Significance
When sound vibrations reach the inner ear, fine protein filaments called “tip links” stretch and open cochlear hair-cell mechanosensitive channels that trigger sensory perception. Similarly, vestibular hair cells use tip links to sense mechanical stimuli produced by head motions. Tip links are formed by cadherin-23 and protocadherin-15, two large proteins involved in hearing loss and balance disorders. Here we present multiple structures, models, and simulations that depict the lower end of the tip link, including the complete protocadherin-15 ectodomain. These models show an essential connection between cadherin-23 and protocadherin-15 with dual molecular “handshakes” and various protein sites that are mutated in inherited deafness. The simulations also reveal how the tip link responds to force to mediate hearing and balance sensing.
Keywords: auditory system, hair cell, tip link, mechanotransduction, PCDH15
Abstract
The vertebrate inner ear, responsible for hearing and balance, is able to sense minute mechanical stimuli originating from an extraordinarily broad range of sound frequencies and intensities or from head movements. Integral to these processes is the tip-link protein complex, which conveys force to open the inner-ear transduction channels that mediate sensory perception. Protocadherin-15 and cadherin-23, two atypically large cadherins with 11 and 27 extracellular cadherin (EC) repeats, are involved in deafness and balance disorders and assemble as parallel homodimers that interact to form the tip link. Here we report the X-ray crystal structure of a protocadherin-15 + cadherin-23 heterotetrameric complex at 2.9-Å resolution, depicting a parallel homodimer of protocadherin-15 EC1-3 molecules forming an antiparallel complex with two cadherin-23 EC1-2 molecules. In addition, we report structures for 10 protocadherin-15 fragments used to build complete high-resolution models of the monomeric protocadherin-15 ectodomain. Molecular dynamics simulations and validated crystal contacts are used to propose models for the complete extracellular protocadherin-15 parallel homodimer and the tip-link bond. Steered molecular dynamics simulations of these models suggest conditions in which a structurally diverse and multimodal protocadherin-15 ectodomain can act as a stiff or soft gating spring. These results reveal the structural determinants of tip-link–mediated inner-ear sensory perception and elucidate protocadherin-15’s structural and adhesive properties relevant in disease.
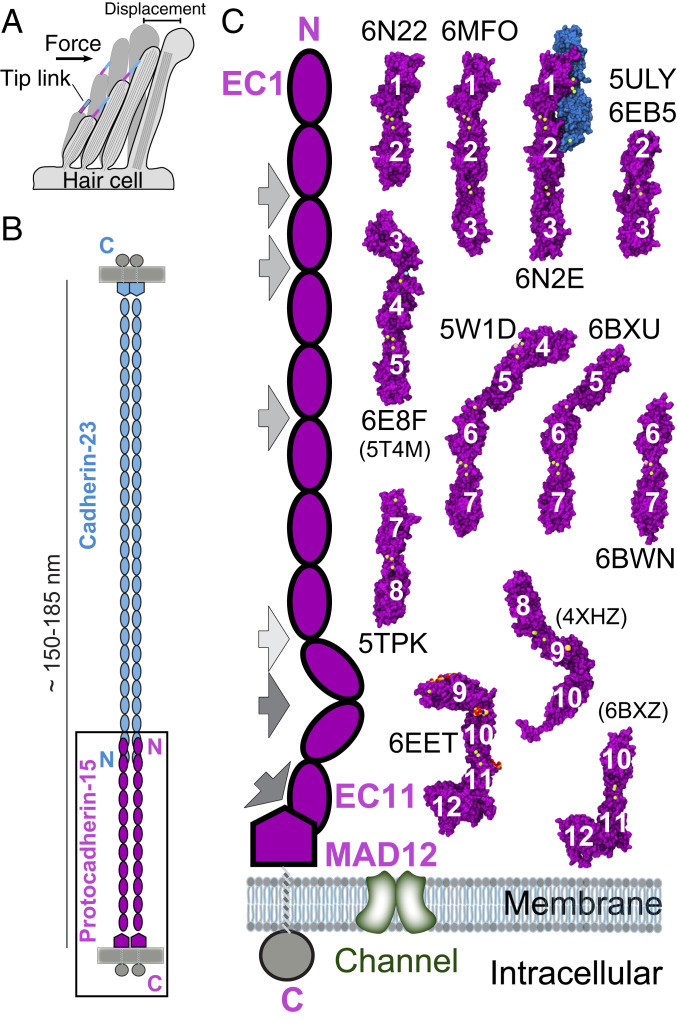
Inner-ear sensory perception begins when hair-cell mechanosensitive ion channels (1–3) are gated by “tip links,” fine protein filaments essential for hearing and balance (Fig. 1A) (4–7). Tip links are 150 to 180 nm long (8–10), their integrity is calcium (Ca2+)-dependent (5, 11), and they are formed by protocadherin-15 (PCDH15; ∼200 kDa) and cadherin-23 (CDH23; ∼370 kDa) (12, 13), two large proteins involved in hereditary deafness (14, 15), audiogenic seizures (16), and progressive blindness (17). Mature tip links are thought to be heterotetramers formed by parallel (cis) homodimers of PCDH15 interacting in an antiparallel trans mode (tip-to-tip) with cis homodimers of CDH23 (13), whereas immature tip links are likely formed by trans interactions of two PCDH15 molecules (18, 19).
Fig. 1.
Inner-ear mechanotransduction and PCDH15 structures. (A) Hair-cell bundle row showing location of tip links. Force from sound or head movements displaces the bundle to activate the transduction apparatus. (B) The tip link is made of CDH23 (blue) and PCDH15 (purple). (C) Schematics of PCDH15 (Left) and structures of PCDH15 fragments (Right) used to build models of the entire PCDH15 ectodomain. PDB codes are indicated for all 11 structures presented here, with codes in parentheses for three additional structures presented elsewhere (28, 31, 32). All structures are in surface representation, with PCDH15 fragments in purple, CDH23 EC1-2 in blue, Ca2+ ions in green, and glycosylation sugars in red and yellow. Gray arrows in the schematics indicate EC linkers with atypical canonical-like linker regions (EC8-9 in light gray), with partial Ca2+-free linker regions (EC2-3, EC3-4, and EC5-6 in gray), and with Ca2+-free linker regions (EC9-10 and EC11-MAD12 in dark gray).
Some details of the PCDH15 and CDH23 interaction are well understood: Immunogold electron microscopy (EM) suggests that tip-link lengths are mostly consistent with the predicted length of the combined proteins interacting tip-to-tip (8, 13); competitive binding of exogenous PCDH15 and CDH23 tip fragments to endogenous proteins blocks regeneration of tip links and associated transduction currents during hair-cell development and after tip-link rupture with a Ca2+ chelator (20); and structures of the complex formed by the tips of PCDH15 and CDH23 engaged in a heterodimeric molecular “handshake” have been obtained and validated in vitro and in vivo (21, 22). Furthermore, mutations that cause deafness in humans and mice have been shown to or are predicted to break the handshake interaction (21–24). In addition, studies of the CDH23 and PCDH15 heterodimer show that its strength can be tuned by PCDH15 isoforms that have distinct N-terminal tips (25). The strength of the heterotetrameric bond can be further diversified when considering combinations of PCDH15 isoforms in parallel. However, little was known about the structure of parallel cis dimers formed by PCDH15 until recently (26–28). While negative-staining transmission EM showed conformationally heterogeneous parallel dimers for tip-link components (13), most extracellular fragments of PCDH15 (and CDH23) studied had been monomeric in solution (26, 29–33). Recent crystal structures of PCDH15 fragments (26–28) and a low-resolution EM-based model suggest two points of dimerization (26, 27), yet a detailed atomistic model of the complete ectodomain is still missing. In addition, how PCDH15 engages in trans homodimers is unclear, and the structural details and strength of the PCDH15 + CDH23 heterotetrameric bond are not known.
The oligomerization states of PCDH15 and CDH23 and the architecture of their heterotetrameric bond are important determinants of tip-link mechanics and inner-ear mechanotransduction (10, 26). Disruption of either the complex between PCDH15 and CDH23 or the oligomerization of PCDH15 impairs transduction (21, 22, 26). The inner-ear transduction channel is gated by a soft element called the “gating spring,” which is either in series with the tip link or the tip link itself (2, 3, 34–36). Whether cadherin tip links are elastic or rigid remains controversial (6, 10, 30, 31). The ultrastructure of tip links suggested a stiff elastic element (6), but the length of tip links varies in situ (8). PCDH15 and CDH23 feature 11 and 27 extracellular cadherin (EC) “repeats” (Fig. 1B), respectively, and initial studies of the structure and simulated dynamics of the CDH23 EC1-2 tip predicted that these canonical repeats and their linker region fully occupied by Ca2+ ions at sites 1, 2, and 3 would be stiff (30). However, while the EC repeats along PCDH15 and CDH23 are predicted to share a common fold, they vary in sequence, which can result in structural heterogeneity, as seen for other long cadherins (37, 38). For example, crystal structures revealed a bent and L-shaped Ca2+-free PCDH15 EC9-10 linker region, with simulations evincing that unbending can provide some elasticity to the tip link (31). Bending and flexibility at this L-shaped EC9-10 linker region are also observed in low-resolution cryo-EM conformations of a cis dimeric PCDH15 fragment encompassing EC8 down to its transmembrane helix (27). These results highlight the diverse mechanical responses of various tip-link cadherin fragments.
Crystal structures and simulations have also shown that other parts of PCDH15 can be structurally distinct. The PCDH15 EC3-4 linker is flexible and binds two Ca2+ ions, instead of three. In addition, this atypical linker binds Ca2+ ions with decreased binding affinity (45 μM for site 3 and >100 μM for site 2) as compared to the canonical Ca2+-binding linker of CDH23 EC1-2 (5 μM for site 3, 44 μM for site 2, and 71 μM for site 1) (30, 32, 39). Occupancy of Ca2+-binding sites at EC linkers will greatly determine the mechanics of tip links and whether unfolding of EC repeats could occur before unbinding of the handshake bond (21). Interestingly, bulk endolymphatic Ca2+ concentration in the cochlea is tightly regulated and varies along its length from ∼20 μM (base) to ∼40 μM (apex) (40, 41). However, Ca2+ concentration near cochlear tip links could be significantly larger (42, 43), and vestibular Ca2+ concentrations range from 90 to 150 μM (41, 44). Yet, the structure and the Ca2+-dependent mechanics of entire heterotetrameric tip links remain undetermined.
