Significance
Hypertrophic cardiomyopathy (HCM) is a common genetic heart disease and a leading cause of sudden cardiac death in young adults. HCM has been linked to mutations in genes encoding sarcomeric proteins, but how different mutations can result in a similar clinical phenotype is unknown. Analysis of surgical heart tissue samples from HCM patients with severe outflow track obstruction using high-resolution mass spectrometry–based top-down proteomics revealed a common pattern of altered sarcomeric proteoforms across HCM tissues compared to non-failing donor heart tissues. Our data suggest that common pathways are associated with clinical phenotypes in patients diagnosed with obstructive HCM, opening the door for the development of interventions that target the HCM phenotype rather than the individual sarcomeric gene mutation.
Keywords: hypertrophic cardiomyopathy, proteoform, posttranslational modifications, phosphorylation, top-down proteomics
Abstract
Hypertrophic cardiomyopathy (HCM) is the most common heritable heart disease. Although the genetic cause of HCM has been linked to mutations in genes encoding sarcomeric proteins, the ability to predict clinical outcomes based on specific mutations in HCM patients is limited. Moreover, how mutations in different sarcomeric proteins can result in highly similar clinical phenotypes remains unknown. Posttranslational modifications (PTMs) and alternative splicing regulate the function of sarcomeric proteins; hence, it is critical to study HCM at the level of proteoforms to gain insights into the mechanisms underlying HCM. Herein, we employed high-resolution mass spectrometry–based top-down proteomics to comprehensively characterize sarcomeric proteoforms in septal myectomy tissues from HCM patients exhibiting severe outflow track obstruction (n = 16) compared to nonfailing donor hearts (n = 16). We observed a complex landscape of sarcomeric proteoforms arising from combinatorial PTMs, alternative splicing, and genetic variation in HCM. A coordinated decrease of phosphorylation in important myofilament and Z-disk proteins with a linear correlation suggests PTM cross-talk in the sarcomere and dysregulation of protein kinase A pathways in HCM. Strikingly, we discovered that the sarcomeric proteoform alterations in the myocardium of HCM patients undergoing septal myectomy were remarkably consistent, regardless of the underlying HCM-causing mutations. This study suggests that the manifestation of severe HCM coalesces at the proteoform level despite distinct genotype, which underscores the importance of molecular characterization of HCM phenotype and presents an opportunity to identify broad-spectrum treatments to mitigate the most severe manifestations of this genetically heterogenous disease.
Hypertrophic cardiomyopathy (HCM) is the most common inherited heart disease and a leading cause of sudden death in young adults (1–4). HCM is highly heterogeneous and has been linked to mutations in the genes that encode proteins of the sarcomere, with over 1,400 HCM-associated mutations identified in at least 11 sarcomeric proteins (1, 5). The sarcomere, the basic contractile apparatus of muscle, consists of alternating actin-based thin filaments and myosin-based thick filaments and is flanked serially by complex protein structures known as Z-disks (6–10). Although the genetic basis of HCM has been identified and continues to be studied, the ability to predict clinical outcomes based on specific mutations in HCM patients is limited (3, 4, 11). Moreover, how mutations in different sarcomeric proteins result in highly similar clinical phenotypes remains unknown. Thus, it is critical to understand the disease at the proteoform level, considering genetic mutations together with alternative splicing and posttranslational modifications (PTMs) (12).
The PTMs of sarcomeric proteins are known to be important mediators of cardiac signaling and exert various effects on contractile function (13–17). Additionally, alternative splicing of sarcomere genes gives rise to variants that also exhibit distinct functionality (18–20). Hence, obtaining a comprehensive, unbiased view of the changes in the sarcomeric proteome is an important first step toward understanding the molecular underpinnings of HCM. “Top-down” mass spectrometry (MS)–based proteomics is increasingly recognized as the premier tool for comprehensive analysis of proteoforms arising from combinatorial PTMs, genetic variation, and alternative messenger RNA splicing (9, 21–25). In contrast to shotgun “bottom-up” proteomics (26, 27), which employs proteolytic digestion of proteins prior to MS analysis introducing a “peptide-to-protein” inference problem, top-down proteomics allows for direct analysis of intact proteins without digestion, thus providing a “bird’s-eye” view of existing proteoforms (9, 12, 21, 23, 28, 29). Subsequently, proteoforms of interest can be fragmented by tandem MS (MS/MS) for sequence characterization and PTM localization (9, 21, 23).
Herein, we sought to determine proteoform-level changes in septal myectomy tissues explanted from patients with obstructive HCM (30) (n = 16) compared to donor tissues (Ctrl, n = 16) to better understand HCM at the protein level and investigate whether different HCM-causing mutations lead to different sarcomeric proteoform profiles. Enabled by a robust top-down proteomics method with efficient chromatography and high-resolution MS, we observed a complex sarcomeric proteoform landscape, arising from alternatively spliced protein variants with combinatorial PTMs, revealing PTM cross-talk among myofilament and Z-disk proteins in the sarcomere. We quantified proteoforms and detected differences in the abundance of sarcomeric isoforms between the HCM and donor groups. Notably, we discovered consistent sarcomeric proteoform alterations in the HCM tissues despite different HCM-causing mutations, suggesting a convergent effect of HCM remodeling that is conserved across HCM genotypes. Our results provide direct evidence supporting the importance of molecular characterization of the HCM proteoform landscape in understanding the specific disease phenotype and underlying altered signaling pathways. If the trend revealed herein holds true for a larger population of HCM patients, this would present new opportunities for therapeutic interventions that broadly spans different HCM genotypes.
Results
A Complex Sarcomeric Proteoform Landscape Revealed by Top-Down Proteomics.
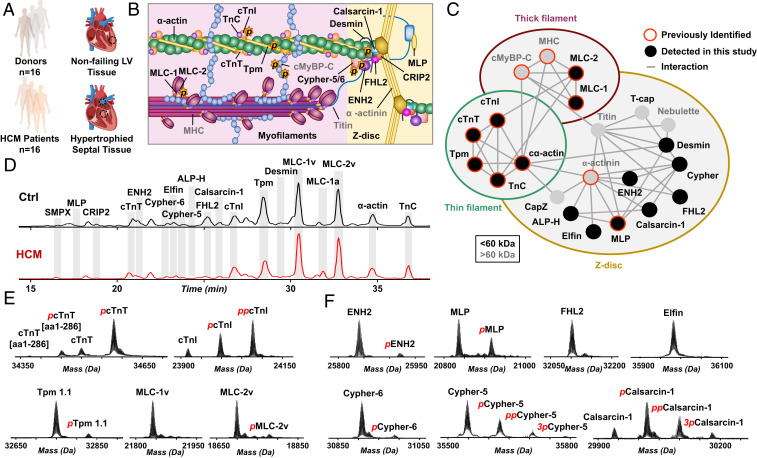
To investigate proteoforms in HCM, we analyzed tissues obtained during septal myectomy procedures (30) in patients who exhibit HCM (n = 16) as compared to left ventricular (LV) tissue from nonfailing donor hearts as controls (Ctrl, n = 16) (Fig. 1A and SI Appendix, Fig. S1). We have shown that LV and interventricular septal tissues in donor hearts (n = 3) have comparable sarcomeric proteoform levels (SI Appendix, Fig. S2 and Table S1), warranting proteoform comparison between donor LV tissues and HCM septal myectomy tissues. For eight of the HCM tissues, the HCM-causing mutations are known (SI Appendix, Table S2). For all but one of these tissues, the mutations were in the gene MYBPC3, which encodes cardiac myosin binding protein C (cMyBP-C), a key thick filament protein serving as an important regulator of myocardial contraction (10, 22). MYBPC3 mutations are also the most common cause of HCM (31). We measured sarcomeric proteoform levels in tissues from donors and HCM patients using a robust liquid chromatography (LC)-MS method (Fig. 1D and SI Appendix, Fig. S1). The method required less than 10 mg of heart tissue and the entire data acquisition process, including sample processing and LC-MS analysis, took less than 3 h per sample. This robust platform is highly reproducible between injections of the same amount of total proteins (SI Appendix, Fig. S3), allowing for a quantitative comparison of sarcomeric proteoforms between tissues from donor hearts and septal myectomy tissues obtained from HCM patients.
