Significance
Parasites are interesting models for studying speciation processes because they have a high potential for specialization, thanks to the intimate ecological association with their hosts and vectors. Yet little is known about the circumstances under which new parasite lineages emerge. Here we studied the genome diversity of parasites of the Leishmania braziliensis species complex that inhabit both Amazonian and Andean biotas in Peru. We identify three major parasite lineages that occupy particular ecological niches and show that these emerged during forestation changes over the past 150,000 y. We furthermore discovered that meiotic recombination between Amazonian and Andean lineages resulted in full-genome hybrids presenting mixed mitochondrial genomes, providing insights into the genetic consequences of hybridization in parasitic protozoa.
Keywords: speciation genomics, ecological speciation, population genomics, vector-borne disease, interspecific hybridization
Abstract
The tropical Andes are an important natural laboratory to understand speciation in many taxa. Here we examined the evolutionary history of parasites of the Leishmania braziliensis species complex based on whole-genome sequencing of 67 isolates from 47 localities in Peru. We first show the origin of Andean Leishmania as a clade of near-clonal lineages that diverged from admixed Amazonian ancestors, accompanied by a significant reduction in genome diversity and large structural variations implicated in host–parasite interactions. Within the Andean species, patterns of population structure were strongly associated with biogeographical origin. Molecular clock and ecological niche modeling suggested that the history of diversification of the Andean lineages is limited to the Late Pleistocene and intimately associated with habitat contractions driven by climate change. These results suggest that changes in forestation over the past 150,000 y have influenced speciation and diversity of these Neotropical parasites. Second, genome-scale analyses provided evidence of meiotic-like recombination between Andean and Amazonian Leishmania species, resulting in full-genome hybrids. The mitochondrial genome of these hybrids consisted of homogeneous uniparental maxicircles, but minicircles originated from both parental species. We further show that mitochondrial minicircles—but not maxicircles—show a similar evolutionary pattern to the nuclear genome, suggesting that compatibility between nuclear-encoded mitochondrial genes and minicircle-encoded guide RNA genes is essential to maintain efficient respiration. By comparing full nuclear and mitochondrial genome ancestries, our data expand our appreciation on the genetic consequences of diversification and hybridization in parasitic protozoa.
Exploring natural genetic variation has been instrumental in understanding how and under which circumstances new species originate. South America encompasses a large fraction of global biodiversity, representing one of the most species-diverse regions on Earth. This is partly because diversification of Neotropical taxa has been influenced by a rich geological and climatic history, including large-scale reconfigurations of the landscape through the Neogene uplift of the Andes (1) and habitat instability through Pleistocene climatic cycling (2). In particular, the Andes—due to a complex interplay of history, geography, and ecology—represent an epicenter for species diversification in birds, reptiles, insects, and plants (3–8). However, little is known about the role of the Andes shaping the evolution of parasitic microorganisms.
Here we used whole-genome sequencing to study the evolutionary history of parasites of the Leishmania braziliensis species complex in Peru. The lowland species L. braziliensis is a zoonotic parasite circulating in a diverse range of wild mammals (9) in Neotropical rainforests. It is one of the major causes of cutaneous leishmaniasis in Latin America and also causes a relatively high frequency of severe mucocutaneous disease (known locally as espundia) where the parasite spreads to mucosal tissue. Human infections appear to be a spillover from this sylvatic life cycle and are probably not important in transmission. In contrast, the montane species Leishmania peruviana is largely endemic to the Pacific slopes of the Peruvian Andes. It is transmitted exclusively in peridomestic xerophytic environments and causes a disease of altitude (1,300 to 2,800 m), known locally as uta, a relatively benign form of cutaneous leishmaniasis. Both Leishmania species were initially shown to be different at only 1 of 16 enzymatic loci (10), but subsequent molecular analyses showed that they correspond to two distinct monophyletic clades of the Viannia subgenus (11–13). Molecular karyotyping further revealed absence of subpopulation structure in Peruvian L. braziliensis in contrast to L. peruviana that was genetically subdivided into different biogeographical regions across and along the Peruvian Andes (14, 15). These species differences persist despite the report of a number of parasites presenting hybrid marker profiles in the eastern Andean valley of the Huánuco region (16, 17).
The L. braziliensis species complex presents an ideal model to study the process of parasite speciation in the Neotropics, as well as understand the genetic consequences of hybridization in parasitic protozoa (18, 19). To this end, we cultured and sequenced cryopreserved Leishmania parasites that were sampled during various studies on the genetics and epidemiology of leishmaniasis in Peru (13, 20–23). The majority of isolates were subject to a small number of passages to reduce potential culture-related biases in parasite genomic characterization (24). Whole-genome sequence data of 67 isolates were aligned against a PacBio assembly including the 35 major chromosomes and the complete mitochondrial maxicircle, and unaligned reads were used to assemble, circularize, and annotate full sets of mitochondrial minicircles. This allowed us to examine the ancestry of a parasitic protozoan based on a joint analysis of the complete nuclear and mitochondrial genome. Our genome-wide survey of population diversity suggests a key role of ecology and climate in parasite diversification and provides unequivocal evidence of mixed mitochondrial genomes in hybrid parasites.
Results
Natural Variation of the L. braziliensis Species Complex.
The genomes of 31 L. peruviana, 23 L. braziliensis, and 13 hybrid L. braziliensis × L. peruviana isolates (SI Appendix, Fig. S1 and Table S1) were sequenced at a median 70× depth (mean = 73×; min = 40×; max = 123×). For comparative purposes, we also included sequencing data of one isolate from the closely related Leishmania panamensis species (SI Appendix, Table S1). All sequence data were aligned against a new long-read assembly of L. braziliensis M2904, and for every isolate, we determined the accessible genome that is characterized by sufficient mapping quality, base quality, and read depth. This revealed that 90.9 to 92.2% of the chromosomal genome (i.e., 29.8 to 30.2 Mb) was accessible for isolates belonging to the L. braziliensis species complex, and 88.1% of the genome (i.e., 28.9 Mb) was accessible for L. panamensis.
Phylogenetic analyses based on 637,821 single-nucleotide polymorphisms (SNPs) revealed a clear distinction between the three Leishmania Viannia species and confirmed that L. peruviana and L. braziliensis correspond to two closely related but distinct monophyletic clades (Fig. 1A). When estimating the number of fixed nucleotide differences between species across their combined accessible genome of 28.3 Mb, we found that L. panamensis showed fixed differences at 301,128 sites (1.06%) from L. braziliensis and 343,175 sites (1.21%) from L. peruviana. In contrast, L. braziliensis and L. peruviana differed at a 10-fold lower number of fixed nucleotide differences (10,368 sites, 0.04%), which is comparable to the 11,228 fixed site differences found between Leishmania infantum and Leishmania donovani (25).
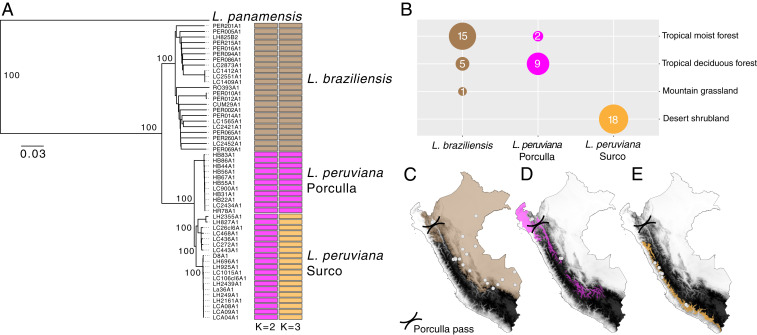
Fig. 1.
(A) Neighbor-joining phylogenetic tree depicting the genetic ancestry of the L. braziliensis species complex (L. braziliensis and L. peruviana) in Peru, including L. panamensis as an outgroup. Colored bar plots show parasite groups as estimated with ADMIXTURE assuming K = 2 and K = 3 populations. (B) Sample sizes of the three major parasite lineages grouped according their originating biome. (C–E) Geographic maps of Peru showing the Andean topography in grayscale, the Porculla pass in the northwest of Peru, the sampling locations of the three major parasite lineages as white dots (C, L. braziliensis; D, L. peruviana Porculla; E, L. peruviana Surco), and their corresponding biomes (C, tropical moist forest [brown]; D, tropical deciduous forest [magenta]; E, desert shrubland [orange]).