To better understand the mechanics of tip links and the first steps of inner-ear mechanotransduction, we have determined 11 X-ray crystal structures of PCDH15 fragments (Fig. 1C and SI Appendix, Table S1), including the structure of a PCDH15 EC1-3 + CDH23 EC1-2 heterotetrameric complex depicting a parallel homodimer of PCDH15 interacting with two molecules of CDH23, and 10 other fragments covering all of PCDH15’s EC repeats, all of its EC linkers, and its membrane-adjacent domain MAD12 (28), also referred to as SEA (45), PICA (26), or EL (27). These structures allowed us to assemble a complete model of the monomeric ectodomain of PCDH15, to suggest models of trans and cis PCDH15 homodimers, and to build a model of the PCDH15–CDH23 bond. In addition, we used steered molecular dynamics (SMD) (46) simulations to predict the Ca2+-dependent mechanics of these models, which suggest a multimodal (stiff or soft) elastic response for PCDH15 and indicates some conditions in which PCDH15 can provide both the elasticity and extensibility typically associated with the hair-cell gating spring.
Results
Sequence analyses of PCDH15 EC repeats and its MAD12 from different species reveal an overall sequence identity of ∼46 ± 9%, with EC1 and MAD12 being the most conserved (∼59%) and EC8 being the least (∼32%) (SI Appendix, Figs. S1 and S2 and Table S2). In contrast, comparison among EC repeats and MAD12 in Homo sapiens (hs) PCDH15 reveals low overall sequence identity (∼21%), suggesting that there is structural variability throughout various parts of the protein’s ectodomain within a single species. Previously published structures of WT Mus musculus (mm) PCDH15 EC1-2 (bound to mm CDH23 EC1-2) (21), mm PCDH15 EC1-3 (26), hs PCDH15 EC3-5 without exon 12a (ex12a−) (32), hs PCDH15 EC8-10 (31), Sus scrofa (ss) PCDH15 EC10-11+MAD12 (EC10-MAD12) (28), and mm PCDH15 EC11-MAD12 (27) reflect this variability, with unique features seemingly adapted for function in heterophilic (EC1-2) and homophilic binding (EC2-3 and EC11-MAD12), and in force communication (EC3-5 and EC8-MAD12). Isoform-specific structural variability of PCDH15’s ectodomain also hints at functionally relevant adaptations (25).
To gain further insights into the function of specific repeats and to build a complete model of PCDH15, we worked with multiple WT (hs NP_001136235.1; mm NP_075604.2) and mutated protein fragments (rationale for mutations are explained throughout the text). Successful expression, purification, crystallization, and structure determination (SI Appendix, Tables S1, S3, and S4) was possible for mm PCDH15 EC1-2 with a biotin acceptor peptide (BAP); hs PCDH15 EC1-3 with mutations p.G16D, p.N369D, and p.Q370N (residue numbering in the text and structures corresponds to processed protein, see SI Appendix, SI Methods), alone and in complex with mm CDH23 EC1-2 carrying the mutation p.T15E, hs PCDH15 EC2-3 (WT and mutant p.V250N), hs PCDH15 EC3-5 with exon 12a (ex12a+), mm PCDH15 EC4-7, mm PCDH15 EC5-7 (p.I582T variant), mm PCDH15 EC6-7, and mm PCDH15 EC7-8 (p.V875A variant). All of these protein fragments were purified from bacterial inclusion bodies under denaturing conditions and refolded (SI Appendix, SI Methods, Note 1, and Table S3). In addition, we solved the structure of the mammalian-expressed, and thus glycosylated, mm PCDH15 EC9-MAD12 fragment. A summary of the protein structures presented is shown in Fig. 1C (see also SI Appendix, Table S1). Below, we discuss overall properties of EC repeats (from N to C terminus) as well as models and simulations of the full-length monomeric and dimeric PCDH15 ectodomain in complex with CDH23 EC1-2 or EC1-3.
PCDH15 N-Terminal Structures Suggest cis and trans Interactions.
Individual EC repeat structures and their relative orientation determine the architecture and shape of entire cadherin ectodomains. While our structures reveal that all PCDH15 repeats (EC1 to EC11) have a typical seven β-strand cadherin Greek-key motif topology (β-strands labeled A to G) (SI Appendix, Fig. S1), there are some significant variations in loops, secondary structure, linker regions, orientation of the EC repeats, and oligomerization that might be important for PCDH15’s function.
The structure of mm PCDH15 EC1-2BAP (Fig. 1C) is similar to other structures in which the same protein fragment has been crystallized in complex with mm CDH23 EC1-2 (21), or as part of mm PCDH15 EC1-3 (26) (core RMSD < 1.5 Å). Size-exclusion chromatography (SEC) experiments suggest that the mm PCDH15 EC1-2BAP fragment is monomeric in solution, and an analysis of the structure’s crystallographic contacts does not reveal possible homophilic interfaces that could mediate PCDH15-PCDH15 trans interactions expected for immature tip links (19). The mm PCDH15 EC1-2BAP linker region is canonical and is fully occupied by Ca2+ ions at sites 1, 2, and 3, as expected for linker regions with the Ca2+-binding motif NTerm-XEX-DXD-D(R/Y)(D/E)-XDX-DXNDN-CTerm (47). In contrast, the EC2-3 linkers in the structures of the hs PCDH15 EC1-3 G16D/N369D/Q370N and hs PCDH15 EC2-3 WT fragments (both dimeric in solution) have only two bound Ca2+ ions (sites 2 and 3) (SI Appendix, Fig. S3A). A couple of modified Ca2+-binding motifs in which DYE is p.200NYE202, and DXNDN is p.236DGDDL240, suggest that impaired Ca2+ binding at site 1 is encoded in the sequence, yet a third structure of a designed mutant hs PCDH15 EC2-3 p.V250N that disrupts cis homodimerization has three bound Ca2+ ions (SI Appendix, Fig. S3B). While we cannot rule out crystallization conditions, including MgCl2 (SI Appendix, Table S4), as the source of this discrepancy in ion occupancy, we do observe the p.E202 side chain now coordinating Ca2+ at sites 1 and 2 (SI Appendix, Fig. S3 A and B). These structures suggest that oligomerization can directly or indirectly alter Ca2+ binding at the EC2-3 linker, an effect that might be relevant to other cadherin complexes.
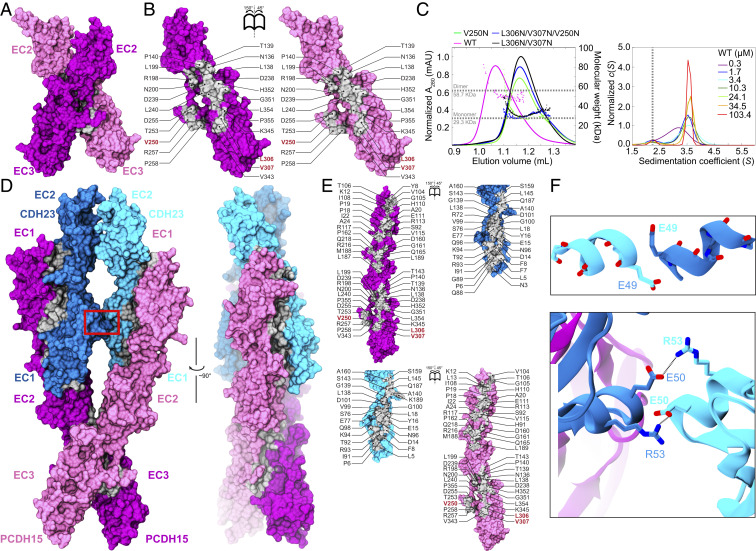
Interestingly, the crystal structures of hs PCDH15 EC1-3 G16D/N369D/Q370N and hs PCDH15 EC2-3 WT reveal crystallographic contacts with arrangements that are compatible with both trans and cis bonds for PCDH15 (Fig. 2A and SI Appendix, Fig. S4). In both cases, the interfaces involve repeats EC2 and EC3. The antiparallel interface observed in hs PCDH15 EC1-3 G16D/N369D/Q370N positions EC2 and EC3 from one subunit opposite to EC3 and EC2 from the other (interface area of 919 Å2 with an aperture angle of 154.5°) (SI Appendix, Fig. S4A), thus forming an antiparallel trans bond similar to that observed for clustered and δ protocadherins where EC1 and EC4 also play a role in the binding interface (48–51). The crystallographic contact observed in the hs PCDH15 EC2-3 WT structure reveals a potential “X” dimerization interface (∼1,052 Å2) (Fig. 2 A and B), where monomers overlap their linker regions with an aperture angle of 58.75°. In this arrangement, the EC2-3 contact forms the hinge of a “molecular scissor” with an estimated KD < 1 μM according to sedimentation velocity (SV) analytical ultracentrifugation (AUC) experiments (Fig. 2 A–C and SI Appendix, Fig. S5A). Both interfaces involve the same sides of EC2 and EC3, suggesting that aperture-angle variations due to some relative displacement of the monomers could result in switching between the antiparallel and X-dimer states, with the latter in principle able to mediate both trans and cis PCDH15–PCDH15 bonds (SI Appendix, Fig. S4 D–F).
Fig. 2.
Dimerization of PCDH15 at the EC2-3 linker and the heterotetrameric tip-link bond. (A) Front view of the hs PCDH15 EC2-3 dimer in surface representation (mauve and purple). Dimer interface is shown in silver. (B) Interaction surfaces exposed and colored as in A with interfacing residues labeled. Red labels indicate sites of mutations used to test interface. (C, Left) SEC-MALS data for hs PCDH15 EC2-3 WT fragment and mutants. Molecular mass is plotted in colored dots. Horizontal dashed lines indicate theoretical molar mass of 29.3 kDa and 58.7 kDa for monomer and dimer, respectively. (Right) c(S) distribution from SV-AUC experiments using hs PCDH15 EC2-3 at various concentrations. Peaks at S ∼3.5 represent dimers. Vertical dashed line indicates expected position for monomer. Data for mutants are shown in SI Appendix, Fig. S5. (D) Front and side views of the heterotetrameric hs PCDH15 EC1-3 G16D/N369D/Q370N + mm CDH23 EC1-2 T15E structure. Molecular surfaces are shown as in A with CDH23 in cyan and blue. Red box indicates CDH23 contacts highlighted in F. (E) Interaction surfaces exposed and colored as in D with interfacing residues labeled. (F) Detail of contacts between CDH23 monomers at their α helices in EC1 (between β-strands C and D; red box in D). (Upper) Residue p.E49 from each helix capping the dipole of the opposite helix. (Lower) p.E50:p.R53 salt bridges “interlocking” the helices of the two CDH23 monomers.
