Significance
How the loss of FMRP affects nonneuronal glial development especially underlying mechanisms remains essentially unexplored. By employing astroglial Fmr1 conditional knockout (cKO) mice and human FMRP-deficient astroglia, in the current study, we showed that the loss of astroglial FMRP cell-autonomously up-regulates brain enriched miR-128-3p in mouse and human FMRP-deficient astroglia, which selectively disrupts developmental astroglial (but not neuronal) mGluR5 signaling. Subsequent transcriptome and proteome profiling from in vivo isolated FMRP-deficient astroglia further indicates that FMRP commonly and preferentially regulates protein expression through posttranscriptional (but not transcriptional) mechanisms (such as miR) in astroglia. Overall, our study defines an FMRP-dependent cell-autonomous miR pathway that alters developmental astroglial mGluR5 signaling, which unveils astroglia-specific molecular mechanisms involved in FXS pathogenesis.
Keywords: astroglia, FMRP, miR-128, mGluR5, fragile X syndrome
Abstract
The loss of fragile X mental retardation protein (FMRP) causes fragile X syndrome (FXS), the most common inherited intellectual disability. How the loss of FMRP alters protein expression and astroglial functions remains essentially unknown. Here we showed that selective loss of astroglial FMRP in vivo up-regulates a brain-enriched miRNA, miR-128-3p, in mouse and human FMRP-deficient astroglia, which suppresses developmental expression of astroglial metabotropic glutamate receptor 5 (mGluR5), a major receptor in mediating developmental astroglia to neuron communication. Selective in vivo inhibition of miR-128-3p in FMRP-deficient astroglia sufficiently rescues decreased mGluR5 function, while astroglial overexpression of miR-128-3p strongly and selectively diminishes developmental astroglial mGluR5 signaling. Subsequent transcriptome and proteome profiling further suggests that FMRP commonly and preferentially regulates protein expression through posttranscriptional, but not transcriptional, mechanisms in astroglia. Overall, our study defines an FMRP-dependent cell-autonomous miR pathway that selectively alters developmental astroglial mGluR5 signaling, unveiling astroglial molecular mechanisms involved in FXS pathogenesis.
The loss of FMRP expression and function, resulting from the CGG expansion at the Fmr1 locus, causes the synaptic connectivity and activity changes that underlie FXS symptoms (1, 2). Extensive studies in cultured neurons and brains of Fmr1 knockout (KO) mice have shown that FMRP primarily functions as an RNA-binding protein to associate with more than 1,000 mRNAs in the brain and potentially regulates (mostly suppresses) their translation (3, 4). Several important intracellular signaling pathways such as mTOR (5), PI3K (6), and GSK3β (7) become elevated in FMRP-deficient mouse or human neurons. Biochemical and genetic evidence suggests that FMRP-mediated translational suppression is closely associated with miRNA (miR) pathways in either regulating miR biogenesis or participating in miR-mediated silencing of target mRNAs through the RNA-induced silencing complex (RISC) in neurons (8). A number of miRs have been identified to be associated with FMRP and regulate synaptic structure and function of hippocampal neurons (9).
In the mammalian central nervous system (CNS), astroglial cells play active and diverse roles in modulating synaptogenesis and synaptic functions, especially during postnatal development. Developing astroglia express Gq protein-coupled receptors (GPCRs), including group I metabotropic glutamate receptors [predominantly mGluR5 with minimal mGluR1 (10, 11)], which is activated by synaptically released glutamate that leads to subsequent release of various gliotransmitters to impact developmental synaptic plasticity and neuronal firing synchronization (12, 13)]. Immature astroglia also secrete multiple factors to promote the number and activity of excitatory synapses (14). Despite the extensive roles that developing astroglia play in modulating synaptogenesis and synaptic functions, how the loss of FMRP affects astroglial development and functions, especially underlying molecular mechanisms, remains essentially unexplored. FMRP expression has been found in nonneuronal glial cells, including astroglia (15, 16). FMRP-deficient astroglia significantly alter neuronal morphology in cocultures (17). Selective deletion of the Fmr1 gene in astroglia leads to reduced expression of astroglial glutamate transporter GLT1 (18). In addition, profiling of FMRP-bound mRNAs from brain tissue has identified a number of mRNAs that are highly and selectively enriched in astroglia, including Slc1a2, Glul, Aldoc, Apoe, and Sparcl1 (4), also indicating a strong molecular involvement of astroglia in FXS pathogenesis. Here we identified altered protein expression in Fmr1 KO astroglia and defined a FMRP-dependent cell-autonomous miR pathway that alters developmental astroglial mGluR5 signaling in human and mouse FXS models.
Results
Up-Regulation of miR-128-3p Levels in Mouse and Human FMRP-Deficient Astroglia.
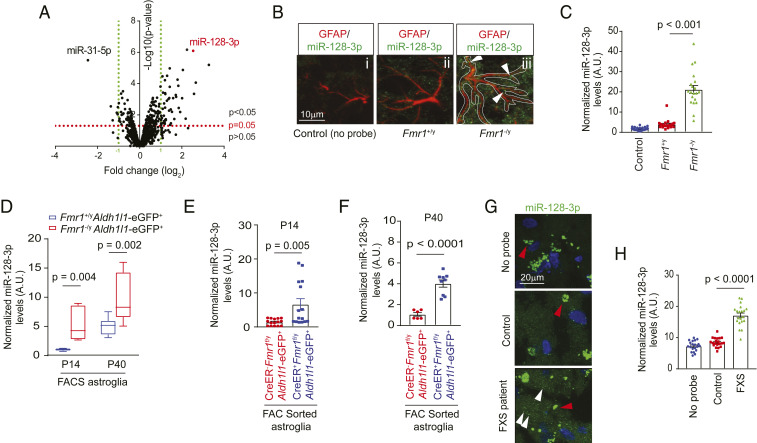
As miRNAs (miRs) play important roles in FMRP-mediated suppression of protein translation (8), to begin exploring the molecular regulators potentially involved in FMRP functions in astroglia, we first defined up- or down-regulated miRs in response to the loss of FMRP in astroglia. Interestingly, in comparison to almost equal up- (5.7%) or down-regulated (4.9%) miRs in FMRP-deficient neurons (SI Appendix, Fig. S1 A and B and Dataset S1), substantially more (29.1%) miRs are up-regulated than down-regulated (11.3%) in astroglia (SI Appendix, Fig. S1 A and B and Fig. 1A and Dataset S2). In addition, significantly altered miRs in Fmr1 KO vs. wild-type (WT) astroglia are distinct from miRs altered in Fmr1 KO vs. WT neurons with only six cochanged miRs (SI Appendix, Fig. S1 A and B and Dataset S3). These results suggest a unique and preferentially more suppressing role of FMRP in regulating miR biogenesis/processing in astroglia than in neurons. Representative top up-regulated miRs (and their primary transcripts) in Fmr1 KO astroglia, let-7c-5p, miR-139-5p, 708-5p, and 128-3p, were further confirmed by qPCR using their specific probes (SI Appendix, Fig. S1 C and D). Reexpression of FMRP in FMRP-deficient astroglia significantly attenuated the up-regulation of these miRs (SI Appendix, Fig. S1 E and F). Interestingly, expression of one of the top up-regulated miRs, miR-128-3p (miRBase accession no. MIMAT0000140) in Fmr1 KO astroglia, is not altered in Fmr1 KO neurons (SI Appendix, Fig. S1 B and G). While the miR-128-3p, a brain enriched miR, has been reported to be involved in neurogenesis (19), formation of fear extinction memory (20), and neuronal excitability (21), our bioinformatic analysis found that miR-128-3p has predicted binding sites on several functional astroglial genes such as Slc1a2 (encoding GLT1), Grm5 (encoding mGluR5), Slc6a1 (encoding GABA transporter), and Slc7a11 (encoding cystine/glutamate exchanger), etc., among others, as summarized in SI Appendix, Fig. S1H. In the brain, miR-128 is predominantly (>80%) transcribed from miR-128-2 but not miR-128-1 locus (21), and mature miR-128-5p was undetected in astroglia in our miR microarray (Fig. 1A).
Fig. 1.
Cell-autonomous up-regulation of miR-128-3p in FMRP-deficient astroglia. (A) Volcano plot of differentially expressed miRs between cultured Fmr1+/y and Fmr1−/y astroglia by miR microarray. n = 3 biological samples per condition. miRs with expression values <23 arbitrary units (A.U.) from the microarray were deemed not expressed. Representative images (B) and quantification (C) of astroglial miR-128-3p expression levels by in situ hybridization in the somatosensory cortex from Fmr1+/y and Fmr1−/y mice at P14. GFAP immunoreactivity was used as a guide to determine astroglial domains for quantifying miR-128 intensity in astroglia. (Scale bar, 10 μm.) White arrows indicate miR-128-3p signal within astroglia. n = 20 individual astroglia/3 mice per condition. P values were determined using one-way ANOVA and post hoc Tukey’s test. (D) Expression levels of miR-128-3p in FAC sorted cortical astroglia from Fmr1+/yAldh1l1-eGFP+ and Fmr1−/yAldh1l1-eGFP+ mice at P14 or P40. n = 3 to 5 biologically independent samples per condition. The data are presented in the box and whisker plot with defined elements, median (Center line), upper and lower quartiles (bounds of box), and highest and lowest values (whiskers). P values were determined using two-tailed unpaired t test. Expression levels of miR-128 in FAC sorted cortical astroglia from control (Slc1a3-CreERT−Fmr1f/yAldh1l1-eGFP+) and astroglial Fmr1 cKO (Slc1a3-CreERT+Fmr1f/yAldh1l1-eGFP+) mice at P14 (E) or P40 (F). n = 3 to 6 biologically independent samples per condition. P values were determined using two-tailed unpaired t test. Representative images (G) and quantification (H) of miR-128-3p expression by in situ hybridization in cortex from human FXS patients and nonneurological disease controls. n = 20 individual astroglia/2 subjects per condition. White arrows: miR-128-3p signals; red arrows: nonspecific fluorescent signals (>0.7 μm in diameter); P values were determined using one-way ANOVA and post hoc Tukey’s test.
