Significance
The evolution of jaws in early vertebrates provided such a predatory advantage that 99% of vertebrate species living today are jawed. What is often overlooked, however, is another structural innovation that happened concurrently and may have been equally critical to the lineage’s success: the evolution of musculoskeletal gill covers to actively drive oxygenated water over the gills. Here, we identify the first essential gene for gill cover formation in modern vertebrates, Pou3f3, and uncover the genomic element that brought Pou3f3 expression into the pharynx more than 430 Mya. Remarkably, small changes in this deeply conserved sequence account for the single large gill cover in living bony fish versus the five separate covers of sharks and their brethren.
Keywords: Pou3f3, gill cover, vertebrate evolution, neural crest, enhancer deletion
Abstract
Whereas the gill chambers of jawless vertebrates open directly into the environment, jawed vertebrates evolved skeletal appendages that drive oxygenated water unidirectionally over the gills. A major anatomical difference between the two jawed vertebrate lineages is the presence of a single large gill cover in bony fishes versus separate covers for each gill chamber in cartilaginous fishes. Here, we find that these divergent patterns correlate with the pharyngeal arch expression of Pou3f3 orthologs. We identify a deeply conserved Pou3f3 arch enhancer present in humans through sharks but undetectable in jawless fish. Minor differences between the bony and cartilaginous fish enhancers account for their restricted versus pan-arch expression patterns. In zebrafish, mutation of Pou3f3 or the conserved enhancer disrupts gill cover formation, whereas ectopic pan-arch Pou3f3b expression generates ectopic skeletal elements resembling the multimeric covers of cartilaginous fishes. Emergence of this Pou3f3 arch enhancer >430 Mya and subsequent modifications may thus have contributed to the acquisition and diversification of gill covers and respiratory strategies during gnathostome evolution.
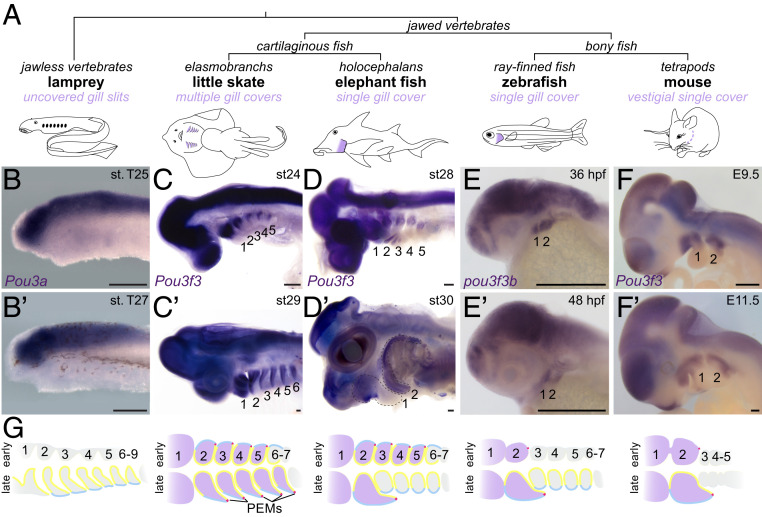
Jawed vertebrates have evolved different anatomical structures to protect and ventilate their gills (Fig. 1 A and G). In bony fishes, the second pharyngeal arch (hyoid) grows caudally over the posterior gill-bearing arches to form a single large gill cover (operculum) supported by intramembranous bones. Although humans and other tetrapods lack functional gills, growth of the hyoid arch over the posterior arches is conserved. The hyoid outgrowth ultimately merges with the trunk to close off the pharyngeal cavity, with defects in this process resulting in branchial cysts and fistulae in humans (e.g., in branchiootorenal syndrome) (1, 2). By contrast, in elasmobranch cartilaginous fishes (sharks, skates, and rays), the hyoid and four posterior arches each undergo posterior-directed growth to generate five separate gill covers supported by cartilaginous rays. In holocephalans, the other extant group of cartilaginous fishes, five separate gill covers also form initially, yet subsequent stalled growth of the four posterior covers results in a single hyoid-derived gill cover (3). Gill cover formation is stimulated by an epithelial signaling center called the posterior ectodermal margin (PEM) that develops along the caudal rim of the hyoid arch in bony fishes and arches 2 to 6 in cartilaginous fishes (later refined to just arch 2 in holocephalans) (Fig. 1G) (2–4). The PEM is a source of Shh, Fgfs, and Bmps (4–7) that are believed to act on the underlying neural crest-derived mesenchyme that generates the gill cover skeleton and connective tissues. Whether this mesenchyme possesses a prepattern for single versus multiple gill covers, or simply receives instructive information from the PEM, had not been examined.
