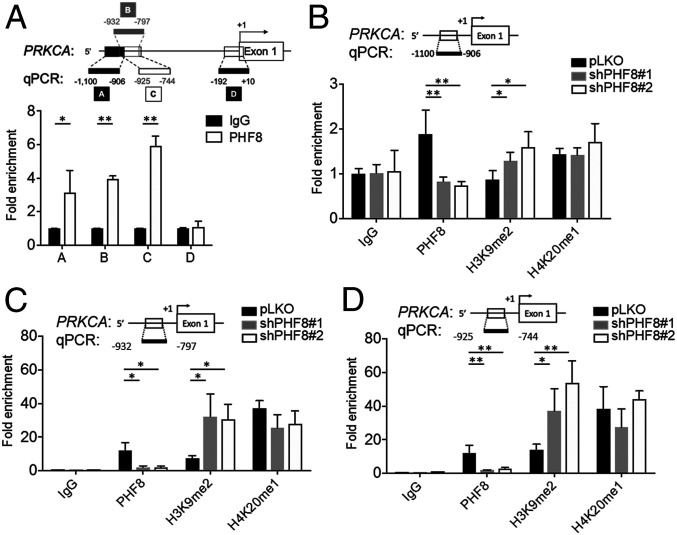
We next evaluated whether PHF8 was directly involved in regulating the expression of the genes identified in the microarray analysis. In a chromatin immunoprecipitation (ChIP) analysis, there was a significantly higher signal of PHF8 than of IgG on the promoter regions of PRKCA and ICAM-1 in MKN28 (Fig. 2C). An additional ChIP analysis with four sets of primers designed across the promoter region of PRKCA in MKN28 revealed positive PHF8-binding signals between −1,100-bp and −744-bp regions, particularly at the site of −925 to −744 bp (Fig. 3A). We further compared the ChIP signals of H3K9me2 and H4K20me1 on the PRKCA locus in cells without or with depletion of PHF8.
Fig. 3.
Analysis of PHF8 occupancy in the promoter region of PRKCA. (A) ChIP analysis of PHF8 enrichment in the promoter region of PRKCA with four sets of primers as indicated. IgG as a control. (B−D) Fold enrichment of PHF8, H3K9me2, and H4K20me1 on the PRKCA promoter in pLKO and shPHF8 (#1 and #2) MKN28 across three regions [−1,100 to −906 (B), −932 to −797 (C), and −925 to −744 (D)]. In A−D, data are presented as the average of three replicates ± SD *P < 0.05, **P < 0.01 (two-tailed Student's t test).
As shown in Fig. 3 B–D, the H3K9me2 signal but not the H4K20me1 one was significantly increased in two independent shPHF8 lines, suggesting that PHF8 might regulate the expression of PRKCA by erasing the repressive H3K9me2 mark on the PRKCA locus.
To substantiate that PRKCA acts as a downstream target of PHF8, we ectopically expressed PKCα in each of the two shPHF8 lines (MKN28, SI Appendix, Fig. S4A and MKN45, SI Appendix, Fig. S5A). Interestingly, the ectopic expression of PKCα in shPHF8 significantly restored the levels of cell proliferation (MKN28, SI Appendix, Fig. S4B and MKN45, SI Appendix, Fig. S5B) and migration (MKN28, SI Appendix, Fig. S4C and MKN45, SI Appendix, Fig. S5C) as compared to those of the pLKO line. Collectively, these data suggest that PHF8 displayed H3K9me2 demethylation activity to up-regulate the expression of PRKCA involved in cell proliferation and migration.
We also evaluated whether PHF8 directly regulated the expression of ICAM-1. SI Appendix, Fig. S6A shows that the depletion of PHF8 reduced the expression of ICAM-1 mRNA in MKN45, consistent with that observed in MKN28 (Fig. 2B). Moreover, we have detected PHF8’s signal in the promoter region of ICAM-1 using a ChIP analysis (SI Appendix, Fig. S6B). The depletion of ICAM-1 led to a decreased degree of migratory effect as compared to the control (SI Appendix, Fig. S6C), indicating that PHF8-regulated expression of ICAM-1 might also contribute to GC progression.
PHF8 Interacts with c-Jun and They Are Corecruited to the PRKCA Locus.