Dual miR-128-3p in situ hybridization and GFAP immunostaining was first performed in Fmr1+/y and Fmr1−/y mice to examine miR-128-3p levels in cortical astroglia. By employing GFAP immunoreactivity as a guide to determine individual astroglial domains (Fig. 1 B, iii), miR-128-3p in situ signals were quantified and showed an approximately fivefold increase in Fmr1−/y cortical astroglia compared to control Fmr1+/y cortical astroglia (Fig. 1C). Cortical astroglia that were acutely sorted through fluorescence activated cell sorting (FACS) at either postnatal day (P) P14 or P40 from Fmr1−/yAldh1l1-eGFP+ mice also showed five- or twofold higher miR-128-3p levels, respectively, than in sorted Fmr1+/yAldh1l1-eGFP+ cortical astroglia (Fig. 1D). In addition, selective deletion of the Fmr1 gene in developing astroglia of astroglial Fmr1 conditional KO (cKO) (Slc1a3-CreER+Fmr1f/y) mice leads to significant reduction of FMRP in astroglia (18) (and also see SI Appendix, Fig. S1I) and similarly up-regulates miR-128-3p at both P14 and P40 in sorted cortical astroglia compared to control Slc1a3-CreER−Fmr1f/y mice (Fig. 1 E and F). These results further confirm a cell-autonomous role of astroglial FMRP in suppressing expression of miR-128-3p in astroglia. To examine whether the observed up-regulation of miR-128-3p in mouse FXS models also occurs in the human FXS condition, we generated human astroglia from astroglial progenitor cells (22) (SI Appendix, Fig. S2A) and silenced FMRP expression using lentiviral FMR1 shRNA (SI Appendix, Fig. S2 B and C). Consistently, silencing of FMRP leads to significantly increased expression of primary (pri)-miR-128-2 (SI Appendix, Fig. S2D), the major transcript to produce mature miR-128-3p, in human astroglia. We also performed in situ hybridization of miR-128-3p on postmortem FXS patient tissues and also observed significantly up-regulated miR-128-3p in FXS patients (white arrows) compared to healthy controls (Fig. 1 G and H).
Astroglial miR-128-3p Cell-Autonomously Regulates Developmental Astroglial mGluR5 Signaling.
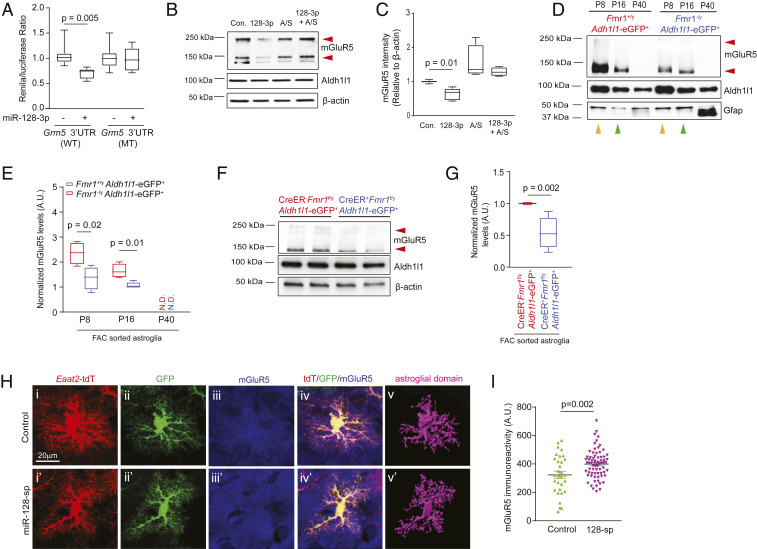
Based on the conserved miR-128-3p predicted binding site on both human and mouse Grm5 mRNAs (SI Appendix, Fig. S3A), we generated luciferase reporters with the Grm5 3′-UTR that contain either the WT or the mutant (MT) miR-128-3p binding site. Transfection of miR-128-3p effectively reduced WT Grm5 3′-UTR luciferase reporter activity but had no effect on the MT Grm5 3′-UTR luciferase reporter activity (Fig. 2A), confirming the specific binding of miR-128-3p with the Grm5 3′-UTR sequence. On the other hand, although miR-128-3p is also predicted to bind to the Slc1a2 3′-UTR (SI Appendix, Fig. S3B), miR-128-3p transfection had no effect in reducing either WT or MT Slc1a2 3′-UTR luciferase reporter activity (SI Appendix, Fig. S3C). Transfection of miR-128-3p is also able to significantly decrease mGluR5 protein levels, which can be fully rescued by miR-128-antisense (A/S) (Fig. 2 B and C). Although mGluR5 in astroglia has been considered a major pathway in mediating glutamatergic neuron-to-astroglial signaling (13, 23), mGluR5 mRNA transcript and its functional activation, measured by downstream Ca2+ increase, is largely present only in developing astroglia and becomes rapidly diminished in adult (cortical and hippocampal) astroglia (24).
Fig. 2.
Direct binding of miR-128-3p to Grm5 mRNA 3′-UTR suppresses developmental expression of astroglial mGluR5 in FXS models. (A) WT and miR-128-3p binding site MT Grm5 3′-UTR luciferase activity in HEK293 cells following miR-128-3p transfection; n = 9 to 12 biological independent samples per condition. P values were determined using one-way ANOVA and post hoc Tukey’s test. Representative mGluR5 immunoblot (B) and quantification (C) from astroglial cultures following miR-128-3p transfection. n = 4 biologically independent experiments per condition. Red arrows: mGluR5 monomer and dimer; P values were determined using one-way ANOVA and post hoc Tukey’s test. Representative mGluR5 immunoblot (D) and quantification (E) in FAC sorted cortical astroglia from Fmr1+/yAldh1l1-eGFP+ and Fmr1−/yAldh1l1-eGFP+ mice at P8, P16, and P40. n = 4 biologically independent samples per condition. Red arrows: mGluR5 monomer and dimer; P values were determined using two-tailed unpaired t test; N.D.: not detected. Representative mGluR5 immunoblot (F) and quantification (G) in FAC sorted cortical astroglia from control and astroglial Fmr1 cKO mice at P14. Red arrows: mGluR5 monomer and dimer; n = 5 to 6 biologically independent samples per condition. P values were determined using two-tailed unpaired t test. (H) Representative mGluR5 immunostaining in control or miR-128-sp expressing Fmr1−/yEaat2-tdT+ astroglia following AAV injection. The astroglial 3D domains were generated using Imaris software based on tdT fluorescence. (Scale bar, 20 μm.) (I) Quantification of mGluR5 immunoreactivity within individual astroglial domains after miR-128 sponge expression in Fmr1−/yEaat2-tdT+ mice. n = 50 to 70 cells/4 mice per group; P value was determined using two-tailed unpaired t test. (Scale bar, 20 μm.) For A, C, E, and G, the data are presented in the box and whisker plot with defined elements, median (Center line), upper and lower quartiles (bounds of box), and highest and lowest values (whiskers).
To determine how mGluR5 expression is altered in FMRP-deficient astroglia during postnatal development, we examined mGluR5 mRNA (at P14) and protein levels (at P8, P16, and P40) in FAC sorted cortical astroglia from Fmr1−/yAldh1l1-eGFP+ and Fmr1+/yAldh1l1-eGFP+ mice. It is clear that mGluR5 protein expression is highest at P8 and becomes undetectable at young adult (P40) in Fmr1+/yAldh1l1-eGFP+ astroglia (Fig. 2D), consistent with its diminishing functional response during development, as previously reported (24). Interestingly, while mGluR5 protein levels are decreased in Fmr1−/yAldh1l1-eGFP+ astroglia compared to Fmr1+/yAldh1l1-eGFP+ astroglia at both P8 (orange arrows) and P16 (green arrows) (Fig. 2 D and E), mGluR5 mRNA levels (at P14) are not altered in Fmr1−/yAldh1l1-eGFP+ compared to Fmr1+/yAldh1l1-eGFP+ cortical astroglia (SI Appendix, Fig. S4A), indicating a posttranscriptional mechanism that abnormally accelerates the developmental down-regulation of mGluR5 protein in FMRP-deficient astroglia. Protein expression of mGluR5 is similarly decreased in sorted cortical astroglia of astroglial Fmr1 cKO (Slc1a3-CreER+Fmr1f/y) mice compared to control Slc1a3-CreER−Fmr1f/y mice at P14 (Fig. 2 F and G). The silencing of FMRP expression using lentiviral FMR1 shRNA in human astroglia also consistently decreases mGluR5 protein levels (SI Appendix, Fig. S4 B and C), confirming the conserved regulatory role of FMRP in mGluR5 expression in both mouse and human models.
To demonstrate that in vivo suppression of miR-128-3p is sufficient to rescue functional expression of mGluR5 in FMRP-deficient astroglia, we generated the AAV-Gfap-GFP/miR-128-sponge (sp) construct and corresponding AAV2/5 virus that express eight tandem and previously validated miR-128-sp sequences (25) downstream of the GFP mRNA (SI Appendix, Fig. S5 A, II), so that miR-128-3p can be selectively and efficiently suppressed in astroglia. As miR-128-3p is abnormally up-regulated in FMRP-deficient astroglia, we injected AAV2/5-Gfap-GFP/miR-128-sp or control AAV2/5-Gfap-GFP (SI Appendix, Fig. S5 A, I and II) intracranially into the brain of P0–P1 pups of Fmr1−/yEaat2-tdT+ mice to selectively express miR-128-sp in astroglia during postnatal development. We have previously shown that the tdTomato (tdT) reporter is selectively expressed in ∼80% of cortical astroglia in Eaat2-tdT mice, allowing for their convenient and reliable identification (11). AAV2/5-mediated expression of miR-128-sp is efficient and specific to cortical astroglia, as indicated by the widespread and selective colocalization of GFP fluorescence with tdT+ astroglia (SI Appendix, Fig. S5B).
We then performed mGluR5 immunostaining on cortical sections of AAV2/5-miR-128-sp-injected Fmr1−/yEaat2-tdT+ mice (P14 age). As mGluR5 is expressed in multiple CNS cell types and its immunostaining is often diffuse (as a surface receptor), we generated astroglial three-dimensional (3D) domains (Fig. 2 H, iii and iii′) from tdT-based confocal images (Fig. 2 H, i and i′) using Imaris software that is able to illustrate the full morphology of individual astroglia, so that mGluR5 immunoreactivity could be specifically and accurately quantified in individual astroglia, as shown in Fig. 2 H, v and v′. Quantification of mGluR5 immunoreactivity from individual AAV2/5-Gfap-GFP/miR-128-sp and control AAV2/5-Gfap-GFP transduced (tdT+GFP+) astroglia revealed that the expression of miR-128-sp significantly increased mGluR5 immunoreactivity by 25% (P = 0.002) in individual astroglia (Fig. 2I). We also generated AAV-miR-128-3p (and scramble control miR) overexpressing constructs and corresponding AAV2/5 viruses (SI Appendix, Fig. S5 A, III and IV) based on a previously characterized AAV-miR vector (26). Consistently, mGluR5 immunoreactivity in individual astroglia of Eaat2-tdT+ mice was reduced by 45% (P < 0.0001, SI Appendix, Fig. S5C) in miR-128-3p overexpressing astroglia compared to that in scramble miR-expressing astroglia following similar intracranial injections (P0–P1) of AAV2/5-miR-128 or control AAV2/5-scramble RNA, respectively. Together with miR-128 up-regulation in Fmr1−/y and Slc1a3-CreER+Fmr1f/y astroglia, these in vivo genetic analyses of the effect of miR-128-3p on astroglial mGluR5 protein expression provide appealing evidence that abnormally up-regulated miR-128-3p, resulting from the cell-autonomous loss of FMRP in astroglia, leads to an accelerated mGluR5 reduction in FMRP-deficient astroglia during development.