The X-dimer interface observed in hs PCDH15 EC2-3 WT is similar to that reported by Dionne et al. (26), where a crystal structure of the glycosylated mm PCDH15 EC1-3 WT fragment (PDB ID code 6CV7) shows an X-dimer with an aperture angle of 59.59° (SI Appendix, Fig. S6A). Overlapping the glycosylated mm PCDH15 EC1-3 WT monomer in the antiparallel configuration of our nonglycosylated hs PCDH15 EC1-3 G16D/N369D/Q370N structure reveals potential steric clashes between sugars at glycosylation site p.N180 and the cysteine-stapled loop in EC3 (with poor density in our structure), thus suggesting that a complete antiparallel trans state is not feasible unless the involved cysteine loop rearranges (SI Appendix, Fig. S4B). It is possible that variations in the angle of the X interface controlled by glycosylation lead to transitions from antiparallel to X-dimer states. Of note, some mutations that disrupt the X-dimer, such as p.V250N and p.V250D (26) in EC2-3, are also predicted to disrupt the PCDH15–PCDH15 antiparallel dimer, and a monomeric state was observed when the p.V250N mutation was incorporated into our glycosylated mm PCDH15 EC1-4 fragment (SI Appendix, Fig. S5B and Table S5). In contrast, the double mutation p.L306N/p.V307N at the EC3–EC3 interface of the X-dimer is not predicted to disrupt the trans antiparallel interface (SI Appendix, Fig. S4A), but the hs PCDH15 EC2-3 p.L306N/p.V307N, hs PCDH15 EC1-3 p.L306N/p.V307N, and hs PCDH15 EC1-4 p.L306N/p.V307N fragments are monomers in solution (SI Appendix, Fig. S5 A and C and Table S5). Thus, the antiparallel trans PCDH15–PCDH15 interface may use the X-dimer interaction as a required intermediate or might be short lived. While the partially deleterious impact of a similar set of mutations (including p.V250D) on hair-cell mechanotransduction highlights the relevance of EC2-3 contacts for PCDH15 function in vivo (26), an evaluation of how these mutations alter PCDH15–PCDH15 tip links (19) is still missing.
Using our structure of hs PCDH15 EC2-3, we asked if its X-dimeric arrangement would be compatible with the trans heterodimeric “handshake” interaction between PCDH15 EC1-2 and CDH23 EC1-2 (21). Alignment of EC2 repeats of PCDH15 from the handshake and X-dimer structures allowed us to build a model of the PCDH15 and CDH23 heterotetramer without steric clashes, suggesting that this is a valid model for the tip-link bond [obtained independently by Dionne et al. (26) using mm PCDH15 EC1-3] (SI Appendix, Fig. S6B). Superposition of the Danio rerio (dr) CDH23 EC1-3 structure (33) onto this complex is also compatible with the heterotetrameric bond conformation in which PCDH15 EC2-3 forms an X-dimer interface (SI Appendix, Fig. S6C), further suggesting that parallel dimerization in CDH23 involves repeats beyond EC3.
Structure of the Heterotetrameric Tip-Link Bond.
To better understand the structural determinants of the heterotetrameric tip-link configuration and to determine the cis configuration of PCDH15 upon complex formation with CDH23, we crystallized higher-affinity variants (24) of tip-link fragments containing repeats EC1 to EC3 of PCDH15 and EC1 to EC2 of CDH23. This structure, comprising two molecules each of hs PCDH15 EC1-3 G16D/N369D/Q370N in complex with mm CDH23 EC1-2 T15E, provides a complete view of the heterotetrameric tip-link bond (Fig. 2D). The two PCDH15 monomers are seen in an X configuration flanking two molecules of CDH23, thereby forming a “double handshake” consistent with the heterotetrameric model deduced from the hs PCDH15 EC2-3 structure and independently proposed by Dionne et al. (26) (Fig. 2 and SI Appendix, Fig. S6 A–C), but with structural variations, as discussed below. The structure is also compatible with longer fragments of CDH23 (SI Appendix, Fig. S6D). Interface areas for the two handshakes in our structures are different, with one of them being larger than all of the previously published heterodimeric handshakes (21) (SI Appendix, Table S6). The aperture angle for the hs PCDH15 dimer is 55.69° in the tetramer, which is smaller than what is observed for the dimeric mm PCDH15 EC1-3 (26) and hs PCDH15 EC2-3 fragments, implying that the “scissor” is less open in the tip-link tetrameric structure (SI Appendix, Fig. S6 A–C).
Interestingly, electron density quality is generally good throughout the heterotetrameric hs PCDH15 EC1-3 G16D/N369D/Q370N + mm CDH23 EC1-2 T15E structure, except for some parts of the CDH23 EC2 repeats, which were not built (β-strands F and G) (SI Appendix, Fig. S7A). Consistently, the PCDH15 EC1 and CDH23 EC1-2 repeats have larger B-factor values, which suggest some inherent flexibility or disorder compared to other regions of the model (SI Appendix, Fig. S7 A–C). Mutations G16D and T15E had been identified as enhancers of equilibrium binding for the mm PCDH15 EC1-2 and mm CDH23 EC1-2 complex (24), and may have helped stabilize a heterotetrameric complex that otherwise requires force or longer CDH23 fragments to form (mutations p.N369D and p.Q370N complete a canonical DXNDN Ca2+-binding motif at the end of PCDH15 EC3 and are not expected to affect the structure in any way). Alternatively, the stoichiometry of the complex might not be uniform across the crystal lattice, yet a composite omit 2mFo–DFc map generated with simulated annealing suggests that both mm CDH23 EC1-2 monomers are present in the crystal (SI Appendix, Fig. S7 B and C).
Some details of the handshake interactions seen in our heterotetrameric hs PCDH15 EC1-3 G16D/N369D/Q370N + mm CDH23 EC1-2 T15E structure (Fig. 2 D and E) are subtly different from those seen in the single handshake mm PCDH15 EC1-2 + CDH23 EC1-2 complex (21) (PDB ID codes 4APX and 4AXW in form I; 4AQ8 in form II). In both PCDH15 monomers of the heterotetrameric structure, the side chain of residue p.R113, known to be important for handshake formation and inner-ear function (13, 20, 21, 52), interacts with CDH23’s p.Q98 rather than with CDH23’s p.E77 (SI Appendix, Fig. S7D). A similar arrangement was seen in the structure of mm PCDH15 EC1-2 isoform CD1-2 bound to mm CDH23 EC1-2 (25) (PDB ID code 4XXW). In addition, the CDH23 EC1 repeat of one of the monomers is closer to its PCDH15 EC2 partner (SI Appendix, Fig. S7 D and E), with PCDH15 EC1-2 and CDH23 EC1-2 interrepeat angles varying across chains and structures (SI Appendix, Fig. S6 A–C), a flexing observed in previously published structures of the mm PCDH15 EC1-2 + CDH23 EC1-2 complex (21).
Importantly, our heterotetramer crystal structure differs from the heterotetramer model generated using our hs PCDH15 EC2-3 dimer (SI Appendix, Fig. S6 B and C) and from the one generated by Dionne et al. (26). The α-helices between β-strands C and D of the CDH23 EC1 repeats in the crystal structure are significantly closer to each other (Fig. 2 D and F) than what is observed in the models. Mismatched species (mouse versus human CDH23 EC1-2 differing at p.R35Q, p.P153Q, p.Q168R, and p.V174T) and engineered mutations are unlikely to explain this difference. Instead, electrostatic interactions among highly conserved charged residues from both CDH23 monomers favor a close contact in this region. Residue p.E49 from one CDH23 monomer “caps” the N terminus of the neighboring CD α-helix in the opposite monomer (Fig. 2F). This conspicuous long-range helix capping (53) appears to stabilize the overall structure of the heterotetramer. Furthermore, p.E50 from one CDH23 monomer forms a salt bridge with p.R53 of the neighboring CDH23 and vice versa (Fig. 2F). Thus, the less open conformation of the PCDH15 “scissor” squeezes the CDH23 monomers together, creating a CDH23–CDH23 interface with favorable contacts, which in turn may stabilize the overall heterotetrameric structure. Taken together, our structures and data strongly support parallel dimerization of PCDH15 at the N-terminal end mediated by EC2-3 repeats and tip-link bonds forming a heterotetramer with two PCDH15 EC1-2 + CDH23 EC1-2 handshakes (Fig. 2 and SI Appendix, Figs. S6 and S7).
PCDH15 Middle Region Structures Reveal Atypical Flexible Linkers.
The middle region of PCDH15, encompassing repeats EC4 to EC8, has been less thoroughly explored than other parts of this protein. A previous structure of hs PCDH15 EC3-5 ex12a− (PDB ID code 5T4M) revealed two bound Ca2+ ions at the EC3-4 linker region and a canonical Ca2+-binding site at the EC4-5 linker region (32). Monomers in the asymmetric unit displayed different orientations of EC3 with respect to EC4, and this flexibility was also evident in simulations of this fragment (32). However, this structure lacked exon 12a, which encodes for a seven-residue insertion p.V(414+1)PPSGVP(414+7) near the EC3-4 linker region. Given that structural variations due to isoform-specific (in-frame) insertions or deletions might be relevant for function, we sought to obtain a structure with exon 12a, present in some of the CD2 isoforms thought to be essential for inner-ear mechanosensation (12, 54, 55). We solved the hs PCDH15 EC3-5 ex12a+ structure, which has three molecules in the asymmetric unit having distinct EC3-4 interrepeat conformations and with two of the linkers with only one bound Ca2+ ion at site 3 (Fig. 3A and SI Appendix, Fig. S8 A–D). The insertion enlarges the EC4 BC loop, which projects away from the repeat without altering its folding (SI Appendix, Figs. S3C and S8A). However, this insertion may affect Ca2+-binding affinity at the nearby EC3-4 linker. Only the most bent conformation of this isoform is compatible with the antiparallel PCDH15 EC1-3 interface (SI Appendix, Fig. S4 A–C), while all conformations observed in the structure are compatible with the PCDH15 X-dimer mediated by EC2-3 (SI Appendix, Fig. S8E). Overall, our mm PCDH15 EC3-5 ex12a+ structure is consistent with enhanced flexibility at the EC3-4 linker.
Fig. 3.
Flexibility of PCDH15 EC3-4 and EC5-6 linker regions. (A) Ribbon representation of hs PCDH15 EC3-5 ex12a+ with monomers (mauve, purple, and magenta) superposed in two views illustrating flexibility at EC3-4. Also superposed are monomers of hs PCDH15 EC3-5 ex12a− (yellow and gray, PDB ID code 5T4M) (32). Black box highlights location of ex12a+ insertion (SI Appendix, Fig. S8A). (B) Ribbon representation of mm PCDH15 EC5-7 with monomers from mm PCDH15 EC4-7 (gray) and from mm PCDH15 EC5-7 I582T (mauve and purple) superposed. (C) Four representative structural conformations of the EC2-3–mediated PCDH15 EC1-7 homodimer from crystallographic conformations of the PCDH15 EC3-5 and EC5-7 linkers observed in our structures hs PCDH15 EC3-5 ex12a+ (colored as in SI Appendix, Fig. S8E), mm PCDH15 EC4-7 (brown), and mm PCDH15 EC5-7 I582T (orange and lime). Distance between the EC7 C termini is indicated for each configuration, some of which are compatible with cis (Left) or trans (Right) dimers (SI Appendix, Fig. S4 D–G).
