Significance
The OAS–RNase L system is an innate immunity pathway activated by viral infection. Viral dsRNA stimulates OAS enzymes to produce short 2′,5′-oligoadenylates (2-5A) that activate RNase L, resulting in cleavage of single-stranded (ss) RNA. We discovered a small-molecule inhibitor of RNase L that rescues the toxic phenotype of cells deficient in the dsRNA-editing enzyme ADAR1. ADAR1 destabilizes dsRNA to prevent OAS activity. ADAR1 mutations are responsible for a subset of cases of Aicardi-Goutières syndrome (AGS), a severe neurodevelopmental and inflammatory genetic disease of children with no effective medical therapy. We posit that an RNase L inhibitor may have utility against cases of AGS in which RNase L is activated and other indications where overactivation of RNase L is harmful.
Keywords: RNase L, valoneic acid dilactone, ADAR1 deficiency, viral infection, small molecule inhibitor
Abstract
The oligoadenylate synthetase (OAS)–RNase L system is an IFN-inducible antiviral pathway activated by viral infection. Viral double-stranded (ds) RNA activates OAS isoforms that synthesize the second messenger 2-5A, which binds and activates the pseudokinase-endoribonuclease RNase L. In cells, OAS activation is tamped down by ADAR1, an adenosine deaminase that destabilizes dsRNA. Mutation of ADAR1 is one cause of Aicardi-Goutières syndrome (AGS), an interferonopathy in children. ADAR1 deficiency in human cells can lead to RNase L activation and subsequent cell death. To evaluate RNase L as a possible therapeutic target for AGS, we sought to identify small-molecule inhibitors of RNase L. A 500-compound library of protein kinase inhibitors was screened for modulators of RNase L activity in vitro. We identified ellagic acid (EA) as a hit with 10-fold higher selectivity against RNase L compared with its nearest paralog, IRE1. SAR analysis identified valoneic acid dilactone (VAL) as a superior inhibitor of RNase L, with 100-fold selectivity over IRE1. Mechanism-of-action analysis indicated that EA and VAL do not bind to the pseudokinase domain of RNase L despite acting as ATP competitive inhibitors of the protein kinase CK2. VAL is nontoxic and functional in cells, although with a 1,000-fold decrease in potency, as measured by RNA cleavage activity in response to treatment with dsRNA activator or by rescue of cell lethality resulting from self dsRNA induced by ADAR1 deficiency. These studies lay the foundation for understanding novel modes of regulating RNase L function using small-molecule inhibitors and avenues of therapeutic potential.
The oligoadenylate synthetase (OAS)–RNase L system is an interferon (IFN)-inducible antiviral pathway in mammalian cells that is activated in the innate immune response to viral infections (1). Upon detection of infection by viruses, cells secrete type I IFNs that bind to the IFNAR1 and IFNAR2 receptor complex present on the surface of the infected cell and the surrounding cells. The JAK-STAT signaling pathway is triggered, causing expression of IFN-stimulated genes (ISGs) that establish an antiviral state (2). OAS proteins, encoded by a subset of ISGs, directly detect viral pathogens by sensing viral double-stranded (ds) RNA (3). dsRNA binding leads to the catalytic activation of OAS isoforms 1 to 3, which utilize ATP to generate the second messenger 2-5A, short oligoadenylates with a 5′-triphosphoryl moiety and unconventional 2′-5′ linkages (1, 4). Polymers with minimal length of three adenylate residues are potent activators of RNase L ribonuclease function (5) through their ability to bind to the N-terminal ankyrin repeat domain of RNase L (6) with subnanomolar affinity (7).
Activation occurs in a manner that induces the parallel back-to-back dimerization of the C-terminal catalytic region of RNase L composed of a pseudokinase domain fused to a ribonuclease domain (8, 9). In conjunction with the constitutive binding of ATP nucleotide to the pseudokinase domain of RNase L, dimerization imposes a productive conformation of the ribonuclease domain by composing composite active sites in trans. The activated RNase L dimer is a metal ion-independent endoribonuclease that indiscriminately cleaves ssRNA preferentially after UU and UA dinucleotide sequence motifs (10, 11), producing RNA fragments with 5′-OH and 2′,3′-cyclic phosphate termini (1). More recently, RNase L was shown to cleave a restricted subset of RNA substrates to arrest protein synthesis (12). Once cleaved by RNase L, target RNAs are further degraded by cellular exonucleases, leading to the inhibition of protein synthesis, disabling the host cell machinery required for viral replication, and elimination of ssRNA viral genomes. Deficiency in the OAS–RNase L pathway leads to a compromised antiviral response in mouse models (13, 14).
A wider role for OAS–RNase L beyond the IFN antiviral state is suggested by human genetic studies implicating RNase L in predisposition toward prostate (15, 16), colorectal (17), and breast (18, 19) cancers. In addition, activation of OAS–RNase L by self dsRNA occurs in the absence of a viral infection when the adenosine deaminase ADAR1 is deficient (20). ADAR1 edits and destabilizes dsRNA (21–23), thereby reducing the activation of OAS by self dsRNA (20). ADAR1 mutations lead to the childhood neurodegenerative and inflammatory disease Aicardi-Goutières syndrome (AGS) in a subset of cases (24). Currently, there is no effective therapy for AGS, but since the genetic ablation of RNase L function can relieve some of the adverse cellular effects caused by the loss of ADAR1 function, it raises the possibility that small-molecule modulators of RNase L could be therapeutically useful in a subset of AGS cases. In addition, RNase L is proinflammatory, and its inhibitors might have utility as antiinflammatory agents (25). These observations highlight the therapeutic potential of a potent small-molecule inhibitor of RNase L.
Small-molecule modulators of RNase L and its closest paralog IRE1 have been discovered that can either potentiate or inhibit ribonuclease activity through a diversity of mechanisms (9, 26, 27). Like RNase L, IRE1 shares a similar domain architecture consisting of an N-terminal sensor domain followed by a catalytic module composed of a protein kinase domain fused to a novel ribonuclease domain. In contrast to RNase L, the N-terminal sensor domain of IRE1 consists of an unrelated globular domain that regulates protein kinase and ribonuclease activities in response to the detection of unfolded proteins in the ER. Two potent inhibitors have been reported for IRE1 with distinct modes of action, including a protein kinase domain binder (28) and a ribonuclease domain binder (27). These molecules provide useful preclinical tools for investigating the therapeutic potential of IRE1 in different disease states.