Fig. 1.
A complex sarcomeric proteoform landscape. (A) Control tissue from left ventricle (LV) of nonfailing donor hearts (Ctrl, n = 16) and HCM tissues procurred via surgical septal myectomy procedure (HCM, n = 16). (B) Schematic representation of cardiac sarcomere, consisting of thin (green) and thick filaments (pink) flanked by Z-disk. (C) Sarcomeric protein interactome showing a complex network of interactions between myofilament and Z-disk proteins. (D) Representative base peak chromatograms showing separation and detection of major sarcomeric proteins by LC-MS for Ctrl and HCM tissues. (E and F) Representative deconvoluted mass spectra showing the proteoforms of (E) myofilament and (F) Z-disk proteins.
In a single LC-MS run, we injected 500 ng of total sarcomeric proteins and detected a panel of sarcomeric proteins in both the Ctrl and HCM samples (Fig. 1D and SI Appendix, Fig. S1). Among the proteins detected were the major myofilament proteins: cardiac troponin T (cTnT), cardiac troponin I (cTnI), troponin C (TnC), tropomyosin (Tpm) isoforms, myosin light chain 1 (MLC-1) isoforms, myosin light chain 2 (MLC-2), and α-actin isoforms (Fig. 1 D and E, and SI Appendix, Fig. S4). We also observed several Z-disk and sarcomere-associated proteins: small muscle protein X-linked (SMPX), muscle LIM protein (MLP), cysteine rich protein 2 (CRIP2), enigma homolog isoform 2 (ENH2), cypher isoforms, elfin, cardiac actinin-associated LIM protein (ALP-H), four and half LIM domains protein 2 (FHL2), calsarcin-1 (also known as myozenin-2), and desmin (Fig. 1 D and F, and SI Appendix, Fig. S5). Many of these proteins localize to the Z-disk of the sarcomere and form a complex interaction network with the myofilament proteins (Fig. 1 B and C). Among the Z-disk and sarcomere-associated proteoforms we identified, isoforms of LIM domain proteins (MLP, CRIP2, ENH2, cypher-6, elfin, cypher-5, ALP-H, and FHL2) were well-represented (Fig. 1F) (32). A full list of sarcomeric proteoforms identified in the donor and HCM tissues can be found in SI Appendix, Table S3.
Importantly, we have obtained a “bird’s-eye” view of sarcomeric proteoform landscape that arises from genetic variants, alternative splicing, and PTMs in the donor and HCM tissues. For instance, proteoforms of ENH2 and elfin resulting from single-nucleotide polymorphisms (SNPs) were detected (SI Appendix, Fig. S6). Interestingly, wild-type elfin and a natural variant, elfin N175S (33), were revealed with evidence of both heterozygous and homozygous expression (SI Appendix, Fig. S7). Moreover, we have shown several sarcomeric proteins expressed as different genetic isoforms, such as the various Tpm isoforms from genes TPM1, TPM2, and TPM3 (SI Appendix, Fig. S8A), or as alternatively spliced isoforms, including cypher-5 and cypher-6 from gene LDB3 (SI Appendix, Fig. S8B). We also detected both skeletal and cardiac isoforms of α-actin (genes ACTA1 and ACTC1, respectively) and the ventricular and atrial isoforms of MLC-1 (genes MYL3 and MYL4, respectively). Additionally, we identified phosphorylated proteoforms for a number of the sarcomeric proteins (e.g., cTnI, cTnT, and ENH2), indicated by a change in mass of +80 Da from the unphosphorylated proteoform. Since most proteoforms are structurally similar (with only small sequence differences or PTMs), most coelute during LC separation and can be quantified together in a single mass spectrum.
Concerted Decrease in cTnI and ENH2 Phosphorylation in HCM.
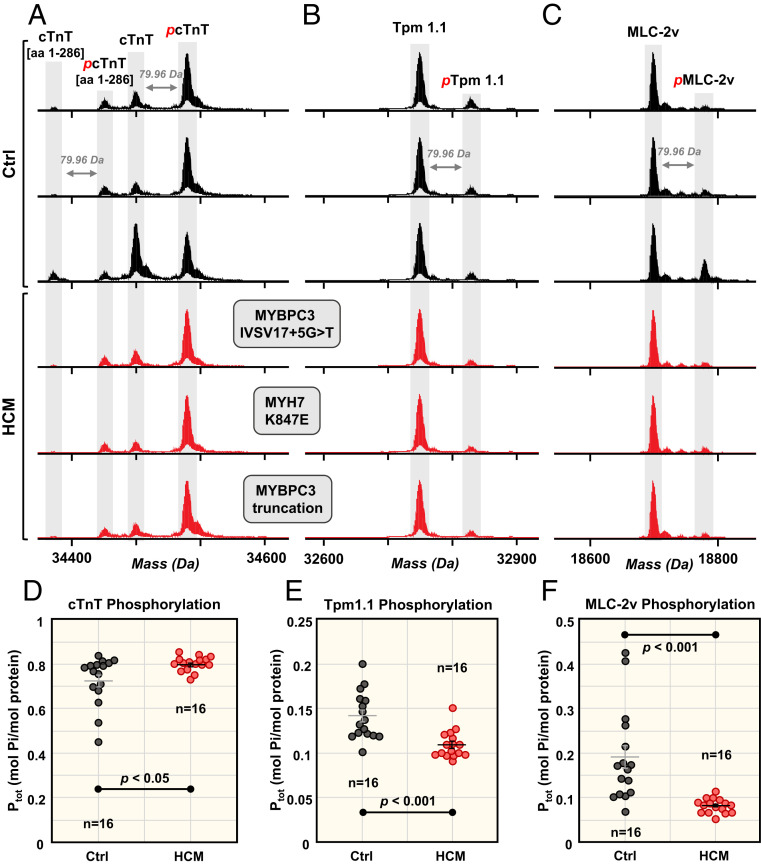
In both donor and HCM tissues, we detected three major cTnI (gene TNNI3) proteoforms: unphosphorylated (cTnI), monophosphorylated (pcTnI), and bis-phosphorylated (ppcTnI) (Fig. 2A). Total phosphorylation levels of cTnI in the donor tissues were between 0.7 and 2.0 mol Pi/mol protein but were significantly decreased (P < 0.001) in the HCM tissues, with levels consistently below 0.6 mol Pi/mol protein (Fig. 2C). To identify the specific sites of cTnI phosphorylation, we performed ultrahigh-resolution MS/MS using electron capture dissociation (ECD), which preserves labile PTMs such as phosphorylation (34). The phosphorylation sites on the ppcTnI were localized to Ser22 and Ser23 (Fig. 2D), the bona fide substrates of cAMP-dependent protein kinase A (PKA), although other kinases can cross-phosphorylate these two sites (13).
Fig. 2.