Another atypical linker region is expected in PCDH15 EC5-6: Sequence alignments show that EC5 has a modified XEX motif (p.498YTD500) and lacks the DRE motif (p.553IVG555) present in canonical Ca2+-binding linker regions (SI Appendix, Figs. S1 and S2 A and B). In addition, the DXNDN linker is p.586PPNNQ590 in EC5-6, although motifs involved in Ca2+ binding at site 3 (top of EC6) are mostly unchanged (DXD is p.621DRE623 and XDX is p.670SDG672). Two of our crystal structures cover the EC5-6 linker, which is seen in three different conformations (one in mm PCDH15 EC4-7 and two in mm PCDH15 EC5-7 I582T) (Fig. 3B and SI Appendix, Fig. S3D). In all cases, only one bound Ca2+ ion is observed at site 3 in EC6, as expected from the modified motifs listed above. Relative orientations of EC6 with respect to EC5 are substantially different (Fig. 3B). In contrast, structures of mm PCDH15 EC6-7 and mm PCDH15 EC7-8 V875A show canonical, rather straight linkers with three bound Ca2+ ions (natural variants I582T and V875A are not expected to alter structures). While the two PCDH15 EC5-7 monomers in the asymmetric unit form a compelling parallel cis dimer in the mm PCDH15 EC5-7 I582T crystal structure (interface area of 1,049.5 Å2) (SI Appendix, Fig. S8 F and G), all of our fragments covering PCDH15 EC4-7, EC5-7, and EC6-7 were monomeric in solution, as also reported for similar fragments by Dionne et al. (26).
Overall, our structures of the middle region of PCDH15 suggest flexibility and altered Ca2+ binding at EC3-4 and EC5-6 linkers. Furthermore, three alternate conformations for each of the EC3-4 and EC5-6 linker regions assembled in an X-dimer mediated by EC2-3 give rise to 45 unique possible combinations of PCDH15 monomers (4 representative conformations are shown in Fig. 3C). Some of these conformations display a remarkable separation between EC7 repeats (up to ∼26.8 nm), compatible with a PCDH15–PCDH15 trans X-dimer (SI Appendix, Fig. S4D). Importantly, given prior simulations of the EC3-4 fragment (32) and the architecture of EC5-6, flexibility at these linker regions will occur even at high Ca2+ concentrations, with a large number of structurally diverse configurations facilitated by the EC2-3 X-dimer.
PCDH15 Ectodomain C-Terminal Structures Reveal Kinks and cis Dimerization.
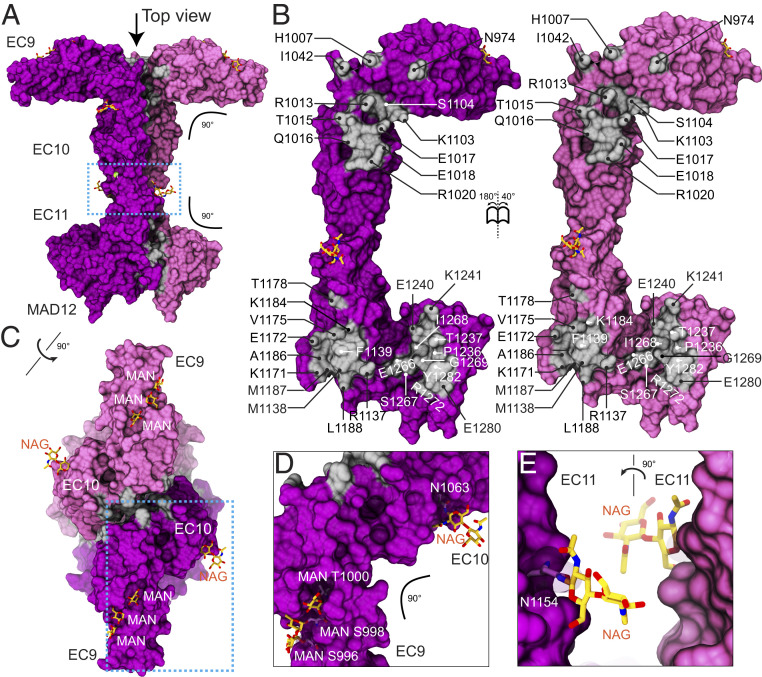
Three previous studies reported high-resolution structures covering parts of the C-terminal end of PCDH15, including EC9 to MAD12 (hs PCDH15 EC8-10, mm PCDH15 EC9-10, ss PCDH15 EC10-MAD12, and mm PCDH15 EC11-MAD12) (27, 28, 31). These structures revealed a semicanonical EC8-9 linker region, a flexible, Ca2+-free and bent EC9-10 linker region, a canonical EC10-11 linker region, and the L-shaped arrangement of a ferredoxin-like MAD12 tucked against EC11, with EC11-MAD12 inducing parallel dimerization (26–28). Our structure of mm PCDH15 EC9-MAD12 (Fig. 4 A and B) displays the bent EC9-10 linker region as observed in the hs PCDH15 EC8-10 and mm PCDH15 EC9-10 structures, albeit with a different azimuthal angle (31). Remarkably, this structure also shows the EC10-MAD12 dimerization domain in a symmetric dimer (one molecule per asymmetric unit). The total dimer interface surface area is 1,267.9 Å2 (including contacts between MAD12 and EC11 and between EC10 repeats) (Fig. 4B), somewhat larger than what is observed in the more asymmetric ss PCDH15 E10-MAD12 structure (28) (∼1,160 Å2, two molecules per asymmetric unit), with the interface area at the EC10–EC10 contact being larger (∼336 Å2 versus ∼185 Å2 for 6BXZ). Glycosylation did not interfere with bending and dimerization (Fig. 4 C–E and SI Appendix, Note 1 and Fig. S9). The mm PCDH15 EC9-MAD12 structure confirms that dimerization is induced by EC11-MAD12 and that it is compatible with a kinked EC9-10.
Fig. 4.
Dimerization of PCDH15 at the EC9-MAD12 region. (A) Molecular surface showing the glycosylated mm PCDH15 EC9-MAD12 parallel dimer in mauve and purple. Dimer interfaces are shown in silver, and glycosylation sugars are shown as yellow sticks. Blue dashed box denotes region highlighted in E. (B) Interaction surfaces exposed and colored as in A with interfacing residues labeled. (C) Top view of the mm PCDH15 EC9-MAD12 parallel dimer, with EC9 repeats pointing in opposite directions. Blue dashed box denotes region highlighted in D. (D and E) Details of glycosylation sites (dashed boxes in A and C).
Intriguingly, bending at the EC9-10 linker region in the context of the dimer positions EC9 repeats pointing in opposite directions, with the projections along their longest principal axis being parallel to a hypothetical membrane plane (SI Appendix, Fig. S4G). Data from SEC-multiple angle light scattering (MALS) confirmed that this protein fragment is dimeric in solution (SI Appendix, Fig. S5B and Table S5), and the EC9-10 “kink” is also consistent with our previous structures (31) and with low-resolution cryo-EM images of a PCDH15 fragment encompassing repeat EC8 down to PCDH15’s transmembrane helix (27). Small-angle X-ray scattering (SAXS) data obtained using bacterially produced mm PCDH15 EC9-MAD12 are also consistent with a dimer in solution, with molecular mass estimates of 98.12 kDa and a 5.6% discrepancy with the sequence-derived molecular mass of 51.98 × 2 = 103.96 kDa (SI Appendix, Note 2, Fig. S10, and Table S7).
Conformational transitions that straighten the EC9-10 linker, observed in simulations (31) and in cryo-EM data (27), strongly suggest that bending and unbending processes are relevant for tip-link assembly and function. The variety of angles that EC9-10 may adopt in solution (27), along with dimerization of PCDH15 at EC2-3 and MAD12, and the asymmetry and flexibility suggested by SAXS data, offer an interesting set of arrangements for PCDH15 + CDH23 filaments that might be adopted by tip links and kinociliary links (8, 56). In addition, flexing at EC3-4 (Fig. 3A), EC5-6 (Fig. 3B), and at EC9-10 (Fig. 4) seems to be a basic geometric requirement for establishing a parallel PCDH15 cis dimer compatible with dimerization points mediated by both EC2-3 and EC11-MAD12.
The structures and models discussed above depict parts of the entire ectodomain of PCDH15, and only small fractions of the entire tip link. Although each of them provides valuable insight into PCDH15 function, the assembly of PCDH15 filaments and force transduction mediated by tip links requires an understanding of the structure, dynamics, and elastic response of the entire PCDH15–CDH23 complex. Therefore, we used our structures to build, simulate, and computationally stretch monomeric PCDH15 EC1-MAD12 models, heterotetrameric PCDH15 EC1-5 + CDH23 EC1-2 or PCDH15 EC1-5 + CDH23 EC1-3 bonds, and two independent complete dimeric PCDH15 EC1-MAD12 models bound to CDH23 EC1-2 or CDH23 EC1-3.
Predicted Elasticity of the Complete PCDH15 Ectodomain Monomer.
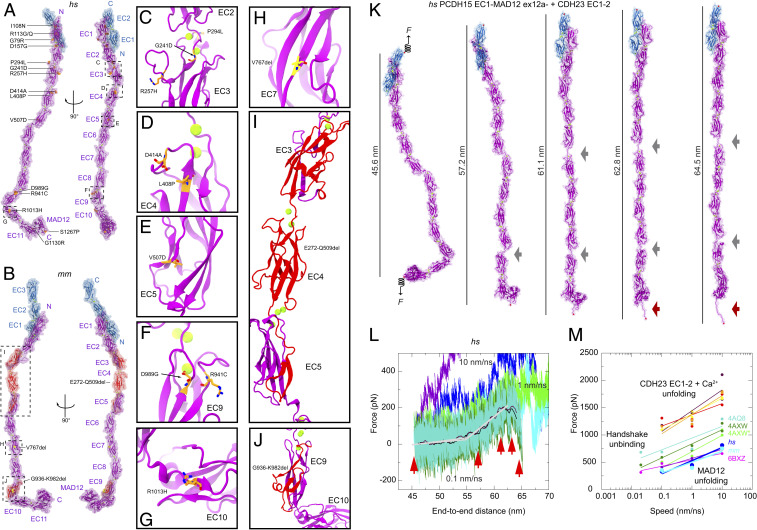
To gain insights into the function and elastic behavior of PCDH15, and as similarly done for parts of CDH23 (33) and the entire ectodomain of the olfactory cell adhesion molecule (57), we used overlapping structures of PCDH15 fragments and built two atomistic models of its entire ectodomain. The first model was built to get the hs PCDH15 EC1-MAD12 ex12a− WT protein ectodomain (p.Q1 to p.I1342), which was coupled to a model of hs CDH23 EC1-2 (p.Q1 to p.D205) using the original handshake interaction (21) as a template (PDB ID code 4APX) (Fig. 5A and SI Appendix, Fig. S11 A–K and Table S8) (∼172 kDa for the complex). The second model was built to get the mm PCDH15 EC1-MAD12 ex12a+ WT protein ectodomain [p.D5 to p.S1341(+7) carrying the p.I582(+7)T and p.V875(+7)A variations] coupled to a model of mm CDH23 EC1-3 (p.V2 to p.D317) using the handshake interaction observed in our hs PCDH15 EC1-3 G16D/N369D/Q370N + mm CDH23 EC1-2 T15E structure (Fig. 5B and SI Appendix, Fig. S11 L–V and Table S8) (∼184 kDa for the complex). These PCDH15 models allowed us to visualize 3 in-frame deletion segments and 15 sites of missense mutations implicated in inherited deafness (Fig. 5 A–J and SI Appendix, Note 3 and Table S9), thus providing a structural framework to propose mechanisms underlying dysfunction of PCDH15.