While effective tool compounds against RNase L are lacking, proof-of-concept studies support the likelihood that RNase L is amenable to small-molecule modulation. For instance, the VEGFR- and PDGFR-specific protein kinase inhibitor sunitinib, used clinically to treat renal cell carcinoma, was recently identified as a cross-reactive inhibitor of RNase L (26). Consistent with its mode of action against VEGFR and PDGFR, it functions against RNase L by competing with ATP for binding to the pseudokinase domain. Sunitinib inhibits the ribonuclease activity of RNase L in vitro with an IC50 of 1.4 μM and the antiviral function of RNase L in cells with an IC50 of 1 μM (26). This is far less potent than the inhibitory activity of sunitinib against VEGFR and PDFGR in vitro (IC50 ∼9 to 10 nM) and in cells (30 to 40 nM) (29), which could limit its utility as a specific biological probe for RNase L.
As a lack of effective tool compounds hampers the investigation of the therapeutic potential of RNase L as a drug target, we set out to identify additional small molecules and scaffolds that may serve as starting points for the development of specific probes. To this end, we performed a biochemical screen against a structurally diverse protein kinase inhibitor library. We expected this screen to yield both activators and inhibitors of RNase L based on our success at identifying both types of modulators against the paralogous kinase-endoribonuclease IRE1 (27, 30, 31). Unexpectedly, we discovered a small molecule that inhibited RNase L ribonuclease activity both in vitro and in cells, but did so without binding the kinase active site. We will further show that the small-molecule inhibitor of RNase L prevented cell death initiated by either exogenously added dsRNA or by accumulation of endogenous self dsRNA resulting from a deficiency in ADAR1.
Results
Screening and Identification of a Small-Molecule Inhibitor of RNase L Ribonuclease Activity.
We screened a 500-compound library composed of a diverse set of known and predicted protein kinase inhibitor scaffolds against the activated form of Sus scrofa (porcine) RNase L. Enzyme activity was monitored in vitro using a real-time fluorescence-based RNA cleavage assay (Fig. 1A). The screen identified ellagic acid (EA), a natural product shown previously to inhibit casein kinase II with 20 nM potency (32, 33), as a top hit (Fig. 1 A and B). We confirmed EA as an inhibitor of RNase L with an IC50 of 73.5 ± 0.2 nM using commercially sourced fresh powder in a 10-point dose–response analysis (Fig. 1C) and also with an orthogonal gel-based RNA cleavage assay (SI Appendix, Fig. S1).
Fig. 1.
Identification of a small-molecule inhibitor of the ribonuclease activity of porcine RNase L. (A) Hit profile against the Ontario Institute for Cancer Research 500-compound protein kinase inhibitor library using an in vitro fluorescence ribonuclease assay. (B) Chemical structure of the RNase L small-molecule inhibitor ellagic acid. (C) Dose–response inhibition profile of ellagic acid against RNase L using the in vitro screening assay. Data represent mean ± SEM of four binding curves.
To investigate structure–activity relationships (SARs), we tested EA and three commercially available analogs for activity against porcine RNase L and its closest functional paralogue, the dual kinase ribonuclease IRE1 (24% identity over the kinase domain and 22% over the entire fused kinase-ribonuclease module; Fig. 2). EA displayed threefold weaker activity against IRE1 (IC50 = 270.6 ± 0.1 nM) than RNase L (IC50 = 73.08 ± 0.16 nM). Substitution of the hydroxyl moiety at position 5 (Fig. 2A shows IUPAC chemical naming convention) with a bulky aryl group to generate valoneic acid dilactone (VAL) resulted in a 100-fold increase in inhibitor potency (0.68 ± 0.09 nM) against RNase L and only a twofold improvement against IRE1 (144.4 ± 0.1 nM; Fig. 2 A and B). This corresponded to an improvement in inhibitor selectivity toward RNase L over IRE1 by a factor of 200.
Fig. 2.
SAR and cross-reactivity analysis of a phenolic analog series (A–D) on the ribonuclease activities of porcine RNase L and murine IRE1: chemical structures (Left) and ribonuclease inhibition profiles (black for RNase L and blue for IRE1; Right). Data represents mean ± SEM of three binding curves. Substituent numbering scheme is shown in A for the primary hit, ellagic acid.
Substitutions of the hydroxyl moieties at positions 4 and 11 with a methoxy group to generate 3,3′-di-O-methylellagic acid abolished all inhibitory activity against IRE1 and rendered the small molecule a weak activator of RNase L (Fig. 2C). Substitution of the hydroxyl moieties at positions 4 and 11 each with an amino group, and eliminating all other ketone and hydroxyl groups at positions 1, 2, 5, 8, 9, and 12 to generate 1,6-diaminopyrene, rendered the molecule ineffective at complete inhibition against both RNase L and IRE1 (Fig. 2D).
Mechanism of Action Analysis.
EA was previously shown to inhibit the kinase activity of CK2 by competitively binding to the ATP binding site of its kinase domain (32). We recapitulated this finding for EA and furthermore showed that VAL was also an effective inhibitor of CK2 kinase activity, with an IC50 of 419 nM and 166 nM, respectively (SI Appendix, Fig. S2 A and B). Thus, we reasoned that EA and its close analog VAL would function similarly by binding to the pseudokinase domain of RNase L. Indeed, precedents for small molecules acting as modulators of ribonuclease activity through binding to the kinase active site of the paralogous protein IRE1 have been established (26, 34, 35). Despite being incompetent for phospho-transfer, the pseudokinase domain is fully competent for binding ATP (9). To test if EA and VAL bind to the pseudokinase domain of RNase L, we examined the ability of EA and VAL to competitively displace the fluorescently labeled ATP analog BODIPY- ATP (Fig. 3A). To this end, we demonstrated that unlabeled ATP could competitively displace BODIPY-ATP from RNase L with an EC50 of 1.85 ± 0.08 μM (Fig. 3 A, ii), a potency comparable to the binding affinity of BODIPY-ATP to RNase L (1.91 ± 0.40 μM; Fig. 3 A, i). Surprisingly, neither EA nor VAL could competitively displace BODIPY-ATP from RNase L (Fig. 3 A, ii). Thus, contrary to expectations, EA and VAL function as inhibitors of RNase L ribonuclease function through a mechanism that does not involve occupancy of the ATP binding site within the pseudokinase domain.