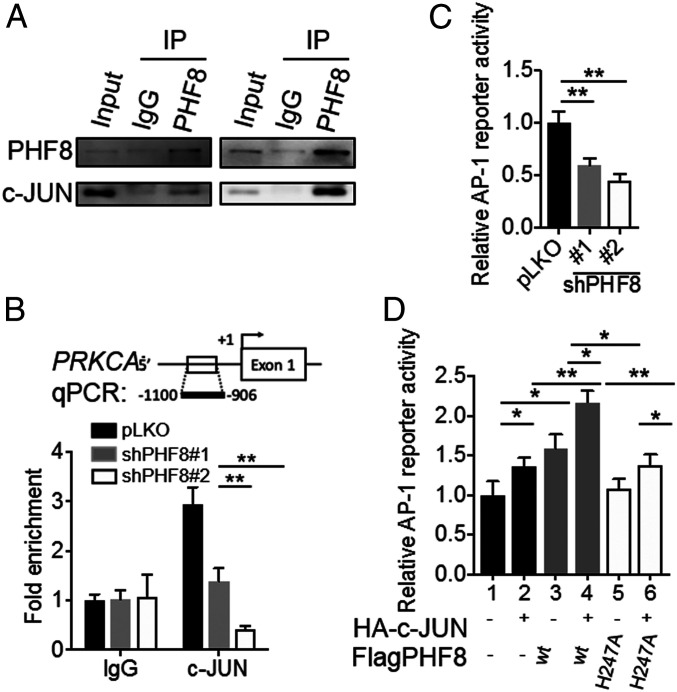
We next identified potential transcription factors that interact with PHF8 using the University of California Santa Cruz Genome Browser (28). We found that c-Jun, a component of the AP-1 transcription factor, shows positive binding peaks in the promoter region of PRKCA in two ChIP-Seq datasets (ChIP-Seq from A549 (ENCLB202COI) (Ab: PHF8): GSM2700325; ChIP-Seq from A549 (ENCLB403GIO) (Ab: c-Jun): GSM2437720). Immunoprecipitation (IP) of lysates from MKN28 or MKN45 using either anti-PHF8 or anti-c-Jun antibodies indeed revealed that endogenous PHF8 is associated with c-Jun (MKN28, Fig. 4A and MKN45, SI Appendix, Fig. S5D). We further determined the crucial region of PHF8 that interacts with c-Jun. We generated full-length PHF8 and various truncated variants fused with a Flag tag (Flag-tagged full-length [FL], N-terminal [ΔN440], and C-terminal [ΔC589] truncated variants). IP analysis revealed that the C-terminal region of PHF8 (residues 441 to 1,024) is most critical for its interaction with c-Jun (SI Appendix, Fig. S7A). We also performed the reciprocal experiment for c-Jun by generating full-length and truncated c-Jun variants fused to an HA-tag (HA-tag FL, N-terminal [ΔN223], and C-terminal [ΔC108] truncated forms). IP analysis showed that c-Jun ΔN223, but not ΔC108, retained the association with PHF8, indicating that the C-terminal region of c-Jun (residues 224 to 331) is critical for the PHF8–c-Jun interaction (SI Appendix, Fig. S7B).
Fig. 4.
PHF8 interacts with c-JUN and regulates AP-1 reporter activity. (A) Endogenous association between PHF8 and c-Jun. MKN28 cell lysates were used for IP assays with IgG, anti-PHF8, or anti-c-Jun. (B) ChIP analysis of c-Jun enrichment on the PRKCA promoter in PHF8-depleted cells. (C) The AP-1 reporter activity of cells (pLKO, shPHF8#1, or shPHF8#2) cotransfected with an AP-1 reporter plasmid and an internal β-galactosidase control plasmid. (D) The AP-1 reporter activity of MKN28 cotransfected with vectors [Flag, wild-type PHF8(WT), or inactive PHF8(H247A), and HA, or HA-c-JUN, plus a β-galactosidase internal control] as indicated. In B−D, data are presented as the average of three replicates ± SD *P < 0.05, **P < 0.01 (two-tailed Student’s t test).
To support the notion that PHF8 acted as a coactivator of c-Jun and regulated the expression of PRKCA, a ChIP analysis was conducted for pLKO and shPHF8 cells using anti-c-Jun and IgG for comparison. Statistically significant enrichment of c-Jun was detected in pLKO cells at the PRKCA locus (MKN28, Fig. 4B and MKN45, SI Appendix, Fig. S5E). Of note, a c-Jun/AP-1 binding site was found in the promoter region of PRKCA based on the ChIP-Seq data (Gene Expression Omnibus [GEO] accession no. GSM2437720). Using the AP-1 reporter activity assay, the depletion of PHF8 significantly reduced the transactivation of AP-1 reporter activity (MKN28, Fig. 4C and MKN45, SI Appendix, Fig. S5F). We next examined whether overexpressing PHF8 and/or c-Jun stimulates AP-1 transcriptional activity. Fig. 4D shows that overexpressing PHF8 or c-Jun alone significantly enhanced the level of AP-1 transactivation. Remarkably, there was an even significantly pronounced increase when overexpressing PHF8 with c-Jun (Fig. 4D). Interestingly, this enhancement was not seen when overexpressing an inactive mutant, PHF8(H247A) (Fig. 4D) (29). These results thus suggest that PHF8 modulates the expression of PRKCA in conjunction with c-Jun through PHF8’s demethylase activity.
The PHF8-PKCa Axis Regulates PTEN Destabilization via Src.