Fig. 5.
Disease-causing mutations and complete model and elasticity of the monomeric PCDH15 ectodomain. (A) Two views of the hs PCDH15 EC1-MAD12 ex12a− + CDH23 EC1-2 model with PCDH15 deafness-causing missense mutations listed. Proteins are shown in purple (PCDH15) and blue (CDH23) ribbon representations with transparent molecular surfaces. Ca2+ ions are shown as green spheres and mutations are highlighted in orange. Dashed boxes and labels correspond to selected missense mutations shown in C–G. (B) The mm PCDH15 EC1-MAD12 ex12a+ + CDH23 EC1-3 model with PCDH15 in-frame deletion mutations that cause deafness highlighted in red (multiresidue deletions) and yellow (single-residue deletions). Dashed boxes and labels correspond to detailed views in H–J. (C–J) Detail of missense mutations and in-frame deletions that cause deafness. (K) Snapshots of the monomeric hs PCDH15 EC1-MAD12 ex12a− + CDH23 EC1-2 system during stretching simulation S1d (0.1 nm/ns) (SI Appendix, Table S10). Stretched C-terminal Cα atoms are shown as red spheres. Springs in first panel indicate position and direction of applied forces. Gray arrows highlight stretching of PCDH15 EC linkers (EC9-10 followed by EC5-6). Dark red arrow indicates unfolding of PCDH15 MAD12’s C-terminal end. (L) Force versus end-to-end distance for constant velocity stretching of hs PCDH15 EC1-MAD12 ex12a− + CDH23 EC1-2 at 10 nm/ns (S1b, purple and blue), 1 nm/ns (S1c, bright green and cyan), and 0.1 nm/ns (S1d, dark green and turquoise; 10-ns running averages shown in black and maroon; gray lines are fits used to determine elasticity of the complex). Red arrowheads indicate time-points for S1d illustrated in K. (M) In silico force peak maxima versus stretching speed for CDH23 EC1-2 unfolding (red, maroon, yellow, and orange) (30), for CDH23 EC1-2 and PCDH15 EC1-2 handshake unbinding (turquiose-4AQ8, dark green-4AXW, and light green-4AXW* [after a 1-μs-long equilibration]) (21, 25), for PCDH15 EC10-MAD12 unfolding (magenta-6BXZ with unfolding at MAD12) (28), for hs PCDH15 EC1-MAD12 ex12a− + CDH23 EC1-2 unfolding (S1b-d, blue with unfolding at MAD12), and for mm PCDH15 EC1-MAD12 ex12a+ + CDH23 EC1-3 unfolding (S2b-d, cyan, with unfolding at MAD12).
Our models of the PCDH15 ectodomain adopt conformations dictated by the relative orientation (tilt and azimuthal angles) of successive EC repeats observed in various crystallographic structures, including the EC9-10 kink observed in hs PCDH15 EC8-10 (31) (used for the hs PCDH15 EC1-MAD12 ex12a− model) and in mm PCDH15 EC9-MAD12 (used for the mm PCDH15 EC1-MAD12 ex12a+ model). These may represent only some of the possible conformations adopted in solution and might not be representative of states observed under physiological tension experienced by hair-cell tip links in vivo (10 to 100 pN) (35, 36, 58, 59). Short equilibrations followed by SMD simulations of the entire hs and mm PCDH15 ectodomain monomers bound to the tips of CDH23 and with all their Ca2+-binding sites occupied allowed us to explore their conformational dynamics and elasticity in silico. Constant-velocity stretching of the hs PCDH15 EC1-MAD12 ex12a− + CDH23 EC1-2 heterodimer complex at 0.1 nm/ns revealed straightening of the structure (Fig. 5K, SI Appendix, Fig. S12A and Table S10, and Movie S1) in a first phase characterized by a soft effective spring constant (k1 ∼3.3 mN/m and ∼10 nm extensibility) (Fig. 5L), followed by stretching of the EC5-6 and EC9-10 linker regions, ensued by stretching of the entire chain (k2 ∼24.4 mN/m and ∼5 nm extensibility). Subsequent unrolling and then unfolding of MAD12 at a force peak of ∼331 pN happened without unbinding of CDH23 EC1-2 from PCDH15. The elastic response of mm PCDH15 EC1-MAD12 ex12a+ + CDH23 EC1-3 at 0.1 nm/ns was similar to that of the human complex (k1 ∼1.5 mN/m and ∼15 nm extensibility, k2 ∼27.5 mN/m and ∼5 nm extensibility, unfolding peak at ∼356 pN) (SI Appendix, Fig. S12 B and C and Movie S2), suggesting that, at the stretching speeds tested, unfolding of MAD12 occurs before unbinding.
The soft elasticity phase observed during stretching simulations of the monomeric PCDH15 EC1-MAD12 + CDH23 EC1-2/3 complexes stems mainly from unbending of the Ca2+-free EC9-10 linker region, as previously predicted (31), and from stretching of EC5-6 and various other linkers. The predicted spring constants at 0.1 nm/ns are an upper limit, as this stretching speed is one order of magnitude faster than physiological speeds of the basilar membrane (60) at loud high-frequency sound and drag might be reduced at slower speeds. Similarly, unfolding forces of MAD12 at 0.1 nm/ns represent an upper bound as simulations and experiments at slower stretching speeds will naturally report smaller forces (61, 62). The elastic response of MAD12 in the context of stretching simulations of the monomeric hs and mm PCDH15 EC1-MAD12 + CDH23 EC1-2/3 complexes is consistent with simulations stretching the dimeric mm PCDH15 EC9-MAD12 (SI Appendix, Fig. S12 D and E and Movie S3) and our previous simulations of ss PCDH15 EC10-MAD12 (28). Our new simulations including the entire monomeric PCDH15 ectodomain predict soft elasticity and unfolding of MAD12 before unbinding of the cadherin handshake at fast stretching speeds.
Predicted Strength of the Heterotetrameric PCDH15–CDH23 Bond.
To better understand the behavior of the handshake bond in the context of the heterotetrameric arrangement predicted from our hs PCDH15 EC2-3 structure and the conformation observed in the hs PCDH15 EC1-3 G16D/N369D/Q370N + mm CDH23 EC1-2 T15E complex, we built two model systems for simulation (SI Appendix, Fig. S13). The first one includes two molecules of hs PCDH15 EC1-5 ex12a− (p.Q1 to p.P587) forming an X-dimer (as seen in the hs PCDH15 EC2-3 structure) coupled to two molecules of hs CDH23 EC1-2 (p.Q1 to p.D205), each arranged as seen in the mouse handshake complex (PDB ID code 4APX) (21) and mutated to match the human sequence (SI Appendix, Fig. S13A). The second system includes two molecules of mm PCDH15 EC1-5 ex12a+ [p.Q1 to p.L585(+7)] forming an X-dimer coupled to two molecules of mm CDH23 EC1-3 (p.Q1 to p.D317) with the cis and trans interactions based on the hs PCDH15 EC1-3 G16D/N369D/Q370N + mm CDH23 EC1-2 T15E tetrameric structure (SI Appendix, Fig. S13C). In both models, referred to here as hs and mm (PCDH15 EC1-5)2 + (CDH23 EC1-2/3)2, all linkers and repeats were in conformations obtained from crystal structures and all Ca2+-binding sites were occupied. To mimic possible low Ca2+ concentration conditions in the cochlea, three additional models of the hs (PCDH15 EC1-5)2 + (CDH23 EC1-2)2 tetramer were built with two, one, or zero Ca2+ ions per linker, respectively (SI Appendix, Table S10). All systems, saturated and not saturated with Ca2+, were equilibrated and then stretched to test their stability and elasticity in silico.
Equilibration trajectories of the hs and mm (PCDH15 EC1-5)2 + (CDH23 EC1-2/3)2 tetramers saturated with Ca2+ revealed a tendency for the PCDH15 EC5 ends to get closer to each other within 10 ns (from ∼16 nm in hs and ∼12 nm in mm to ∼10 nm in both). The hs CDH23 EC1-2 monomers also got close to each other, mimicking the arrangement seen in the hs PCDH15 EC1-3 G16D/N369D/Q370N + mm CDH23 EC1-2 T15E structure, where β-strands A and G of EC2 from one CDH23 monomer are closer to the same strands in EC2 from the adjacent monomer. However, the p.E50:p.R53 salt bridge between adjacent CDH23 monomers (as seen in the crystal structure of the heterotetrameric bond) (Fig. 2F) did not form in the hs complex. The PCDH15 p.R113 salt bridge with CDH23 p.E77, absent in the mm model, was formed throughout its equilibration trajectory. While these rearrangements of specific bonds highlight the dynamic nature of the heterotetrameric complex, key features—like the double handshake bond and the PCDH15 EC2-3 interface for parallel dimerization—remained stable during these short equilibrations.
Systems with fewer Ca2+ ions at all linkers exhibited two distinct behaviors in our equilibrium simulations. The system with two Ca2+ ions per linker was stable, with the two PCDH15 EC5 repeats coming as close as ∼9 nm in a configuration that is more compatible with a parallel dimer than the one observed in the system with fully saturated linkers (three Ca2+ ions per linker, except for PCDH15 EC2-3 and EC3-4 with two Ca2+ ions each). More dramatic conformational changes, including bending and twisting of linkers, were observed for systems with one and zero Ca2+ ions per linker, especially at the PCDH15 EC3-4 linker regions. The conformational changes observed with less Ca2+, resulting in overall shrinkage of the end-to-end distance of the complex, are consistent with results from simulations of tip-link cadherins (21, 30, 32) that in the absence of bound Ca2+ ions exhibited enhanced hinge-like flexibility at their linker regions. These results are also consistent with EM images showing curled and collapsed tip-link cadherins in the absence of Ca2+ (13).