Fig. 3.
Enzymatic and biophysical characterization of the mechanism of action of EA and VAL on porcine RNase L. (A, i) Binding of BODIPY-ATP to RNase L as assessed by monitoring the fluorescence polarization signal of BODIPY-ATP in the presence of increasing concentrations of RNase L. Kd represents mean ± SEM of three binding curves. (A, ii) In vitro competition binding profiles of EA, VAL, and ATP to RNase L using a BODIPY-ATP fluorescent probe as assessed by fluorescence polarization. EC50 represents mean ± SEM of three displacement curves. (B) Effect of enzyme concentration on the dose–response inhibition profiles of VAL against RNase L ribonuclease activity. IC50 represents mean ± SEM of three inhibition profiles. (C) Effect of RNA substrate concentration on the dose–response inhibition profiles of VAL against RNase L ribonuclease activity. A total of 0.4 nM of RNase L was used for the reactions. IC50 values represent mean ± SEM of three inhibition profiles.
To gain further insight into inhibitor mechanism of action, we performed dose–response analyses with VAL in reactions using three different concentrations of RNase L (1.6 nM, 3 nM, and 8 nM; Fig. 3B). As enzyme concentration increased, we observed a concomitant increase in IC50 values for VAL (0.56 nM ± 0.07, 2.80 ± 0.08 nM, and 23.15 ± 0.14 nM, respectively). This finding that inhibitor potency tracked closely with enzyme concentration was consistent with a mode of action in which VAL binds directly to RNase L and not solely to the RNA substrate. We next performed dose–response analyses using fixed RNase L concentrations but with three different concentration of RNA substrate (50 nM, 150 nM, and 450 nM). We observed no significant change in IC50 values as RNA substrate concentration was varied (Fig. 3C). This behavior is also consistent with a mode of action in which VAL does not simply sequester RNA substrate and suggests that VAL binds to RNase L in a manner that does not compete with RNA substrate.
To confirm direct binding of VAL to RNase L and to discern any dependency for binding on the activators 2-5A and ATP/MgCl2, we exploited the sensitivity of solution NMR, which can detect binding events at substochiometric levels (reviewed in ref. 36). As evidenced by the dose-dependent attenuation of VAL resonance peaks at 7.46, 7.38, and 6.66 ppm in Carr-Purcell-Meiboom-Gill (CPMG) analyses and by the appearance of the same peaks in positive saturation transfer difference (STD) analyses, RNase L in isolation was sufficient for binding to VAL (Fig. 4A and SI Appendix, Fig. S3A). Addition of the 40-pM-affinity (7) ligand 2-5A in molar excess to RNase L had negligible effect on the CPMG and STD binding signals of VAL (Fig. 4B and SI Appendix, Fig. S3B), while addition of the 1.2-µM-affinity ligand ATP/MgCl2 (9) in molar excess to RNase L (Fig. 4C and SI Appendix, Fig. S3C), or the combination of both 2-5A and ATP/MgCl2 (Fig. 4D and SI Appendix, Fig. S3D), reduced CPMG and STD signals to a small degree. Further analysis revealed that 100 μM MgCl2 alone was sufficient to reduce CPMG and STD signals of RNase L to VAL, suggesting that the reduction in NMR signals is not related specifically to competition with nucleotide (SI Appendix, Fig. S4A). Demonstrating the specificity of the CPMG and STD NMR binding experiments, VAL showed no evidence of binding to four control proteins under comparable conditions to those performed with RNase L (SI Appendix, Fig. S4B).
Fig. 4.
Binding of VAL to porcine RNase L assessed by NMR and SPR. (A–D) Dose-dependent CPMG-NMR spectra for VAL recorded in the presence of the indicated concentrations of RNase L, 2-5A, and ATP/MgCl2. (E–G) Steady-state binding analysis of VAL to immobilized RNase L using SPR. Representative plots are shown for one of three replicate experiments. Kd represents the average for n = 3 ± SD.
To corroborate our NMR findings, we employed surface plasmon resonance (SPR). VAL demonstrated saturable binding to immobilized apo-RNase L with a Kd of 0.372 ± 0.017 µM (Fig. 4E and SI Appendix, Fig. S3E). Consistent with our NMR findings, preincubation of RNase L with 2-5A or the combination of 2-5A/ATP-MgCl2 had negligible effect on the binding of VAL to RNase L (Kd = 0.304 ± 0.015 µM and 0.299 ± 0.016 µM, respectively; Fig. 4 F and G and SI Appendix, Fig. S3 F and G). Analysis of the less potent ribonuclease inhibitor sunitinib (SU) revealed overall weaker binding affinity for RNase L (SI Appendix, Fig. S5). While nonsaturable binding kinetics precluded accurate estimate of the binding affinity of sunitinib to apo RNase L (SI Appendix, Fig. S5A), Kd values of 2.53 ± 0.09 µM and 2.06 ± 0.09 µM were determined for RNase L preequilibrated with 2-5A and 2-5A+ATP-MgCl2, respectively (SI Appendix, Fig. S5 B and C). Although the binding affinity of VAL for RNase L was unexpectedly shifted relative to its inhibitory properties in vitro, both NMR and SPR findings confirm the ability of VAL to bind directly to RNase L.