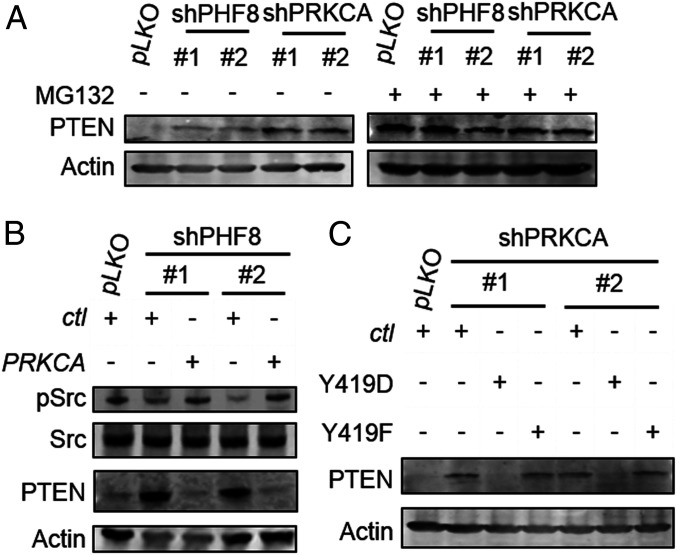
PRKCA encodes PKCα, a serine/threonine protein kinase that serves as a signaling molecule activated by Ca2+ and phospholipids (30); accordingly, we explored the signaling pathway mediated by the PHF8-PKCα axis. We used a Western microarray analysis to evaluate the patterns of 96 antibodies simultaneously in pLKO, shPHF8, and shPRKCA cells. SI Appendix, Fig. S8 shows that two pathways were altered substantially: PI3K and MAPK. In particular, the tumor suppressor PTEN/pPTEN had the highest signal intensities in both shPHF8 and shPRKCA cells. Western blotting analyses of pLKO and shPHF8 lines confirmed that PTEN was clearly up-regulated by the depletion of PHF8 (MKN28, Fig. 5A and MKN45, SI Appendix, Fig. S9A). Moreover, the level of PTEN was substantially higher in each of the two shPRKCA lines than in pLKO, indicating that PHF8-PKCα signaling led to PTEN destabilization (MKN28, Fig. 5A and MKN45, SI Appendix, Fig. S9A). We thus asked whether PTEN was regulated at the level of transcriptional silencing or translational modification (31, 32) in the context of the PHF8-PKCα axis. We first evaluated whether treatment with MG132, a proteasome inhibitor could restore the PTEN signal, since PTEN is quite susceptible to proteasomal degradation, particularly upon posttranslational modifications (31, 32). The protein level of PTEN was indeed rescued in MG132-treated pLKO cells (MKN28, Fig. 5B and MKN45, SI Appendix, Fig. S9A).
Fig. 5.
The PHF8-PKCα axis regulates PTEN destabilization by SRC activation. (A) Immunoblotting analysis of PTEN expression in shPHF8 (#1 and #2) and shPRKCA (#1 and #2) MKN28 with or without MG132 treatment. (B) Abundance of Src and pSrc (Y419) in pLKO or shPHF8 (#1 or #2) transfected with a control (ctl) or a PRKCA-expressing vector. (C) Detection of PTEN in shPRKCA (#1 and #2) cells transfected with a kinase-active Src (Y419D) or a kinase-dead Src (Y419F) vector.
We next asked whether Src kinase serves as a downstream effector of PKCα based on two reports: 1) Tatin et al., who clearly define the PKCα-Src-CDC42 signaling cascade (33); and 2) Lu et al., who show that Src activation promotes the degradation of PTEN (34). Analyses of the abundance of Src and activated Src (pY419) in pLKO and two shPHF8 lines, interestingly, revealed that there was indeed a reduced level of activated Src (pY419) in PHF8-depleted cells (MKN28, Fig. 5B and MKN45, SI Appendix, Fig. S9B). Furthermore, complementation with PKCα using a PRKα-expressing vector in each of the two shPHF8 lines restored the level of activated Src (MKN28, Fig. 5B and MKN45, SI Appendix, Fig. S9B), supporting the notion that Src served as a downstream effector of PKCα. We further generated a kinase-dead Src variant (Y419F) and a constitutively active Src variant (Y419D). The introduction of Y419D, but not Y419F, greatly diminished the abundance of PTEN (MKN28, Fig. 5C and MKN45, SI Appendix, Fig. S9C). Together, these results suggested that PHF8 negatively regulates PTEN destabilization via the PKCα-Src-induced signaling pathway.
Targeting the PKCα-Src Pathway in Metastatic GC.