Genotyping across the accessible genome of the 67 L. braziliensis species complex isolates disclosed a total of 389,259 SNPs and 114,082 small insertions/deletions (INDELs) called against the reference. SNPs and variable sites (i.e., excluding SNPs fixed in a given species) were evenly distributed across the 35 major chromosomes, but L. peruviana showed a 2.6-fold lower density in SNPs (4.1/kb vs. 10.6/kb) and a 9.5-fold lower density in variable sites (1/kb vs. 9.5/kb) compared to L. braziliensis, despite our slightly large sample of L. peruviana (SI Appendix, Fig. S2). While the allele frequency spectrum of L. braziliensis was dominated by rare variants, the large majority of SNP loci were entirely fixed (67%) and homozygous (89%) in L. peruviana (SI Appendix, Fig. S3). Several of these fixed SNP mutations were virtually absent in L. braziliensis and deleterious to genes coding for an ion transporter protein, kinesin-C (26), and the subunit 2 of the class I transcription factor A complex (27, 28) (SI Appendix, Table S2).
Both L. peruviana and L. braziliensis showed variation in chromosome copy numbers, but somy profiles were not species-specific (SI Appendix, Fig. S4, grayscale boxes). Chromosome 31 was tetrasomic for 59 isolates, a consistent observation for all Leishmania species (29, 30). Chromosomes 19, 26, 27, 32, and 34 were disomic for all isolates (SI Appendix, Fig. S4). A total of 164 gene copy number variations were identified in L. peruviana (51 deletions and 113 amplifications). Six deletions and five amplifications were shared among all L. peruviana isolates, eight of which encompassed genes encoding cell surface glycoproteins such as the gp63 leishmanolysin gene family and the ∂-amastin surface glycoproteins (Table 1 and SI Appendix, Fig. S5). Specifically, the gp63 gene array on chromosome 10 was reduced from an average 64 copies in L. braziliensis to 37 copies in L. peruviana (Table 1 and SI Appendix, Fig. S5). ∂-amastin genes increased from an average 17 copies in L. braziliensis to an average 38 copies in L. peruviana (Table 1). However, two ∂-amastin genes on chromosome 8 were entirely lost in L. peruviana, reducing the overall ∂-amastin gene repertoire in this species (Table 1). Other major differences encompass putative proteins encoding kinesin, autophagy protein ATG8 ubiquitin, and a glycerol uptake protein (Table 1 and SI Appendix, Fig. S5).
Table 1.
Ancestral deletions and amplifications in Andean L. peruviana species
| Type | Orthologous group or gene IDs | Product | Chromosomes | L. peruviana | L. braziliensis |
| Amplification | OG5_143904 | ∂-amastin-like surface protein | 20 | 22 (8–34) | 1 (0–3) |
| Amplification | OG5_183241 | ∂-amastin-like protein | bin | 16 (12–21) | 9 (8–12) |
| Amplification | OG5_137181 | Autophagy protein Atg8 ubiquitin like, putative | 19 | 12 (8–16) | 5 (3–7) |
| Amplification | OG5_136713 | Kinesin motor domain containing protein, putative | 20, bin | 18 (15–24) | 12 (10–16) |
| Deletion | OG5_126749 | GP63, leishmanolysin | 10, bin | 37 (23–50) | 64 (53–79) |
| Deletion | OG5_128109 | glycerol uptake protein, putative | 19 | 3 (3–4) | 9 (5–12) |
| Deletion | LbrM.20.2.206380.1–LbrM.20.2.206460.1 | Amastin surface glycoprotein, putative | 20 | 0 (0–1) | 5 (3–7) |
| Deletion | OG5_130987 | ∂-amastin-like protein | 8 | 0 (0–1) | 4 (3–5) |
| Deletion | OG5_130729 | Amastin-like surface protein | 20 | 1 (0–2) | 4 (3–6) |
| Deletion | OG5_132980 | ∂-amastin | 8 | 0 (0–1) | 3 (1–5) |
The numbers in the two species columns reflect the mean predicted copy numbers with the minimum and maximum copy numbers given in parentheses.
Comparative Population Genomics of Lowland and Montane Leishmania Parasites.
Analyses of population structure using unsupervised clustering with ADMIXTURE (Fig. 1A) revealed three major groups of parasites, each corresponding to a particular biome. The first group was composed of the lowland L. braziliensis parasites that were largely found within tropical moist forests at a median altitude of 631 m (Fig. 1 B and C). The second and third groups were composed of the montane L. peruviana parasites found within two different biomes. The Porculla lineage was found at an average altitude of 1,985 m within tropical deciduous forests that span the northeastern slopes of the Peruvian Andes and the Porculla pass (Fig. 1 B and D), which is an east–west pass through the Andes that forms a biogeographic connection between the lowland forests of the Pacific coast and the Amazon basin. The Surco lineage was exclusively found within desert shrubland along the Pacific slopes of the Peruvian Andes, at an average altitude of 2,769 m (Fig. 1 B and E). The Surco lineage was further subdivided into three differentiated sublineages, hereafter referred to as Surco North (SUN), Surco Central (SUC), and Surco Central/South (SUCS) (SI Appendix, Fig. S6). In contrast, clustering analyses failed to identify subpopulations in L. braziliensis, and the L. braziliensis phylogenetic network featured long branches that separate most isolates with little clade structure (SI Appendix, Fig. S7), suggesting relatively high gene flow within the Peruvian L. braziliensis lineage.
To make predictions about relative recombination rates, Hardy–Weinberg equilibrium (HWE) was tested by estimating FIS per SNP locus in L. peruviana and L. braziliensis taking into account Wahlund effects (Materials and Methods). The FIS distribution was skewed toward negative FIS for L. peruviana (mean FIS = −0.54) with almost half of the SNP loci showing FIS = −1, suggesting heterozygote excess, as would be predicted for a population experiencing predominantly clonal propagation (SI Appendix, Fig. S8 A and B and Table S3). In contrast, L. braziliensis displayed a unimodal distribution centered around zero (mean = −0.11), suggesting that the population is close to HWE and that L. braziliensis may experience relatively high recombination rates (SI Appendix, Fig. S8C and Table S3). Despite strong genetic differentiation and reduced recombination rates in L. peruviana, there were signals of historical hybridization events, in particular among the Surco subpopulations (SI Appendix, Fig. S9). Specifically, we found that SUN isolates and SUC isolate LH827 contained genomic windows with elevated proportions of heterozygous sites compared to the other L. peruviana isolates. Phylogenetic networks of SNPs in several such heterozygous regions revealed signals of introgressive hybridization from the SUC/SUCS populations into the SUN population (SI Appendix, Fig. S9). The occurrence of hybridization among the Surco populations was also highlighted by isolate LH741 that showed a mixed ancestry between the SUN and SUCS populations (SI Appendix, Fig. S6).
Late Pleistocene Origin of Montane Leishmania Parasites.
A time-resolved phylogeny calibrated based on an estimated molecular clock of 0.81% per My for maxicircles (95% highest posterior density [HPD] interval 0.5 to 1.2%; Materials and Methods) suggested that the common ancestor of L. peruviana and L. braziliensis lived ∼126 kya (CI 59 to 225 kya) (Fig. 2A) during the Last Interglacial (LIG; 130 to 115 kya). Subsequent diversification of L. peruviana occurred at several occasions during the Last Glacial Period which lasted from 115 to 11.7 kya (Fig. 2A). These results suggest that the (sub)diversification of L. peruviana may have been promoted by extensive climatic cycling and vegetational shifts.