Coordinated decrease in cTnI and ENH2 phosphorylation in HCM tissues. Representative deconvoluted mass spectra for (A) cTnI and (B) ENH2 from donor hearts (black) and HCM tissues (red). Mono- and bis-phosphorylation are denoted by red p and pp, respectively. (C) Total protein phosphorylation (Ptot) calculated by mol Pi/mol protein for cTnI and ENH2 in Ctrl (n = 16) and HCM (n = 16). Horizontal bars represent the mean of the group and error bars represent SEM in gray for Ctrl and black for HCM. Groups were considered significantly different at P < 0.05. (D) Localization of cTnI phosphorylation to Ser22/23 and (E) localization of ENH2 phosphorylation to Ser118 by ECD. (F) Linear correlation between cTnI phosphorylation and ENH2 phosphorylation (R2 = 0.9276).
Interestingly, we also observed significantly decreased phosphorylation (P < 0.001) of ENH2 (gene PDLIM5, isoform 2) in HCM tissues compared to donor tissues (Fig. 2 B and C). We detected three ENH2 proteoforms in the samples analyzed: unphosphorylated (ENH2), monophosphorylated (pENH2), and bis-phosphorylated (ppENH2). Total ENH2 phosphorylation in donor tissues ranged from 0.4 to 1.1 mol Pi/mol protein, whereas total phosphorylation for HCM tissues was consistently below 0.3 mol Pi/mol protein. The total phosphorylation for cTnI and ENH2 across the Ctrl and HCM cohorts exhibited a strong linear correlation, with an R2 value of 0.9276 (Fig. 2F). This suggests the phosphorylation of ENH2 and cTnI are coregulated by the same kinase(s) or that a cross-talk mechanism exists between the kinases regulating the phosphorylation of these two proteins. Moreover, ultrahigh-resolution ECD tandem mass spectra of pENH2 unambiguously localized a phosphorylation site to Ser118 (Fig. 2E), which is consistent with the phosphorylation site for swine pENH2 (8). Because the ENH2 phosphorylation site at Ser118 exists within a consensus PKA substrate sequence (RRXS/T), we hypothesized that ENH2 can be phosphorylated by PKA. To test this hypothesis, we incubated bacterially expressed recombinant human ENH2 with PKA in vitro. Intact mass spectra of ENH2 before and after incubation with PKA unambiguously demonstrated that PKA indeed phosphorylates ENH2 in vitro, evident by the nearly complete conversion to pENH2 proteoform after incubation with PKA (SI Appendix, Fig. S9 A and B). We next determined that Ser118 was phosphorylated by PKA in vitro using MS/MS (SI Appendix, Fig. S9D). Importantly, regardless of HCM-causing mutation, the total phosphorylation levels for both cTnI and ENH2 were consistently decreased in the HCM tissues compared to the donor tissues (Fig. 2C), and unphosphorylated cTnI and ENH2 are consistently the most abundant proteoforms detected in the HCM tissues (SI Appendix, Fig. S10).
Altered Phosphorylation of cTnT, Tpm1.1, and MLC-2v in HCM.
We observed multiple proteoforms for cTnT (gene TNNT2, isoform 6), including full-length cTnT (cTnT) and truncated cTnT with the C-terminal lysine cleaved (cTnT [amino acids (aa) 1 to 286]), and monophosphorylated forms of both (pcTnT and pcTnT [aa 1 t 286], respectively). In all samples, the monophosphorylated full-length cTnT (pcTnT) was the most abundant proteoform observed, with less than one-third of cTnT observed to be unphosphorylated (Fig. 3A). We quantified phosphorylation of both cTnT (aa 1 to 286) and the full-length cTnT. On average, cTnT (aa 1 to 286) and cTnT total phosphorylation increased by 8% and 10% in the HCM tissues compared to donor tissues (Fig. 3D and SI Appendix, Fig. S11). While an apparent increase in total cTnT and cTnT (aa 1 to 286) phosphorylation is observed in the HCM tissues compared to donor tissues, only the difference in total cTnT phosphorylation is significant between Ctrl and HCM groups (P < 0.05).
Fig. 3.
Altered phosphorylation of cTnT, Tpm1.1, and MLC-2v. Representative deconvoluted mass spectra from donor hearts (black) and HCM tissues (red) for (A) cTnT, (B) Tpm1.1, and (C) MLC-2v. Monophosphorylation is denoted with red p. Total phosphorylation (Ptot) calculated by mol Pi/mol protein for (D) cTnT, (E) Tpm1.1, and (F) MLC-2v for Ctrl (n = 16) and HCM (n = 16). Horizontal bars represent the mean of the group and error bars represent SEM (gray, Ctrl; black, HCM). Groups were considered statistically different at P < 0.05.
In contrast to cTnT, decreased levels of total phosphorylation were observed for the myofilament proteins Tpm1.1 (gene TPM1, isoform 1) and MLC-2v (gene MYL2) in HCM tissues compared to donor tissues (Fig. 3 B and C). Unmodified (Tpm1.1 and MLC-2v) and monophosphorylated (pTpm1.1 and pMLC-2v) proteoforms were detected for both. Tpm1.1 total phosphorylation levels were relatively low (less than 0.2 mol Pi/mol protein) in all of the tissues analyzed (Fig. 3E). On average, Tpm1.1 phosphorylation decreased by 23% in the HCM myectomy tissues compared to the donor tissues (P < 0.001). MLC-2v phosphorylation decreased by 57% in the HCM myectomy tissues compared to donor tissues (P < 0.001) (Fig. 3F). Total phosphorylation levels of cTnT, Tpm1.1, and MLC-2v were consistent among the HCM myectomy tissues analyzed, regardless of HCM-causing mutations (Fig. 3 D–F and SI Appendix, Fig. S12).
Differential Expression of Tropomyosin Isoforms and MLC-1 Isoforms in HCM.
While proteoforms of Tpm1.1 (gene TPM1, isoform 1, α-Tpm) were most abundant in both donor and HCM tissues, we also detected lower-abundance Tpm isoforms such as Tpm1.2 (gene TPM1, isoform 6, κ-Tpm), Tpm2.2 (gene TPM2, isoform 1, β-Tpm), and Tpm3.12 (gene TPM3, isoform 1, γ-Tpm) (Fig. 4 A and B) (35, 36). Subsequently, we quantified the abundance of Tpm2.2 and Tpm3.12 proteoforms relative to the total abudance of Tpm1.1. There was an apparent decrease in Tpm2.2 levels in HCM tissues compared to donor tissues, with Tpm2.2/Tpm1.1 proteoform ratios ranges of 0.06 to 0.01 and 0.04 to 0.01 in donor and HCM tissues, respectively. Furthermore, we detected the unphosphorylated and monophosphorylated proteoforms of Tpm3.12 (Tpm3.12 and pTpm3.12, respectively) in eight HCM myectomy tissues (with a range of 0.04 to 0.003 Tpm3.12/Tpm1.1), whereas none of the control samples had detectable levels of Tpm3.12. Nonetheless, it is difficult to accurately quantify the levels of Tpm2.2 and Tpm3.12 due to its low abundance (Fig. 4B and SI Appendix, Fig. S13).
Fig. 4.
Differential tropomyosin isoform expression in HCM. (A) Representative deconvoluted spectra showing Tpm proteoforms Tpm1.2, Tpm1.1, and pTpm1.1 proteoforms. (B) Zoom-in to baseline of deconvoluted mass spectra to show lower abundance Tpm proteoforms, Tpm2.2, Tpm3.12, and pTpm3.12. Monophosphorylation denoted by red p.