Fig. 1.
Arch Pou3f3 expression correlates with gill cover formation. (A) Vertebrate phylogeny highlighting the gill cover type of each lineage. (B–F and B′–F′) In situ hybridizations at two stages for Pou3f3 homologs in lamprey, little skate, elephant fish, zebrafish, and mouse show conserved expression in the CNS but divergent patterns in the pharyngeal arches. Note the later suppression of Pou3f3 in the posterior arches of the elephant fish. The dotted lines in D′ outline the first and second arches. (Scale bars, 250 μm.) (G) Schematized frontal sections through the pharyngeal arches (numbered) at early (Top) and late (Bottom) stages, with arch Pou3f3 expression coded in purple, PEMs in red, neural crest-derived mesenchyme in gray, endoderm in yellow, and ectoderm in blue. E, embryonic day; st, stage.
Results and Discussion
Pharyngeal Arch Pou3f3 Expression Correlates with Gill Cover Formation.
POU Class 3 Homeobox 3 (POU3F3), also known as BRN1, is a proneural transcription factor with widespread expression in the developing central nervous system (CNS) (8) as well as in dorsal pharyngeal arch mesenchyme. We noted that zebrafish (9), mouse (10), and frog (11) Pou3f3 homologs are atypical among arch-enriched genes in that they are expressed only in the mandibular (first) and hyoid arches and not in the more posterior gill-forming arches (Fig. 1 E and F; compare with dlx2a, dlx6a, and hand2 in SI Appendix, Fig. S1 A and B). At later stages, the two zebrafish paralogs, pou3f3a and pou3f3b, are restricted to the growing gill cover (SI Appendix, Fig. S1 C and D). This restriction to the forming gill cover prompted us to examine the expression of Pou3f3 orthologs in species with divergent gill cover patterns. In the sea lamprey (Petromyzon marinus), a jawless vertebrate lacking gill covers, we detected robust expression of the three Pou3 homologs Pou3a, Pou3b, and Pou3c in the brain but not the pharyngeal arches (Fig. 1B and SI Appendix, Fig. S2). Diffuse Pou3c expression was detected throughout the pharyngeal region at later stages, although it was not enriched in arch mesenchyme. In the little skate (Leucoraja erinacea), an elasmobranch with five gill covers, Pou3f3 is expressed in the brain and arches 1 to 6 (Fig. 1C). In the elephant fish (Callorhinchus milii), a holocephalan, Pou3f3 initially displayed expression in arches 1 to 5 and the brain but later became restricted to the hyoid and mandibular arches, consistent with the secondary regression of the posterior covers (3) (Fig. 1D). These patterns collectively indicate ancestral expression of Pou3f3 homologs in the brain, with cooption of Pou3f3 into arch mesenchyme in jawed vertebrates tightly correlating with the presence, number, and developmental progression of gill covers across diverse species.
A Deeply Conserved Pou3f3 Enhancer Drives Differential Expression in Bony and Cartilaginous Fish.