Stretching of the equilibrated hs (PCDH15 EC1-5)2 + (CDH23 EC1-2)2 tetramer saturated with Ca2+ at the constant speeds tested (10, 1, and 0.1 nm/ns) revealed unbinding of the CDH23 EC1-2 monomers from PCDH15 without unfolding of EC repeats (SI Appendix, Fig. S13A and Movie S4). At the slowest stretching speed (0.1 nm/ns), the hs (PCDH15 EC1-5)2 + (CDH23 EC1-2)2 complex straightened, with repeats EC4-5 of PCDH15 establishing close contacts in a parallel arrangement. The “scissor” conformation of the PCDH15 EC1-3 dimer closed itself as the stretching proceeded, thereby squeezing the CDH23 EC1-2 monomers together. As a result, an intricate network of hydrogen bonds developed between conserved CDH23 EC1 residues. Furthermore, a rotation of the CDH23 EC1-2 monomers (SI Appendix, Fig. S13A) preceded unbinding at large force (∼1,080 pN, simulation S3d), which was roughly three times as large as the force observed during unfolding of MAD12 in the monomeric hs PCDH15 EC1-MAD12 + CDH23 EC1-2 complex (∼330 pN, simulation S1d), and ∼1.5 times larger than the force predicted for the unbinding of two noninteracting parallel handshakes (∼720 pN) (21). Similarly, stretching of the mm (PCDH15 EC1-5)2 + (CDH23 EC1-3)2 complex at 0.1 nm/ns resulted in unbinding at a large force peak (∼800 pN, simulation S7d) (SI Appendix, Fig. S13D), with squeezing of the CDH23 EC1-3 monomers. Thus, force-induced “locking” of CDH23 may make the tip-link bond more resistant against unbinding. Remarkably, two force peaks were observed at the slowest stretching speed (0.02 nm/ns, simulation S7e), suggesting the existence of an intermediate state before complete unbinding (SI Appendix, Fig. S13E, and Movie S5), which occurred after forces reached ∼730 pN. The PCDH15 EC2-3 X-dimer interface was distorted but not lost after unbinding. These results suggest that the mechanical strength of the tetrameric tip-link bond is bolstered by the PCDH15 EC2-3 X-dimer.
Stretching of the equilibrated hs (PCDH15 EC1-5)2 + (CDH23 EC1-2)2 tetramers not saturated with Ca2+ at all linker regions revealed a more complex set of events during unbinding (SI Appendix, Fig. S13B). SMD simulations of the system with two Ca2+ ions per linker at 0.1 nm/ns revealed unbinding without unfolding (force peak at ∼737 pN, simulation S4d) with complete separation of the PCDH15 EC2-3 X-dimer interface. SMD simulations of the system with one Ca2+ ion per linker region at 0.1 nm/ns revealed sequential unbinding (main force peak at ∼833 pN, simulation S5d) with severe stretching of linker regions. SMD simulations of the system with 0 Ca2+ ions per linker region at 0.1 nm/ns revealed minor partial unfolding, rupture of the EC2-3 X-dimer, and unbinding (force peak at 853 pN, simulation S6d) (Movie S6). The starting states for SMD simulations of systems with 1 and 0 Ca2+ ions per linker region were shorter due to bending of ECs with respect to each other, resulting in a longer stretching phase (<25 nm to ∼27.5 nm in SI Appendix, Fig. S13B) at low force with rather soft elasticity (k1Ca ∼20.5 mN/m and k0Ca ∼21.7 mN/m) dominated by unbending and straightening of the repeats. Extension of the linkers between EC repeats at higher force resulted in a displacement of the force peak to longer end-to-end distances.
Overall, our simulations of the heterotetrameric PCDH15–CDH23 bond suggest that parallel dimerization and Ca2+ ions modulate the tip-link strength, which might in turn determine whether unbinding happens before unfolding of EC repeats.
Predicted Elasticity of the Complete PCDH15 Ectodomain Dimer.
Our structures and biochemical data obtained using various PCDH15 fragments indicate that there are two possible points of parallel dimerization. The first one involves PCDH15 EC2-3 and the second involves PCDH15 EC11-MAD12. These results are consistent with data presented by Dionne et al. (26) on various fragments of PCDH15, and also with structures of the PCDH15 ectodomain tail reported by Ge et al. (27). Additional MALS experiments with the full-length ectodomain of mm PCDH15 EC1-MAD12 confirm that it is a dimer in solution that can withstand mild Ca2+-chelation with EDTA (SI Appendix, Fig. S5 D and E). However, atomistic models of the entire PCDH15 ectodomain seem to be incompatible with an arrangement in which both points of dimerization exist simultaneously (SI Appendix, Fig. S4G), even when the PCDH15 EC9-10 linker unbends (SI Appendix, Fig. S14 A–E). This incompatibility might result from the constraints imposed by limited crystallographic conformations sampled by fragments containing only a few EC repeats, rather than the entire PCDH15 EC1-MAD12 protein. To model a parallel dimeric PCDH15 complex as observed in tip links (6, 13), we systematically searched for conformations obtained from equilibrium and SMD simulations that could be assembled into structures compatible with both points of dimerization (Fig. 6 A and B, SI Appendix, Fig. S14 E–G, and Movies S7 and S8).
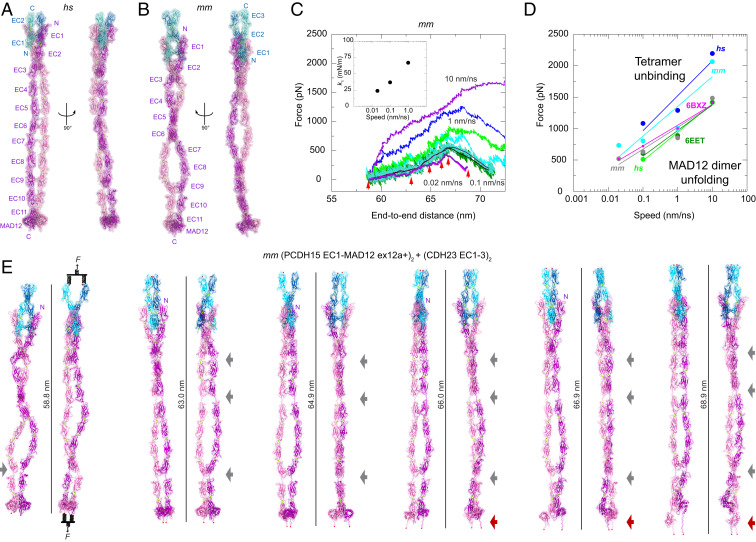
Fig. 6.
Models and mechanics of the PCDH15 ectodomain as part of an heterotetrameric complex with CDH23. (A and B) Two views of the (A) hs (PCDH15 EC1-MAD12 ex12a−)2 + (CDH23 EC1-2)2 and (B) mm (PCDH15 EC1-MAD12 ex12a+)2 + (CDH23 EC1-3)2 heterotetrameric systems before simulation. (C) Force applied to slabs versus protein separation (SI Appendix, SI Methods, Fig. S15, and Table S10) for constant velocity stretching simulations of the mm (PCDH15 EC1-MAD12 ex12a+)2 + (CDH23 EC1-3)2 system at 10 nm/ns (S9b, purple and blue), 1 nm/ns (S9c, bright green and cyan), 0.1 nm/ns (S9d, dark green and turquoise with 10-ns running averages in black and maroon), and 0.02 nm/ns (S9e, 10-ns running averages in magenta and indigo). Red arrowheads indicate time-points for simulation S9e, illustrated in E. Inset shows k1 at different stretching speeds. (D) In silico force peak maxima versus stretching speed for hs (PCDH15 EC1-5 ex12a−)2 + (CDH23 EC1-2)2 (S3b-d, blue), for mm (PCDH15 EC1-5 ex12a+)2 + (CDH23 EC1-3)2 (S7b-e, cyan), for PCDH15 EC10-MAD12 unfolding (magenta-6BXZ with unfolding at MAD12) (28), for mm PCDH15 EC9-MAD12 dimer unfolding (S11b-d, dark green with unfolding at MAD12), for hs (PCDH15 EC1-MAD12 ex12a−)2 + (CDH23 EC1-2)2 unfolding (S8b-d, bright green with unfolding at MAD12), and for mm (PCDH15 EC1-MAD12 ex12a+)2 + (CDH23 EC1-3)2 unfolding (S9b-e, gray with unfolding at MAD12). (E) Snapshots of the heterotetrameric mm (PCDH15 EC1-MAD12 ex12a+)2 + (CDH23 EC1-3)2 system during stretching simulation S9e (0.02 nm/ns). Stretched C-terminal Cα atoms are shown as red spheres. Stretching using slabs was carried out by attaching two slabs to springs that were in turn attached to the terminal ends of each protein monomer. Slabs were moved in opposite directions at constant speed through individual springs. Gray arrows highlight stretching of PCDH15 EC linkers (EC9-10 followed by EC5-6 and EC3-4). Dark red arrow indicates unfolding of PCDH15 MAD12’s C-terminal end.
The first tetramer model has the hs (PCDH15 EC1-MAD12 ex12a−)2 + (CDH23 EC1-2)2 complex (p.Q1 to p.I1342 and p.Q1 to p.D205 per monomer; ∼343 kDa), while the second contains the mm (PCDH15 EC1-MAD12 ex12a+)2 + (CDH23 EC1-3)2 complex [p.Q1 to p.S1341(+7) carrying the p.I582(+7)T and p.V875(+7)A variations for PCDH15 and p.Q1 to p.D317 for CDH23; ∼369 kDa] (SI Appendix, Table S8). All EC repeats and linker regions in these hs and mm (PCDH15 EC1-MAD12)2 + (CDH23 EC1-2/3)2 tetrameric models were obtained from crystallographic conformations or coordinates from simulations of crystal structures (SI Appendix, Fig. S14 F and G). In equilibrium simulations of the hs heterotetrameric model, in which the PCDH15 EC9-10 segment is initially straight (Fig. 6A and SI Appendix, Fig. S14F), we observed rebending at this linker region, with one of the PCDH15 monomers more kinked than the other (SI Appendix, Fig. S15A, Left). A similar asymmetric bent conformation is maintained in an equilibration of the mm model (Fig. 6 B and E). These results are consistent with simulations of the isolated PCDH15 EC8-10 fragment (31) and suggest that the EC9-10 kink can exist within the entire PCDH15 homodimer.