We next investigated the levels of MLC-1 isoforms in the donor and HCM tissues, since partial replacement of MLC-1v with the atrial isoform of myosin light chain 1 (MLC-1a) has been previously observed in hypertrophic myocardium (37). We detected MLC-1v as the most abundant MLC-1 isoform and MLC-1a at low levels in both donor and HCM tissues. The percent MLC-1a (MLC-1a/total MLC-1 content) was increased in HCM tissues compared to donor tissues, with levels ranging from 0.7 to 11.3% in donor tissues and 1.1 to 13.9% in HCM tissues (SI Appendix, Fig. S14). Marston and coworkers found twice as much MLC-1a, on average, in a small cohort of HCM tissues (n = 3) compared to donor tissues, using two-dimensional electrophoresis (2DE) and sypro ruby staining (37). However, we analyzed a larger HCM cohort of tissues (n = 16) and MS is capable of high-sensitivity measurements compared to 2DE.
Top-Down Characterization of Cypher Proteoforms in HCM.
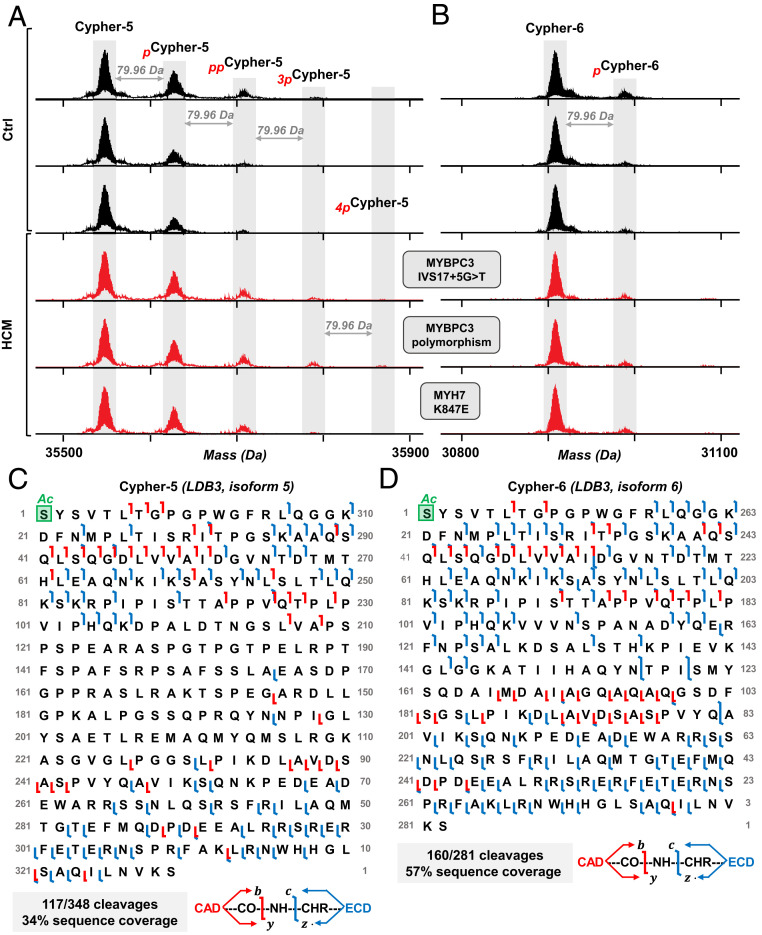
In addition to ENH2, we identified another important Z-disk protein, cypher, belonging to the LIM domain protein family (32). Cypher is encoded by the gene LDB3 and expressed in striated muscle tissue with many splicing isoforms (18, 38). Here, we identified two isoforms from the LDB3 gene, cypher-5 (isoform 5) and cypher-6 (isoform 6), both of which contain an N-terminal α-actinin–associating PDZ domain and lack the C-terminal LIM domains that are thought to recruit protein kinase C to the Z-disk (38). We detected multiple phosphorylated species of both isoforms (Fig. 5 A and B). Mono- and bis-phosphorylated cypher-5 (pcypher-5 and ppcypher-5) were observed in both donor and HCM tissues, whereas the HCM tissues also exhibited tris- and tetra-phosphorylated proteoforms (3pcypher-5 and 4pcypher-5) (Fig. 5A and SI Appendix, Fig. S15A). The total phosphorylation of cypher-5 ranged from 0.29 to 0.56 and 0.26 to 0.71 mol Pi/mol protein for Ctrl and HCM, respectively (Dataset S1). We detected two proteoforms of cypher-6, unphosphorylated (cypher-6) and monophosphorylated (pcypher6). Cypher-6 was found predominantly unphosphorylated in all tissue samples, with 0.02 to 0.18 mol Pi/mol protein in donor tissues and 0.00 to 0.10 mol Pi/mol protein in HCM tissues (Fig. 5B and SI Appendix, Fig. S15B). In contrast to the apparent increase in cypher-5 phosphorylation in the HCM tissues, cypher-6 phosphorylation appeared to be reduced in the HCM tissues. Some HCM samples were found to have low or undetectable levels of pcypher-6. However, accurate quantification of the low-abundance proteoforms remains difficult.
Fig. 5.
Top-down MS characterization of cypher proteoforms in HCM. Representative deconvoluted mass spectra for (A) cypher-5 and (B) cypher-6 from donor hearts (black) and HCM tissues (red). Sequence tables showing MS/MS cleavages from CAD and ECD for (C) cypher-5 and (D) cypher-6. Mono, bis-, tris, tetra-phosphorylation indicated by red p, pp, 3p, “4p”, respectively. Schematic showing protein backbone cleavages is shown, with fragments produced by CAD and ECD shown in red and blue, respectively.
Given that the intact endogenous cypher-5 and cypher-6 had not been detected in human tissue previously, we next determined the sequences of these isoforms using ultrahigh-resolution Fourier transform ion cyclotron resonance (FTICR) MS/MS analysis (Fig. 5 C and D), obtaining direct sequence evidence for both the N- and C-terminal sides of both cypher isoforms. We observed fragments corresponding to 117/348 protein backbone bond cleavages for the unphosphorylated cypher-5, for 34% sequence coverage by combining ECD and collisionally activated dissociation (CAD) experiments (Fig. 5C). With combined ECD and CAD experiments, we obtained 57% sequence coverage for the unphosphorylated cypher-6, accounting for 160/281 protein backbone cleavages for the DNA-predicted sequence with the initiator methinonine cleaved and N-terminal acetylation (Fig. 5D). The overall low abundance of cypher-5 and cypher-6 phosphorylated proteoforms in the sample led to low fragment ion signal-to-noise ratio in the MS/MS, which made definitive localization of the phosphorylation sites challenging.
Meanwhile, another important Z-disk protein, calsarcin-1, was identified in the sarcomeric protein-enriched fractions with unphosphorylated, monophosphorylated, bis-phosphorylated, and triphosphorylated proteoforms (SI Appendix, Fig. S16A). Ultrahigh-resolution FTICR MS analysis of the calsarcin-1 proteoforms measured the masses with high accuracy and confirmed the sequence through isolation of the monophosphorylated proteoform (SI Appendix, Fig. S16B). ECD fragmentation of the monophosphorylated and presence of phosphorylated z·-ions suggests that a phosphorylation site may be located at Thr107 or Thr111, phosphorylated residues that have been previously confirmed at the peptide level (39, 40). Monophosphorylated calsarcin-1 was the predominant proteoform observed in almost all of the tissue samples for both groups, but no statistically significant difference between total phosphorylation of calsarcin-1 in donor and HCM heart tissues was found (P = 0.49) (SI Appendix, Fig. S17).
Discussion
Converging Sarcomeric Proteoform Profiles in HCM.