To assess binding of VAL to RNase L in a more biological/cellular context, we performed cellular thermal shift assay (CETSA) analyses. Here, the binding of a ligand to a target within a cellular lysate is detected by the stabilization of the target as assessed by Western analysis following incubations performed at different temperatures. We observed a weak but reproducible stabilization of overexpressed Flag-RNase L in cellular lysates exposed to 100 μM VAL relative to exposure to a control DMSO only (SI Appendix, Fig. S6 A–C). Specifically, VAL-dependent stabilization of Flag-RNase L was observed in cellular lysates treated at 45 °C, 49 °C, 51 °C, and 55 °C (SI Appendix, Fig. S6A, compare lanes 5 and 6, 9 and 10, 11 and 12, and 15 and 16, respectively; SI Appendix, Fig. S6 B and C, are a triplicate). We note that the absence of a strong protein stabilization signal for VAL was not unexpected, since marginal stabilization of RNase L by VAL was observed in thermal denaturation experiments performed with purified proteins (SI Appendix, Fig. S7). In these experiments, VAL, like 2-5A and RNA, caused no appreciable stabilization of RNase L on their own, while a weak stabilization signal was observed for VAL on RNase L in the presence of 2-5A, ATP/MgCl2, and RNA. In comparison, ATP/MgCl2 caused a pronounced stabilization of RNase L on its own (ΔTm = 6 ± 0.2 °C; SI Appendix, Fig. S7A). Together, our biophysical studies support the notion that VAL exerts its inhibitory effect on the ribonuclease activity of RNase L by binding directly to RNase L, to a site distinct from ATP and likely from 2-5A.
Functional Characterization of VAL and EA in Cells.
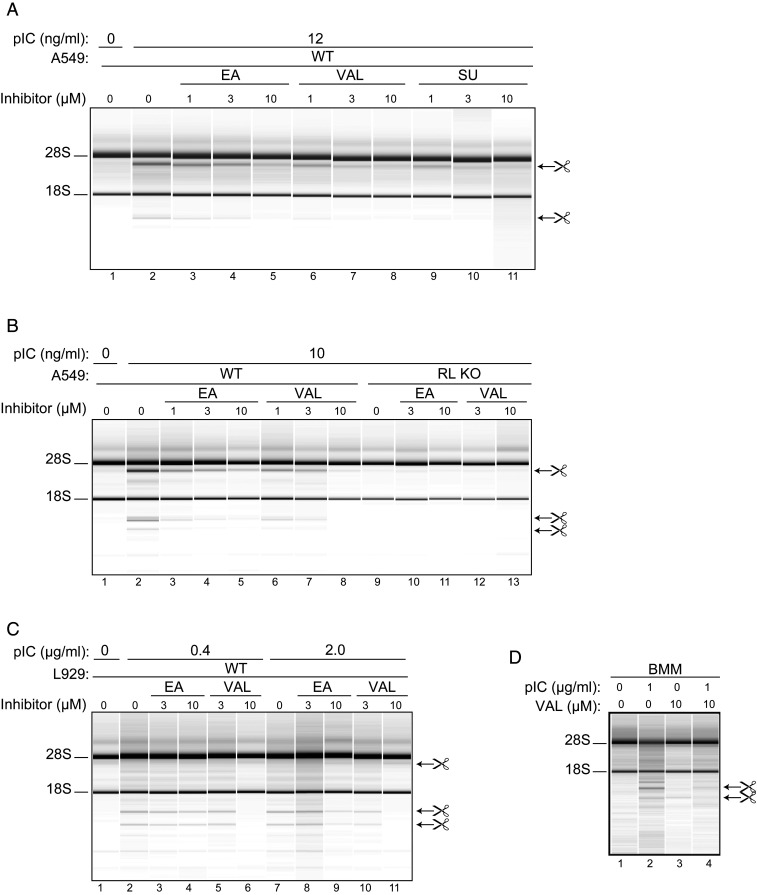
To determine if VAL and EA can function as inhibitors of RNase L in cells, we examined their effect on the induced rRNA cleavage pattern of a wild type human A549 cell line and matched RNase L−/− knockout (KO) cell line transfected with synthetic polyribonucleotide dsRNA duplex poly(rI):poly(rC) (denoted pIC). Transfection of A549 cells with 10 to 12 ng/mL of pIC led to the activation of RNase L as detected by monitoring highly specific and characteristic discrete cleavage products of 28S and 18S rRNA from ribosomes (37) (Fig. 5 A and B, compare lanes 1 vs. 2). This RNA cleavage pattern was due to the activity of RNase L, as the RNase L KO cell line was not affected by transfection with pIC (Fig. 5B, compare lanes 2 vs. 9).
Fig. 5.
Inhibition of RNase L from different species by VAL in intact cell lines and in primary cells. (A) Human A549 cells were incubated with different concentrations (as indicated) of EA, VAL, and sunitinib (SU) for 3 h before transfection with 12 ng/mL pIC for 2 h; (B) WT and RNase L (RL) KO A549 cells were incubated with EA or VAL for 3 h and then transfected with 10 ng/mL of pIC for 2 h; and (C) mouse WT L929 cells were incubated with or without EA or VAL for 3 h and then transfected with 0.4 or 2 µg/mL of pIC for 2 h or mock-transfected without removing compounds. (D) Mouse bone marrow macrophages (BMMs) were treated with 10 µM VAL for 3 h and then transfected with 1 µg/mL of pIC for 5 h or mock-transfected without removing VAL. Two (A and D) or three (B and C) biological replicates were done. Total RNA isolated from the cells was separated on RNA chips as described previously (9).