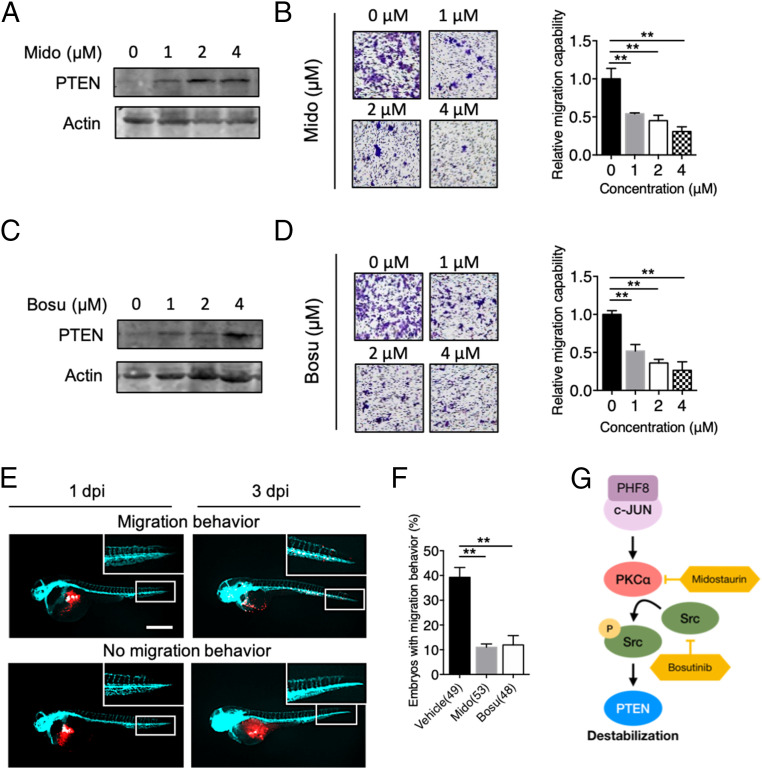
We next tested whether targeting the PHF8-PKCα-Src-PTEN axis using pharmacological inhibitors curbs GC metastasis. To do so, we utilized midostaurin (35, 36), a PKCα inhibitor, and bosutinib (37, 38), a Src inhibitor (FDA-approved since 2017). Treatment with midostaurin indeed led to an elevated level of PTEN in a dose-dependent manner in MKN28 (Fig. 6A) and in MKN45 (SI Appendix, Fig. S10A). Importantly, the degree of cell migration was also significantly suppressed (MKN28, Fig. 6B and MKN45, SI Appendix, Fig. S10B). Likewise, the inhibition of Src by bosutinib resulted in an increased level of PTEN expression in both MKN28(Fig. 6C) and MKN45 (SI Appendix, Fig. S10C). As such, there was also a reduced degree of cell migration (MKN28, Fig. 6D and MKN45, SI Appendix, Fig. S10D).
Fig. 6.
Pharmacological inhibition of PHF8-PKCα-Src-PTEN blocks GC progression in vitro and in vivo. (A) Analysis of PTEN protein level in MKN28 treated with 0, 1, 2, or 4 μM of midostaurin (Mido). (B) Detection of migration activity of MKN28 treated with Mido by using Transwell migration assay. (C) Analysis of PTEN protein level in MKN28 treated with 0, 1, 2, or 4 μM of bosutinib (Bosu). (D) Detection of migration activity of MKN28 treated with Bosu by using Transwell migration assay. In B and D, data are presented as the average of three replicates ± SD **P < 0.01 (two-tailed Student’s t test). (E and F) Zebrafish xenotransplantation assays using Mido or Bosu. MKN28 labeled with CM-Dil (red fluorescence dye in the membrane) were ectopically injected into the yolk-sac parts of 2-d-old Tg(fli1:EGFP) (fish with green fluorescence in blood vessels) zebrafish embryos, followed by immersion in solutions containing 1 μM midostaurin or bosutinib (a sublethal dose) at 1 dpi. Fluorescence microscopic analysis was conducted at 1 dpi and 3 dpi. Representative fluorescence images of a zebrafish embryo displaying cell dissemination (Upper) or no migration (Lower) at 3 dpi (E). Cyan, blood vessel. (Scale bar, 100 µm.) Quantification of embryos with migration behavior in vehicle or drug-treated groups (F). Total number of embryos used in vehicle, Mido, and Bosu is shown in the bracket. Data were obtained from three independent studies. **P < 0.01 (two-tailed Student’s t test). (G) A schematic diagram shows that the PHF8-c-Jun complex contributes to GC progression through activation of PKCα-Src-dependent signaling to suppress PTEN. Targeting the PHF8-c-Jun-PKCα-Src-PTEN axis represents a prognostic/therapeutic target in advanced GC.
We next corroborated this finding using a zebrafish xenotransplantation model. Cells labeled with Vybrant CM-DiI (CM-Dil) (red fluorescence dye) were injected into the embryos of Tg(fli1:EGFP) (fish with green fluorescence in blood vessels), followed by immersion in solutions containing 1 μM midostaurin or bosutinib (a sublethal dose) at 1 dpi. Implantation of MKN28 or MKN45 cells in embryos often resulted in cell dissemination; cells clearly metastasized to distal parts of the body (MKN28, Fig. 6E and MKN45, SI Appendix, Fig. S10E). A large proportion of MKN28-injected embryos treated by midostaurin or bosutinib had reduced levels of widespread dissemination and invasion (39.3% in the vehicle group, 11% in the midostaurin group, 11.95% in the bosutinib group) (Fig. 6F). MKN45 injection also resulted in a significantly higher levels of metastatic activity in the vehicle group (34.25%) than in the two inhibitor groups (13.88% in the midostaurin group and 18.08% in the bosutinib group) (SI Appendix, Fig. S10F).