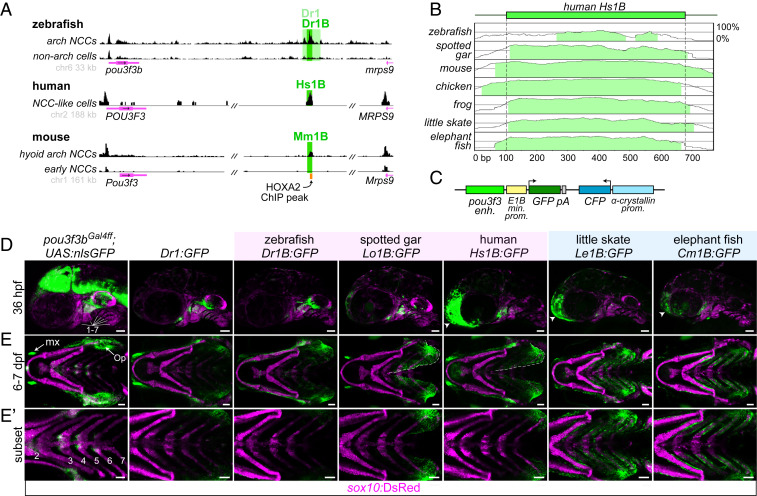
We next sought to identify cis-regulatory elements that could explain the distinct Pou3f3 expression patterns. Assay for transposase-accessible chromatin sequencing (ATAC-seq) on sorted zebrafish sox10:DsRed+; fli1a:EGFP+ arch mesenchyme versus control double-negative cells revealed differentially accessible regions downstream of pou3f3a (“Dr4”; 380 base pairs [bp] in length; 11,919 bp 3′ to the transcription start site [TSS]) and pou3f3b (“Dr1”; 2,033 bp in length; 21,728 bp 3′ to the TSS) (Fig. 2A and SI Appendix, Fig. S3A and Table S1). These elements have no sequence homology with each other and do not appear to be paralogs of an ancestral regulatory element present before the teleost whole genome duplication. In transgenesis assays, both pou3f3b-Dr1 and pou3f3a-Dr4 drove GFP expression in neural crest-derived mesenchyme of the dorsal mandibular and hyoid arches at 36 h post fertilization (hpf), with limited or no expression in the posterior arches (Fig. 2 C and D and SI Appendix, Fig. S3C). Neither Dr1 nor Dr4 drove CNS expression, in contrast to a pou3f3bGal4ff; UAS:nlsGFP knock-in reporter line that recapitulates endogenous pou3f3b expression in the CNS as well as the dorsal first and second arches (Fig. 2D and SI Appendix, Fig. S4 A–C). At 6 to 7 d post fertilization (dpf), pou3f3b-Dr1 but not pou3f3a-Dr4 continued to drive robust expression in the operculum (Fig. 2E and SI Appendix, Fig. S3C), consistent with the broader endogenous expression of pou3f3b during opercular outgrowth (SI Appendix, Fig. S1 C and D). We focused on pou3f3b-Dr1 for further analysis, as pou3f3a-Dr4 is conserved only in the teleost fish species most closely related to zebrafish (e.g., cavefish, catfish, and piranha) (SI Appendix, Fig. S5). Within pou3f3b-Dr1, we identified a 515-bp 5′ fragment (“Dr1A”) that drove expression in only the maxillary domain, similar to pou3f3a-Dr4 (SI Appendix, Fig. S3 B–D). We also identified a central 610-bp sequence (“Dr1B”) that drove robust expression in the dorsal mandibular and hyoid arches starting at 36 hpf and the growing operculum at 7 dpf (Fig. 2 A, D, and E). Whereas Dr1A is found only in teleost fish (n = 43 of 45 genomes; SI Appendix, Fig. S5), Dr1B is exceptionally well conserved across gnathostomes (Fig. 2B). Homologous sequences were recovered in syntenic regions of all mammalian (n = 20), avian (n = 5), reptilian (n = 5), amphibian (n = 1), and cartilaginous fish (n = 4) genomes evaluated (SI Appendix, Table S1). Zebrafish Dr1B was the most divergent in an alignment of 11 vertebrate 1B sequences, and no 1B sequence could be retrieved in 13 of 45 teleosts (SI Appendix, Figs. S5 and S6), in line with previous work showing rapid evolution of regulatory sequences following the teleost whole-genome duplication ∼300 Mya (12). We also did not identify homologous 1B sequences in lamprey or two invertebrate chordates (amphioxus and tunicate) that lack gill covers. In addition, we independently predicted the human “Hs1B” and mouse “Mm1B” sequences as enhancers by virtue of their open chromatin and 25-state epigenomic annotation in published datasets for human and mouse neural crest-derived cells (Fig. 2A) (13–16).