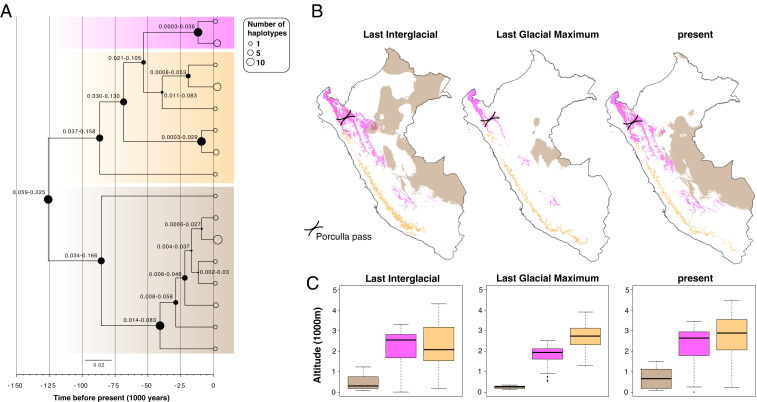
Fig. 2.
(A) Time-calibrated phylogenetic tree for the L. braziliensis species complex based on maxicircle gene alignments. At each node, the size of the filled circles reflects the posterior density ranging between 0.15 and 1, while the numbers represent the 95% highest posterior density of the divergence time estimates. Thick transparent boxes mark the maxicircle haplotypes of L. braziliensis (brown), L. peruviana Porculla (magenta), and L. peruviana Surco (orange). The sizes of the open circles at the tips of the branches reflect the number of haplotype sequences (legend on the top right). (B) Geographic maps of Peru showing the modeled distribution and (C) average altitude of the predicted habitat patches, for each of the three major Leishmania lineages during the LIG, LGM, and present (see A for color codes).
Environmental niche modeling (ENM) using present and past bioclimatic variables predicted the putative range of each Leishmania lineage (L. braziliensis, L. peruviana Porculla, and L. peruviana Surco) during the Last Glacial Maximum (LGM; 21 kya) and the LIG (130 kya). The bioclimatic variables that contributed most to the model results were temperature and precipitation seasonality. Present-day predicted areas included the species’ known distributions in Peru and largely matched the biogeographic regions (tropical moist forest, tropical deciduous forest, and desert shrubland) associated to each Leishmania population (Fig. 2B and SI Appendix, Fig. S10). During the LGM, there was a strong contraction, fragmentation, and isolation of suitable habitat for all Leishmania populations (Fig. 2B and SI Appendix, Fig. S10), resulting in a pronounced difference in altitude range between L. braziliensis and the montane L. peruviana lineages (Fig. 2C). During the LIG, the potential range of L. peruviana was similar to the current range, but the range of L. braziliensis was slightly shifted to the north of Peru, resulting in a spatial range overlap between the two Andean biotas and the Amazonian biota near the Porculla pass (Fig. 2B and SI Appendix, Fig. S10). Altogether, these results show that fluctuations in bioclimatic variables over the past 150,000 y affected the predicted habitat range of the major Peruvian Leishmania lineages.
Meiotic-Like Recombination Between Lowland and Montane Leishmania Parasites.
We also included 13 hybrid L. peruviana × L. braziliensis isolates from the Huánuco region where both Leishmania species and their hybrids occur sympatrically (16, 17). Earlier molecular work suggested the presence of four zymodemes and seven microsatellite genotypes within the hybrid population (17), and here we sequenced isolates from each zymodeme. Of a total 149,735 SNPs that were identified in the hybrid population, 61,804 sites (41.3%) were fixed homozygous, and 73,375 sites (49%) were fixed heterozygous, leaving 14,556 sites (9.7%) at which individual isolates varied in genotype. Heterozygous sites were evenly distributed across the genome and rarely interrupted by homozygous stretches (SI Appendix, Fig. S11). The genome-wide frequency distribution of allelic read depths at heterozygous sites was centered around 0.5 for all hybrids, as would be predicted for diploid Leishmania parasites (29). Hybrids were near-identical, each pair differing by a median 1,245 heterozygous sites and 1 homozygous site (SI Appendix, Table S4). Exceptions were isolates PER011 and LC2520 that showed 167 homozygous SNP differences (SI Appendix, Table S4), but close inspection revealed that 165 SNPs were located within a telomeric 63-kb window on chromosome 32 that was homozygous for a different parental allele in the two isolates (SI Appendix, Fig. S11).
Principal component analyses (PCA) based on genome-wide SNPs showed that hybrids occupied a tight central position between L. braziliensis and the L. peruviana SUCS population (Fig. 3A), suggesting that all hybrids are first-generation offspring. Estimates of raw nucleotide differences revealed a similar genetic distance between the hybrids and the L. braziliensis isolates LC2551, LC1409, and LC1412 on one hand and the L. peruviana SUCS population on the other hand. To investigate the ancestry of the hybrids in more detail, we focused on the 113,266 SNPs that were shared between the 13 hybrids and representative isolates of each parent group (LH925 for L. peruviana SUCS and LC1412 for L. braziliensis). SNPs were counted when homozygous for the reference allele (R/R), homozygous for the alternate allele (A/A), or heterozygous (A/R). Of the 54,734 SNPs that were homozygous (A/A) in one parent and absent (R/R) in the other, an average 54,038 SNPs (99.5%) were heterozygous (A/R) in the 13 hybrids. Of the 14,025 SNPs that were homozygous in one parent (A/A) and heterozygous in the other parent (A/R), an average 7,399 SNPs (53%) were heterozygous (A/R), and 6,596 SNPs (47%) were homozygous (A/A) in the hybrid offspring. Of the 41,424 SNPs that were heterozygous (A/R) in one parent and absent (R/R) in the other, an average 21,053 SNPs (50.8%) were heterozygous (A/R), and 20,295 SNPs (48.9%) were absent (R/R) in the hybrid offspring. These results confirm that hybrid parasites inherited parental alleles in a 1:1 ratio, as would be predicted for first-generation offspring of a Mendelian cross.
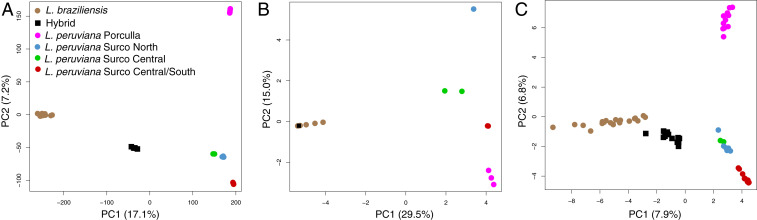
Fig. 3.
Principal component analyses of the L. braziliensis species complex in Peru based on (A) 389,259 genome-wide SNPs, (B) 89 SNPs of the mitochondrial maxicircle coding region, and (C) sequence similarity of 950 mitochondrial minicircle sequence classes. Hybrid isolates were projected onto the PCA space of L. braziliensis and L. peruviana, where the first axis separates L. braziliensis and L. peruviana, and the second axis distinguishes the main L. peruviana populations.
Mixed Ancestry of Mitochondrial Genomes in Hybrid Parasites.
We also examined the ancestry of the L. braziliensis × L. peruviana hybrids based on sequence data of the complete mitochondrial genome. The mitochondrial DNA of trypanosomatids shows extraordinary complexity, consisting of a giant network of thousands of heterogeneous minicircles (0.5 to 2.5 kb in size, depending on the species) interlaced with 20 to 50 homogeneous maxicircles (20 to 30 kb) (31). For maxicircles, we focused on SNPs identified within the coding region after aligning reads to the M2904 maxicircle reference sequence. For minicircles, we developed the Python package Kinetoplast Genomics (KOMICS) (SI Appendix, SI Methods and Fig. S12) to assemble and circularize the minicircle sequences for each of the 67 isolates. This resulted in a combined number of 9,003 high-quality minicircle contigs, of which 6,949 (77%) circularized (SI Appendix, SI Results A, Table S5, and Fig. S13). Minicircle complexity and ancestry was studied using a clustering approach to find sets of very similar minicircle sequences (hereafter referred to as minicircle sequence classes [MSCs]). Assuming 97% identity (SI Appendix, SI Results A, and Figs. S14 and S15), a significantly lower number of MSCs were found per isolate in L. peruviana (mean = 81 MSCs per isolate) compared to L. braziliensis (mean = 147 MSCs per isolate; P < 0.0001).