We next tested whether midostaurin or bosutinib impedes tumor growth in vivo by using a MKN28 xenograft model. SI Appendix, Fig. S11 shows that both drugs significantly impaired tumor growth as compared to vehicle group. Collectively, these results suggested that the inhibition of PKCα or Src is an effective strategy to curb tumor progression in vivo.
Immunohistochemical Analyses of PHF8, PKCα, and PTEN in GC Subjects.
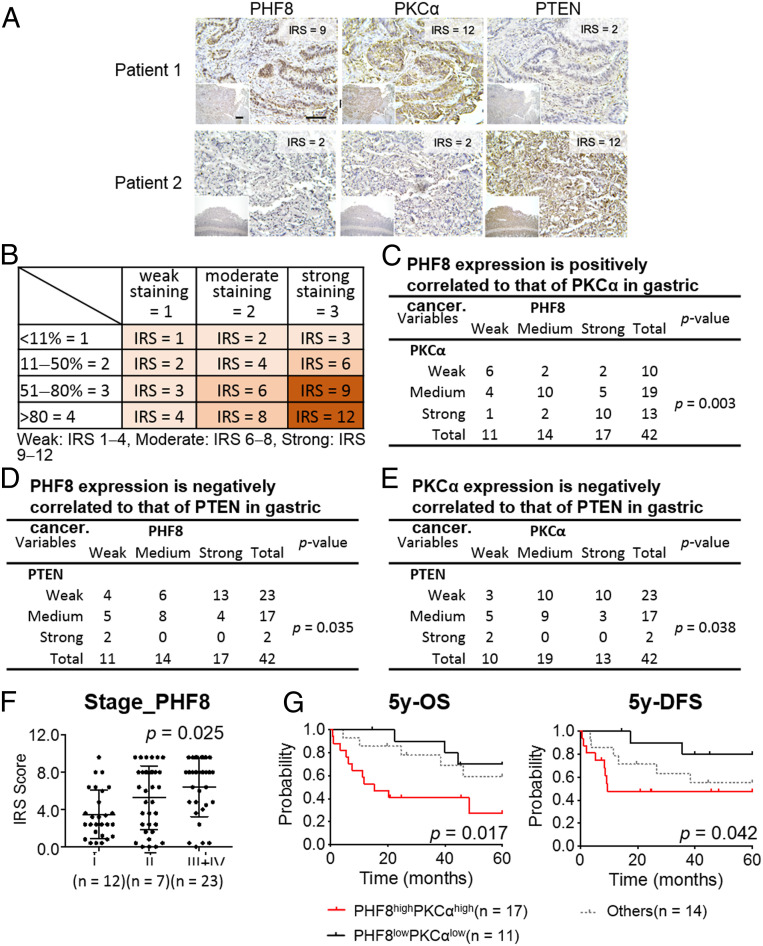
Given the identified PHF8-PKCα-PTEN axis that contributes to GC progression using two GC cell models, we further evaluated the clinical relevance of these markers. We conducted an immunohistochemistry (IHC) analysis of PHF8, PKCα, and PTEN for patients with GC. Three consecutive paraffin-embedded human GC biopsies (n = 42) were obtained from Chang Gung Memorial Hospital (CGMH), Taoyuan, Taiwan (Fig. 7A). Additionally, the tissue array ST1505 (US Biomax; n = 50) was characterized (SI Appendix, Fig. S12A). IHC results were scored based on two parameters: the intensity grade (score: 1to 3) and the proportion of positive tumor cells (score: 1 to 4). The immunoreactive score (IRS) was obtained by multiplying the intensity grade by the positive proportion score (Fig. 7B). Remarkably, there were significant correlations between these markers (Fig. 7 C–E and SI Appendix, Fig. S12 B–D), i.e., a positive correlation between PHF8 and PKCα (CGMH, P = 0.003; tissue assay ST1505, P = 0.037) and negative correlations between PHF8 and PTEN (CGMH, P = 0.035; tissue assay ST1505, P = 0.019) and between PKCα and PTEN (CGMH, P = 0.038; tissue assay ST1505, P = 0.018). Of note, PHF8 abundance was significantly positively correlated with tumor stage (CGMH, P = 0.025; tissue assay ST1505, P = 0.046) (Fig. 7F and SI Appendix, Fig. S12E). We then evaluated the clinical outcome in CGMH patients with available follow-up clinical data (n = 42). Interestingly, the PHF8highPKCαhigh group (n = 20) was significantly associated with a poor 5-y OS (P = 0.017) and 5-y disease-free survival (DFS) (P = 0.042) as compared with the PHF8lowPKCαlow group (n = 12) (Fig. 7G).
Fig. 7.