In the presence of EA, VAL, or SU, the RNA cleavage pattern in human A549 cells induced by treatment of cells with pIC was progressively reduced in a concentration-dependent manner (Fig. 5A, compare lanes 3 to 5, 6 to 8, and 9 to 11, respectively). Consistent with its superior potency in vitro, VAL was a more potent inhibitor of RNA cleavage than EA in these human cells. A similar trend of potency was observed with a mouse L929 cell line using higher amounts of activator pIC (Fig. 5C, compare lanes 4 and 6 and lanes 9 and 11, for example). (The specific rRNA cleavage products produced by RNase L are distinct and different in human and mouse cells; ref. 37.) We note, however, that the potency of VAL (IC50 = 1 μM) and EA in both A549 (human) and L929 (mouse) cell lines was drastically reduced (∼1,000-fold) relative to the potency observed in vitro against porcine RNase L. Only part of this potency shift (28-fold change) could be attributed to species variation (IC50 of 1.2 ± 0.1 nM vs. 33.9 ± 0.1 nM for porcine and human RNase L, respectively; SI Appendix, Fig. S8), suggesting other factors were at play. To determine if VAL is able to enter and inhibit RNase L in a primary cell type, bone marrow macrophages (BMMs) were isolated from mice. RNase L-mediated rRNA cleavage products were clearly visible by RNA chip analysis in response to pIC transfection (Fig. 5D, compare lanes 1 and 2). However, there was near-complete inhibition of RNase L activation when BMMs were pretreated with VAL for 3 h and then transfected with 1 μg/mL of pIC (Fig. 5D, compare lanes 2 and 4).
As the potency of VAL on RNase L in cells was unexpectedly similar to that reported previously for sunitinib (Fig. 5A) (26), we investigated whether VAL would display comparable cytotoxic properties in cells. As shown in Fig. 6 A and B, VAL and EA were not cytotoxic to A549 cells up to the 25-μM concentration tested (25-fold higher than the IC50 for RNase L cleavage activity in cells) over 30 h. In comparison, A549 cells treated with 3 to 25 μM SU displayed considerable cytotoxicity over the concentrations required for the repression of RNase L cleavage activity, typically 5 μM (26) (Fig. 6C). As A549 cells are expected to be tolerant to the loss of RNase L under the conditions tested (38), these results led us to conclude that VAL is a more effective and useful molecular probe to dissect RNase L function in intact cells than sunitinib.
Fig. 6.
VAL lacks cytotoxicity and prevents cell death from dsRNA or ADAR1 deficiency. A549 cells were cultured in the absence or presence of the indicated concentrations of VAL (A), EA (B), or sunitinib (SU; C). Percent cell survival was measured by real-time imaging of dye-labeled cells (Methods). Data represent the mean ± SD of three survival profiles. (D) Effect of VAL on pIC-induced cell death in A549. A549 cells were preincubated with or without 10 µM VAL for 2 h. The cells were then either mock-transfected with Lipofectamine 2000 (Invitrogen) alone (number of replicates, n = 3) or transfected with 20 ng/mL pIC (number of replicates, n = 9). Percentage of cell survival was determined by real-time imaging (Methods). Data are averages ± SD. (E) MHV NS2 expressing ADAR1 WT or ADAR KO A549 cells were treated with siRNAs against NS2 (siNS2-1, siNS2-2) or nonspecific control siGFP. At the time of transfection, cells were treated with 20 μM of VAL or mock-treated, and, at 96 h post transfection, cells were fixed and stained with crystal violet. (F) Western blots of A549 cells transfected with control siRNA or siRNA against ADAR1 for 48 h probed with anti-ADAR1 or anti–β-actin antibody as a loading and transfer control. Lanes 1 to 3 and lanes 4 to 7 are replicates. (G) Effect of VAL on siRNA-mediated knockdown of ADAR1 in A549 cells. A549 cells (5 × 104) were plated in 24-well plates. After 12 h, the cells were preincubated with or without 10 µM VAL for 2 h before siRNA transfection with 30 nM of control siRNA or ADAR1-siRNA (Santa Cruz Biotechnology) with DharmaFECT 1 (ThermoFisher). Phase-contrast images of control siRNA- or ADAR siRNA-treated cells in the presence and absence of VAL are shown at the indicated times post transfection. (H) Quantification of percent cell survival in cells treated as indicated in G was determined by real-time imaging (Methods). Data are averages ± SD from six identically treated replicates. P values calculated with two-way ANOVA using GraphPad Prism are indicated.
We next investigated the function of VAL under cellular conditions expected to activate RNase L RNA cleavage activity. Transfection of A549 cells with 20 ng/mL pIC, a potent activator of OAS enzymes, caused rapid death over 48 h (Fig. 6D). Previously, we showed that RNase L knockout renders cells insensitive to cell death by pIC (20). Strikingly, the pretreatment of cells with 10 µM VAL prevented cell death in response to pIC transfection.
The finding that VAL can inhibit the ribonuclease activity of RNase L in cells induced by pIC treatment led us to investigate the utility of VAL in a more disease-relevant cellular context. Specifically, we aimed to determine whether VAL could rescue the cell-lethal phenotype of ADAR1 deficiency caused by the hyperactivation of RNase L (20). For this analysis, we employed an ADAR1 KO A549 lung cancer cell line, deficient in the ability to catalyze the conversion of adenosine to inosine in cellular dsRNAs (20). This activity is critical for the destabilization of dsRNA structures in the cell; therefore, absence of ADAR1 results in the accumulation of unnaturally high levels of dsRNA, which are detected by OAS, leading to the induction of an apoptotic response through RNase L. Viability of the ADAR1 KO cell line can be restored by the knockout of the RNase L gene locus or by antagonizing RNase L function by the expression of the murine coronavirus NS2 accessory protein (20). NS2 is a phosphodiesterase that subverts the activation of the OAS–2-5A–RNase L pathway by degradation of the intermediate 2-5A second messenger (39).
We employed the A549 cells in which ADAR1 is present (ADAR1 WT) or has been knocked out with CRISPR-Cas9 (ADAR1 KO) (20). (Fig. 6E). To maintain viability of the ADAR1 KO cell line, NS2 was stably expressed in both cell lines. Transfection of the ADAR1 KO cell line but not the parental ADAR WT cell line with siRNA against NS2 selectively killed the ADAR1 KO cell line (Fig. 6E, compare columns 3 and 4), as would be expected since NS2 is required for viability of this cell line. Previously, we showed the effectiveness of shRNA to NS2 by Western analysis (20). However, treatment with 10 μM VAL (Fig. 6E, columns 1 and 2) suppressed the cell death effect of NS2 siRNA knockdown (Fig. 6E, compare columns 1 vs. 3).