Close examination of 89 SNPs identified within the coding region of the mitochondrial maxicircle revealed that all hybrids were identical to the putative L. braziliensis parent isolates LC2551, LC1409, and LC1412. This uniparental inheritance of the maxicircle may also explain the discordant pattern between the genome (Fig. 3A) and the mitochondrial maxicircle (Fig. 3B) in the observed population genomic structure of L. peruviana. In contrast, PCA based on minicircle sequence similarity (Fig. 3C) showed an identical pattern as seen with genome-wide SNPs (Fig. 3A), with the first axis separating L. braziliensis from L. peruviana and the second axis dividing the main L. peruviana populations. Interestingly, hybrids did not cluster with either parental species as would be predicted for a uniparentally inherited kinetoplast but occupied an intermediate position between L. braziliensis and the L. peruviana SUCS population (Fig. 3C). To investigate this in more detail, we quantified the number of MSCs shared between each hybrid isolate and representative isolates of each parent group (LH925 for L. peruviana SUCS and LC1412 for L. braziliensis). While both putative parents shared 14 MSCs, hybrids shared on average 34 MCSs (29%) with the putative L. peruviana parent and 58 MCSs (50%) with the putative L. braziliensis parent (Table 2), clearly evidencing biparental inheritance of minicircles. The 21% lower number of L. peruviana (29%) versus L. braziliensis (50%) MSCs in hybrid parasites seems to roughly reflect the difference (18%) in minicircle complexity between the putative parents (73 MCSs or 41% [=73/180] in L. peruviana versus 107 MCSs or 59% [=107/180] in L. braziliensis), suggesting hybrid parasites inherited minicircles in a 1:1 ratio. Finally, hybrids contained an elevated number of MSCs (average 118 MSCs) (Table 2) compared to the putative parents (73 MCSs in L. peruviana and 107 MSCs in L. braziliensis).
Table 2.
Total number of MSCs for the 12 L. braziliensis × L. peruviana hybrids for which minicircles were successfully assembled
| Number of MSCs | L. peruviana | L. braziliensis | |
| LC2877A1 | 94 | 29 (31%) | 52 (55%) |
| HR410A1 | 113 | 34 (31%) | 57 (50%) |
| LC2435A1 | 105 | 33 (31%) | 54 (51%) |
| LC2520A1 | 104 | 28 (27%) | 57 (55%) |
| LC1407A1 | 114 | 36 (32%) | 61 (52%) |
| LC1408A1 | 127 | 38 (30%) | 66 (52%) |
| LC1418A1 | 118 | 37 (31%) | 56 (48%) |
| LC1419A1 | 111 | 40 (36%) | 58 (53%) |
| LH1099A1 | 112 | 27 (24%) | 56 (50%) |
| PER011A1 | 211 | 33 (16%) | 69 (33%) |
| LC2851A1 | 99 | 28 (28%) | 52 (53%) |
| HR80A1 | 110 | 39 (35%) | 52 (47%) |
| Average | 118 | 34 (29%) | 58 (50%) |
Percentages indicate the proportion of MSCs that were found in the putative L. braziliensis or L. peruviana parent; the remaining MSCs were unassigned.
The minicircles encode guide RNA (gRNA) genes that are responsible for directing an elaborate U-indel RNA editing process that generates translatable maxicircle-encoded transcripts (31). Correct RNA editing is essential for parasite viability (32) and depends on a functionally complete set of minicircles (33). Putative guide RNA genes (gRNAs) were identified by aligning minicircle and maxicircle sequences to predicted edited mRNA sequences, allowing for G-U base pairs (34). This was done for four isolates representing L. braziliensis, L. peruviana Porculla, L. peruviana SUCS, and a hybrid L. braziliensis × L. peruviana parasite (SI Appendix, Fig. S16). A total of 19 to 21 gRNAs were identified within the maxicircle of L. peruviana and L. braziliensis (SI Appendix, SI Results B, and Table S6), a number that far exceeded the seven maxicircle-encoded gRNAs (Ma-gRNAs) reported for Leishmania tarentolae (35) and Crithidia fasciculata (36). A total of 123 minicircle-encoded gRNAs (Mi-gRNAs) were identified in L. peruviana Surco, 151 Mi-gRNAs in L. peruviana Porculla, 154 Mi-gRNAs in L. braziliensis, and 157 Mi-gRNAs in hybrid L. peruviana × L. braziliensis (SI Appendix, SI Results B, and Table S7). The distribution and ancestry of the predicted Ma-gRNAs and Mi-gRNAs was examined across three extensively edited genes (GR4, ND8, and ND9), revealing two major results (SI Appendix, Fig. S17). First, many of the novel Ma-gRNAs covered editing sites that were not covered by minicircle-encoded candidates (SI Appendix, Fig. S17, black stars), suggesting that these Ma-gRNAs may be essential to prevent a break of the 3′–5′ editing cascade. Second, L. peruviana × L. braziliensis hybrids showed a mosaic ancestry of Mi-gRNAs originating from both parental species, with L. peruviana-specific Mi-gRNAs aligning in locations where there were no L. braziliensis-specific Mi-gRNAs (SI Appendix, Fig. S17, blue stars). This result shows that in addition to mixed ancestry of Mi-gRNAs, complete editing in hybrid kinetoplasts may be dependent on Mi-gRNAs from both parents.
Discussion
We have presented population genomic analyses based on the complete nuclear and mitochondrial genome of natural Leishmania isolates that provide comprehensive insights into the origin, evolution, and subsequent hybridization of these Neotropical parasites.
Andean L. peruviana parasites demonstrated stable long-term genetic diversification, evolving as near-clonal lineages that emerged from admixed Amazonian ancestors. The origin of the Andean lineages was accompanied by a strong population bottleneck, as evidenced by a genome-wide fixation of SNP polymorphisms. Several of these fixed SNP mutations were deleterious to genes involved in the parasite’s biosynthetic capability or cytokinesis, which may explain the slower growth rate in vitro of L. peruviana compared to L. braziliensis (37, 38). In addition, all L. peruviana isolates shared eight structural variants encompassing genes involved in host–parasite interactions. Most notable was a 51.1-kb deletion spanning the gp63 leishmanolysin gene family, important for amastigotes to subvert the macrophage immune response (39); reduced copy numbers at this locus may explain the lower virulence of L. peruviana amastigotes in vivo (38). Interestingly, hybrid L. peruviana × L. braziliensis parasites have a gp63 copy number comparable to L. braziliensis, which may explain why these hybrids could develop a similar pathogenicity as L. braziliensis (17). Beside reduced gp63 copy numbers in L. peruviana, we also observed a simultaneous loss and expansion of ∂-amastin surface proteins that are essential for parasite–macrophage interactions and the viability of intracellular amastigotes (40). Further experimental work is needed to verify the relatively role of these structural variations on the loss virulence in L. peruviana.
Beside a clear dichotomy in population genomic structure between the Andean and Amazonian Leishmania species, parasite genomic diversity was principally partitioned by ecotype. The strict ecological association of the major Leishmania lineages in Peru suggests that reproductive isolation is primarily driven by colonization of host–vector communities specific to each ecotype. Indeed, different vector Lutzomyia sand flies and reservoir host mammals are implicated in the transmission of Andean versus Amazonian leishmaniasis, and many of these show a fragmented distribution across the Peruvian Andes (41, 42). The ability of parasites to occupy novel environmental niches may often be accompanied by host (or vector) switches (43) and facilitated by climatological variation and ecological perturbation. Here we show that the history of diversification of Andean lineages is limited to the Late Pleistocene and consistent with predicted changes in habitat range. We propose that spatial overlap of Andean and Amazonian ecotypes at the Porculla pass during the LIG may have facilitated the intrusion of Amazonian Leishmania parasites west of the Andes. Subsequent habitat and altitudinal range contractions during last glacial episodes resulted in the fragmentation of Leishmania-specific ecotypes, a process that may have fueled the diversification of Leishmania in Peru.