Fig. 2.
A deeply conserved Pou3f3 enhancer differentially regulates arch expression in bony and cartilaginous fish. (A) A conserved regulatory element downstream of Pou3f3 genes. ATAC-seq data from zebrafish fluorescence-activated cell sorting-purified fli1a:EGFP; sox10:DsRed double-positive arch neural crest cells (NCCs) and double-negative non-arch cells (Top), human in vitro-derived neural crest-like cells (data from ref. 15) (Middle), and mouse hyoid arch cells versus early neural crest progenitors (data from ref. 14) (Bottom). Data were visualized with the UCSC Genome Browser, and analyzed peaks are indicated. (B) mVista alignment (60, 61) of conserved 1B sequences relative to Hs1B. (C) Structure of reporter construct. (D) Compared with a pou3f3b knock-in line (Left), Dr1 and 1B reporter lines show primarily arch-restricted GFP expression at 36 hpf (lateral perspective; arches are numbered), with an additional domain of expression in the anterior head (arrowheads) as indicated. (E and E′) At 6 to 7 dpf (ventral perspective), Dr1 and bony vertebrate 1B sequences drive robust expression in the hyoid opercular mesenchyme (Op), with nominal expression in the more posterior arches, while cartilaginous fish 1B reporters induce expression evenly across the hyoid and posterior gill-bearing arches. Expression expands into the ventral operculum in the human and gar reporters (dotted line). Images in E′ are cropped subsets highlighting the posterior arches. sox10:DsRed labels neural crest-derived and otic cells at 36 hpf and chondrocytes at 7 dpf. enh., enhancer; kb, kilobase pairs; min. prom., minimal promoter; mx, maxillary expression; prom., promoter. (Scale bars, 50 µm.)
We next examined the ability of 1B sequences from diverse vertebrates to drive arch expression patterns corresponding to their gill cover patterns. To do so, we cloned 1B sequences from three bony vertebrates (zebrafish, spotted gar, and human) and two cartilaginous fishes (little skate and elephant fish) and tested their abilities to drive GFP expression in zebrafish stable transgenesis assays. At 36 hpf, 1B sequences from all five species directed GFP expression in hyoid and mandibular arch mesenchyme, with few or no GFP+ cells in the posterior arches (Fig. 2D). However, these patterns conspicuously diverged by 7 dpf. Whereas the zebrafish, gar, and human enhancers drove expression primarily in the operculum, skate and elephant fish enhancers drove strong expression in the operculum and all posterior gill-bearing arches (Fig. 2E). The zebrafish but not gar or human enhancers also recapitulated the clear restriction of GFP expression to the dorsal half of the operculum seen in the knock-in line, suggesting divergence within bony fishes of the sequences conferring ventral repression. These experiments demonstrate that small shifts in the 1B enhancer, likely occurring after bony and cartilaginous fish diverged ∼430 Mya (17), can explain, at least in part, the unique arch expression patterns of vertebrate Pou3f3 homologs.
Pou3f3 Is Required for Formation of the Entire Gill Cover Skeleton in Zebrafish.