Equilibrated states of both hs and mm (PCDH15 EC1-MAD12)2 + (CDH23 EC1-2/3)2 tetrameric complexes were used as starting points for SMD simulations at various stretching speeds (Fig. 6 C–E, SI Appendix, Fig. S15, and Movies S9 and S10). In the slowest simulations (0.1 nm/ns) of the hs complex, straightening by unbending of various linker regions (most notably the EC9-10 kink) was followed by elongation of the EC3-4, EC5-6, and EC9-10 linkers and subsequent partial unrolling and unfolding of both MAD12s at the C-terminal end. Unbinding of CDH23 from PCDH15 was not observed at the slowest stretching speeds used. The initial straightening phase had an effective spring constant of k1 ∼45 mN/m (extensibility ∼5 nm), while unrolling and unfolding occurred at ∼510 pN, with a recoil reflected in rebending at the EC9-10 linker region (SI Appendix, Fig. S15 A and B). A similar series of events was observed for the mm complex when stretched at 0.1 nm/ns, with an initial straightening phase that had an effective spring constant of k1 ∼37 mN/m (extensibility ∼5 nm) and that was followed by MAD12 unrolling and unfolding at ∼630 pN (SI Appendix, Fig. S15 C and D). The effective spring constant decreased to k1 ∼24 mN/m (extensibility ∼5 nm) (Fig. 6 C, Inset) at a stretching speed of 0.02 nm/ns, with subsequent asymmetric unrolling and unfolding of MAD12s occurring at a force peak of ∼520 pN without unbinding (Fig. 6 C–E and Movie S11). Simulations thus predict that the complete PCDH15 ectodomain dimer is significantly stiffer than the monomeric PCDH15 ectodomain. Unfolded MAD12s, however, could provide soft elasticity and extreme extensibility (∼40 nm).
Discussion
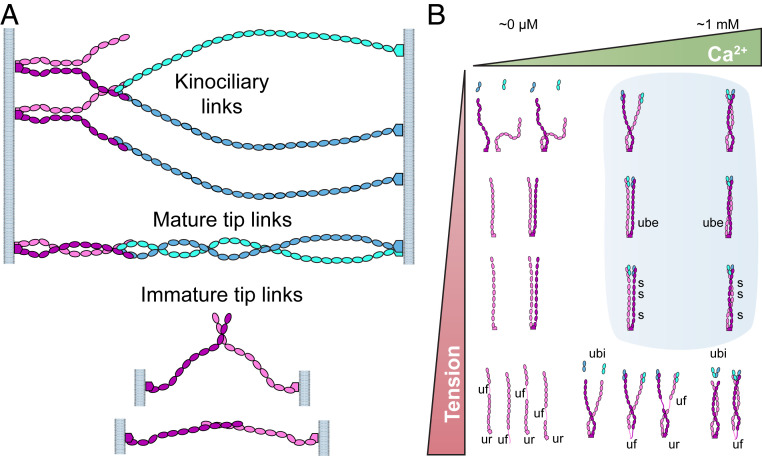
Our structures, biochemical data, and simulations provide an integrated atomistic view of the entire PCDH15 ectodomain and its possible modes of homophilic and heterophilic interaction (Fig. 7A). We found two points of parallel dimerization for PCDH15 (at EC2-3 and EC11-MAD12) and provide a detailed and unique view of the two trans PCDH15-CDH23 handshakes in the heterotetrameric bond facilitated by the PCDH15 EC2-3 X-dimer. Simulations suggest that the two handshakes in the heterotetramer are squeezed together by closing of the X-dimer scissor when tension that mimics physiological stimuli is applied. This squeezing strengthens the heterotetrameric bond with unbinding forces predicted to be on average ∼1.3 times larger than the sum of unbinding forces for independent handshakes, reminiscent of catch-bond behavior where the dissociation lifetime of a bond is increased by tensile forces (63). The dual handshake may also facilitate, through avidity, a strengthened mechanical response and extended lifetime of the tip-link bond (64).
Fig. 7.
Interaction modes and elastic behavior of PCDH15. (A) Potential homophilic and heterophilic interactions of PCDH15 (mauve and purple) and CDH23 (cyan and blue) suggested by structural analyses. Top arrangement could be adopted by kinociliary links (56), while the middle heterophilic complex is compatible with the heterotetrameric tip link (13, 21). The two remaining homophilic arrangements shown at the bottom are possible homophilic trans interactions in immature tip links (18, 19). (B) Structures and simulations indicate that the PCDH15 ectodomain can have distinct elastic responses that are tuned by tension and Ca2+. Phase diagram shows possible hypothetical states for PCDH15’s ectodomain. At very low Ca2+ concentrations (Upper Left) PCDH15 will not interact with CDH23 and its linkers will be flexible and easily stretchable under tension. At high tension and very low Ca2+ concentrations (Lower Left), the Ca2+-free linkers will be weak and unfolding (uf) at any of the EC repeats will ensue. Under normal physiological conditions (blue shade), PCDH15 will form a cis dimer that interacts with CDH23 and that can exhibit soft elasticity mediated by bending and unbending transitions of Ca2+-free linker regions EC5-6 and EC9-10. As tension increases, unbending (ube) results in stiffening of PCDH15. Stretching of linkers (s) and unrolling of MAD12 (ur) at higher tension may soften the elastic response of PCDH15 for the subsequent stimulus. Under more extreme tension (Lower Right) unbinding of PCDH15 from CDH23 (ubi) or unfolding of PCDH15 (uf) may occur. Glycosylation and specifics of PCDH15 splice isoforms may also influence the tip-link state and elastic response. The effect of resting tension (58, 59) is further discussed in SI Appendix, Fig. S16.
Our structures also unmasked three points of Ca2+-independent flexibility for PCDH15 at linker regions EC3-4, EC5-6, and EC9-10. Simulations of the entire monomeric PCDH15 ectodomain bound to CDH23 EC1-2/3 revealed a soft elastic response dominated by these flexible points, while the heterotetrameric complex (dimeric PCDH15 bound to two CDH23 monomers) was stiffer yet still presented kinks at the PCDH15 EC9-10 linker region that could provide some limited extensibility. Intriguingly, our simulations predict that PCDH15’s MAD12 unrolls and then unfolds before CDH23 unbinding at the stretching speeds (down to 0.02 nm/ns). Unfolding of MADs in PCDH15 and CDH23 (28) could explain uncompromised hair-cell mechanotransduction under extreme stimuli that would require large tip-link extensibility (∼100 nm) (65, 66). These results highlight the potential complexities of the tip-link mechanical response and provide a structural framework to both compare and interpret complementary experimental results (26, 27, 59, 64, 67–69) and to understand the function of PCDH15 as a key component of the tip links that open inner-ear transduction channels (SI Appendix, Note 4).
In vivo conditions for tip links vary significantly across organs and species. For example, Ca2+ concentration near tip links will greatly depend on the organ in which hair cells are located. The vestibular endolymph Ca2+ concentration is 90 to 150 μM, while the bulk cochlear endolymph Ca2+ concentration varies from 20 to 40 μM (40, 41). Intriguingly, Ca2+ concentrations near hair bundles in the cochlear subtectorial space might be significantly higher than previously thought (>300 μM), both because of the action of stereocilia Ca2+ pumps functioning in a restricted space and because of the buffering effect of the tectorial membrane (42, 43). Similarly, resting tension and physiological forces will vary within and across organs, where glycosylation and differential expression of various tip-link PCDH15 isoforms may also be diverse. The complexity of PCDH15’s ectodomain revealed by our structures and simulations and the various sets of environments in which it functions in mechanotransduction indicate that PCDH15 might be a versatile and multimodal protein that can be tuned to display distinct elastic responses (Fig. 7B and SI Appendix, Fig. S16). While under some conditions PCDH15 may display the soft elasticity and extensibility attributed to gating springs, including significant extensibility through MAD12 unfolding, directly determining the exact in vivo conditions in which PCDH15 functions remains a necessary and challenging step required to fully comprehend the role played by tip links in inner-ear mechanotransduction.
PCDH15 is also found in auditory cortex interneurons (16), eye photoreceptors (52, 70), and various other tissues (15), where it may play a role in cell–cell adhesion and tissue development and maintenance. PCDH15’s interaction with CDH23 seems to be essential for auditory cortex wiring (16), while its specific role in photoreceptor function is less clear. Our PCDH15 structures and biochemical assays probing mutations that impair heterophilic binding to CDH23, cis dimerization, and Ca2+ binding provide data that can inform the exploration of PCDH15’s function beyond inner-ear mechanotransduction.
Materials and Methods
Detailed procedures for all methods are provided in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Michael Hammel and Daniel Rosenberg for data collection and initial size-exclusion chromatography–small-angle X-ray scattering data analysis; Marina Bakhtina for assistance with analytical ultracentrifugation experiments; Benjamin Donovan for assistance with cloning; Michelle E. Gray for maintenance of the Expi293 cell line; and members of the M.S. research group for their feedback, training, and discussions. This work was supported by the Ohio State University, by the National Institutes of Health–National Institute on Deafness and Other Communication Disorders (R01 DC015271), and by the National Science Foundation through Extreme Science and Engineering Discovery Environment (XRAC MCB140226). Simulations were performed at the Texas Advanced Computing Center-Stampede, Pittsburgh Supercomputing Center-Bridges, and Ohio Supercomputer Center-Owens (PAS1037) supercomputers. Use of Advanced Photon Source Northeastern Collaborative Access Team beamlines was supported by the NIH (P30 GM1241653 and S10 RR029205) and the Department of Energy (DE-AC02-06CH11357) through Grants GUP 40277, 49774, and 59251. Small-angle X-ray scattering data collection at the Advanced Light Source SIBYLS beamline was funded through the Department of Energy Biological and Environmental Research Integrated Diffraction Analysis Technologies program and NIH Grants NIGMS P30 GM124169 ALS-ENABLE and S10OD018483. B.L.N. was supported by a 2017 Cellular, Molecular, and Biochemical Sciences Program training grant fellowship. P.D.-l.-T. and R.A.-S. were Pelotonia fellows. L.N.W. received summer support from The Ohio State University Mayer’s Research scholarship.
Footnotes
Competing interest statement: M.S. is coinventor on a patent application that describes the use of modified PCDH15 proteins for gene therapy.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920444117/-/DCSupplemental.
Data Availability.
Coordinates for all structures are available in the Protein Data Bank (IDs 6N22, 6MFO, 6N2E, 5ULY, 6EB5, 6E8F, 5W1D, 6BXU, 6BWN, 5TPK, and 6EET; see SI Appendix for additional details). Other data are available upon request.