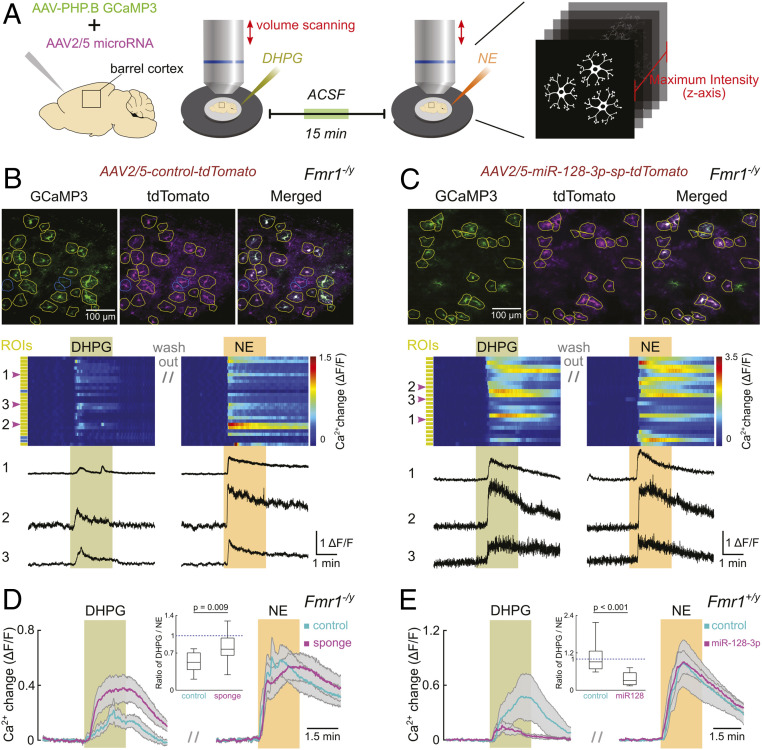
To further explore whether abnormally up-regulated miR-128-3p indeed impairs cortical astroglial mGluR5-mediated signaling, we conducted volume two-photon Ca2+ imaging of barrel cortex astroglia (Fig. 3A) in acute brain slices (P13–P16) from Fmr1−/y mice that were intracranially injected (at P0–P1) with a mixture of AAV-PHP.B-gfaABC1D-GCaMP3 (customized from refs. 27, 28) and AAV2/5-Gfap-tdT/miR-128-sp or control AAV2/5-Gfap-tdT (SI Appendix, Fig. S6 A, I, II, and V), respectively. Many astroglia were transduced by both AAV viruses, indicated by the coexpression of GCaMP3 and tdT reporters (Fig. 3B). Each slice was exposed to bath-applied (RS)-3,5-dihydroxyphenylglycine (DHPG), an mGluR1 and mGluR5 selective agonist, in the presence of tetrodotoxin (TTX) to minimize circuit effects, followed by exposure to norepinephrine (NE) to obtain a similar Gq-coupled receptor-mediated reference Ca2+ response (29) as endogenous control. Consequently, only astroglia with detectable response to NE were analyzed (yellow regions of interest [ROIs] in Fig. 3B).
Fig. 3.
miR-128-3p expression essentially and sufficiently regulates astroglial mGluR5-mediated Ca2+ response in adolescent mice. (A) Graphical representation of the experimental design. Acute slice preparation of mouse barrel cortex was performed on P13 to P16 mice following P0–P1 injection of AAVs. Two-photon volume scanning was applied to compare individual astroglial responsiveness to DHPG and NE. Ca2+ dynamics were quantified following a collapse of volume data into planar data. Upper, representative Ca2+ maximum intensity projection images from Fmr1−/y mouse during exposure to NE; green: astroglial GCaMP3 expression; magenta: expression of AAV-control-tdT (B) or AAV-miR-128-sp-tdT (C). Yellow circles: ROIs of individual tdT+ astroglia responding to NE. Blue circles: ROIs of individual tdT+ astroglia not responding to NE. Middle, pseudocolor plot represents Ca2+ dynamics of individual astroglia following AAV-control-tdT (B) or AAV-miR-128-sp-tdT (C) injections. Yellow or blue rectangles on the side indicate corresponding type of ROI in Upper. Green bar highlights DHPG (50 μM) application; orange bar highlights NE (30 μM) application. Lower, corresponding Ca2+ change traces from representative astroglia marked as 1, 2, and 3. (D) Quantitative analysis of Ca2+ responses from all examined Fmr1−/y mouse astroglia expressing either control (cyan) or miR128-sp (magenta). Each colored trace with the respective gray shading represents mean ± SEM of Ca2+ dynamics of astroglia from all slices. Colored bars indicate application of DHPG and NE, respectively. The Inset boxplot demonstrates the ratio of peak Ca2+ response induced by DHPG to that induced by NE. n = 13 slices (171 astroglia)/3 mice (control) and 13 slices (226 astroglia)/4 mice (sp). (E) Quantitative analysis of Ca2+ responses from all examined Fmr1+/y mouse astroglia expressing either control (cyan) or miR-128-3p (magenta). n = 10 slices (181 astroglia)/3 mice (control) and 10 slices (168 astroglia)/4 mice (miR-128-3p). P values were calculated from the Student’s t test. For Inset plot in D and E, the data are presented in the box and whisker plot with defined elements, median (Center line), upper and lower quartiles (bounds of box), and highest and lowest values (whiskers).
FMRP-deficient cortical astroglia that were transduced with control AAV2/5-Gfap-tdT responded to DHPG with peak Ca2+ increases that were approximately half as large as their responses to NE (DHPG/NE response ratio range: 0.207 to 0.776 with a median of 0.522) (Fig. 3 B and D). Since cortical astroglia from less than 3-wk-old mice express mGluR5, but not mGluR1 (10, 11), responses to DHPG can be attributed to mGluR5 (but not mGluR1) activation on astroglia. In contrast, when AAV2/5-Gfap-tdT/miR-128-sp was overexpressed to suppress miR-128-3p, cortical astroglia responded more similarly to DHPG and NE (DHPG/NE response ratio range: 0.290 to 1.295 with a median of 0.768) (Fig. 3 C and D), indicating a rescued and increased responsiveness to DHPG rather than an altered responsiveness to NE (SI Appendix, Fig. S6 B and C). Consistently, our bioinformatic analysis also found no predicted miR-128-3p binding sites on Adra1 a, b, or d mRNAs that encodes different alpha1-adrenergic receptor isoforms for NE activation. On the other hand, cortical astroglia from Fmr1+/y littermates at the same age responded similarly to DHPG as to NE (DHPG/NE response ratio range: 0.586 to 2.178 with a median of 0.912) (control, Fig. 3E), confirming that mGluR5 functional response is reduced in FMRP-deficient astroglia (control, Fig. 3D), consistent with its diminished protein levels (Fig. 2). Interestingly, when miR-128-3p was overexpressed in Fmr1+/y cortical astroglia through similar intracranial injections to mimic the increased miR-128-3p in Fmr1−/y astroglia, responses to DHPG were less than a third of the amplitude of responses to NE (DHPG/NE response ratio range: 0.139 to 0.722 with a median of 0.302) (Fig. 3E and SI Appendix, Fig. S6D), while the responsiveness to NE was not altered (Fig. 3E and SI Appendix, Fig. S6E). Together these findings suggest that increased expression of miR-128-3p in adolescent Fmr1−/y mice is necessary and sufficient for an accelerated functional down-regulation of mGluR5 signaling in cortical astroglia.
FMRP-Dependent Posttranscriptional Regulation of Protein Expression in Astroglia.
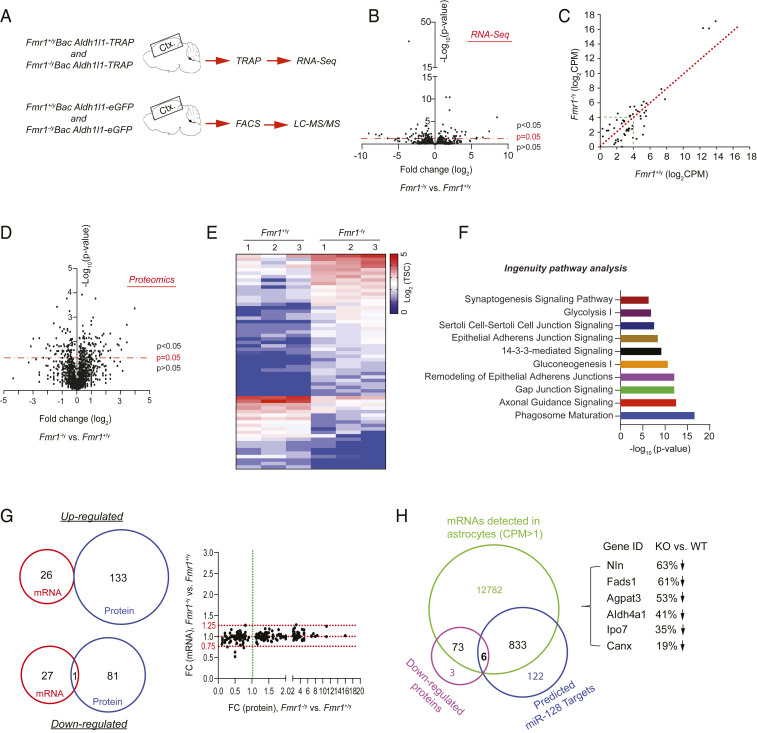
Encouraged by the observation that FMRP alters miR-128-3p levels to regulate developmental astroglial mGluR5 signaling, we set out to examine the genome-wide mRNA and protein expression profiles in cortical astroglia in Fmr1+/y and Fmr1−/y mice to determine whether FMRP-dependent posttranscriptional regulation is commonly observed in astroglia. How the loss of FMRP affects genome-wide mRNA and protein expression in astroglia (especially from in vivo conditions) has not been previously explored. We isolated cortical astroglial ribosome-bound RNA (including translating mRNA) with the translating ribosome affinity purification (TRAP) approach from Fmr1+/yAldh1l1-TRAP and Fmr1−/yAldh1l1-TRAP mice (P40) and performed RNA-seq (Fig. 4A), as we previously described (30). The TRAP approach preferentially profiles translating mRNA in cells, thus allowing a more similar match to the protein content than the total RNA profiling approaches (31). In addition, we isolated in vivo cortical astroglia by FACS from Fmr1+/yAldh1l1-eGFP and Fmr1−/yAldh1l1-eGFP mice (P40) and performed liquid chromatography tandem mass spectrometry (LC-MS/MS)-based proteomic analysis (Fig. 4A). The astroglia specificity of RNA sequencing (RNA-seq) and proteomic data were confirmed with known CNS cell type specificity genes (SI Appendix, Fig. S7 A and B). RNA-seq results showed that overall translating mRNA levels in cortical astroglia of Fmr1−/y mice is highly similar to that in Fmr1+/y mice (Fig. 4B and Dataset S4) with 54 differentially expressed genes (DEGs, P < 0.05). Similar to the qPCR results in SI Appendix, Fig. S4A, we observed no significant difference in mGluR5 mRNA levels in Fmr1+/y (mean counts per million mapped reads [cpm]: 45) and Fmr1−/y (mean cpm: 41) mice. Many of these DEGs have either low expression levels (<24 cpm) or only show modest fold changes (FCs), indicated by being close to the equality line (Fig. 4C and Dataset S5), further indicating that mRNA profiles are only modestly altered in Fmr1 KO astroglia.