Clinical relevance of PHF8, PKCα, and PTEN in 42 GC subjects obtained from CGMH. (A) Representative image of immunohistochemical profiles. PHF8, PKCα, and PTEN were immunostained for each of gastric tissue specimens (n = 42). (Scale bar, 100 μm.) (B) IRS score of GC samples. (C−E) The correlation of IHC signals for two-group comparisons: PHF8 and PKCα (C), PHF8 and PTEN (D), PKCα and PTEN (E). Statistical significance was evaluated using the χ2 test. (F) PHF8 expression is significantly correlated with tumor stage in patients with GC. Statistical calculation is conducted using one-way ANOVA analysis. (G) Five-year OS and 5-y DFS analysis according to the level of PHF8 and PKCα expression in patients with GC (n = 42). High, IRS ≥ 8; low, IRS ≤ 6. Statistical significance (PHF8highPKCαhigh vs. PHF8lowPKCαlow) was determined by log-rank test.
Discussion
Although the comprehensive profiling of altered driver/passenger genes across GC genomes offers a classification scheme for the identification of potential therapeutic targets and predictive/prognostic markers (8, 9), the dysregulated epigenetic landscape of GC remains largely elusive. In this study, a large-scale in silico analysis of histone lysine demethylases revealed that PHF8 is a potential oncogenic KDM associated with poor prognosis in HER2-negative GC. Our analysis of two metastatic GC cell lines indeed revealed that the depletion of PHF8 significantly reduced cell proliferation and migration in vitro and in vivo. Interestingly, PHF8 is often overexpressed in several malignancies, including prostate cancer (39), esophagus cancer (40), lung cancer (41), laryngeal and hypopharyngeal squamous cell carcinoma (42), acute lymphoblastic leukemia (43), and GC (44). These results together suggest that PHF8 serves as a potential oncogenic epigenetic regulator.
PHF8 has previously been identified as a causative gene of X-linked mental retardation due to the loss-of-activity mutation F279S (45–47). PHF8 regulates the expression of cytoskeleton-related genes and neurite elongation (48). Consistent with these previous results, we show that PHF8 regulates the expression of genes related to cell migration/motility. Above all, we demonstrate that PHF8 interacts with c-Jun and functions as a coactivator to directly regulate a crucial motility-related gene PRKCA, possibly by H3K9me2 demethylation activity. c-Jun, a major component of the AP-1 complex involved in invasiveness, is triggered by diverse stimuli, including growth factors, cytokines, and extracellular stresses (49, 50). In GC, c-Jun directly regulates FOXK1 expression to promote cancer progression (51). We have also shown that c-Jun contributes to the up-regulation of IL-8 via JNK signaling under H. pylori challenge (16). Together, these results suggest that the PHF8-c-Jun axis plays a crucial role in stress-induced chromatin remodeling and that c-Jun functions as an important determinant of GC progression.
Strikingly, we show that the abundance of PTEN, a key tumor suppressor with frequent gene mutations or deletions in tumors (52), is negatively regulated by the PHF8-c-Jun-PKCα pathway. The dysfunction of PTEN, either by mutations, deletions, or nongenomic silencing, causes hyperactivated PI3K-AKT signaling, a crucial event that drives carcinogenesis in many cancers (31). The frequency of genetic alterations in PTEN varies among geographic regions (Asian: 2.52% vs. Caucasian: 9.05%, P = 0.008) (53). The loss or reduced expression of PTEN occurs at the stage of dysplasia and is highly frequent in GC (54). PTEN loss is more frequent in advanced than in early-stage GCs (55). Here, we further show that PKCα suppresses PTEN by Src activation. Our results provide an epigenetic silencing mechanism by the PHF8-c-Jun-PKCα-Src axis that leads to a PTEN deficiency and support the application of PHF8/PKCα as a biomarker for a subpopulation of PI3K-driven tumors.
The inhibition of PKCα using midostaurin (IC50: 22 nM) (35), a multikinase inhibitor, significantly suppressed migratory activity in vitro and in vivo. Of note, midostaurin was approved for the treatment of adult patients with aggressive systemic mastocytosis, systemic mastocytosis with associated hematological neoplasm, or mast cell leukemia in 2017 (36). We also show that the pharmacological inhibition of Src using bosutinib (IC50: 1.2 nM) (37), a Src tyrosine kinase inhibitor approved for chronic myeloid leukemia (38, 56), effectively blocks migratory activity in vitro and in vivo. Our results provide proof-of-concept evidence that targeting PKCα/Src is a feasible approach for PI3K-driven GC harboring a PTEN deficiency via the PHF8-PKCα pathway.
Interestingly, two-thirds of those carrying high-IRS PHF8-PKCα subjects (n = 42) exhibited low or no expression of PTEN (n = 28). Notably, the survival and recurrence-free rates were significantly poorer for this subgroup. We have previously participated in the 2012 international clinical (CLASSIC) trial to evaluate the survival benefit of adjuvant chemotherapy for GC after curative D2 gastrectomy (57). After a careful follow-up assessment, the survival rate was superior in the adjuvant capecitabine plus oxaliplatin group than in the placebo group (the 3- and 5-y OS improved by 10%). Given the results of this study, the patient population harboring PHF8/PKCα may benefit from PI3K pathway inhibitors as a combined chemotherapy; this should be a focus of further research.