References
- 1.Fettiplace R., Kim K. X., The physiology of mechanoelectrical transduction channels in hearing. Physiol. Rev. 94, 951–986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hudspeth A. J., Integrating the active process of hair cells with cochlear function. Nat. Rev. Neurosci. 15, 600–614 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Corey D. P., Ó Maoiléidigh D., Ashmore J. F., Mechanical Transduction Processes in the Hair Cell (Springer, Cham, 2017), pp. 75–111. [Google Scholar]
- 4.Pickles J. O., Comis S. D., Osborne M. P., Cross-links between stereocilia in the guinea pig organ of Corti, and their possible relation to sensory transduction. Hear. Res. 15, 103–112 (1984). [DOI] [PubMed] [Google Scholar]
- 5.Assad J. A., Shepherd G. M., Corey D. P., Tip-link integrity and mechanical transduction in vertebrate hair cells. Neuron 7, 985–994 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Kachar B., Parakkal M., Kurc M., Zhao Y., Gillespie P. G., High-resolution structure of hair-cell tip links. Proc. Natl. Acad. Sci. U.S.A. 97, 13336–13341 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu A., Lagier S., Vologodskaia M., Fabella B. A., Hudspeth A. J., Direct mechanical stimulation of tip links in hair cells through DNA tethers. eLife 5, e16041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackney C. M., Furness D. N., The composition and role of cross links in mechanoelectrical transduction in vertebrate sensory hair cells. J. Cell Sci. 126, 1721–1731 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Pepermans E., Petit C., The tip-link molecular complex of the auditory mechano-electrical transduction machinery. Hear Res 330, 10–17 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Jaiganesh A., Narui Y., Araya-Secchi R., Sotomayor M., Beyond cell-cell adhesion: Sensational cadherins for hearing and balance. Cold Spring Harb. Perspect. Biol. 10, 029280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y., Yamoah E. N., Gillespie P. G., Regeneration of broken tip links and restoration of mechanical transduction in hair cells. Proc. Natl. Acad. Sci. U.S.A. 93, 15469–15474 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed Z. M., et al. , The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci. 26, 7022–7034 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazmierczak P., et al. , Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Di Palma F., et al. , Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat. Genet. 27, 103–107 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Alagramam K. N., et al. , Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum. Mol. Genet. 10, 1709–1718 (2001). Correction in: Hum. Mol. Genet.10, 2603 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Libé-Philippot B., et al. , Auditory cortex interneuron development requires cadherins operating hair-cell mechanoelectrical transduction. Proc. Natl. Acad. Sci. U.S.A. 114, 7765–7774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman T. B., Schultz J. M., Ahmed Z. M., Tsilou E. T., Brewer C. C., Usher syndrome: Hearing loss with vision loss. Adv. Ortohinolaryngol. 70, 56–65 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Rzadzinska A. K., Steel K. P., Presence of interstereocilial links in waltzer mutants suggests Cdh23 is not essential for tip link formation. Neuroscience 158, 365–368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Indzhykulian A. A., et al. , Molecular remodeling of tip links underlies mechanosensory regeneration in auditory hair cells. PLoS Biol. 11, e1001583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lelli A., Kazmierczak P., Kawashima Y., Müller U., Holt J. R., Development and regeneration of sensory transduction in auditory hair cells requires functional interaction between cadherin-23 and protocadherin-15. J. Neurosci. 30, 11259–11269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sotomayor M., Weihofen W. A., Gaudet R., Corey D. P., Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature 492, 128–132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geng R., et al. , Noddy, a mouse harboring a missense mutation in protocadherin-15, reveals the impact of disrupting a critical interaction site between tip-link cadherins in inner ear hair cells. J. Neurosci. 33, 4395–4404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdi S., et al. , Diversity of the genes implicated in Algerian patients affected by Usher syndrome. PLoS One 11, e0161893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhary D., Kumar A., Magliery T. J., Sotomayor M., Using thermal scanning assays to test protein-protein interactions of inner-ear cadherins. PLoS One 12, e0189546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narui Y., Sotomayor M., Tuning inner-ear tip-link affinity through alternatively spliced variants of protocadherin-15. Biochemistry 57, 1702–1710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dionne G., et al. , Mechanotransduction by PCDH15 relies on a novel cis-dimeric architecture. Neuron 99, 480–492.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge J., et al. , Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5. eLife 7, e38770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De-la-Torre P., Choudhary D., Araya-Secchi R., Narui Y., Sotomayor M., A mechanically weak extracellular membrane-adjacent domain induces dimerization of protocadherin-15. Biophys. J. 115, 2368–2385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elledge H. M., et al. , Structure of the N terminus of cadherin 23 reveals a new adhesion mechanism for a subset of cadherin superfamily members. Proc. Natl. Acad. Sci. U.S.A. 107, 10708–10712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sotomayor M., Weihofen W. A., Gaudet R., Corey D. P., Structural determinants of cadherin-23 function in hearing and deafness. Neuron 66, 85–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Araya-Secchi R., Neel B. L., Sotomayor M., An elastic element in the protocadherin-15 tip link of the inner ear. Nat. Commun. 7, 13458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powers R. E., Gaudet R., Sotomayor M., A partial calcium-free linker confers flexibility to inner-ear protocadherin-15. Structure 25, 482–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaiganesh A., et al. , Zooming in on cadherin-23: Structural diversity and potential mechanisms of inherited deafness. Structure 26, 1210–1225.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corey D. P., Hudspeth A. J., Kinetics of the receptor current in bullfrog saccular hair cells. J. Neurosci. 3, 962–976 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howard J., Hudspeth A. J., Compliance of the hair bundle associated with gating of mechanoelectrical transduction channels in the bullfrog’s saccular hair cell. Neuron 1, 189–199 (1988). [DOI] [PubMed] [Google Scholar]
- 36.Cheung E. L. M., Corey D. P., Ca2+ changes the force sensitivity of the hair-cell transduction channel. Biophys. J. 90, 124–139 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin X., et al. , Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers. Proc. Natl. Acad. Sci. U.S.A. 109, E127–E134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukasaki Y., et al. , Giant cadherins Fat and Dachsous self-bend to organize properly spaced intercellular junctions. Proc. Natl. Acad. Sci. U.S.A. 111, 16011–16016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazra J. P., et al. , Broken force dispersal network in tip-links by the mutations at the Ca2+-binding residues induces hearing-loss. Biochem. J. 476, 2411–2425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosher S. K., Warren R. L., Very low calcium content of cochlear endolymph, an extracellular fluid. Nature 273, 377–378 (1978). [DOI] [PubMed] [Google Scholar]
- 41.Salt A. N., Inamura N., Thalmann R., Vora A., Calcium gradients in inner ear endolymph. Am. J. Otolaryngol. 10, 371–375 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Yamoah E. N., et al. , Plasma membrane Ca2+-ATPase extrudes Ca2+ from hair cell stereocilia. J. Neurosci. 18, 610–624 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strimbu C. E., Prasad S., Hakizimana P., Fridberger A., Control of hearing sensitivity by tectorial membrane calcium. Proc. Natl. Acad. Sci. U.S.A. 116, 5756–5764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mammano F., Bortolozzi M., Ortolano S., Anselmi F., Ca2+ signaling in the inner ear. Physiology (Bethesda) 22, 131–144 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Pei J., Grishin N. V., Expansion of divergent SEA domains in cell surface proteins and nucleoporin 54. Protein Sci. 26, 617–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isralewitz B., Gao M., Schulten K., Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 11, 224–230 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Nagar B., Overduin M., Ikura M., Rini J. M., Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature 380, 360–364 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Goodman K. M., et al. , Structural basis of diverse homophilic recognition by clustered α- and β-protocadherins. Neuron 90, 709–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicoludis J. M., et al. , Structure and sequence analyses of clustered protocadherins reveal antiparallel interactions that mediate homophilic specificity. Structure 23, 2087–2098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper S. R., Jontes J. D., Sotomayor M., Structural determinants of adhesion by protocadherin-19 and implications for its role in epilepsy. eLife 5, e18529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Modak D., Sotomayor M., Identification of an adhesive interface for the non-clustered δ1 protocadherin-1 involved in respiratory diseases. Commun. Biol. 2, 354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed Z. M., et al. , PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum. Mol. Genet. 12, 3215–3223 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Aurora R., Rose G. D., Helix capping. Protein Sci. 7, 21–38 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb S. W., et al. , Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development 138, 1607–1617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pepermans E., et al. , The CD2 isoform of protocadherin-15 is an essential component of the tip-link complex in mature auditory hair cells. EMBO Mol. Med. 6, 984–992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodyear R. J., Forge A., Legan P. K., Richardson G. P., Asymmetric distribution of cadherin 23 and protocadherin 15 in the kinocilial links of avian sensory hair cells. J. Comp. Neurol. 518, 4288–4297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kulahin N., et al. , Structural model and trans-interaction of the entire ectodomain of the olfactory cell adhesion molecule. Structure 19, 203–211 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Jaramillo F., Hudspeth A. J., Displacement-clamp measurement of the forces exerted by gating springs in the hair bundle. Proc. Natl. Acad. Sci. U.S.A. 90, 1330–1334 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tobin M., Chaiyasitdhi A., Michel V., Michalski N., Martin P., Stiffness and tension gradients of the hair cell’s tip-link complex in the mammalian cochlea. eLife 8, e43473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robles L., Ruggero M. A., Mechanics of the mammalian cochlea. Physiol. Rev. 81, 1305–1352 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee E. H., Hsin J., Sotomayor M., Comellas G., Schulten K., Discovery through the computational microscope. Structure 17, 1295–1306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rico F., Gonzalez L., Casuso I., Puig-Vidal M., Scheuring S., High-speed force spectroscopy unfolds titin at the velocity of molecular dynamics simulations. Science 342, 741–743 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Rakshit S., Sivasankar S., Biomechanics of cell adhesion: How force regulates the lifetime of adhesive bonds at the single molecule level. Phys. Chem. Chem. Phys. 16, 2211–2223 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Mulhall E. M., et al. , The dynamic strength of the hair-cell tip link reveals mechanisms of hearing and deafness. bioRxiv:10.1101/763847 (12 September 2019). [Google Scholar]
- 65.Shepherd G. M., Corey D. P., The extent of adaptation in bullfrog saccular hair cells. J. Neurosci. 14, 6217–6229 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holt J. R., Corey D. P., Eatock R. A., Mechanoelectrical transduction and adaptation in hair cells of the mouse utricle, a low-frequency vestibular organ. J. Neurosci. 17, 8739–8748 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bartsch T. F., et al. , Elasticity of individual protocadherin 15 molecules implicates tip links as the gating springs for hearing. Proc. Natl. Acad. Sci. U.S.A. 116, 11048–11056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oroz J., et al. , Nanomechanics of tip-link cadherins. Sci. Rep. 9, 13306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang Y.-Q., et al. , Ankyrin is an intracellular tether for TMC mechanotransduction channels. Neuron 107, 112–125.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahly I., et al. , Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. J. Cell Biol. 199, 381–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates for all structures are available in the Protein Data Bank (IDs 6N22, 6MFO, 6N2E, 5ULY, 6EB5, 6E8F, 5W1D, 6BXU, 6BWN, 5TPK, and 6EET; see SI Appendix for additional details). Other data are available upon request.