Fig. 4.
Genome-wide profiling of translating mRNAs and proteins in Fmr1+/y and Fmr1−/y cortical astroglia. (A) Schematic diagram of TRAP/RNA-seq and FACS/LC-MS/MS to profile ribosome-bound mRNA and proteomics from cortical astroglia of Bac Aldh1l1-TRAP or Bac Aldh1l1-eGFP mice (P40), respectively. (B) Volcano plot of expressed mRNAs in cortical astroglia identified by TRAP/RNA-seq from Fmr1+/yAldh1l1-TRAP and Fmr1−/yAldh1l1-TRAP mice. n = 3 biologically independent samples per condition; somatosensory cortex sections (1 mm) were used in TRAP isolation. (C) Scatterplot of averaged expression values (log2 cpm) of DEGs between Fmr1+/yAldh1l1-TRAP and Fmr1−/yAldh1l1-TRAP cortical astroglia. (D) Volcano plot of proteins identified by LC-MS/MS from FAC sorted cortical astroglia of Fmr1+/yAldh1l1-eGFP and Fmr1−/yAldh1l1-eGFP mice. n = 3 biologically independent samples per condition. (E) Heatmap of top DEPs (FC >2 fold) between cortical astroglia of Fmr1+/yAldh1l1-eGFP and Fmr1−/yAldh1l1-eGFP mice. TSC: total spectrum counts. (F) Top 10 signal pathways clustered from DEPs using IPA. (G) Essentially no overlap between altered mRNAs and proteins between Fmr1+/y and Fmr1−/y astroglia illustrated by Venn diagrams and scatterplot. Scatterplot shows fold changes of altered proteins and their corresponding mRNAs. (H) Identification of down-regulated proteins in Fmr1−/y astroglia whose mRNAs have predicted miR-128-3p binding sites. Venn diagram showing the number of expressed mRNAs (cpm > 1) and down-regulated proteins, as well as predicted miR-128-3p mRNA targets, respectively. There are six mRNAs having miR-128-3p binding targets and also encoding for proteins that are down-regulated in Fmr1−/y astroglia compared to Fmr1+/y astroglia. These six genes and their protein expression changes in Fmr1−/y vs. Fmr1+/y astroglia are shown.
In contrast, 215 differentially expressed proteins (DEPs, P < 0.05) were identified in FAC sorted cortical astroglia from Fmr1+/y and Fmr1−/y mice (Fig. 4D and Dataset S6) with the top DEPs (FC > 2, P < 0.05) illustrated in the heat map (Fig. 4E). As expected, mGluR5 protein was not found in our proteomic datasets, because its protein expression in astroglia has significantly diminished at P40 (Fig. 2D). Many of these DEPs are enzymes, transporters, and protein kinases (SI Appendix, Fig. S7C) that are significantly clustered in important astroglial pathways and astroglial interaction with neurons, including phagocytosis, gap junctions, axonal guidance, and synaptogenesis signaling based on Ingenuity Pathway Analysis (IPA) (Fig. 4F and Dataset S7). Surprisingly, the protein changes induced by the loss of FMRP are essentially nonoverlapping with the mRNA changes in Fmr1 KO astroglia (Venn diagrams in Fig. 4G), clearly suggesting posttranscriptional (but not transcriptional) mechanisms for protein changes in Fmr1−/y astroglia. Subsequent examination of altered proteins and their corresponding mRNAs confirmed an overall small fold change (FC, <0.25 fold) of most of these mRNAs between WT and Fmr1 KO astroglia, while the FC of altered proteins between WT and Fmr1 KO astroglia has a much greater range (0.1- to 16-folds, scatterplot in Fig. 4G and Dataset S8).
To further determine whether miRs are potentially involved in FMRP-dependent protein changes in Fmr1−/y astroglia, we compared miR-128-3p predicted mRNA targets, expressed mRNAs in astroglia (cpm > 1), and down-regulated proteins in Fmr1−/y astroglia (P < 0.05) and found that six of them have predicted miR-128-3p binding sites (Fig. 4H and Dataset S9). Previous studies have identified many mRNAs that are associated with FMRP, presumably FMRP targets, in developing mouse brain using the high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) approach (4), we then compared miR-128-3p predicted mRNA targets that encode proteins altered in proteomic analysis between WT and Fmr1 KO astroglia with previously identified FMRP-associated mRNAs. We found only three such mRNAs (Agpat3, Slc6a17, and Syt1). In addition to miR-128-3p, we also overlapped predicted mRNA targets of miR-31-5p, one of the most down-regulated miRs with up-regulated proteins in Fmr1−/y astroglia and also found three genes with significantly increased protein but not mRNA levels in Fmr1 KO astroglia (SI Appendix, Fig. S7D and Dataset S9), further supporting common miR-mediated posttranscriptional mechanisms in regulating FMRP-dependent protein expression in astroglia. By comparing DEGs or DEPs with previously identified FMRP-associated mRNAs, we found that there is only one DEG (Lars2) that is also associated with FMRP. However, there are 29 DEPs (Dataset S13) whose encoding mRNAs are also associated with FMRP, further suggesting that FMRP is able to directly regulate protein translation of these mRNAs in astroglia.
Discussion
Our current results demonstrated a FMRP-dependent and miR-mediated posttranscriptional mechanism in regulating the developmental function of important astroglial mGluR5 signaling. This mechanism appears conserved, as we made consistent observations in both rodent and human FMRP-deficient astroglia in vitro and in vivo. Our transcriptomic and proteomic profiling from WT and Fmr1 KO astroglia also identified altered mRNAs and proteins in astroglia in response to the loss of FMRP. These results suggest that the loss of FMRP preferentially induces widespread changes in protein expression by posttranscriptional (but not transcriptional) mechanisms in Fmr1 KO astroglia. Our results provide interesting insights into the genome-wide molecular changes of astroglia in FXS conditions and begin to unveil the role and mechanism of FMRP in regulating protein expression in astroglia.
Altered synaptic signaling and enhanced neuronal excitability have been widely observed in mouse and human FXS models to play a key role in developing FXS phenotypes (1). FMRP ablation resulted in dysregulation of the maturation of glutamatergic signaling, especially the developmental switching of the NMDA/AMPA ratio during the critical period in Fmr1 KO mice (32). Enhanced neuronal excitability and circuit level hyperexcitability have also been considered one of the central mechanisms underlying FXS-related behavioral hyperactivity and seizures (33). Activation of astroglial mGluR5 (and other Gq GPCRs) is known to evoke intracellular Ca2+ elevations and induce subsequent release of various gliotransmitters that modulate synaptic signaling and neuronal excitability (12). In addition, mGluR5 activation contributes to developmental induction of the predominant astroglial glutamate transporter GLT1 and developmental dynamics of astroglial processes (11, 34). Selective deletion of mGluR5 in astroglia significantly reduces GLT1 expression and pharmacological inhibition of mGluR5 reduced synaptic activity-evoked peripheral astroglial process (PAP) motility (11, 34). It is well established that astroglial GLT1-mediated glutamate uptake and PAP coverage of synapses actively and significantly impact synaptic transmission (35). Thus, premature decrease of astroglial mGluR5 by FMRP deficiency-induced up-regulation of miR-128-3p would alter key astroglial functions from gliotransmission to glutamate uptake and synaptic coverage, contributing to altered synaptic signaling and enhanced neuronal excitability in FXS conditions, as we recently observed in synaptic signaling and behaviors of astroglial Fmr1 cKO mice (18, 36). It is important to note that while our results define a miR-based molecular pathway in down-regulating mGluR5 in FMRP-deficient astroglia, mGluR1/5 expression is not altered in FMRP-deficient neurons despite the observation of enhanced group I mGluR-mediated synaptic plasticity in Fmr1 KO mice (37, 38). These contrasting observations in neurons and astroglia point to a cell type-specific mechanism, presumably differential coordination between FMRP and miRs (such as miR-128-3p) in regulating mRNA translation (like mGluR5).
In contrast to well-established and expanding functions of FMRP in neurons, the functions of FMRP, especially in the regulation of protein expression in glia, including astroglia, remain very little known. It is also unexplored how mRNAs and proteins in astroglia are altered in response to the loss of FMRP. Our current proteomic analysis of FAC sorted WT and Fmr1 KO astroglia begins to provide a first glimpse of protein expression changes in Fmr1 KO astroglia. Interestingly, although we employed the TRAP/RNA-seq approach which preferentially profiles translating but not total mRNAs thus allowing more direct comparison with protein expression changes in WT vs. Fmr1 KO astroglia, we only observed drastic changes in protein expression with essentially no changes of corresponding mRNAs. Our results are in contrast to evident mRNA changes in FMRP-deficient hippocampal CA1 neurons or adult neural stem cells using either RiboTag or ribosome profiling approaches (39–41). The differential and cell type-specific mRNA expression changes in the FMRP-deficiency condition support the notion that there are preferential FMRP-dependent posttranscriptional mechanisms, such as miRs, in regulating protein expression in astroglia. These results are consistent with astroglia specific and FMRP-dependent miR-128-3p regulation of mGluR5 expression. Our miR microarray analysis from FMRP-deficient neurons or astroglia also suggest that FMRP appears to preferentially suppress miR biogenesis/processing in astroglia.
Although FMRP has been implicated in regulating miR biogenesis at the precursor-miRNA (pre-miR) to mature miR level by interacting with Dicer (42, 43), the up-regulation of mature miRs and their corresponding pri-miRs in FMRP-deficient astroglia (SI Appendix, Fig. S1 C and D) suggests that FMRP may preferentially suppress transcription of miR genes in astroglia. Indeed, loss of FMRP has been shown to induce widespread changes in chromatin regulation (44) that may selectively promote transcription of certain (miR) genes. Whether this is the mechanism that results in preferential up-regulation of miR expression in FMRP-deficient astroglia remains to be investigated. In addition, we are able to identify additional down-regulated proteins whose encoding mRNAs have predicted miR-128-3p binding sites, or inversely up-regulated proteins whose encoding mRNAs have predicted binding sites of miR-31-5p, one of the top down-regulated miRs in Fmr1 KO astroglia. Whether these FMRP-dependent posttranscriptional mechanisms involve direct binding to mRNAs and how specific miRs regulate FMRP-dependent protein expression in astroglia remain to be investigated in future studies, which will provide new insights into astroglial mechanisms in understanding the pathogenesis of FXS.