The eastern Andean valley of the Huánuco region is characterized by Andean tropical deciduous forest but surrounded by Amazonian tropical moist forest. It is assumed that migration since 1975 in and around the Huánuco valley has promoted infections by multiple Leishmania species from lowland and highland areas, leading to the establishment of a hybrid population in this region (16, 17, 44). Here we show clear signs of meiotic-like recombination between Andean and Amazonian Leishmania species in the Huánuco valley. Our observations are consistent with recent genomic evidence of meiosis-like sexual recombination in experimental hybrids (19, 45). All L. peruviana × L. braziliensis hybrids contained both parental alleles, providing evidence that hybrids inherited a full set of chromosomes from each parent and are thus full-genome hybrids. The observation of near-identical genomes and identical mitochondrial maxicircles sampled over a period of 11 y suggest that hybrids arose from a rare and relatively recent mating event between L. peruviana and L. braziliensis, although we cannot exclude the possibility of multiple mating events between closely related parental parasites (46, 47). The high number of heterozygous sites across the genome, the virtual absence of homozygous regions, and the central position of the hybrids between either parental species in the PCA space provide clear evidence that they are natural first-generation hybrids that propagated mitotically since the initial cross. Rare non-Mendelian observations of homozygous tracts in telomeric regions are likely due to gene conversion events during meiosis or during mitotic divisions of the progeny clones (19), rather than due to backcrossing, inbreeding, or selfing that would result in a genome-wide mosaic of ancestry from the two parents (19, 48, 49). The absence of genomic signatures of backcrossing or inbreeding suggest that these L. peruviana × L. braziliensis hybrids may be sterile, as shown experimentally for interspecies Leishmania hybrids (19). Hybrid sterility among closely related species is the most common form of postzygotic reproductive isolation (50–52) and may be one of the factors that contributed to the speciation of L. peruviana.
The mitochondrial minicircles were inherited from both parental species, a phenomenon that has been described previously in trypanosomatids, although those studies could not reveal the extent of exchange (53–55). In contrast, the mitochondrial maxicircles demonstrated clear and consistent uniparental inheritance from the L. braziliensis parent. While these observations would suggest differential inheritance of maxicircles and minicircles, the most likely explanation is complete fusion of the entire parental kinetoplast DNA networks during genetic exchange into a single hybrid network. Subsequent mitotic divisions then resulted in homogenization of the maxicircle population, as has been suggested for Trypanosoma brucei (56). Assuming a stochastic segregation model, it was estimated that fixation in experimental T. brucei clones would complete in 139 generations (∼35 d) for 50 maxicircles and 27,726 generations (∼19 y) for 10,000 minicircles (56). In Saccharomyces cerevisiae, the mitochondrial genome homogenizes in only 20 generations (57). Rapid stochastic loss of maxicircles following mating could explain why there is currently no evidence for maxicircle heteroplasmy in natural or experimental Leishmania hybrids (19). Nevertheless, the presence of minicircles from both parental species increased the complexity of mitochondrial genomes in hybrid parasites, as evidenced by a larger number of minicircle sequence classes within hybrid isolates (average 118 classes) compared to either putative parents (73 classes in L. peruviana and 107 classes in L. braziliensis). This number is still much lower than the expected ∼180 sequence classes assuming kinetoplasts simply fused during genetic exchange, but we believe that mitotic divisions since the initial cross many generations ago have resulted in a loss of minicircle classes.
The most obvious benefits of biparental inheritance of mitochondria in trypanosomatids would be to restore gRNA redundancy to compensate for the inevitable loss of minicircles during life cycle stages where not all mitochondrial genes are under selective pressure. Mathematical models in L. tarentolae showed that nonessential minicircle classes were lost within a few hundred generations in the absence of recombination, i.e., mixing different minicircle populations (58). In addition, there is a lack of editing of several respiratory complex I NADH dehydrogenase genes in axenic cultures of Leishmania promastigotes (35, 59), suggesting that—in the absence of recombination—there is a loss of minicircle-encoded gRNAs for genes not required for life in culture. This loss may be especially efficient in Leishmania species that contain a single gRNA gene per minicircle class. Here the number of minicircle classes was significantly lower within the near-clonal and bottlenecked Andean parasites when compared to the admixed Amazonian Leishmania populations. Within this context, an interesting observation is the concordant population structure as observed with genome-wide SNPs and mitochondrial minicircles, while the mitochondrial maxicircle revealed different ancestries. This observation clearly suggests that minicircles follow a similar evolutionary path to the nuclear genome, which may indicate that compatibility between nuclear-encoded mitochondrial genes and minicircle-encoded guide RNA genes is essential to maintain efficient respiration. Here we have shown that complete editing in hybrid mitochondria may be dependent on gRNAs from both parents. This suggests that besides replenishing the minicircle repertoire, biparental inheritance of minicircles may present parasites the opportunity to incorporate novel combinations of guide RNA genes, some of which might provide more efficient editing than others, and allow for selection in recovering optimal mitonuclear interactions.
In summary, our data describe genome-wide diversity of Neotropical Leishmania parasites across and along the Peruvian Andes and show that the major parasite lineages in this group occupy particular ecological niches. This strict ecological association presents an opportunity to use this group as a model system to gain a refined understanding of the emergence and spread of leishmaniasis in the face of ecological perturbations due to human intervention and global warming, with implications for understanding these processes in other vector-borne parasitic diseases. In addition, our analyses of Leishmania hybrids suggests that genetic exchange may be crucial to the long-term survival of these parasites in the wild as it replenishes the genetic repertoire of the mitochondrial genome. This reinvigorates the need to study the biological and epidemiological significance of genetic exchange in parasitic protozoa.
Material and Methods
Parasite Collection and DNA Sequencing.
We included a total of 31 L. peruviana isolates from Peru, originating from the regions of Piura (n = 9), Huánuco (n = 2), Ancash (n = 6), Lima (n = 6), and Ayacucho (n = 6), that largely reflect the distribution of Andean cutaneous leishmaniasis (CL) (SI Appendix, Fig. S1 and Table S1), with 1 Peruvian isolate of unknown origin. We also included 23 L. braziliensis isolates from Peru (n = 21), Bolivia (n = 1), and Brazil (n = 1). In Peru, L. braziliensis isolates originated from the regions of Huánuco (n = 8), Cajamarca (n = 1), Cusco (n = 3), Madre de Dios (n = 3), Loreto (n = 2), Junín (n = 1), Pasco (n = 1), and Ucayali (n = 2) (SI Appendix, Fig. S1 and Table S1). We also included 13 isolates previously characterized as hybrid L. braziliensis × L. peruviana (16, 17) from the Huánuco region where both Leishmania species occur sympatrically. For comparative purposes, we also included L. panamensis isolate REST417. Parasites were grown in liquid culture medium for 3 to 4 d at the Antwerp Institute of Tropical Medicine or the London School of Hygiene and Tropical Medicine, and their DNA was extracted using either the QIAGEN QIAamp DNA Mini Kit or a phenol-chloroform extraction. At the Wellcome Sanger Institute, genomic DNA was sheared into 400 to 600 base pair fragments by focused ultrasonication (Covaris Inc.), and amplification-free Illumina libraries were prepared (60). One hundred base pair paired end reads were generated on the HiSeq 2000, and 150 base pair paired end reads were generated on the HiSeq ×10 according to the manufacturer’s standard sequencing protocol. Sequence data are available at Sequence Read Archive (SRA) BioProject ERP118170.
Mapping, Variant Calling and Assembly of the Mitochondrial Minicircle Genome.