HCM is a genetically heterogeneous disease clinically characterized by abnormal thickening of the myocardium, which imposes a mechanical burden on the heart (1). Despite the prevalence of HCM, therapeutic options for patients with obstructive HCM are few, with the first-line medical therapy including β-blockers, nondihydropyridine Ca2+ channel blockers, and disopyramide (1). Because these medications often fail to improve symptoms, septal myectomy can be pursued to reduce LV tract obstruction and ameliorate the symptoms (1, 30). In this study, all of the septal myectomy tissues analyzed are from patients with obstructive HCM, presenting the opportunity to measure proteoforms from HCM patients with severe obstruction at a similar stage during disease presentation.
HCM is an autosomal dominant disease with over 1,400 mutations identified in at least 11 genes that encode the protein constituents of sarcomeres; hence, HCM is also known as “the disease of the sarcomere” (4, 41). Although genetic mutations in sarcomeric proteins are generally considered as the causes of HCM, a knowledge gap exists regarding how these mutations lead to cardiac hypertrophy and heart failure. Moreover, genotype alone is not sufficient to predict clinical outcomes in HCM patients, given the multifactorial nature of the disease (3, 11, 42). Unlike genomics, proteomics more precisely reflects cellular responses and disease phenotypes (21, 27, 28). Thus, a major question we sought to address in this study was the effects of different HCM-causing mutations on the sarcomeric proteoform landscape. Our results reveal intriguing HCM-associated alterations in sarcomeric proteoforms, providing potential insights into the specific signaling pathways involved in pathologic cardiac hypertrophy in obstructive HCM. The study provides direct evidence supporting the potential value of molecular characterization of the HCM phenotype at the proteoform level, which will ultimately contribute to a deeper understanding of this genetically heterogenous disease.
Notably, we discovered that, regardless of the specific HCM-causing mutation, uniform proteoform changes were present between the Ctrl and HCM groups, particularly in the phosphorylation changes for cTnI, ENH2, cTnT, Tpm1.1, and MLC-2v (SI Appendix, Table S4). Furthermore, despite the complexity and heterogeneity of these HCM samples, including variabilities in age, gender, geographic locations, comorbidity, and medications for symptom management (SI Appendix, Table S2 and Dataset S2), we have observed consistent alterations in the sarcomeric protein PTM changes and isoform expression in HCM patients. The similarities observed in low phosphorylation levels of cTnI among the HCM tissues analyzed could result from the commonality of pressure overload, which is secondary to obstructive HCM rather than a direct consequence of individual genetic mutation. This is supported by the recent study by Marston and coworkers, which demonstrated that pressure overload was the most important stimulus leading to low phosphorylation levels of cTnI and cMyBP-C (43). In contrast, we generally observed a wider spread of total phosphorylation levels for donor tissues compared to the HCM group (Figs. 2 C and F and 3 D–F). This reflects the heterogeneous nature of the proteoform landscape across donor samples which is likely due to considerable differences in the donor characteristics (SI Appendix, Table S2 and Dataset S2).
While the initial impact of the primary disease-causing mutation may have unique features, the convergence of the HCM phenotype suggests common pathways that may be mediated by PTMs or alternative splicing to influence the function of sarcomeric proteins during hypertrophic remodeling. The observed convergence at the proteoform level might be activated via a common pathway, presenting an opportunity to develop therapeutic approaches with the potential to be broadly effective in treating this genetically heterogeneous disease. Importantly, our finding that proteoform profiles converge for the HCM tissues analyzed in this study highlights the importance of characterization of molecular phenotype. Note that we have captured only a small subset of the HCM genotypes, and thus it is reasonable to conclude that our data suggest a convergence to a common molecular pathway among the obstructive HCM patients with mutations in either MYH7 or MYBPC3 as analyzed in this study, but independent of the specific mutation type.
Admittedly, here we only investigated surgical tissues from HCM patients who exhibited with severe outflow track obstruction and underwent septal myectomy procedures. Since it is impossible to obtain tissue samples from early-stage HCM patients with mild symptoms, this portion of the HCM population was not represented in our study. Thus, it remains unclear whether the sarcomeric protein alterations are features intrinsic to the late-stage obstructive HCM patients as a reflection of the obstructive physiology and thus caution needs to be taken on how to extrapolate these data to the entire HCM patient population.
Top-Down Proteomics for Effective Mapping Sarcomeric Proteoform Landscape.
Enabled by top-down proteomics, we have identified a complex sarcomeric proteoform landscape including many myofilament and Z-disk proteins in the donor and HCM tissues (SI Appendix, Table S3). Specifically, we have mapped nearly all of the myofilament proteins including cTnI, cTnT, TnC, Tpm, and α-actin of the thin filament and MLC-1 and MLC-2 of the thick filament. Moreover, we have identified a large number of Z-disk proteins such as ENH2, MLP, cypher, calsarcin-1, elfin, ALP-H, FHL2, and CRIP2. It remains challenging to detect and quantify large heart proteins, such as cMyBP-C (141 kDa) (37, 44, 45) and MHC (223 kDa), because of the exponential decay in MS signal-to-noise ratio observed with the increasing molecular weight (46), combined with coelution of low-molecular-weight or high-abundance proteins. Size-based protein fractionation is essential to separate the larger proteins from the smaller ones for the top-down MS analysis of large proteins (47, 48). Although we have recently developed a 2DLC method coupling serial size-exclusion chromatography with reversed-phase chromatography that enabled the detection and identification of larger heart proteins, including cMyBP-C and MHC (48, 49), it required much larger amount of tissues than the HCM septal myectomy tissues we obtained here. Importantly, we have discovered many PTMs and alternatively spliced isoforms and polymorphism of these important Z-disk proteins such as phosphorylation of cypher-5 and cypher-6, MLP, and calsarcin-1, and CRIP2. In addition, we detected proteoforms arising from SNPs (elfin and ENH2) in donor and HCM tissues. Furthermore, we identified the endogenous short splicing isoforms of cypher (gene LDB3, isoforms 5 and 6) from human tissue, which contain only the N-terminal PDZ domains and lack the C-terminal LIM domains. We characterized cypher-5 and cypher-6 sequences using ultrahigh-resolution FTICR MS/MS (Fig. 5). We also identified calsarcin-1 and characterized its sequence using ultrahigh-resolution FTICR MS/MS (SI Appendix, Fig. S16). Unphosphorylated, mono-, bis-, and tris-phosphorylated proteoforms of calsarcin-1 were identified in all samples and did not exhibit a change in total phosphorylation between Ctrl and HCM groups (SI Appendix, Fig. S17). Other phosphorylated Z-disk proteins identified in this study, such as MLP, CRIP2, and ALP-H, were too low abundance across the samples to be reliably quantified.