To validate these findings in real time, siRNA depletion of ADAR1 was used as an alternative to CRISPR-Cas9–mediated knockout of ADAR1. The efficacy of ADAR1 depletion was shown by Western blot analysis (Fig. 6F). ADAR1 depletion by siRNA oligonucleotides caused A549 cell death as determined by real-time cell imaging (Fig. 6 G and H). A large increase in numbers of dead cells was observed after 72 h of ADAR1 siRNA treatment, whereas relatively few dead cells were observed in siRNA-treated cells in the presence of VAL. The much more modest reduction in cell survival by control siRNA (Fig. 6H), also inhibited by VAL, is likely due to nonspecific induction of the IFN system (40) with limited OAS–RNase L activation. These data are fully consistent with VAL functioning in an on-target manner to inhibit the ribonuclease activity of RNase L in cells and highlights the potential promise of the therapeutic utility of an RNase L-selective inhibitor in the treatment of Aicardi-Goutières syndrome characterized by genetic lesions in ADAR1 gene locus, and possibly against other conditions in which there is excessive RNase L activity.
Discussion
Our studies have identified VAL and EA as potent low-nanomolar inhibitors of RNase L ribonuclease activity in vitro (in cell-free systems) and at low micromolar levels when added to intact cells. Importantly, VAL and EA lacked cytotoxicity at concentrations that were effective in the inhibition of RNase L activity.
VAL and EA belong to a class of polyphenol natural products found in pomegranates, chestnut, tree bark, and walnuts (41–43) with poorly defined cellular functions and activities. For example, EA displays antibacterial activity in vitro and in mice (44) and relieves hepatic oxidative stress and insulin resistance in diabetic rats (45) and oxidative stress in heart tissue of rats (46). More limited studies on VAL have revealed antibacterial activity in vitro (47) and inhibition of the cytotoxicity of the DNA methyltransferase inhibitor 5-azacytidine (48). For all cellular activities characterized to date, no molecular mechanisms for EA or VAL have been described.
VAL and EA display 212- and 4-fold specificity for RNase L over its closest paralog, IRE1, which shares 22% sequence identity over the dual catalytic region encompassing the fused protein kinase and endoribonuclease domains. EA was identified previously as a potent inhibitor of casein kinase IIalpha (CK2α; IC50 = 0.04 μM) (32) and a weak inhibitor of glycogen phosphorylase b (IC50 = 3.2 μM) (49). In the case of CK2α, cocrystal structure analysis revealed that EA functions by binding to the kinase domain in a manner competitive with ATP (32). Contrary to expectation based on the mode of action of EA on CK2α, VAL and EA do not act by binding to the ATP binding site of the kinase domain of RNase L. EA was shown to exert an inhibitory effect on the matrix metalloproteinases-2 by chelating zinc (50). However, since RNase L is not a metallo-dependent nuclease, and the magnesium required for ATP activator binding to the kinase domain is present in our studies in vast excess, it is unlikely that a chelator function of EA would account for the inhibitory effect of EA and VAL on RNase L. Our NMR and SPR studies showed that VAL binds to RNase L without a requirement for 2-5A or ATP/MgCl2, indicating that the dimerization status or the activation state of RNase L are not critical determinants for VAL inhibitory function. Last, we note that a sizable shift (∼400-fold) between the binding affinity of VAL for RNase L determined by SPR relative to its inhibitory properties toward RNase L in vitro suggests a level of complexity we do not fully understand. Thus, the question of how precisely VAL exerts its inhibitory effect on RNase L ribonuclease activity remains an open question. Interestingly, the lack of a robust thermal stabilization (ΔTm) signal for VAL on RNase L in purified systems parallels a surprisingly weak thermal stabilization signal for 2-5A in contrast to ATP/MgCl2. As the primary interaction site of 2-5A and ATP/MgCl2 are the Ankyrin domain and kinase domain, respectively, the weak thermal stabilization by VAL hints that it too might engage the Ankyrin domain, albeit with a nonoverlapping binding mode. This inference would also imply that the thermal denaturation of RNase L is dominated by the unfolding kinetics of the kinase domain more so than the Ankyrin repeat domain. A detailed understanding of the mechanism of action of RNase L awaits a high-resolution structure of the inhibitor in action.
Our studies also show that VAL is an effective inhibitor of RNase L in human and mouse cells, although with a 1,000-fold decrease in potency than that observed in vitro. This difference is due in part to the species variation between the two experimental systems tested, as porcine RNase L proved to be 28-fold more sensitive toward VAL than human RNase L when tested side by side in vitro. Other potential contributing factors that await further characterization include plasma binding, metabolic stability, membrane permeability, and efflux characteristics.
Despite reduced potency in cells, VAL still displays superior binding and inhibitory functionality compared with sunitinib, a previously described biological inhibitor of RNase L (26). While the two molecules display comparable low-micromolar potencies against RNase L in human cells, VAL does not display the cytotoxicity observed with sunitinib. Since RNase L-knockout cells are viable (13, 38), the cytotoxicity displayed by sunitinib treatment is likely due to the targeting of other protein kinases. This is not unexpected, since the effective concentration of sunitinib required for comparable inhibition of RNase L is 100-fold higher than that required for inhibition of sunitinib’s clinically defined receptor tyrosine kinase targets VEGFR1-3, PDGFR-β, C-Kit, and Flit-3 (IC50s in cells range from 2 to 14 nM; ref. 51).
A subset of AGS cases are caused by ADAR1 mutations that lead to high levels of IFN production and IFN-stimulated gene expression. While the specific pathways and mechanisms underlying the pathology of AGS in cases involving ADAR1 mutations are not known, MDA5, PKR (52), and OAS–RNase L are implicated (20). However, genetic evidence for the role of either PKR or RNase L in the pathophysiology of AGS has not been shown. In our prior studies, we showed that loss of ADAR1 activity in A549 cells leads to RNase L activation and subsequent apoptotic cell death (20). Thus, we tested VAL in an A549 ADAR1-knockout or -knockdown cellular background and observed that VAL selectively inhibited RNase L to suppress cell death. By demonstrating that inhibiting RNase L ribonuclease activity can attenuate ADAR1-associated apoptosis, these results indicate that RNase L is a viable therapeutic target in the context of select cases of AGS with ADAR1 mutations (53). Furthermore, because VAL inhibits RNase L, it is reasonable to expect that, during viral infections, VAL treatment will result in fewer RNA cleavage products to stimulate RIG-I/MDA5 and NLRP3 (25, 54). Therefore, RNase L inhibitors may also be used in the future to mitigate overactive inflammatory responses to viral infections, which have contributed to a tragically high death toll for COVID-19 patients in the global pandemic in 2019 to 2020 (55–58).