Materials and Methods
Mice.
The Fmr1f/f mice were generated as previously described (45). Bac Slc1a3-CreERT transgenic mice (C57 BL/6J background; stock no. 012586) and Fmr1 KO mice (FVB background; stock no. 003024) were obtained from The Jackson Laboratory. Astroglial Fmr1 cKO mice were previously generated using Bac Slc1a3-CreERT and Fmr1f/f mice (18). Bac Aldh1l1-TRAP and Bac Aldh1l1-eGFP mice were obtained from the GENSAT project (The Rockefeller University). Eaat2-tdT mice were previously generated in the laboratory (11, 46). Only male mice were used from Fmr1 KO, astrocyte Fmr1 cKO mice in both in vitro and in vivo experiments, because the Fmr1 locus is on the X chromosome and males are more severely affected than females in FXS mouse models and patients. All mice were maintained on a 12-h light/dark cycle with food and water available ad libitum. Care and treatment of animals in all procedures strictly followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Guidelines for the Use of Animals in Neuroscience Research and the Tufts University Institutional Animal Care and Use Committee.
Drug Administration.
Tamoxifen [(Z)-4-hydroxytamoxifen/4-OHT; Sigma-Aldrich; H7904] was resuspended at 20 mg/mL in 100% ethanol and diluted in sunflower seed oil (Spectrum; S1929) to a final concentration of 2 mg/mL. For astrocyte Fmr1 cKO mice, a daily i.p. injection of 20 μL 4-OHT was administered from P4 to P9 for a total dose of 0.24 mg. All control mice received the same dose of 4-OHT injections.
RNA Isolation and miR qPCR.
Total RNA was extracted from cell pellets, or FAC-sorted cells using TRIzol reagent (Thermo Fisher Scientific; 15596018), by following the manufacturer’s instructions. To remove DNA contamination, 1 μg RNA samples were treated with 1 unit of DNase I at 37 °C for 30 min. DNase I was later inactivated by adding 1 μL 25 mM ethylenediaminetetraacetic acid (EDTA) and incubating at 65 °C for 10 min. The quantity and quality of isolated RNA was determined using the Agilent BioAnalyzer according to the manufacturer’s instructions. The miR qPCR was performed based on TaqMan miR assay protocol. Briefly, the 10 ng of total RNA samples were converted to cDNA using the TaqMan miR Reverse Transcription Kit (Applied Biosystems) with specific primers for each individual miR and control U6 small nuclear (sn) RNA (Applied Biosystems). For pri-miRNAs, Grm5, Slc1a2, and Actb random primers, provided in High Capacity cDNA Reverse Transcription Kit (Invitrogen; 4368814), were used for the reverse transcription. Specific primers for Grm5, Slc1a2, and Actb (chosen from PrimerBank: https://pga.mgh.harvard.edu/primerbank/) were used in qPCR. Actb was used as an endogenous control for normalization of pri-miRNA and mRNA. For mouse pri-miR-128-2 qPCR, the Taqman pri-miR-128-2 kit (Mm03307278_pri) was used; For human pri-miR-128-2 qPCR, total RNA was similarly isolated using the TRIzol reagent. cDqPCRNA was synthesized using PrimeScript RT Reagent Kit (Takara, catalog no. RR037A) with random 6-mers. qPCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) on a StepOnePlus Real-Time PCR cycler (Applied Biosystems). Gapdh gene was utilized as an endogenous gene. Primers (5′ to 3′) used for qPCR are listed below: Actb-F: ggctgtattcccctccatcg, Actb-R: ccagttggtaacaatgccatgt; Slc1a2-F: acaatatgcccagcaggtaga, Slc1a2-R: gacaccaaacacagtcagtga; Grm5-F: acaatatgcccagcaggtaga, Grm5-R: gacaccaaacacagtcagtga; pri-miR-128-2-F: tgtgcagtgggaaggggg, pri-miR-128-2-R: gacacagtagggaaagagaccg; Fmr1-F: gcaaatgtgtgccaaagaggc, Fmr1-R:ctccgaaagtgcatgtcaatcag; and Gapdh-F: caaattccatggcaccgtca, Gapdh-R: ttggagggatctcgctcctgg.
miR Microarray and Analysis.
Primary cultured cortical astroglial and neuronal RNA from Fmr1+/y and Fmr1−/y mice were hybridized to GeneChip miRNA (Affymetrix, version 4.0 array) at the Boston University microarray facility (n = 3 per group). Raw Affymetrix CEL files of miRNA microarrays were normalized to produce probe set-level expression values across mouse and control probe sets using the Affymetrix Expression Console (version 1.4.1.46) with robust multiarray average (RMA) normalization and detection above background (DABG). Analysis was limited to the 1,908 mouse miRs interrogated by the array. Principal component analysis (PCA) was performed using the prcomp R function with expression values that had been normalized across all samples to a mean of zero and a SD of one. All samples had similar quality metrics, including mean relative log expression (RLE) values and percent present calls (% P), indicating that all samples were of similar quality. Pairwise differential expression of microRNAs was assessed using the moderated (empirical Bayesian) t test implemented in the limma package (version 3.14.4). Correction for multiple hypothesis testing was accomplished using the Benjamini–Hochberg false discovery rate (FDR). Human homologs of mouse genes were identified using HomoloGene (version 68). All microarray analyses were performed using the R Environment for Statistical Computing (version 2.15.1).
Preparation of Cell Suspension and FACS.
Animals were deeply anesthetized with ketamine (100 mg/kg) + xylazine (10 mg/kg) in saline by i.p. injection and perfused intracardially with Hanks’ balanced salt solution (HBSS) (Thermo Fisher Scientific; 14175103). The brain cortices were immediately dissected in cold HBSS solution. Cell suspension was prepared using the Neural Tissue Dissociation Kit (Miltenyi Biotech, 130-092-628) by following the manufacturer’s instructions. The brain cortices were cut into small pieces and treated with papain enzymatic mix at 37 °C for 15 min, and then digested with DNase I at 37 °C for 10 min. Cell mixtures were filtered through a cell strainer (70 μm) and resuspended in cold HBSS solution for sorting. Cells were sorted using a MoFlo MLS high-speed cell sorter (Beckman Coulter) with Summit version 4.3 software at the Tufts FACS facility. FAC sorted cells were spun down at 13,000 × g for 5 min and kept at −80 °C until RNA and protein extraction.
Affinity Purification of Translating RNA, Sequencing, and Analysis.
Ribosome-bound RNAs were isolated as described previously (47). Briefly, somatosensory cortical sections (1 mm) from Fmr1+/yAldh1l1-TRAP and Fmr1−/yAldh1l1-TRAP mice were prepared on a mouse brain matrix. The anti-eGFP antibody (mouse clone HtzGFP-19C8, Memorial Sloan Kettering Cancer Center) was coupled to magnetic beads (Dynabeads, Invitrogen) as described in the kit instructions. Tissue homogenates were prepared and mixed with eGFP antibody-coupled beads for immunoprecipitation. Following immunoprecipitation, RNA was isolated using the TRIzol reagent and precipitated with isopropanol. Three biological replicates were separated and prepared for each experimental condition. Preparation of RNA sequencing libraries and sequencing was carried out by the Tufts University Core Facility (TUCF) Genomics facility. The TrUSeq Stranded Total RNA with RiboZero Gold (Illumina) was used in library preparation. The 100-bp paired-end sequencing was carried out on an Illumina HiSEq. 2500 sequencer. RNA-seq libraries were assigned randomly to sequencing lanes to avoid lane bias. The FastQC program was used to examine the quality of generated reads. For mouse samples, we usually obtained 20 to 30 million reads for each library with a Phred quality score >32. The total number of mapped reads for each sample was at least 60 million, sufficient for reliable calculations of cpm values and subsequent differential gene expression analysis. For RNA-seq analysis, reads were aligned to the reference mouse genome (GRCm38/mm10) using STAR (v 2.6.0) software. RSEM (v 1.3) was used to calculate cpm values. The genes with an averaged cpm >1 were considered detectable transcripts. EdgeR (3.5) was used to identify significant changes in mRNA expression among different samples using the biological replicates with a default q-value (FDR-adjusted P value) cutoff of 0.05 (q < 0.05).
In Situ Hybridization of miR-128-3p.
A locked-nucleic-acid (LNA) fluorescent miR-128-3p probe was used according to the manufacturer’s instructions. First, animals were anesthetized with an i.p. injection of a ketamine/xylazine mixture (100/10 mg/kg) and intracardially perfused with cold phosphate-buffered saline (PBS). Brain tissues from freshly perfused mice and frozen human brain samples were embedded in optimal cutting temperature (OCT)-Compound Tissu-Tek (VWR; 25608-930). Human postmortem brain (FXS and nonneurological disease controls) cortical tissues were obtained from the National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank. Cortical sections (20 μm) were prepared using a Leica HM525 cryostat and mounted on glass SuperFrost+ slides (Thermo Fisher Scientific, 1255015). Slides were washed with PBS for 10 min and fixed with freshly made 4% paraformaldehyde (PFA) at room temperature (RT) for 2 min. After a 10-min incubation with proteinase K at 37 °C, the slides were hybridized with denatured FAM-labeled miR-128-3p LNA probe (Exiqon) diluted to 40 nM overnight at 55 °C. miR-128-3p probes were denatured at 90 °C for 4 min. After stringent washes with saline-sodium citrate (SSC) buffer the immunostaining procedure was performed. Slides were incubated in blocking solution [1% bovine serum albumin (BSA), 5% goat serum, and 0.2% Triton-X 100 in PBS] for 1 h and then incubated with anti-FAM rabbit antibody (1:250; Sigma-Aldrich: A889) overnight to amplify the LNA probe signal. The following day, after washing the slides three times in PBS, a Alexa 488 goat anti-rabbit secondary antibody (1:2,000; Life Technologies: A11008_9205179) was added for 1 h at RT. The sections were then washed three times in PBS. Following this, immunostaining with anti-GFAP mouse antibody and Alexa 555 goat anti-mouse secondary antibody was performed to label astrocytes following standard immunostaining procedures. The slides were then mounted with ProLong Gold antifade reagent with DAPI (Invitrogen, P36931).
Generation of Luciferase Reporters and Luciferase Assay.