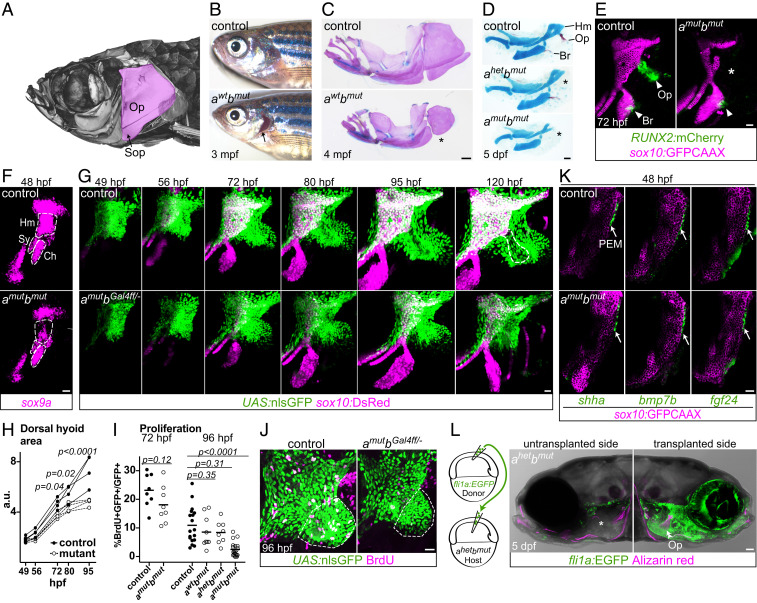
Given the strong correlation of Pou3f3 expression with gill cover pattern, we next assessed the requirement for Pou3f3 in gill cover development. We generated zebrafish pou3f3ael489 and pou3f3bel502 frame-shift alleles with TALENs and confirmed total loss of protein expression in double mutants with a pan-Pou3f3 antibody (SI Appendix, Fig. S7 A and B). The fan-shaped opercle and adjacent subopercle intramembranous bones form the primary skeleton of the operculum in teleost fishes (Fig. 3A). Whereas adult pou3f3a mutants appear normal, adult pou3f3b−/− and pou3f3a−/−; pou3f3b+/− mutants have variably truncated gill covers associated with reduced opercles and missing or fused subopercle bones (Figs. 3 B and C and 4B). The opercle is entirely absent in compound pou3f3b mutants lacking one or both copies of pou3f3a (Fig. 3D), which develop cardiac edema and die at ∼6 to 7 dpf. By contrast, the branchiostegal ray bone, derived from more ventral hyoid arch cells that do not express pou3f3a/b (18), develops normally in double mutants, as assessed by a RUNX2:mCherry osteoblast reporter line and endogenous runx2b expression at 3 dpf (Fig. 3E and SI Appendix, Fig. S7C), despite a failure of mineralization that is likely secondary to edema. Double mutants also display reductions in the hyomandibular cartilage (to which the opercle bone attaches) and its sox9a+ progenitors (Fig. 3 D–F), as well as loss of the specialized joint cells (trps1:GFP+) that connect the opercle to the hyomandibula and disorganization of opercular muscles (SI Appendix, Fig. S7 D and E). A requirement for Pou3f3 in bone and cartilage development in the dorsal (i.e., proximal) mandibular and hyoid arches is also conserved in mammals, as Pou3f3−/− mice lack squamosal and jugal bones (mandibular derivatives) and display an abnormal stapes cartilage (hyoid derivative) (10).
Fig. 3.