Materials and Methods
Plasmids and Protein Expression and Purification.
Expression constructs, plasmids, cloning, and full description of protein expression and purification are detailed in the SI Appendix.
In Vitro Biotinylation of AviTagged RNase L.
In vitro biotinylation reaction procedure using recombinant RNase L WT-AviTag is described in the SI Appendix.
In Vitro Endoribonuclease and Fluorescence Polarization Assays.
RNase L endoribonuclease assay.
Fluorescence-based endoribonuclease cleavage assays were performed as previously described (9) and detailed in the SI Appendix.
IRE1 endoribonuclease assay.
Fluorescence-based endoribonuclease cleavage assays were performed using purified IRE1 protein or RNase L and RNA substrate as described previously (9) and detailed in the SI Appendix.
Ribonuclease Activity Inhibitor Screen.
A 500-compound library (OICR-L100, Medicinal Chemistry Platform at the Ontario Institute for Cancer Research) assembled from previously reported protein kinase inhibitors and close analogs was used for an RNase L activity inhibitor screen. The full description of the procedure is provided in the SI Appendix.
Inhibitor Binding Analysis Using NMR.
NMR sample preparation and binding experiments are described in the SI Appendix.
Inhibitor Binding Analysis Using Surface Plasmon Resonance (SPR).
A complete description of the SPR analysis of biotinylated C-terminal AviTagged RNase L is included in the SI Appendix.
Inhibitor-Binding Analysis Using Differential Scanning Fluorimetry.
Differential scanning fluorimetry (thermal melt) was performed with recombinant RNase L H680N protein. A detailed protocol is provided in the SI Appendix.
In Vitro Inhibition of CK2α by VAL or EA.
Inhibitor activity of VAL and EA against CK2α was determined by Eurofins. The detailed method is described in the SI Appendix.
Cell Culture, siRNA Transfection, PolyI:polyC (pIC) Transfections, and Antibodies for Western Blot.
Human lung epithelial A549 cells, mouse L929 cells, human embryonic kidney HEK293T (293T) cells, and mouse bone marrow macrophages (BMMs) were maintained according to standard procedures. Cell culture, cell transfection, and Western blotting were done as described in the SI Appendix.
Cellular Thermal Shift Assay (CETSA).
CETSA experiments were conducted as described in ref. 59 and detailed in the SI Appendix.
Monitoring Specific rRNA Cleavage Products as an Index of RNase L Activity in Intact Cells.
The cell-based assay for rRNA cleavages in intact cells was performed as described previously (9, 37).
Cell Survival Assays and ADAR1 KO Cell-Based Assays.
Cell transfection, compound treatment, and cell survival assays were done as described previously (20) and detailed in the SI Appendix.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the Canadian Cancer Society (Impact Grant 704116 to F.S.), Canadian Institute for Health Research (FDN143277 to F.S.), National Institute of Allergy and Infectious Disease of the NIH under award numbers R01AI104887 (to S.R.W. and R.H.S.) and R01AI135922 (to R.H.S), the National Natural Science Foundation of China (21778008 to H.H.), and the Shenzhen Science and Technology Innovation Committee (KQJSCX20170330154900 to H.H.). All NMR experiments were performed at the Beijing NMR Center and the NMR facility of National Center for Protein Sciences at Peking University with the assistance of Dr. Xiaogang Niu. S.D. is a recipient of the Banting Postdoctoral Fellowship. We thank Dr. David Uehling from the Ontario Institute for Cancer Research for coordinating experiments performed by Eurofins.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006883117/-/DCSupplemental.
Data Availability.
All of the data and reagents that support the findings of this study are available within the main text or the SI Appendix.
References
- 1.Cooper D. A., Jha B. K., Silverman R. H., Hesselberth J. R., Barton D. J., Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic Acids Res. 42, 5202–5216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D., How cells respond to interferons. Annu. Rev. Biochem. 67, 227–264 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Kristiansen H., Gad H. H., Eskildsen-Larsen S., Despres P., Hartmann R., The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 31, 41–47 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Kerr I. M., Brown R. E., pppA2“p5”A2“p5”A: An inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. U.S.A. 75, 256–260 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong B. et al., Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J. Biol. Chem. 269, 14153–14158 (1994). [PubMed] [Google Scholar]
- 6.Dong B., Silverman R. H., A bipartite model of 2-5A-dependent RNase L. J. Biol. Chem. 272, 22236–22242 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Silverman R. H. et al., Purification and analysis of murine 2-5A-dependent RNase. J. Biol. Chem. 263, 7336–7341 (1988). [PubMed] [Google Scholar]
- 8.Han Y. et al., Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343, 1244–1248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H. et al., Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol. Cell 53, 221–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wreschner D. H., McCauley J. W., Skehel J. J., Kerr I. M., Interferon action–sequence specificity of the ppp(A2’p)nA-dependent ribonuclease. Nature 289, 414–417 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Floyd-Smith G., Slattery E., Lengyel P., Interferon action: RNA cleavage pattern of a (2′–5′)oligoadenylate–Dependent endonuclease. Science 212, 1030–1032 (1981). [DOI] [PubMed] [Google Scholar]
- 12.Donovan J., Rath S., Kolet-Mandrikov D., Korennykh A., Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou A. et al., Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16, 6355–6363 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samuel M. A. et al., PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80, 7009–7019 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpten J. et al., Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 30, 181–184 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Silverman R. H., Implications for RNase L in prostate cancer biology. Biochemistry 42, 1805–1812 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Krüger S. et al.; German HNPCC Consortium , The additive effect of p53 Arg72Pro and RNASEL Arg462Gln genotypes on age of disease onset in Lynch syndrome patients with pathogenic germline mutations in MSH2 or MLH1. Cancer Lett. 252, 55–64 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Madsen B. E. et al., Germline mutation in RNASEL predicts increased risk of head and neck, uterine cervix and breast cancer. PLoS One 3, e2492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen-Dumont T. et al.; ABCFR , Is RNASEL:p.Glu265* a modifier of early-onset breast cancer risk for carriers of high-risk mutations? BMC Cancer 18, 165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y. et al., Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. eLife 6, e25687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George C. X., John L., Samuel C. E., An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1). J. Interferon Cytokine Res. 34, 437–446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomaselli S., Galeano F., Locatelli F., Gallo A., ADARs and the balance game between virus infection and innate immune cell response. Curr. Issues Mol. Biol. 17, 37–51 (2015). [PubMed] [Google Scholar]