The oligos of miR-128-binding sites on the Grm5 and Slc1a2 mRNA 3′-UTR luciferase reporter constructs were synthesized by Integrated DNA Technologies as follows: wild-type Grm5: 5′-ttttatactgtgaaaaaaatacag-3′, mutant Grm5: 5′-ttttataaaaaatacag-3′; and wild-type Slc1a2: 5′-gctggcaaactgtgaatctaatt-3′, mutant Slc1a2: 5′-gctggcaaatctaatt-3′. Synthesized oligos were cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, E1330). Constructs were confirmed by sequencing. For HEK293 cell transfection, HEK293 cells (ATCC, CRL-1573) were seeded 1 × 104 per well in 96-well plates and transfected at 70 to 90% confluency with 0.1 μg luciferase reporter constructs alone or luciferase reporter construct with miR-128-3p mimics (100 nM, Dharmacon) using Dharma FECT Reagent. The pmirGLO vector alone was used as the mock transfection. Luciferase activity was measured 48 h after transfection of HEK293 cells using the Dual-Glo luciferase kit (Promega, E2920). Coexpressed Renilla luciferase on the pmirGLO vector was used as an internal control to normalize the firefly luciferase activity.
Primary Neuronal and Astrocyte Culture and Transfection.
For cortical astrocyte cultures, P0–P3 mouse pups were decapitated, and cerebral cortices were removed and transferred into 37 °C prewarmed astrocyte growth culture medium (Dulbecco minimum essential medium [DMEM[ [Gibco, 11995], supplemented with 10% fetal bovine serum [FBS] [Sigma-Aldrich, F4135], and 1% penicillin/streptomycin [P/S]) for dissection. Olfactory bulbs and cerebellum were removed, and the meninges were stripped. Cortices were finely chopped and placed into 0.05% Trypsin-EDTA solution (Gibco, 25300-054) for 10 min at 37 °C. The enzymatic reaction was stopped by addition of astrocyte culture medium. The tissue was gently dissociated by trituration with a firepolished Pasteur pipette. The dissociated cells were centrifuged for 10 min at 300 × g at RT. Supernatant was removed and cells were resuspended in fresh astrocyte growth medium. Dissociated cells were filtered through a 70-μm cell strainer (BD Falcon Nylon Cell Strainer, 32350) to collect a clear astrocyte cell suspension. After counting the cells, astrocytes were plated on six-well plates (1 × 106 cells/well). For neuronal primary cultures, cortical neuronal cells were isolated from embryonic days 14 to 16 (E14 to E16) mouse brains. The neuron culture medium was composed of neurobasal medium (Gibco, 21103-049), 2% B27 serum-free supplement (Invitrogen, 17504044), 1% of GlutaMAX (Gibco, 35050-061), and 1% P/S. The brain dissociation procedure was similar to the astrocyte isolation procedure described above. Freshly prepared neurons were then plated on six-well plates (1 × 106 cell/well). Primary astrocyte transfection with luciferase or Fmr1 cDNA (2.5 μg/well) in a six-well plate was performed with Lipofectamine 3000 reagent (Invitrogen, L3000001). Transfection of the miR, miR-A/S (antisense), or a mixture of miRs and miR-A/S (25 nM) was performed with DharmaFECT dual transfection reagent (Thermo Fisher Scientific) following the manufacturer’s instructions.
Human Cell Culture and Astrocyte Differentiation.
Human embryonic stem cell (hESC) line H1 (WA01) was obtained from WiCell. Pluripotent stem cells were cultured on mouse embryonic fibroblast (MEF) feeder layers (WiCell) with a daily change of hESC medium of DMEM/F12 (Thermo Fisher Scientific), 20% knockout serum replacement (Thermo Fisher Scientific), 0.1 mM 2-mercaptoethanol (Sigma), 1× l-glutamine (Thermo Fisher Scientific), and 6 ng/mL FGF-2 (Waisman Biomanufacturing). Cells were passaged using 6 U/mL of dispase (Thermo Fisher Scientific) in hESC medium, washed, and replated at a dilution of 1:5 to 1:10. Neural induction was carried out using a dual SMAD method (48) with modifications. In brief, 5 d after hPSCs were passaged onto MEFs, neural differentiation was induced by switching hESC medium to neural induction medium (NIM) of DMEM/F12:Neurobasal 1:1, 1× N2 supplement [Waisman Center induced pluripotent stem cells (iPSC) Service], 1× l-glutamine, 1× antibiotic-antimycotic (Gibco), 10 μM SB432542 (Selleck), 100 nM LDN193189 (Selleck), and 5 μM XAV-939 (Selleck). Cells were cultured in NIM for 9 d with a daily medium change. The cells were then dissociated with dispase and cultured as neurospheres in astrocyte progenitor cell (APC) medium of neurobasal medium (Thermo Fisher Scientific), 1× GlutaMAX (Thermo Fisher Scientific), 1× N2, 0.5× B27 without vitamin A (Thermo Fisher Scientific), 1× antibiotic-antimycotic, 5 ng/mL FGF-2, 10 ng/mL EGF. Astrocyte progenitor cells were propagated and terminally differentiated into mature human astrocytes as previously described (22, 49). Briefly, 7-mo astrocyte progenitor spheres were dissociated using Accutase and plated in a six-well plate precoated with polyethyleneimine (PEI) and Matrigel. Astrocyte progenitors were plated 5 × 105 cells per well and allowed to differentiate for 1 wk in astrocyte differentiation media (ADM) of neurobasal medium, 1× GlutaMAX, 1× N2, 0.5× B27 without vitamin A, 1× antibiotic-antimycotic, 10 ng/mL BMP4 (R&D Systems), and 10 ng/mL ciliary neurotrophic factor (CNTF) (R&D Systems).
Lentiviral FMR1 Knockdown.
Plasmid constructs encoding scrambled control (shSC: 5′-gcttcgcgccgtagtctta-3′) or shFMR1 (5′-gcatgtgatgcaacttaca-3′) were generated by insertion into the pCDH-CMV-MCS-Ef1α-copGFP lentivector (System Biosciences, CD511B-1) with the GFP reporter replaced with mCherry reporter and insertion of the hUBC promoter amplified from pHAGE-UBC-NLS-HA-PCP-GFP construct (Addgene, 64539). All plasmid constructs were verified by DNA sequencing. shSC-mCherry and shFMR1-mCherry lentiviral production was performed as described previously (50). Briefly, lentiviral DNA was cotransfected with packaging plasmids pCMV-VSV-G and psPAX2 into HEK293 T cells using the calcium phosphate method. The medium containing lentivirus was collected at 24- and 60-h posttransfection, pooled, and centrifuged to remove cellular debris. Human astrocytes were infected 1 wk after plating with lentivirus encoding shFMR1-mCherry or shSC-mCherry and collected 3 d postinfection.
Immunoblot.
FAC sorted or cultured astrocytes were homogenized, and the total protein amount was determined by the Bradford protein assay. A total of 10 μg or 20 μg of cell lysate was loaded on 4 to 15% gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) gels, and the protein mixtures were separated by size using gel electrophoresis. Separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (0.22 μm, Bio-Rad) at constant 2.5 Amp for 10 min using the Bio-Rad Trans-Blot Turbo Transfer System. The membrane was blocked with 3% nonfat milk in TBST (Tris buffer saline with 0.1% Tween 20) then incubated with the appropriate primary antibody overnight at 4 °C. On the following day, the membrane was incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:5,000) diluted in TBST for 1 h at RT. Bands were visualized by ECL Plus chemiluminescent substrate (Thermo Fisher Scientific) the Bio-Rad ChemiDoc MP imaging system. The antibodies used were: Anti-mGluR5 (1:1,000, Millipore, rabbit), anti-GFAP (1:2,000, Dako, rabbit), anti-FMRP (1:20, mouse monoclonal antibody 2F5, Developmental Studies Hybridoma Bank or 1:1,000, Thermo Fisher Scientific, mouse), anti-ALDH1L1 (1:20, mouse monoclonal antibody, Developmental Studies Hybridoma Bank), anti-GAPDH (1:2,500, Thermo Fisher Scientific, mouse; 1:2,500, Thermo Fisher Scientific, rabbit) and anti-β-actin (1:1,000; Sigma-Aldrich, mouse).
Immunohistochemistry and Confocal Image Acquisition.
Animals were deeply anesthetized with ketamine (100 mg/kg) + xylazine (10 mg/kg) in saline by i.p. injection and perfused intracardially with 4% paraformaldehyde (PFA) in PBS. The brains were dissected and kept in 4% PFA overnight at 4 °C, then cryoprotected by immersion in 30% sucrose. Brains were embedded and frozen in OCT-Compound Tissu-Tek (VWR, 25608-930). Coronal sections (20 μm) were prepared with a cryostat (Leica HM525) and mounted on glass SuperFrost+ slides (Thermo Fisher Scientific, 1255015). Slides were rinsed three times in PBS, then treated with blocking buffer (1% BSA, 10% goat-serum, and 0.2% Triton X-100 in PBS) for 30 min at RT. Primary antibodies for GFAP (1:1,000, Thermo Fisher Scientific, IF03L, mouse), mGluR5 (1:1,000, Millipore, AB5675, rabbit), and S100β (1:1,000, DakoCytomation, A5110, rabbit) were incubated overnight at 4 °C. After washing slides three times in PBS, the corresponding secondary antibody (1:1,000, Life Technologies, A21070) was added for 1 h at RT. The sections were rinsed three times in PBS before mounting. Low magnification images were taken using the Zeiss Axio Imager with ApoTome. Confocal images were taken using the Nikon A1R or Leica SPE confocal laser scanning microscope (15- to 20-μm Z-stack with 0.5-μm step) magnified with ×40 (numerical aperture 0.8) or ×63 (numerical aperture 1.0) objectives. Images were taken under optimized settings which were consistent between control group and viral transduced group.
Adeno-Associated Viral (AAV) Vector Cloning and AAV Preparation.