Paired sequence data were aligned against a long-read assembly of the M2904 reference genome containing the 35 major chromosomes that cover 32.73 Mb and a complete circularized maxicircle sequence of 27.69 kb (available on https://tritrypdb.org/tritrypdb/app as LbraziliensisMHOMBR75M2904_2019). The major difference with the previous Illumina-based M2904 reference genome is that chromosome 20 (a fusion of chromosome 20 and 34 in L. braziliensis) was successfully scaffolded into a single chromosome. Mapping was done using SMALT v0.7.4 (https://www.sanger.ac.uk/science/tools/smalt-0), whereby the hash index was built with words of 13 base pair length (k = 13) that are sampled every other position in the genome (s = 2). Duplicate reads were tagged using MarkDuplicates as implemented in Picard tools v1.92 (https://broadinstitute.github.io/picard/). Reads that did not align to the reference genome or to the maxicircle were used for assembly of the mitochondrial minicircles. To this end, we implemented a bioinformatics pipeline in Python to automate the assembly and circularization of minicircle sequences independently for all isolates. The pipeline named KOMICS is described in SI Appendix, SI Methods, and an overview is given in SI Appendix, Fig. S12. The program is available at https://frebio.github.io/komics. Minicircle sequences were clustered based on sequence similarity using VSEARCH v2.14.1 (61).
SNP and small INDEL calling within the 35 chromosomes and the maxicircle was done in GATK v4.0.2 (62). More specifically, we used GATK’s HaplotypeCaller to produce genotype VCF files for every isolate, CombineGVCFs to merge the genotype VCF files of all isolates, GenotypeGVCFs to perform joint genotyping, and finally, SelectVariants to separate SNPs and INDELs. Low-quality SNPs were excluded using VariantFiltration when QUAL < 500, DP < 5, QD < 2.0, FS > 60.0, MQ < 40.0, MQRankSum < −12.5, or ReadPosRankSum < −8.0 or when SNPs occurred within SNP clusters (clusterSize = 3 and clusterWindowSize = 10). Low-quality INDELs were excluded when QD < 2.0, FS > 200.0, or ReadPosRankSum < −20.0. Our SNP and INDEL datasets were further refined by running GATK CallableLoci on each sample BAM file to determine genomic intervals that are callable in each sample with the following parameters: –minDepth 5 –minBaseQuality 25 –minMappingQuality 25. BEDOPS (63) –intersect was used to identify the callable genomic regions common to all isolates, and only variants within the callable genome were retained for downstream analyses. The final set of SNPs and INDELs were annotated using the L. braziliensis M2904 annotation file with SNPEFF v4.3 (64).
Chromosome and Gene Copy Number Variation.
Chromosome and local copy number variations were calculated based on haploid read depths. Per site coverages were obtained with SAMTOOLS v1.0 (65). Assuming diploidy, aneuploidy was estimated as two times the haploid chromosomal read depths, which was obtained by normalizing the median chromosomal read depths by the median genome-wide read depth. L. peruviana-specific local copy number variations were detected by comparing haploid read depths in nonoverlapping 10-kb windows or across protein-coding genes between L. peruviana and L. braziliensis. Haploid copy numbers were obtained by normalizing the median read depths per 10-kb window or per gene by the median chromosomal read depth. The haploid copy numbers of genes were summed up per orthologous gene group. In order to identify structural variations specific to L. peruviana, we subtracted the haploid copy numbers per L. peruviana isolate by the average haploid copy number observed in L. braziliensis, yielding a normal distribution centered around zero for each L. peruviana isolate. Local copy number variations were then defined where the z score was lower than −3 (deletions) or larger than 3 (amplifications). The haploid copy numbers of consecutive deletions/amplifications in 10-kb windows were summed up to obtain an estimate of the total haploid copy number within a given genomic region.
Population Genomic and Phylogenomic Analyses.
Population genomic structure was examined using phylogenetic network analyses in SPLITSTREE v4 (66), PCA in ADEGENET (67) as implemented in R v3.5.1 (68), and genotype-based clustering analyses in ADMIXTURE v1.3.0 (69). Weir and Cockerham’s FST were estimated in nonoverlapping 50-kb windows using VCFtools v0.1.12 (70). Neighbor-joining trees were built with the R package APE (71) based on raw nucleotide distances (for genome-wide SNPs) or Euclidean distances (for minicircles; see below).
To test for HWE, FIS was calculated in R using the formula 1 − (Ho/Hs), where Ho is the observed proportion of heterozygous genotypes and Hs the expected proportion of heterozygous genotypes assuming HWE (i.e., 2pq with p and q the frequency of the reference and alternate alleles, respectively). To control as much as possible for any spatiotemporal Wahlund effects, analyses of HWE were focused on four L. peruviana isolates sampled in 1989 to 1990 in the Piura region (HB22, HB31, HB55, and LC900), five L. peruviana isolates sampled in 1990 in the Ayacucho region (La36, LCA04, LCA08, LCA09, and LH249), and six L. braziliensis isolates sampled between 1991 and 1995 in the Huánuco region (LC2551, LC1409, LC1412, LC2873, LC2421, and LC2452).
Mitochondrial phylogenetic analyses were specifically focused on five maxicircle genes that are never edited (COI, ND1, ND2, ND4, and ND5) in order to guarantee robust alignments devoid of INDELs and with clear start and end positions. GATK’s FastaAlternateReferenceMaker was used to incorporate strain-specific SNPs into the mitochondrial gene sequences of each isolate. Note that we could not find any heterozygous SNP, suggesting absence of heteroplasmy and indicating that all L. peruviana isolates contained a single maxicircle sequence type. Gene alignments were concatenated and identical sequences (i.e., haplotypes) were removed using the R package APE, resulting in a dataset of 17 unique haplotypes. In addition, maxicircle coding sequences were downloaded for 15 Old and New World Leishmania species in the National Center for Biotechnology Information nucleotide database (accession nos. BK010875-80, BK010882-84, 1036_mexFOS1, CP022652, MK514111, and MK514113-5), from which the five never-edited maxicircle genes were extracted and concatenated. This resulted in a final alignment of 32 maxicircle sequences of length 6,105 bp. A dated phylogeny was obtained in BEAST v1.8.2 (72) with a strict molecular clock, a GTR + I + G4 substitution model, and a Yule tree prior. For molecular clock analyses, we could not resort to tip dating as mitochondrial mutation rates are too slow to generate sufficient mutations in our sampling time range. We therefore estimated molecular clocks for maxicircles using various divergence time estimates for the split of Leishmania major and L. infantum. Examination of the literature including studies using various genetic datasets and calibration assumptions revealed two groups of divergence times, either 20 to 25 Mya (group A) or 12 to 16 Mya (group B) (SI Appendix, Table S8). We therefore estimated molecular clocks in BEAST using two different models including a lognormal distribution prior (either with mu = 0.001, sigma = 0.8, and offset = 11.5 for group A or mu = 0.001, sigma = 1, and offset = 20 for group B) for the taxon set including L. major, Leishmania arabica, Leishmania tropica, Leishmania aethiopica, L. donovani, and L. infantum. This revealed a molecular clock for maxicircles of 0.99% (95% HPD interval = 0.83 to 1.16%) for group A and 0.59% (95% HPD interval = 0.5 to 0.68%) for group B. Finally, we ran BEAST using only a maxicircle gene alignment for the 16 haplotypes belonging to the L. braziliensis species complex, a GTR + I + G4 substitution model, a Yule tree prior, and a strict molecular clock with a uniform distribution between 0.5% (the lower 95% HPD estimate for group B) and 1.2% (the higher 95% HPD estimate for group A). The uniform prior distribution was specifically chosen to reflect the uncertainty in molecular clocks as estimated using various genetic datasets and biogeographic hypotheses (SI Appendix, Table S8).
Ecological Niche Modeling.
Species distribution models of the three studied taxa were generated using the program MAXENT v.3.3.3 (73), which estimates the potential distribution based on presence-only data. MAXENT is particularly well suited for species with few data records (74). Environmental layers consisted of 19 temperature and precipitation variables downloaded from the WorldClim dataset (http://www.worldclim.org) and past climate reconstructions at a scale of 30 arc-seconds (ca. 1 km2) for current and LIG (120 to 140 kya) scenarios and 2.5 arc-minutes (c. 5 km2) for LGM (21 kya).