- 23.Patterson J. B., Samuel C. E., Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice G. I. et al., Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakrabarti A. et al., RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe 17, 466–477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jha B. K. et al., Inhibition of RNase L and RNA-dependent protein kinase (PKR) by sunitinib impairs antiviral innate immunity. J. Biol. Chem. 286, 26319–26326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanches M. et al., Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors. Nat. Commun. 5, 4202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrington P. E. et al., Unfolded protein response in cancer: IRE1α inhibition by selective kinase ligands does not impair tumor cell viability. ACS Med. Chem. Lett. 6, 68–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendel D. B. et al., In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: Determination of a pharmacokinetic/pharmacodynamic relationship. Clin. Cancer Res. 9, 327–337 (2003). [PubMed] [Google Scholar]
- 30.Wiseman R. L. et al., Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol. Cell 38, 291–304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkmann K. et al., Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J. Biol. Chem. 286, 12743–12755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekiguchi Y. et al., Structural insight into human CK2alpha in complex with the potent inhibitor ellagic acid. Bioorg. Med. Chem. Lett. 19, 2920–2923 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Cozza G. et al., Identification of ellagic acid as potent inhibitor of protein kinase CK2: A successful example of a virtual screening application. J. Med. Chem. 49, 2363–2366 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Korennykh A. V. et al., The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L. et al., Divergent allosteric control of the IRE1α endoribonuclease using kinase inhibitors. Nat. Chem. Biol. 8, 982–989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lepre C. A., Moore J. M., Peng J. W., Theory and applications of NMR-based screening in pharmaceutical research. Chem. Rev. 104, 3641–3676 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Silverman R. H., Skehel J. J., James T. C., Wreschner D. H., Kerr I. M., rRNA cleavage as an index of ppp(A2’p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 46, 1051–1055 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y. et al., Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U.S.A. 113, 2241–2246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L. et al., Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11, 607–616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sledz C. A., Holko M., de Veer M. J., Silverman R. H., Williams B. R., Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5, 834–839 (2003). [DOI] [PubMed] [Google Scholar]
- 41.García-Villalba R. et al., Validated method for the characterization and quantification of extractable and nonextractable ellagitannins after acid hydrolysis in pomegranate fruits, juices, and extracts. J. Agric. Food Chem. 63, 6555–6566 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Vekiari S. A., Gordon M. H., García-Macías P., Labrinea H., Extraction and determination of ellagic acid contentin chestnut bark and fruit. Food Chem. 110, 1007–1011 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Li L. et al., Polyphenolic profiles and antioxidant activities of heartnut (Juglans ailanthifolia Var. cordiformis) and Persian walnut (Juglans regia L.). J. Agric. Food Chem. 54, 8033–8040 (2006). [DOI] [PubMed] [Google Scholar]
- 44.De R. et al., Antimicrobial activity of ellagic acid against Helicobacter pylori isolates from India and during infections in mice. J. Antimicrob. Chemother. 73, 1595–1603 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Polce S. A. et al., Ellagic acid alleviates hepatic oxidative stress and insulin resistance in diabetic female rats. Nutrients 10, 351 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yüce A., Ateşşahin A., Ceribaşi A. O., Aksakal M., Ellagic acid prevents cisplatin-induced oxidative stress in liver and heart tissue of rats. Basic Clin. Pharmacol. Toxicol. 101, 345–349 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Khan I. et al., Punica granatum peel extracts: HPLC fractionation and LC MS analysis to quest compounds having activity against multidrug resistant bacteria. BMC Complement. Altern. Med. 17, 247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee S. et al., OAS-RNase L innate immune pathway mediates the cytotoxicity of a DNA-demethylating drug. Proc. Natl. Acad. Sci. U.S.A. 116, 5071–5076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kyriakis E. et al., Natural flavonoids as antidiabetic agents. The binding of gallic and ellagic acids to glycogen phosphorylase b. FEBS Lett. 589, 1787–1794 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Huang S. T., Yang R. C., Wu H. T., Wang C. N., Pang J. H., Zinc-chelation contributes to the anti-angiogenic effect of ellagic acid on inhibiting MMP-2 activity, cell migration and tube formation. PLoS One 6, e18986 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aparicio-Gallego G. et al., New insights into molecular mechanisms of sunitinib-associated side effects. Mol. Cancer Ther. 10, 2215–2223 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Chung H. et al., Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sase S., Takanohashi A., Vanderver A., Almad A., Astrocytes, an active player in Aicardi-Goutières syndrome. Brain Pathol. 28, 399–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malathi K., Dong B., Gale M. Jr., Silverman R. H., Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448, 816–819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehta P. et al.; HLH Across Speciality Collaboration, UK , COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi Y. et al., COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 27, 1451–1454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C., Wu Z., Li J. W., Zhao H., Wang G. Q., Cytokine release syndrome in severe COVID-19: Interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents 55, 105954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedersen S. F., Ho Y. C., SARS-CoV-2: A storm is raging. J. Clin. Invest. 130, 2202–2205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jafari R. et al., The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100–2122 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data and reagents that support the findings of this study are available within the main text or the SI Appendix.