In conclusion, we provide evidence for a role of the PHF8-PKCα-Src axis in GC driven by hyperactivated PI3K signaling via PTEN deficiency. Our results suggest that PHF8/PKCα is a potential prognostic marker and can be used to identify patient subsets who may benefit from therapies targeting PKCα and Src. In support of this proposal, midostaurin and bosutinib inhibited tumor migration in vitro and in vivo. More comprehensive profiling analyses, including synthetic lethality experiments, are needed.
Materials and Methods
MKN28 and MKN45 were cultured in RPMI medium 1640 supplied with 10% fetal bovine serum at 37 °C in 5% CO2. PHF8 knockdown MKN28 and MKN45 were generated based on lentivirus-mediated shPHF8s. Detailed information is provided in SI Appendix, SI Materials and Methods. ChIP assay was performed with the use of the Magna ChIP assay kit (Millipore). Enrichment of specific gene occupancy onto promoter was detected by qRT-PCR as described in SI Appendix, SI Materials and Methods and detailed protocols for the reagents, cell proliferation assay, migration assays, immunoblot, immunoprecipitation assay, luciferase activity assay, and IHC analysis are given in detailed descriptions in SI Appendix.
Supplementary Material
Acknowledgments
A special thanks to Dr. Pien-Chien Huang and Dr. Ru-Chih Huang (Johns Hopkins University) for their valuable discussions and critical reading of the manuscript. We thank Dr. Guan-Ying Tseng and Dr. Hsiao-Bei Yang (Ton-Yen General Hospital, Hsinchu, Taiwan) for the pilot samples. We also thank Taiwan Bioinformatics Institute Core Facility for assistance on using Oncomine (National Core Facility Program for Biotechnology). This work was supported by Ministry of Science and Technology (MOST), National Health Research Institutes (NHRI), Chang Gung Memorial Hospital (CGMH), and National Tsing Hua University (NTHU) in Taiwan (MOST-107-2321-B-007-005, MOST-107-2314-B-007-001, MOST-108-2321-B-007-004, MOST-109-2327-B-007-001, MOST 108-2319-B-400-001, NHRI-EX107-10517BI, NHRI-EX109-10917BI, CORPG3I0091, CORPG3J0351, and CGMH-NTHU105N717CV7).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919766117/-/DCSupplemental.
Data Availability.
Global gene expression analyses performed in MKN28 pLKO vs. shPHF8#1 have been deposited in the GEO database under accession number GSE117980.
References
- 1.Sitarz R. et al., Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 10, 239–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waddell T. et al., Gastric cancer: ESMO-ESSO-ESTRO clinical practice guidelines for diagnosis, treatment and follow-up. Eur. J. Surg. Oncol. 40, 584–591 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H., Gastric cancer. Lancet 388, 2654–2664 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Wagner A. D. et al., Chemotherapy for advanced gastric cancer. Cochrane Database Syst. Rev., CD004064 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Bernards N. et al., No improvement in median survival for patients with metastatic gastric cancer despite increased use of chemotherapy. Ann. Oncol. 24, 3056–3060 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Bang Y. J. et al.; ToGA Trial Investigators , Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 376, 687–697 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Van Cutsem E. et al., HER2 screening data from ToGA: Targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer 18, 476–484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network , Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristescu R. et al., Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 21, 449–456 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Flavahan W. A., Gaskell E., Bernstein B. E., Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg A. P., Koldobskiy M. A., Göndör A., Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaminska K. et al., Prognostic and predictive epigenetic biomarkers in oncology. Mol. Diagn. Ther. 23, 83–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varier R. A., Timmers H. T., Histone lysine methylation and demethylation pathways in cancer. Biochim. Biophys. Acta 1815, 75–89 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Zeng J. et al., The histone demethylase RBP2 Is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology 138, 981–992 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Han F. et al., JMJD2B is required for Helicobacter pylori-induced gastric carcinogenesis via regulating COX-2 expression. Oncotarget 7, 38626–38637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M. C. et al., KDM4B is a coactivator of c-Jun and involved in gastric carcinogenesis. Cell Death Dis. 10, 68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuai W. X. et al., Interleukin-8 associates with adhesion, migration, invasion and chemosensitivity of human gastric cancer cells. World J. Gastroenterol. 18, 979–985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortschegger K., Shiekhattar R., Plant homeodomain fingers form a helping hand for transcription. Epigenetics 6, 4–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosammaparast N., Shi Y., Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Rhodes D. R. et al., Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X. et al., Data from “Variation in gene expression patterns in human gastric cancers ” (Chen Gastric 2003). Mol. Biol. Cell 14, 3208–3215, https://www.oncomine.org/ (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Errico M. et al., Data from “Genome-wide expression profile of sporadic gastric cancers with microsatellite instability” (D’Errico Gastric 2009). Eur. J. Cancer 45, 461–469, https://www.oncomine.org/ (2009). [DOI] [PubMed] [Google Scholar]