Current climatic data from each occurrence point were extracted using the program Quantum-GIS v2.18 (https://www.qgis.org/en/site/). For L. braziliensis, we only included samples from the tropical moist forest below 700 m. Highly correlated variables (R > 0.7) were removed to avoid overfitting the data. The 19 bioclimatic variables were reduced to 6 variables for L. braziliensis, 2 variables for L. peruviana Porculla, and 6 variables for L. peruviana Surco. The models were run with the following parameters: quadratic, product, threshold and hinge, 500 iterations, regularization multiplier equal 1, and 10 replicates subsampled. We used the average prediction from all of the model replicates to construct the ENM species distribution maps. Present-day ENMs were projected into bioclimatic variables predicted for two different past scenarios, LIG (120 to 140 kya) and LGM (21 kya) from Model of Interdisciplinary Research on Climate and Community Climate System Model. A jackknife procedure was performed to measure the percentage of contribution and the importance of the variables to the models.
Prediction of Guide RNA Genes on Mitochondrial Maxicircles and Minicircles.
Putative maxicircle contigs were generated with SPADES v3.13 (75) using a multiple kmer strategy (21, 33, 55, 77, 99, and 127), identified by BLAST, and annotated with RATT (76) using the L. tarentolae annotation file. This resulted in an 18,775-bp maxicircle fragment for L. peruviana HR78, 18,314 bp for L. peruviana LCA04, and 19,988 bp for L. braziliensis LC1412, all covering the maxicircle coding region.
In order to obtain species-specific differences in edited maxicircle mRNA sequences, A, C, and G residues in published sequences of L. tarentolae (77–80) and/or Leishmania mexicana amazonensis LV78 (81) were corrected based on the assembled maxicircle sequences. Illumina reads from whole-cell RNA sequencing were then aligned to these manually edited sequences using SMALT, and the alignments were carefully inspected for indications of potential differences from the published editing patterns.
Canonical gRNAs were then predicted as follows. Coding and template strands of maxicircles and template strands of minicircles were aligned (not permitting gaps) to edited mRNA to predict canonical gRNAs. Each strand was split into 120-nt fragments with each fragment overlapping by 60 nt. Given that gRNA genes are about 40 nt long, an overlap of 60 nt was deemed sufficient to capture all gRNAs. Each fragment was then aligned to all edited mRNA sequences. All unique alignments which met the following criteria were recorded: 1) contained from 25 (for minicircles) and 40 (for maxicircles) to 60 noncontiguous matches (Watson–Crick or G-U base pairs), 2) contained no more than two contiguous mismatches, 3) had an anchor duplex of at least 6WC base pairs or 5WC + 1GU + 1WC bps or 4WC + 1GU + 2WC bps, and 4) covered at least one U insertion or deletion event.
Plots of predicted gRNA positions on minicircles (SI Appendix, Fig. S16) revealed that highly likely gRNAs (i.e., those longer than 40 nt with low frequency of mismatches) occurred at a well-defined position of between 450 and 525 nt downstream from the start of CSB3. All gRNAs falling into this region were assumed to be canonical gRNAs. This does mean that some minicircles encode more than one predicted canonical gRNA. In such cases, without transcriptomic data, it is impossible to determine which predicted gRNAs are transcribed.
Supplementary Material
Acknowledgments
F.V.d.B. was supported by the Department of Economy, Science and Innovation in Flanders and by the Research Foundation Flanders (Grants 1226120N and 1528117N). This work also received financial support from the European Commission (Contracts TS2-CT90-0315 and TS3-CT92-0129) and Directie-Generaal Ontwikkelingssamenwerking en Humanitaire Hulp (DGD) (Belgian cooperation). J.A.C. and M.B. acknowledge support from Wellcome (Grant 206194). A.S. is supported by the UK Medical Research Council Fellowship MR/L019701/1. S.C. was supported by a joint PhD program funded equally by the UK Biotechnology and Biological Sciences Research Council, and the UK Engineering and Physical Sciences Research Council.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920136117/-/DCSupplemental.
Data Availability.
Sequence data have been deposited in the European Nucleotide Archive under accession number PRJEB35158.
References
- 1.Hoorn C. et al., Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330, 927–931 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Nevado B., Contreras-Ortiz N., Hughes C., Filatov D. A., Pleistocene glacial cycles drive isolation, gene flow and speciation in the high-elevation Andes. New Phytol. 219, 779–793 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Beckman E. J., Witt C. C., Phylogeny and biogeography of the New World siskins and goldfinches: Rapid, recent diversification in the Central Andes. Mol. Phylogenet. Evol. 87, 28–45 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Castroviejo-Fisher S., Guayasamin J. M., Gonzalez-Voyer A., Vilà C., Neotropical diversification seen through glassfrogs. J. Biogeogr. 41, 66–80 (2014). [Google Scholar]
- 5.De-Silva D. L., Elias M., Willmott K., Mallet J., Day J. J., Diversification of clearwing butterflies with the rise of the Andes. J. Biogeogr. 43, 44–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebel E. R. et al., Rapid diversification associated with ecological specialization in Neotropical Adelpha butterflies. Mol. Ecol. 24, 2392–2405 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Hughes C., Eastwood R., Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. U.S.A. 103, 10334–10339 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagomarsino L. P., Condamine F. L., Antonelli A., Mulch A., Davis C. C., The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). New Phytol. 210, 1430–1442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roque A. L. R., Jansen A. M., Wild and synanthropic reservoirs of Leishmania species in the Americas. Int. J. Parasitol. Parasites Wildl. 3, 251–262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arana M., Evans D. A., Zolessi A., Cuentas A. L., Arevalo J., Biochemical characterization of Leishmania (Viannia) braziliensis and Leishmania (Viannia) peruviana by isoenzyme electrophoresis. Trans. R. Soc. Trop. Med. Hyg. 84, 526–529 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Bañuls A. L. et al., Is Leishmania (Viannia) peruviana a distinct species? A MLEE/RAPD evolutionary genetics answer. J. Eukaryot. Microbiol. 47, 197–207 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Garcia A. L. et al., American tegumentary leishmaniasis: Antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect. Genet. Evol. 5, 109–116 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Odiwuor S. et al., Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect. Genet. Evol. 12, 1994–2002 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Dujardin J. C. et al., From population to genome: Ecogenetics of Leishmania (Viannia) braziliensis and L. (V.) peruviana. Ann. Trop. Med. Parasitol. 89 (suppl. 1), 45–53 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Dujardin J. C. et al., Molecular epidemiology and diagnosis of Leishmania: What have we learnt from genome structure, dynamics and function? Trans. R. Soc. Trop. Med. Hyg. 96 (suppl. 1), S81–S86 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Dujardin J. C. et al., Putative Leishmania hybrids in the eastern Andean valley of Huanuco, Peru. Acta Trop. 59, 293–307 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Nolder D., Roncal N., Davies C. R., Llanos-Cuentas A., Miles M. A., Multiple hybrid genotypes of Leishmania (viannia) in a focus of mucocutaneous Leishmaniasis. Am. J. Trop. Med. Hyg. 76, 573–578 (2007). [PubMed] [Google Scholar]
- 18.Ramírez J. D., Llewellyn M. S., Reproductive clonality in protozoan pathogens—Truth or artefact? Mol. Ecol. 23, 4195–4202 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Inbar E. et al., Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania. PLoS Genet. 15, e1008042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dujardin J. C. et al., Molecular karyotype variation in Leishmania (Viannia) peruviana: Indication of geographical populations in Peru distributed along a north-south cline. Ann. Trop. Med. Parasitol. 87, 335–347 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Dujardin J. C. et al., Karyotype plasticity in neotropical Leishmania: An index for measuring genomic distance among L. (V.) peruviana and L. (V.) braziliensis populations. Parasitology 110, 21–30 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Yardley V. et al., American tegumentary leishmaniasis: Is antimonial treatment outcome related to parasite drug susceptibility? J. Infect. Dis. 194, 1168–1175 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Rougeron V. et al., Extreme inbreeding in Leishmania braziliensis. Proc. Natl. Acad. Sci. U.S.A. 106, 10224–10229 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domagalska M. A. et al., Genomes of Leishmania parasites directly sequenced from patients with visceral leishmaniasis in the Indian subcontinent. PLoS Negl. Trop. Dis. 13, e0007900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franssen S. U. et al., Global genome diversity of the Leishmania donovani complex. eLife 9, e51243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L., Hu H., Li Z., A kinetoplastid-specific kinesin is required for cytokinesis and for maintenance of cell morphology in Trypanosoma brucei. Mol. Microbiol. 