The U6-miR-128/scrambled miRNA-CMV-GFP AAV constructs were generated by replacing the mCherry reporter in the AAV-CMV-mCherry/U6-mcs construct (University of Iowa viral vector core, G0774) with the eGFP reporter amplified from AAV-Gfap-eGFP plasmid (Addgene, 50473) and inserting miR-128 hairpin/scrambled sequence (synthesized by IDT) into multiple cloning sites (mcs) in the AAV-CMV-eGFP/U6-mcs vector with SpeI and XhoI digestions. Primers for eGFP amplification: Egfp-F (NheI): 5′tatatagctagcatggtgagcaagggcgagg3′; Egfp-R (SacI): 5′tatatagagctcctacttgtacagctcgtccat3′; AAV-Gfap-GFP/miR-128-sponge (sp) construct was generated by replacing the Synapsin promoter and miR-124-sp in the AAV-Synapsin-GFP/miR-124-sp plasmid (a gift from Dr. Johan Jakobsson, Lund University, Lund, Sweden) with the Gfap promoter from the pGfa2-nLac plasmid (Addgene, 53126) and the miR-128-sp sequence synthesized from IDT, respectively. Primers for amplifying the Gfap promoter: Gfap-F (ApaI): 5′-tatatagggcccgcggccgcacgcgtgatctaacat-3′; Gfap-R (AgeI): 5′-tatataaccggtggtggcgtcgactctagaccccg-3′; For constructs used for calcium imaging, miR-128 hairpin sequence or scramble sequence were cloned into the AAV-CMV-mCherry/U6-mcs construct with SpeI and XhoI digestions. The GFP reporter in AAV5-Gfap-GFP/miR-128-sp was replaced with the tdTomato (tdT) reporter amplified by PCR from the tdT N1 plasmid (Addgene, 54642). Primers for amplifying tdT: tdt-F (AgeI): 5′-tatataaccggtcgccaccatggtga-3′; tdt-R (FseI): 5′-tatataggccggccttacttgtacagctcgtccat-3′; miR-128 sp (8× tandem sponge sequences): 5′-aagagaccaaccactgtgagaatatgaaaagagaccaaccactgtgaagatataaagagaccaaccactgtgattgaaaaaagagaccaaccactgtgagtaataaaagagaccaaccactgtgaaagtataaagagaccaaccactgtgagtaataaaagagaccaaccactgtgaaagtataaagagaccaaccactgtga-3′; miR-128 hairpin sequence (for miR-128-3p overexpression): 5′-ctcgagtgagcgccacagtgaaccggtctctttactgtaaagccacagatgggtaaagagaccggttcactgtgacgcctactagt-3′; Scrambled sequence: 5′-ctcgagtgagcgcagcgaacgacttacgcgtttactgtaaagccacagatgggtaaacgcgtaagtcgt
tcgctacgcctactagt-3′. All AAV2/5 viruses except pAAV-Gfap-GCaMP3 (see below) were generated by the Boston Children’s Hospital Viral Core. In the AAV-Gfap-mKate2.5f plasmid (a gift from Viviana Gradinaru, California Institute of Technology, Pasadena, CA; Addgene: 99129) we replaced mKate2.5f with GCaMP3 which was PCR amplified out of the Zac2.1 gfaABC1D-Cyto-GCaMP3 plasmid (a gift from Baljit Khakh, UCLA, Los Angeles, CA; Addgene: 44331) by adding a 5′-AflII and a 3′-BamHI restriction digestion site. AAV-Gfap-GCaMP3 was then packaged into AAV-PHP.B capsid (a gift from Viviana Gradinaru; Addgene: 103002) by the University of Pennsylvania Vector core. The final product is called AAV-PHP.B-gfaABC1D-GCaMP3 henceforth.
Sample Preparation and LC-MS/MS Analysis.
LC-MS/MS and a total spectrum count (TSC)-based quantification method was performed at Beth Israel Deaconess Medical Center Mass Spectrometry Core. Briefly, FAC sorted cortical astroglia were homogenized and cell lysates (10 µg) were separated on SDS/PAGE as described above. The gel was washed with clean H2O for 5 min twice and fixed with 40% methanol, 10% acetic acid, and 50% H2O for 1 h. The gel was then stained with Coomassie blue solution for 45 min and subsequently destained overnight with 40% methanol, 10% acetic acid, and 50% H2O. Gel bands were then excised from the gel and washed twice in a tube with 50% acetonitrile. In-gel digestion with trypsin was performed and the peptide mixtures were analyzed by microcapillary LC-MS/MS using a 5500 QTRAP hybrid triple quadrupole mass spectrometer (AB/SCIEX) with amide hydrophilic interaction liquid chromatography (HILIC) chromatography (Waters).
Bioinformatic Analysis of miR Target mRNAs and IPA Functional Analysis.
The miR-128-3p binding targets in human or mouse were predicted using Targetscan Mouse (version 7.2). The miR-128-3p target mRNAs with at least one conserved site (including three types of seed-matched sites: conserved 8mer sites, conserved 7mer-m8 sites, conserved 7mer-A1 sites) of miR-128-3p were selected and compared in human and mouse. Identified human and mouse conserved miR-128-3p target mRNAs were then cross-examined with their expression levels [fragments per kilobase of transcript per million mapped reads (FPKM) > 10] in published human and mouse astrocyte RNA-seq datasets (51) to identify mRNA targets that are clearly expressed in human and mouse astrocytes. Identified miR-128-3p target mRNAs that are expressed in human and mouse astrocytes were analyzed in IPA (version 01–08) for their functional characterization, and the output was plotted as a pie chart.
Intracranial Injection of Neonatal Mouse Pups.
Viral vectors were delivered through free-hand intracranial injection under hypothermia anesthesia by ice at P0–P3, as described previously (52). For mGluR5 immunostaining, AAV2/5-Gfap-GFP/control (8.5 × 1014 genome copies [GCs]) or AAV2/5-Gfap-GFP/miR-128-sp (1.9 × 1013 GCs) were separately injected into Fmr1−/y mice; AAV2/5-CMV-eGFP/U6-scramble (7.9 × 1013 GCs) or AAV2/5-CMV-eGFP/U6-miR-128 (1.4 × 1013 GCs) were separately injected into Fmr1+/y mice; AAV-PHP.B-gfaABC1D-GCaMP3 (1.25 μL, 1.7 × 109 GCs) was coinjected in combination with one of the following: 1) AAV2/5-Gfap-tdT-control (1.25 μL, 1.7 × 1010 GCs) or AAV2/5-Gfap-tdT-128sp (1.25 μL, 5.3 × 1010 GCs) into Fmr1−/y mice; 2) AAV2/5-CMV-mCherry/U6-miR-128 (1.25 μL, 2.6 × 1010 GCs) or AAV2/5-CMV-mCherry/U6-scramble (1.25 μL, 1.2 × 1011 GCs) into Fmr1+/y mice. The injection micropipette made from borosilicate glass capillaries (1B150-4, World Precision Instruments) using a glass puller (PC-10, Narishige International USA, Inc.) was attached to a 3-mL syringe and front filled with virus solution. Following disinfection of the head skin using 70% ethanol, the injection was conducted at an inclination of 30° to the horizontal plane through the frontal bone in the middle between the center of the right eye and midline. The accuracy of the virus delivery into the right lateral ventricle was monitored by adding 0.5 μL sterile-filtered 1% Fast Green (Sigma-Aldrich, F7252) in saline to the injection solution. Following delivery to the right lateral ventricle the solution was diffused to the left lateral ventricle. To minimize any effect of the injection trauma, slice physiology experiments were restricted to the left hemisphere. Animals were placed on a warming pad following injection for recovery (active movement) before being returned to their dam.
Slice Preparation.
Mice were anesthetized by isoflurane inhalation and decapitated at P13 to P16. The forebrain from the left hemisphere was glued on 2% agarose gel with tissue adhesive (1469SB, 3M Vetbond) mounted on the flat specimen holder of a vibratome (Leica VT1200, Leica Biosystems). During slice preparation the brain tissue was submerged in saturated 95% O2 and 5% CO2 ice-cold cutting-slicing solution containing (in mM, pH 7.4): 135 C7H17NO5 NMDG (N-methyl-D-glucamine), 1 KCl, 1.2 KH2PO4, 1.5 MgCl2, 0.5 CaCl2, 20 C5H14NO∙HCO3 (choline bicarbonate), 11 C6H12O6 (dextrose). Parasagittal forebrain slices were cut at 250-μm thickness and collected at 37 °C in oxygenated (95% O2, 5% CO2) artificial cerebrospinal fluid (aCSF) containing (in mM, pH 7.4): 119 NaCl, 2.5 KCl, 1 NaH2PO4, 1.3 MgCl2, 2 CaCl2, 26.2 NaHCO3, 11 C6H12O6 (dextrose). After completion of slicing, the beaker with collected slices was transferred to room temperature while the solution continued to be bubbled with 95% O2 and 5% CO2. Before being transferred to a recording chamber, the respective slice was incubated for ∼30 min in aCSF (constantly bubbled with 95% O2 and 5% CO2) containing 1 μM TTX (Tocris Bioscience, 1069).
Two-Photon Ca2+ Imaging in Acute Brain Slices.
For all astrocyte Ca2+ imaging, a resonant scanner-based two-photon movable objective microscope (MOM) (Sutter Instruments) was used with a 16×, 0.80 NA water-immersion objective (Nikon). A pulsed Ti:Sapphire laser beam at 920 nm, 80-MHz repetition rate, 120-fs pulse width (Insight DS+, Spectra-Physics MKS Instruments Light & Motion) was focused on the barrel cortex. Emitted light was detected using photomultiplier tubes (H10770PA-40; Hamamatsu Photonics). The laser beam tuned to 920 nm for GCaMP3 signals and 1,040 nm for red fluorescence of either tdT or mCherry, were focused to visualize as many astrocytes labeled by both fluorophores as possible within the 400 μm by 400 μm (512 pixels by 512 pixels) field of view. However, the 1,040-nm laser was only used at the beginning of each experiment for identifying cells transduced by AAVs manipulating miR-128 and respective controls. Laser power used for GCaMP3 imaging ranged from 2 to 12 mW at the front objective aperture, while the laser power for imaging tdT or mCherry was in the range 10 to 60 mW. The microscope was controlled by a Dell personal computer [Intel(R) Core(TM) i7-5930K CPU @ 3.50 GHz, 16 GB of RAM] running ScanImage (v5.5; Vidrio Technologies, LLC) software within MATLAB R2019a (Mathworks). All data were acquired using volumetric scanning (PD72Z4CAA, Physik Instrumente) with a step size of 10 µm, covering 40 µm in the z-plane, at a frequency of 5 volumes/s. The slices in the recording chamber were continuously superfused at a flow rate of 2 mL/min with aCSF at room temperature containing 1 μM TTX. (RS)- DHPG, 50 μM (19641-83-9, Tocris Bioscience) and DL-NE, 30 μM (A7256-1G, Sigma-Aldrich) were bath applied sequentially on the same slice. DHPG was applied for 2 min following 2 min of baseline recordings in aCSF solution. After 15 min of DHPG washout, NE was applied for 2 min. Both DHPG and NE testing solutions contained 1 μM TTX. We maintained the same agonist application order (first DHPG, then NE) since we determined ROIs based on individual astrocytes’ responsiveness to NE.
Quantification of mGluR5 Immunoreactivity and miR-128-3p In Situ Signals.