- 23.Cui J. et al., Data from “An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer” (Cui Gastric 2011). Nucleic Acids Res. 39, 1197–1207, https://www.oncomine.org/ (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho J. Y. et al., Data from “Gene expression signature-based prognostic risk score in gastric cancer” (Cho Gastric 2011). Clin. Cancer Res. 17, 1850–1857, https://www.oncomine.org/ (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szász A. M. et al., Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7, 49322–49333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motoyama T., Hojo H., Watanabe H., Comparison of seven cell lines derived from human gastric carcinomas. Acta Pathol. Jpn. 36, 65–83 (1986). [DOI] [PubMed] [Google Scholar]
- 27.Letrado P., de Miguel I., Lamberto I., Díez-Martínez R., Oyarzabal J., Zebrafish: Speeding up the cancer drug discovery process. Cancer Res. 78, 6048–6058 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Kent W. J. et al., The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Z. et al., PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 20, 794–801 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Rosse C. et al., PKC and the control of localized signal dynamics. Nat. Rev. Mol. Cell Biol. 11, 103–112 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Dillon L. M., Miller T. W., Therapeutic targeting of cancers with loss of PTEN function. Curr. Drug Targets 15, 65–79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M. S. et al., PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat. Commun. 6, 7769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatin F., Varon C., Génot E., Moreau V., A signalling cascade involving PKC, Src and Cdc42 regulates podosome assembly in cultured endothelial cells in response to phorbol ester. J. Cell Sci. 119, 769–781 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Lu Y. et al., Src family protein-tyrosine kinases alter the function of PTEN to regulate phosphatidylinositol 3-kinase/AKT cascades. J. Biol. Chem. 278, 40057–40066 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Fabbro D. et al., PKC412–a protein kinase inhibitor with a broad therapeutic potential. Anticancer Drug Des. 15, 17–28 (2000). [PubMed] [Google Scholar]
- 36.Kasamon Y. L. et al., FDA approval summary: Midostaurin for the treatment of advanced systemic mastocytosis. Oncologist 23, 1511–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boschelli D. H. et al., Optimization of 4-phenylamino-3-quinolinecarbonitriles as potent inhibitors of Src kinase activity. J. Med. Chem. 44, 3965–3977 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Vultur A. et al., SKI-606 (bosutinib), a novel Src kinase inhibitor, suppresses migration and invasion of human breast cancer cells. Mol. Cancer Ther. 7, 1185–1194 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Q. et al., The histone demethylase PHF8 promotes prostate cancer cell growth by activating the oncomiR miR-125b. OncoTargets Ther. 8, 1979–1988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun X. et al., Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS One 8, e77353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Y., Pan X., Zhao H., The histone demethylase PHF8 is an oncogenic protein in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 451, 119–125 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Zhu G. et al., Elevated expression of histone demethylase PHF8 associates with adverse prognosis in patients of laryngeal and hypopharyngeal squamous cell carcinoma. Epigenomics 7, 143–153 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Fu Y. et al., The histone demethylase PHF8 promotes adult acute lymphoblastic leukemia through interaction with the MEK/ERK signaling pathway. Biochem. Biophys. Res. Commun. 496, 981–987 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Li S. et al., Histone demethylase PHF8 promotes progression and metastasis of gastric cancer. Am. J. Cancer Res. 7, 448–461 (2017). [PMC free article] [PubMed] [Google Scholar]
- 45.Laumonnier F. et al., Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 42, 780–786 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi H. H. et al., Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleine-Kohlbrecher D. et al., A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38, 165–178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asensio-Juan E., Gallego C., Martínez-Balbás M. A., The histone demethylase PHF8 is essential for cytoskeleton dynamics. Nucleic Acids Res. 40, 9429–9440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mechta-Grigoriou F., Gerald D., Yaniv M., The mammalian jun proteins: Redundancy and specificity. Oncogene 20, 2378–2389 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Ozanne B. W., Spence H. J., McGarry L. C., Hennigan R. F., Transcription factors control invasion: AP-1 the first among equals. Oncogene 26, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Peng Y. et al., Direct regulation of FOXK1 by C-jun promotes proliferation, invasion and metastasis in gastric cancer cells. Cell Death Dis. 7, e2480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey M. H. et al., Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia F. et al., Discordance of somatic mutations between asian and caucasian patient populations with gastric cancer. Mol. Diagn. Ther. 21, 179–185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L. et al., PTEN encoding product: A marker for tumorigenesis and progression of gastric carcinoma. World J. Gastroenterol. 9, 35–39 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu X. et al., Loss and reduced expression of PTEN correlate with advanced-stage gastric carcinoma. Exp. Ther. Med. 5, 57–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isfort S., Brümmendorf T. H., Bosutinib in chronic myeloid leukemia: Patient selection and perspectives. J. Blood Med. 9, 43–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bang Y. J. et al.; CLASSIC trial investigators , Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): A phase 3 open-label, randomised controlled trial. Lancet 379, 315–321 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Global gene expression analyses performed in MKN28 pLKO vs. shPHF8#1 have been deposited in the GEO database under accession number GSE117980.