83, 565–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandenburg J. et al., Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 26, 4856–4866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen T. N., Nguyen B. N., Lee J. H., Panigrahi A. K., Günzl A., Characterization of a novel class I transcription factor A (CITFA) subunit that is indispensable for transcription by the multifunctional RNA polymerase I of Trypanosoma brucei. Eukaryot. Cell 11, 1573–1581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers M. B. et al., Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 21, 2129–2142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imamura H. et al., Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. eLife 5, e12613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukes J. et al., Kinetoplast DNA network: Evolution of an improbable structure. Eukaryot. Cell 1, 495–502 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnaufer A. et al., An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291, 2159–2162 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Cooper S. et al., Assembly and annotation of the mitochondrial minicircle genome of a differentiation-competent strain of Trypanosoma brucei. Nucleic Acids Res. 47, 11304–11325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum B., Bakalara N., Simpson L., A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60, 189–198 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Simpson L., Douglass S. M., Lake J. A., Pellegrini M., Li F., Comparison of the mitochondrial genomes and steady state transcriptomes of two strains of the trypanosomatid parasite, leishmania tarentolae. PLoS Negl. Trop. Dis. 9, e0003841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Spek H. et al., Conserved genes encode guide RNAs in mitochondria of Crithidia fasciculata. EMBO J. 10, 1217–1224 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torrico M. C., De Doncker S., Arevalo J., Le Ray D., Dujardin J. C., In vitro promastigote fitness of putative Leishmania (Viannia) braziliensis/Leishmania (Viannia) peruviana hybrids. Acta Trop. 72, 99–110 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Cortes S. et al., In vitro and in vivo behaviour of sympatric Leishmania (V.) braziliensis, L. (V.) peruviana and their hybrids. Parasitology 139, 191–199 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Contreras I. et al., Leishmania-induced inactivation of the macrophage transcription factor AP-1 is mediated by the parasite metalloprotease GP63. PLoS Pathog. 6, e1001148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Paiva R. M. C. et al., Amastin knockdown in Leishmania braziliensis affects parasite-macrophage interaction and results in impaired Viability of intracellular amastigotes. PLoS Pathog. 11, e1005296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashiguchi Y. et al., Andean cutaneous leishmaniasis (Andean-CL, uta) in Peru and Ecuador: The vector Lutzomyia sand flies and reservoir mammals. Acta Trop. 178, 264–275 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Caceres A. G. et al., Epidemiology of Andean cutaneous leishmaniasis: Incrimination of Lutzomyia ayacuchensis (Diptera: Psychodidae) as a vector of Leishmania in geographically isolated, upland valleys of Peru. Am. J. Trop. Med. Hyg. 70, 607–612 (2004). [PubMed] [Google Scholar]
- 43.Messenger L. A. et al., Ecological host fitting of Trypanosoma cruzi TcI in Bolivia: Mosaic population structure, hybridization and a role for humans in Andean parasite dispersal. Mol. Ecol. 24, 2406–2422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato H., Cáceres A. G., Hashiguchi Y., First evidence of a hybrid of Leishmania (Viannia) braziliensis/L. (V.) peruviana DNA detected from the phlebotomine sand fly Lutzomyia tejadai in Peru. PLoS Negl. Trop. Dis. 10, e0004336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louradour I., Ferreira T. R., Ghosh K., Shaik J., Sacks D., In vitro generation of Leishmania hybrids. Cell Rep. 31, 107507 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Rougeron V., De Meeûs T., Bañuls A.-L., A primer for Leishmania population genetic studies. Trends Parasitol. 31, 52–59 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Akopyants N. S. et al., Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324, 265–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tihon E., Imamura H., Dujardin J.-C., Van Den Abbeele J., Van den Broeck F., Discovery and genomic analyses of hybridization between divergent lineages of Trypanosoma congolense, causative agent of Animal African Trypanosomiasis. Mol. Ecol. 26, 6524–6538 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Rogers M. B. et al., Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet. 10, e1004092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greig D., Reproductive isolation in Saccharomyces. Heredity 102, 39–44 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Maheshwari S., Barbash D. A., The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45, 331–355 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Ouyang Y., Zhang Q., Understanding reproductive isolation based on the rice model. Annu. Rev. Plant Biol. 64, 111–135 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Gibson W., Garside L., Kinetoplast DNA minicircles are inherited from both parents in genetic hybrids of Trypanosoma brucei. Mol. Biochem. Parasitol. 42, 45–53 (1990). [DOI] [PubMed] [Google Scholar]
- 54.Gibson W., Crow M., Kearns J., Kinetoplast DNA minicircles are inherited from both parents in genetic crosses of Trypanosoma brucei. Parasitol. Res. 83, 483–488 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Rusman F. et al., Elucidating diversity in the class composition of the minicircle hypervariable region of Trypanosoma cruzi: New perspectives on typing and kDNA inheritance. PLoS Negl. Trop. Dis. 13, e0007536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner C. M. R., Hide G., Buchanan N., Tait A., Trypanosoma brucei: Inheritance of kinetoplast DNA maxicircles in a genetic cross and their segregation during vegetative growth. Exp. Parasitol. 80, 234–241 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Birky C. W., Relaxed cellular controls and organelle heredity. Science 222, 468–475 (1983). [DOI] [PubMed] [Google Scholar]
- 58.Savill N. J., Higgs P. G., A theoretical study of random segregation of minicircles in trypanosomatids. Proc. Biol. Sci. 266, 611–620 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simpson L., Thiemann O. H., Savill N. J., Alfonzo J. D., Maslov D. A., Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. U.S.A. 97, 6986–6993 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozarewa I. et al., Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6, 291–295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rognes T., Flouri T., Nichols B., Quince C., Mahé F., VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKenna A. et al., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neph S., Reynolds A. P., Kuehn M. S., Stamatoyannopoulos J. A., Operating on genomic ranges using BEDOPS. Methods Mol. Biol. 1418, 267–281 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Cingolani P. et al., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Jombart T., Ahmed I., Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R Development Core Team , R: A Language and Environment for Statistical Computing, (Foundation for Statistical Computing, Vienna, Austria, 2018). [Google Scholar]
- 69.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danecek P. et al.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phillips S. J., Anderson R. P., Schapire R. E., Maximum entropy modeling of species geographic distributions. Ecol. Modell. 190, 231–259 (2006). [Google Scholar]
- 74.Wisz M. S. et al., Effects of sample size on the performance of species distribution models. Divers. Distrib. 14, 763–773 (2008). [Google Scholar]
- 75.Bankevich A. et al., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Otto T. D., Dillon G. P., Degrave W. S., Berriman M., RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 39, e57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaw J. M., Feagin J. E., Stuart K., Simpson L., Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell 53, 401–411 (1988). [DOI] [PubMed] [Google Scholar]
- 78.Shaw J. M., Campbell D., Simpson L., Internal frameshifts within the mitochondrial genes for cytochrome oxidase subunit II and maxicircle unidentified reading frame 3 of Leishmania tarentolae are corrected by RNA editing: Evidence for translation of the edited cytochrome oxidase subunit II mRNA. Proc. Natl. Acad. Sci. U.S.A. 86, 6220–6224 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sturm N. R., Simpson L., Partially edited mRNAs for cytochrome b and subunit III of cytochrome oxidase from Leishmania tarentolae mitochondria: RNA editing intermediates. Cell 61, 871–878 (1990). [DOI] [PubMed] [Google Scholar]
- 80.Bhat G. J., Koslowsky D. J., Feagin J. E., Smiley B. L., Stuart K., An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell 61, 885–894 (1990). [DOI] [PubMed] [Google Scholar]
- 81.Maslov D. A., Complete set of mitochondrial pan-edited mRNAs in Leishmania mexicana amazonensis LV78. Mol. Biochem. Parasitol. 173, 107–114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in the European Nucleotide Archive under accession number PRJEB35158.