For quantification of astroglial mGluR5, a 3D reconstruction of astroglial domains was performed using the original confocal Z-stack images in Imaris software (Bitplane). The surface tool was used to build the astroglial domain based on Eaat2-tdT fluorescence signal. The sensitivity threshold (absolute intensity) of tdT fluorescence was manually adjusted so that the generated astroglial domain in the 3D image matched with the tdT fluorescence in the original confocal images. Astrocytes that were only partially included in the confocal and 3D images were excluded from analysis. The immunoreactivity of mGluR5 within individual astrocytes can be directly measured from the generated 3D domain in Imaris. For the quantification of miR-128-3p in situ hybridization, images were taken with the Zeiss Imager Apotome function (63× oil-immersion objective lens). The intensity of miR-128 in each astrocyte was measured using ImageJ. For the quantification of miR-128 immunoreactivity in mouse cortical slices, a maximum intensity projection of the GFAP immunoreactivity was generated. Then the individual astrocyte was outlined based on GFAP signal using the freehand tool in ImageJ and saved as a ROI. The mean intensity of miR-128-3p within the individual astrocytes was measured using ImageJ. For the quantification of miR-128-3p in situ signals in human samples, miR-128-3p in situ signals were quantified from selected regions (14.6 mm × 8.7 mm) where GFAP immunoreactivity is evident using the ROI tool in ImageJ. Nonspecific fluorescence signals were defined as large size (>0.7 μm in diameter) puncta. Regions with no miR-128-3p signals were used as the image background during the quantification.
Quantification of Astroglial Ca2+ Signals on Brain Slices.
Slice imaging data were saved in ScanImage v5.5 as tiff files, imported to MATLAB R2019a and stored as mat files. A maximum intensity projection was applied to each scanned volume to obtain a more complete representation of individual astrocytes’ Ca2+ dynamics at five measurements/s. ROIs were drawn surrounding astrocytes that expressed GCaMP3 as well as tdT or mCherry. For each ROI the green fluorescence detector offset that was determined with the laser turned off was subtracted from the fluorescence time series, which was determined as mean fluorescence signal within the ROI within each consecutive maximum intensity projection. GCaMP3 fluorescence dynamics within individual ROIs were then expressed as ΔF/F, calculated as (each maximum intensity projection fluorescence value − median fluorescence value during baseline [first 600 volumes/2 min])/median fluorescence value during baseline. We then determined whether ROI-defined astrocytes were responsive to NE. Astrocytes were considered responsive if their ΔF/F time series during NE application had at least two consecutive values exceeding 3× SD of the entire baseline time series. Astrocyte Ca2+ responses to bath application of DHPG or NE were slow, usually lasting more than 1 min. To compare the relative responsiveness of individual astrocytes to DHPG and NE we calculated the ratio of the respective peak responses, peak ΔF/FDHPG/peak ΔF/FNE with each peak value representing the maximum response during the 2-min drug application or within 2 min following drug application.
Statistical Analysis.
For Ca2+ imaging data, statistical analyses were performed using MATLAB R2016a (MathWorks). For each group of a dataset the Lilliefors test was applied to test for Gaussian distribution. If all groups followed a Gaussian distribution and two groups were compared, we applied two-tailed Student’s t test, otherwise the Kruskal–Wallis test was performed. All values are presented as the mean ± SEM unless otherwise stated, with n indicating the number of slices. The corresponding statistical approach used for each experiment is described in each method section and in the figure legends. All other analyses were performed using GraphPad Prism 7. All values were plotted as individual values or box and whisker plot with all defined elements, including median (center line), upper, and lower quartiles (bounds of box), and the highest and lowest values (whiskers). For graphs with error bars, error bars were presented as SEM. For multiple groups (>2), one-way analysis of variance (ANOVA) was used to analyze the variance, followed by a Tukey’s post hoc test to compare multiple groups. For two-group comparison, two-tailed unpaired Student’s t test was used. Statistical significance was tested at a 95% (P < 0.05) confidence level and P values are shown in each graph.
Supplementary Material
Acknowledgments
We thank Dr. Julia Yelick’s initial analysis of the miR-128 binding site on the Slc1a2 gene, Alessandra Tamashiro Orrego for performing 4-OHT injections, Leona Tu for help in preparing tissues for histology, Rachel Jarvis for proofreading of the manuscript, Dr. Johan Jakobsson (Lund University) for the gift of the original miR sponge construct, Dr. David Nelson (Baylor College of Medicine) for providing Fmr1f/f mice, Dr. Hye-Young Lee for making Fmr1 KO mice available at University of Texas Health Science Center at San Antonio (UTHSCSA), Raehum Paik from the UTHSCSA Mouse Genome Engineering and Transgenic Facility for support with the AAV-GCaMP3 construct, and Dr. Meng Li (University of Wisconsin–Madison) for preparing human astroglial progenitor cultures. This work was supported by NIH grant MH106490 (Y.Y.), R01MH116582 (X.Z.), R21NS105339 (X.Z.), NIH grant MH113780 (M.P.), the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (M.P.), and a Fragile X Syndrome Research Foundation (FRAXA) Research Foundation postdoc fellowship (Y.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014080117/-/DCSupplemental.
Data Availability.
RNA-seq data and miRNA microarray data have been deposited in Gene Expression Omnibus (GEO), accession nos. GSE157723 and GSE157724. All study data are included in the article and supporting information.
References
- 1.Telias M., Molecular mechanisms of synaptic dysregulation in fragile X syndrome and autism spectrum disorders. Front. Mol. Neurosci. 12, 51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verkerk A. J. et al., Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Bassell G. J., Warren S. T., Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 60, 201–214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darnell J. C. et al., FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma A. et al., Dysregulation of mTOR signaling in fragile X syndrome. J. Neurosci. 30, 694–702 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross C. et al., Increased expression of the PI3K enhancer PIKE mediates deficits in synaptic plasticity and behavior in fragile X syndrome. Cell Rep. 11, 727–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Portis S., Giunta B., Obregon D., Tan J., The role of glycogen synthase kinase-3 signaling in neurodevelopment and fragile X syndrome. Int. J. Physiol. Pathophysiol. Pharmacol. 4, 140–148 (2012). [PMC free article] [PubMed] [Google Scholar]
- 8.Jin P. et al., Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7, 113–117 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Edbauer D. et al., Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller S., Romano C., Cotman C. W., Growth factor upregulation of a phosphoinositide-coupled metabotropic glutamate receptor in cortical astrocytes. J. Neurosci. 15, 6103–6109 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morel L., Higashimori H., Tolman M., Yang Y., VGluT1+ neuronal glutamatergic signaling regulates postnatal developmental maturation of cortical protoplasmic astroglia. J. Neurosci. 34, 10950–10962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halassa M. M., Fellin T., Haydon P. G., The tripartite synapse: Roles for gliotransmission in health and disease. Trends Mol. Med. 13, 54–63 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Bazargani N., Attwell D., Astrocyte calcium signaling: The third wave. Nat. Neurosci. 19, 182–189 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Allen N. J., Eroglu C., Cell biology of astrocyte-synapse interactions. Neuron 96, 697–708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacey L. K., Doering L. C., Developmental expression of FMRP in the astrocyte lineage: Implications for fragile X syndrome. Glia 55, 1601–1609 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Higashimori H. et al., Astroglial FMRP-dependent translational down-regulation of mGluR5 underlies glutamate transporter GLT1 dysregulation in the fragile X mouse. Hum. Mol. Genet. 22, 2041–2054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs S., Doering L. C., Astrocytes prevent abnormal neuronal development in the fragile x mouse. J. Neurosci. 30, 4508–4514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashimori H. et al., Selective deletion of astroglial FMRP dysregulates glutamate transporter GLT1 and contributes to fragile X syndrome phenotypes in vivo. J. Neurosci. 36, 7079–7094 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W. et al., MiRNA-128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex. eLife 5, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Q. et al., The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat. Neurosci. 14, 1115–1117 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Tan C. L. et al., MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342, 1254–1258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krencik R., Weick J. P., Liu Y., Zhang Z. J., Zhang S. C., Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 29, 528–534 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Ascenzo M. et al., mGluR5 stimulates gliotransmission in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 104, 1995–2000 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun W. et al., Glutamate-dependent neuroglial calcium signaling differs between young and adult brain. Science 339, 197–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McSweeney K. M. et al., Inhibition of microRNA 128 promotes excitability of cultured cortical neuronal networks. Genome Res. 26, 1411–1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boudreau R. L., Monteys A. M., Davidson B. L., Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA 14, 1834–1844 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigetomi E. et al., Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J. Gen. Physiol. 141, 633–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deverman B. E. et al., Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paukert M. et al., Norepinephrine controls astroglial responsiveness to local circuit activity. Neuron 82, 1263–1270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morel L. et al., Molecular and functional properties of regional astrocytes in the adult brain. J. Neurosci. 37, 8706–8717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heiman M., Kulicke R., Fenster R. J., Greengard P., Heintz N., Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9, 1282–1291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harlow E. G. et al., Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron 65, 385–398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Contractor A., Klyachko V. A., Portera-Cailliau C., Altered neuronal and circuit excitability in fragile X syndrome. Neuron 87, 699–715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernardinelli Y. et al., Activity-dependent structural plasticity of perisynaptic astrocytic domains promotes excitatory synapse stability. Curr. Biol. 24, 1679–1688 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Tzingounis A. V., Wadiche J. I., Glutamate transporters: Confining runaway excitation by shaping synaptic transmission. Nat. Rev. Neurosci. 8, 935–947 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Jin S. H. H., et al. , Astroglial FMRP modulates synaptic signaling and behavior phenotypes in FXS mouse model. bioRxiv:10.1101/2020.02.10.941971 (11 February 2020). [DOI] [PubMed]
- 37.Huber K. M., Gallagher S. M., Warren S. T., Bear M. F., Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl. Acad. Sci. U.S.A. 99, 7746–7750 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todd P. K., Mack K. J., Malter J. S., The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc. Natl. Acad. Sci. U.S.A. 100, 14374–14378 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ceolin L. et al., Cell type-specific mRNA dysregulation in hippocampal CA1 pyramidal neurons of the fragile X syndrome mouse model. Front. Mol. Neurosci. 10, 340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu B. et al., Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, E11397–E11405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson S. R. et al., Cell-type-specific translation profiling reveals a novel strategy for treating fragile X syndrome. Neuron 95, 550–563.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plante I. et al., Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J. Biomed. Biotechnol. 2006, 64347 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheever A., Ceman S., Phosphorylation of FMRP inhibits association with Dicer. RNA 15, 362–366 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korb E. et al., Excess translation of epigenetic regulators contributes to fragile X syndrome and is alleviated by Brd4 inhibition. Cell 170, 1209–1223.e1220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mientjes E. J. et al., The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiol. Dis. 21, 549–555 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Yang Y. et al., Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia 59, 200–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doyle J. P. et al., Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers S. M. et al., Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones J. R. et al., Mutations in GFAP disrupt the distribution and function of organelles in human astrocytes. Cell Rep. 25, 947–958.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li M., Rossi J. J., Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol. Biol. 433, 287–299 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y. et al., Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broekman M. L., Comer L. A., Hyman B. T., Sena-Esteves M., Adeno-associated virus vectors serotyped with AAV8 capsid are more efficient than AAV-1 or -2 serotypes for widespread gene delivery to the neonatal mouse brain. Neuroscience 138, 501–510 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data and miRNA microarray data have been deposited in Gene Expression Omnibus (GEO), accession nos. GSE157723 and GSE157724. All study data are included in the article and supporting information.