Almost all of the Z-disk proteins (with the exception of calsarcin-1) identified in our study belong to a family of LIM domain proteins, which represent an important class of proteins expressed in mammalian hearts and have been implicated in heart development and diseases (32). Z-discs anchor adjacent thick and thin filaments, allowing for sliding of the thick filaments relative to the thin filaments to drive muscle contraction (6, 50). Recently the structural and functional importance of Z-disk has been increasingly recognized as it serves as an interface between biomechanical sensing and signaling in cardiac and skeletal muscle functions and diseases (51). ENH2 is a short, alternatively spliced isoform of ENH that belongs to the PDZ-LIM protein family and colocalizes to the Z-disk (8). MLP comprises two LIM domains and is tethered to the Z-disk via its interactors, α-actinin2 and T-cap, playing an essential role in mechanical stretch sensor machinery (52). Cypher (also known as ZASP) (38, 53, 54) has been identified as a novel A-kinase anchoring protein serving as important sarcomeric signaling scaffold in regulating the phosphorylation of contractile proteins in addition to its structural role for the sarcomeric integrity (55). Calsarcin-1 is a sarcomeric calcineurin-binding protein located at the Z-disk protein and the only member of the calsarcin family expressed in the adult heart which interacts with α-actinin2 and T-cap, as well as Cypher/ZASP, playing a pivotal role as modulators of calcineurin signaling (51, 56). Elfin (also known as CLP36, CLIM1), encoded by PDLIM1, is another PDZ-LIM family member and predominantly expressed in the heart and skeletal muscle (51). FHL2 is a LIM-only protein colocalized to the Z-disk in the cardiac and skeletal muscles which is believed to negatively regulate the hypertrophic signaling (51). ALP-H is encoded by the gene PDLIM3, associates α-actinin2, and anchors actin filaments to the Z-disk (32, 57). CRIP2 is a LIM domain containing protein closely related to MLP and highly expressed in the heart during development and at adult stage but the roles of CRIP2 in cardiac function remain unexplored (58).
Differential Regulation of Sarcomeric Protein Proteoforms in HCM.
Our robust LC-MS platform allowed for a “bird’s-eye” view of sarcomeric proteoforms and unambiguous quantification of sarcomeric proteoforms in HCM tissues compared to donor tissues. Specifically, we discovered a significant decrease in phosphorylation of cTnI, ENH2, MLC-2v, and Tpm1.1, with a significant increase in the phosphorylation of cTnT in HCM tissues compared to donor tissues (SI Appendix, Table S4). Previous studies have investigated changes in the sarcomeric protein isoform expression and changes to PTMs in HCM primarily using sodium dodecyl sulfate polyacrylamide gel electrophoresis, immunoblotting, and MS-based shotgun proteomics (37, 44, 59–61). However, antibody-based approaches heavily rely on prior knowledge of protein targets. While MS-based techniques provide information without a priori knowledge, shotgun bottom-up proteomics has intrinsic limitations due to the “peptide-to-protein” inference problem and is, therefore, unable to provide proteoform-specific information (12, 27).
We discovered that cTnT phosphorylation increases in HCM tissues (∼10%) compared to donor tissues (Figs. 3 A and C), which gives rise to the possibility of cTnT phosphorylation involvement in hypertrophic signaling and the cause of functional abnormality previously determined by Bayliss et al. (61). The phosphorylation of the cTnT (aa 1 to 286) proteoform increased by 8% in the HCM tissues compared to donor tissues (SI Appendix, Fig. S11), though not significantly. cTnT is a major component of the Ca2+-regulating troponin complex, functional abnormality of which has been determined as the cause of the uncoupling of cTnI phosphorylation and myofibriliar Ca2+ sensitivity in HCM myofibrils (61).
Coordinated Decrease in cTnI and ENH2 Phosphorylation Suggests Dysregulation in PKA-Mediated Phosphorylation in HCM.
The decrease in cTnI phosphorylation at Ser22/23 observed in this study (Figs. 2 A, C, and E) is consistent with previous reports which investigated HCM (59, 61, 62). The troponin complex, composed of cTnI, cTnT, and TnC, is the Ca2+ regulatory protein complex in the cardiac sarcomere. Myofibrilar Ca2+ sensitivity is regulated by the PKA-mediated phosphorylation of cTnI at Ser22/23, with the phosphorylation of cTnI shown to decrease the Ca2+ sensitivity of the myofilament in normal hearts (63, 64). However, despite a dramatic reduction in the phosphorylation of observed consistently for cTnI in HCM, the Ca2+ binding affinity of troponin was unchanged (61), suggesting a reduction in the ability of cTnI phosphorylation to regulate myofilament calcium sensitivity in HCM. Furthermore, manipulation of the cTnI phosphorylation did not alter thin filament Ca2+ sensitivity in human HCM myofibrils, unlike in the thin filaments of donor hearts where increased cTnI phosphorylation led to decreased Ca2+ sensitivity (61). These data suggest that cTnI phosphorylation is uncoupled from myofilament Ca2+ sensitivity in the HCM phenotype.
Here we discovered a coordinated decrease in the phosphorylation of both cTnI and ENH2 (Fig. 2) in HCM tissues which indicates the potential dysregulation of PKA signaling in HCM. ENH is a family of PDZ-LIM domain proteins that localize to Z-disk and serve as scaffolds for signaling complexes (65). Recently we provided evidence that the ENH protein influences tension redevelopment kinetics in mouse myocardium (66). Here we have determined the phosphorylation site in pENH2 and provide evidence that PKA can phosphorylate ENH2 at Ser118. Nevertheless, the specific functional role of ENH2 and its phosphorylation in the regulation of contractility remain unclear. Although the Z-disk is traditionally viewed as a structural component of the sarcomere, emerging evidence indicates Z-disks are epicenters for signaling in the sarcomere with a number of different kinases and phosphatases localized to the Z-disk (6, 50). Here we provide direct evidence of PKA-regulated phosphorylation of a Z-disk protein and its alteration in human HCM. Moreover, the significant linear correlation between ENH2 and cTnI phosphorylation suggests that ENH2 and cTnI may be regulated by the same PKA signaling pathway, implying a PTM cross-talk between myofialment and Z-disk proteins.
Conclusion
Here, we employed a robust LC-MS–based top-down proteomics method for identification and quantification of cardiac sarcomeric proteoforms from HCM and donor human heart tissues. We unveiled a complex proteoform landscape and report proteoform-level evidence for important Z-disk proteins such as FHL2, ALP-H, elfin, cypher-5, cypher-6, and calsarcin-1. Moreover, we used ultrahigh-resolution top-down FTICR MS to characterize the sequences of endogenous cypher-5, cypher-6, and calsarcin-1. Our analysis revealed differential phosphorylation of sarcomeric proteins in HCM septal myectomy tissues compared to donor heart tissues. Importantly, we observed consistently altered sarcomeric proteoforms in the myocardium of HCM patients, regardless of disease-causing mutations and other confounding factors. There was a strong linear correlation in the decreased phosphorylation of cTnI and ENH2 in HCM tissues compared to donor tissues, suggesting dysregulation of PKA signaling in HCM and PTM cross-talk between myofilament and Z-disk proteins. Taken together, our data indicate that convergent pathways could be activated despite different sarcomeric gene mutations in MYH7 or MYBPC3, resulting in consistent alterations in sarcomeric proteoforms aligned with similar clinical phenotypes in the obstructive HCM patients. This study underscores the importance of molecular characterization of HCM at the proteoform level to understand the phenotypic manifestations and underlying dysfunctional signaling pathways. If our findings hold true across a larger population of HCM patients, this would provide new opportunities for novel therapeutic interventions that broadly target the obstructive HCM phenotype rather than specific genotypes. We envision that future proteomics studies covering a wide range of HCM phenotypes will hold great promise to help define disease progression and prognosis based on the proteoform landscape.
Materials and Methods
Detailed materials and methods are outlined in SI Appendix, Supplemental Methods.
Reagents and Chemicals.
All reagents were purchased from Sigma-Aldrich, Inc. unless otherwise noted. High-performance LC-grade water, acetonitrile, and ethanol were purchased from Fischer Scientific.
Human Cardiac Tissue Collection and Preservation.
LV myocardium from nonfailing hearts from brain-dead donors with no history of heart diseases but unsuitable for heart transplant were used as control tissues in this study (Ctrl, n = 16). The donor hearts LV tissues were obtained either from the University of Wisconsin Organ and Tissue Donation (67–69) or from the Sydney Heart Bank. Interventricular septal myocardium from patients with obstructive HCM undergoing septal myectomy surgery (30) for relief of symptoms was analyzed in this study (HCM, n = 16). Septal myectomy tissues from either the University of Wisconsin Hospital or the Imperial College of London were collected and processed in a highly uniform manner. The procedures for the collection of human donor heart and septal myectomy tissues were approved by the Institutional Review Board of the University of Wisconsin–Madison, University of Sydney, and the Imperial College of London, respectively. Available clinical data for the deidentified tissues used in this study can be found in SI Appendix, Table S2 and Dataset S2.
Sarcomeric Protein Extraction from Cardiac Tissue.
Sarcomeric proteins were extracted as previously reported, with minor modifications (67). To minimize artificial protein modifications and oxidation, tissue homogenization was performed at 4 °C. The tissue (7 to 10 mg) was first homogenized rapidly in 50 µL Hepes buffer and following centrifugation, the remaining pellets were further homogenized in 50 µL trifluoroacetic acid solution. The sarcomeric protein-enriched fractions were diluted 20-fold for a Bradford protein assay, and all of the samples were normalized to 100 ng/µL for LC-MS analysis.
LC-MS Analysis and Top-Down MS Characterization.
LC-MS analysis was carried out using a NanoAcquity ultra-high pressure LC system (Waters) coupled to a high-resolution Impact II quadrupole time-of-flight mass spectrometer (Bruker Daltonics) (67). Five hundred nanograms of total sarcomeric protein (per sample) were separated using a home-packed PLRP column (PLRP-S, 1,000 Å pore size, 10-µm particle size, 500-µm inner diameter) using an organic gradient of 5 to 95% mobile phase B (mobile phase A: 0.1% formic acid in water; mobile phase B: 0.1% formic acid in 50:50 acetonitrile:ethanol) at a constant flow rate of 14 µL/min. Individual protein fractions were collected following reversed-phase LC separation for ultrahigh-resolution top-down MS using a 7-T linear ion trap (LTQ)/FTICR mass spectrometer (Thermo Scientific) or 12-T solariX FTICR mass spectrometer (Bruker Daltonics) equipped with an automated chip-based nano electrospray ionization source (Triversa NanoMate; Advion Bioscience). Proteoforms of interest were first isolated in the gas phase and fragmented by ECD and CAD.
Data Analysis.
All LC-MS data were processed and analyzed using the DataAnalysis software (version 4.3; Bruker Daltonics). The relative abundance (A) of a particular proteoform is reported as the ratio of the peak intensity of the proteoform to the summed peak intensities of all proteoforms of the same protein. Tandem mass spectra (MS/MS) were analyzed using the MASH Suite Pro (70) software which was developed in-house. All of the program-processed data were manually validated to obtain accurate sequence and PTM information.
Statistical Analysis.
A Wilcoxon rank sum test was performed to evaluate the statistical significance of differences between the Ctrl (n = 16) and HCM (n = 16) groups. This test was selected because not all data exhibit a normal distribution based on a Shapiro–Wilk test for normality. Differences between sample means were considered statistically significant at P < 0.05 (SI Appendix, Table S5).
Supplementary Material
Acknowledgments
Financial support was provided by NIH R01 HL096971 (to Y.G.). Y.G. also acknowledges R01 GM117058, GM125085, HL109810, and S10 OD018475. T.T. acknowledges support from the NIH Chemistry-Biology Interface Training Program T32 GM008505. W.C. acknowledges American Heart Association predoctoral fellowship 17PRE33660224. S.D.M. acknowledges support from NIH Training Grant T32 GM008688. T.J.K. acknowledges NSF Grant EEC-1648035 and NIH grants R01 HL129798 and U01 HL134764. R.L.M. acknowledges support of NIH Grant R01 HL139883. P.G.V. and S.B.M. acknowledge the British Heart Foundation (PG/17/5/32705). We thank James Anderson, the Surgical Recovery and Organ Preservation Manager, and Carrie Sparks, the Data Coordinator for Organ and Tissue Donation Office at University of Wisconsin, for their assistance in compiling donor data. We also thank Samantha Knott for assistance with graphics for our paper.
Footnotes
Competing interest statement: T.J.K. is a consultant for Fujifilm Cellular Dynamics Incorporated.
This article is a PNAS Direct Submission. J.E.V.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006764117/-/DCSupplemental.
Data Availability.
All study data are included in the paper and supporting information.
References
- 1.Maron B. J.; Clinical Course and Management of Hypertrophic Cardiomyopathy , Clinical course and management of hypertrophic cardiomyopathy. N. Engl. J. Med. 379, 655–668 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Yotti R., Seidman C. E., Seidman J. G., Advances in the genetic basis and pathogenesis of sarcomere cardiomyopathies. Annu. Rev. Genomics Hum. Genet. 20, 129–153 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Landstrom A. P., Ackerman M. J., Mutation type is not clinically useful in predicting prognosis in hypertrophic cardiomyopathy. Circulation 122, 2441–2449, discussion 2450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frey N., Luedde M., Katus H. A., Mechanisms of disease: Hypertrophic cardiomyopathy. Nat. Rev. Cardiol. 9, 91–100 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Seidman C. E., Seidman J. G., Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: A personal history. Circ. Res. 108, 743–750 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang P. M., Sykes B. D., Targeting the sarcomere to correct muscle function. Nat. Rev. Drug Discov. 14, 313–328 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Yuan C., Solaro R. J., Myofilament proteins: From cardiac disorders to proteomic changes. Proteomics Clin. Appl. 2, 788–799 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Peng Y. et al., Top-down proteomics reveals concerted reductions in myofilament and Z-disc protein phosphorylation after acute myocardial infarction. Mol. Cell. Proteomics 13, 2752–2764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai W., Tucholski T. M., Gregorich Z. R., Ge Y., Top-down proteomics: Technology advancements and applications to heart diseases. Expert Rev. Proteomics 13, 717–730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kensler R. W., Craig R., Moss R. L., Phosphorylation of cardiac myosin binding protein C releases myosin heads from the surface of cardiac thick filaments. Proc. Natl. Acad. Sci. U.S.A. 114, E1355–E1364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tower-Rader A., Desai M. Y., Phenotype-genotype correlation in hypertrophic cardiomyopathy: Less signal, more noise? Circ. Cardiovasc. Imaging 10 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Smith L. M., Kelleher N. L., Proteoforms as the next proteomics currency. Science 359, 1106–1107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Layland J., Solaro R. J., Shah A. M., Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc. Res. 66, 12–21 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Oliveira S. M. et al., AMP-activated protein kinase phosphorylates cardiac troponin I and alters contractility of murine ventricular myocytes. Circ. Res. 110, 1192–1201 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Chung H. S. et al., Transient receptor potential channel 6 regulates abnormal cardiac S-nitrosylation in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. U.S.A. 114, E10763–E10771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marston S. B., Walker J. W., Back to the future: New techniques show that forgotten phosphorylation sites are present in contractile proteins of the heart whilst intensively studied sites appear to be absent. J. Muscle Res. Cell Motil. 30, 93–95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez-Correa G. A. et al., O-linked GlcNAc modification of cardiac myofilament proteins: A novel regulator of myocardial contractile function. Circ. Res. 103, 1354–1358 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C. et al., Characterization and in vivo functional analysis of splice variants of cypher. J. Biol. Chem. 278, 7360–7365 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Lau E. et al., Splice-junction-based mapping of alternative isoforms in the human proteome. Cell Rep. 29, 3751–3765.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan C. C. et al., Sarcomeric perturbations of myosin motors lead to dilated cardiomyopathy in genetically modified MYL2 mice. Proc. Natl. Acad. Sci. U.S.A. 115, E2338–E2347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siuti N., Kelleher N. L., Decoding protein modifications using top-down mass spectrometry. Nat. Methods 4, 817–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge Y., Rybakova I. N., Xu Q., Moss R. L., Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc. Natl. Acad. Sci. U.S.A. 106, 12658–12663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B., Brown K. A., Lin Z., Ge Y., Top-down proteomics: Ready for prime time? Anal. Chem. 90, 110–127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aebersold R. et al., How many human proteoforms are there? Nat. Chem. Biol. 14, 206–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ntai I. et al., Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk. Proc. Natl. Acad. Sci. U.S.A. 115, 4140–4145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Fonslow B. R., Shan B., Baek M. C., Yates J. R. 3rd, Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 113, 2343–2394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aebersold R., Mann M., Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Gregorich Z. R., Chang Y. H., Ge Y., Proteomics in heart failure: Top-down or bottom-up? Pflugers Arch. 466, 1199–1209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown K. A. et al., A photocleavable surfactant for top-down proteomics. Nat. Methods 16, 417–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura R. A., Holmes D. R. Jr., Clinical practice. Hypertrophic obstructive cardiomyopathy. N. Engl. J. Med. 350, 1320–1327 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Marston S. et al., Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ. Res. 105, 219–222 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Li A., Ponten F., dos Remedios C. G., The interactome of LIM domain proteins: The contributions of LIM domain proteins to heart failure and heart development. Proteomics 12, 203–225 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Ota T. et al., Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36, 40–45 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Zubarev R. A. et al., Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 72, 563–573 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Peng Y. et al., In-depth proteomic analysis of human tropomyosin by top-down mass spectrometry. J. Muscle Res. Cell Motil. 34, 199–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geeves M. A., Hitchcock-DeGregori S. E., Gunning P. W., A systematic nomenclature for mammalian tropomyosin isoforms. J. Muscle Res. Cell Motil. 36, 147–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacques A. M. et al., The molecular phenotype of human cardiac myosin associated with hypertrophic obstructive cardiomyopathy. Cardiovasc. Res. 79, 481–491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheikh F., Bang M. L., Lange S., Chen J., “Z”eroing in on the role of Cypher in striated muscle function, signaling, and human disease. Trends Cardiovasc. Med. 17, 258–262 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulsson A. K. et al., Post-translational regulation of calsarcin-1 during pressure overload-induced cardiac hypertrophy. J. Mol. Cell. Cardiol. 48, 1206–1214 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P., Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang J., Wakimoto H., Seidman J. G., Seidman C. E., Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science 342, 111–114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marian A. J., van Rooij E., Roberts R., Genetics and genomics of single-gene cardiovascular diseases: Common hereditary cardiomyopathies as prototypes of single-gene disorders. J. Am. Coll. Cardiol. 68, 2831–2849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copeland O. et al., Pressure overload is associated with low levels of troponin I and myosin binding protein C phosphorylation in the hearts of patients with aortic stenosis. Front. Physiol. 11, 241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Copeland O. et al., Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J. Mol. Cell. Cardiol. 49, 1003–1011 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Hoskins A. C. et al., Normal passive viscoelasticity but abnormal myofibrillar force generation in human hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 49, 737–745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Compton P. D., Zamdborg L., Thomas P. M., Kelleher N. L., On the scalability and requirements of whole protein mass spectrometry. Anal. Chem. 83, 6868–6874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran J. C., Doucette A. A., Gel-eluted liquid fraction entrapment electrophoresis: An electrophoretic method for broad molecular weight range proteome separation. Anal. Chem. 80, 1568–1573 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Cai W. et al., Top-down proteomics of large proteins up to 223 kDa enabled by serial size exclusion chromatography strategy. Anal. Chem. 89, 5467–5475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaffer L. V., Tucholski T., Shortreed M. R., Ge Y., Smith L. M., Intact-mass analysis facilitating the identification of large human heart proteoforms. Anal. Chem. 91, 10937–10942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pyle W. G., Solaro R. J., At the crossroads of myocardial signaling: The role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 94, 296–305 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Frank D., Kuhn C., Katus H. A., Frey N., The sarcomeric Z-disc: A nodal point in signalling and disease. J. Mol. Med. (Berl.) 84, 446–468 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Knöll R. et al., The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111, 943–955 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Zhou Q., Ruiz-Lozano P., Martone M. E., Chen J., Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to α-actinin-2 and protein kinase C. J. Biol. Chem. 274, 19807–19813 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Zheng M. et al., Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum. Mol. Genet. 18, 701–713 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin C. et al., Cypher/ZASP is a novel A-kinase anchoring protein. J. Biol. Chem. 288, 29403–29413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frey N. et al., Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 10, 1336–1343 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Zheng M., Cheng H., Banerjee I., Chen J., ALP/Enigma PDZ-LIM domain proteins in the heart. J. Mol. Cell Biol. 2, 96–102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei T. C., Lin H. Y., Lu C. C., Chen C. M., You L. R., Expression of Crip2, a LIM-domain-only protein, in the mouse cardiovascular system under physiological and pathological conditions. Gene Expr. Patterns 11, 384–394 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Messer A. E., Gallon C. E., McKenna W. J., Dos Remedios C. G., Marston S. B., The use of phosphate-affinity SDS-PAGE to measure the cardiac troponin I phosphorylation site distribution in human heart muscle. Proteomics Clin. Appl. 3, 1371–1382 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Schulz E. M. et al., Decreasing tropomyosin phosphorylation rescues tropomyosin-induced familial hypertrophic cardiomyopathy. J. Biol. Chem. 288, 28925–28935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayliss C. R. et al., Myofibrillar Ca(2+) sensitivity is uncoupled from troponin I phosphorylation in hypertrophic obstructive cardiomyopathy due to abnormal troponin T. Cardiovasc. Res. 97, 500–508 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Messer A. E. et al., Mutations in troponin T associated with Hypertrophic Cardiomyopathy increase Ca(2+)-sensitivity and suppress the modulation of Ca(2+)-sensitivity by troponin I phosphorylation. Arch. Biochem. Biophys. 601, 113–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Solaro R. J., Moir A. J., Perry S. V., Phosphorylation of troponin I and the inotropic effect of adrenaline in the perfused rabbit heart. Nature 262, 615–617 (1976). [DOI] [PubMed] [Google Scholar]
- 64.Zhang R., Zhao J., Mandveno A., Potter J. D., Cardiac troponin I phosphorylation increases the rate of cardiac muscle relaxation. Circ. Res. 76, 1028–1035 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Cheng H. et al., Loss of enigma homolog protein results in dilated cardiomyopathy. Circ. Res. 107, 348–356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregorich Z. R. et al., Deletion of Enigma Homologue from the Z-disc slows tension development kinetics in mouse myocardium. J. Gen. Physiol. 151, 670–679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai W. et al., Temperature-sensitive sarcomeric protein post-translational modifications revealed by top-down proteomics. J. Mol. Cell. Cardiol. 122, 11–22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J. et al., Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure. J. Proteome Res. 10, 4054–4065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai W. et al., An unbiased proteomics method to assess the maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 125, 936–953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai W. et al., MASH suite Pro: A comprehensive software tool for top-down proteomics. Mol. Cell. Proteomics 15, 703–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the paper and supporting information.