Pou3f3 is required for formation of the opercular skeleton in zebrafish. (A) Micro-computed tomography of an adult zebrafish skull, with the opercle (Op) and subopercle (Sop) bones pseudocolored pink. (B and C) Reduction or loss (*) of Op and Sop bones exposes the gills (black arrow) in adult pou3f3b mutants. In C, dissected jaw skeletons were stained with Alcian blue (cartilage) and Alizarin red (bone), and the ceratohyal and branchiostegal ray series were removed for clarity. (D) In larval mutants, the Op and supporting hyomandibula cartilage (Hm) are progressively reduced (*) with decreasing Pou3f3 dosage. (E) Loss of the Op bone in double mutants is preceded by loss of RUNX2:mCherry+ preosteoblasts in this domain (*) but not the branchiostegal ray (Br) bone domain at 3 dpf. (F) Double mutants show a reduction of sox9a expression in the Hm precartilage condensation but not the neighboring symplectic (Sy) or ceratohyal (Ch) domains. (G) Opercular outgrowth initiates but is not sustained in pou3f3a; pou3f3b mutants. Control pou3f3a+/−; pou3f3bGal4ff/+ and mutant pou3f3a−/−; pou3f3bGal4ff/− larvae carrying UAS:nlsGFP and sox10:DsRed (cartilage) transgenes were repeatedly imaged between 49 and 120 hpf. Compaction of mesenchyme around the forming opercle bone (outlined in Upper Right) was not evident in mutants. (H) Quantification of total dorsal hyoid arch area in individually tracked control and pou3f3a−/−; pou3f3bGal4ff/− siblings (repeated-measures ANOVA: genotype P = 0.0069; time: P < 0.0001; genotype × time: P < 0.0001). (I) A trend toward moderately lower rates of proliferation in double mutants at 72 hpf becomes significant at 96 hpf (unpaired t test). Horizontal lines denote the mean. (J) Representative BrdU-labeled control and mutant samples, with the quantified opercular region outlined. (K) PEM markers shha, bmp7b, and fgf24 are expressed at normal levels in pou3f3a; pou3f3b double mutants at 48 hpf (white arrows). sox10:GFPCAAX labels arch mesenchyme. (L) Unilateral transplantation of fli1a:EGFP donor neural crest cells into a pou3f3a+/−; pou3f3b−/− host rescued Op formation. Images in G, J, and L are maximum-intensity projections; single optical sections are presented in K. (Scale bars: C, 500 μm; D and L, 50 μm; E–G, J, and K, 20 μm.)
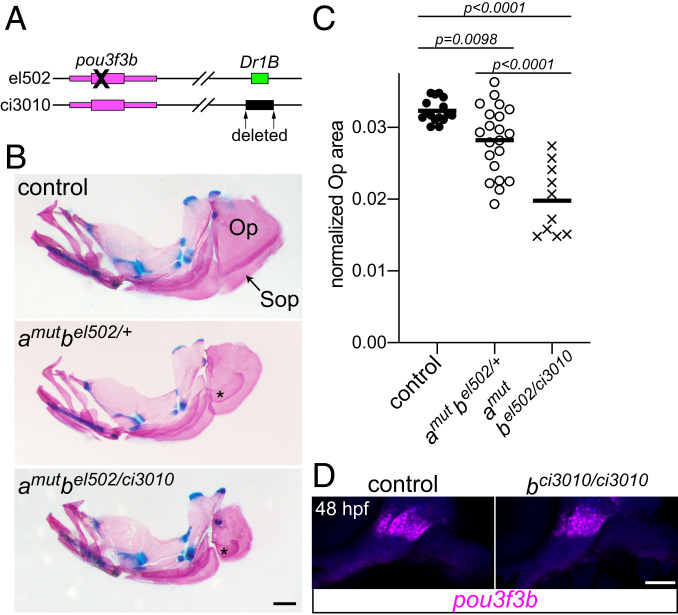
Fig. 4.
Requirement of the Dr1B enhancer for gill cover growth. (A) Schematic of two mutant alleles in the D. rerio pou3f3b locus. el502 is a 2-bp deletion in the pou3f3b-coding sequence, and ci3010 is a 1,025-bp deletion of the Dr1B enhancer. (B) Representative adult mutants stained with Alcian blue and Alizarin red show reduced opercles (Op) and subopercles (Sop) compared with the wild-type control. Fused Sop bones are indicated with asterisks. All fish were 19-mm standard length. (Scale bar, 500 μm.) (C) Opercle area normalized to standard length, compared by one-way ANOVA with Tukey’s multiple-comparisons test. Horizontal lines denote the mean. (D) In situ hybridization for pou3f3b (magenta) in the Dr1B enhancer mutant relative to a sibling control reveals no pronounced loss of expression at 48 hpf. Background fluorescence (blue) shows the general structure of the arches. (Scale bar, 50 μm.)
We next sought to uncover the cellular basis of gill cover defects in Pou3f3 mutants by tracking the behaviors of Pou3f3-expressing cells in living fish. To accomplish this, we inserted a Gal4ff cassette into the endogenous pou3f3b 5′ untranslated region (UTR) sequence and crossed this line with a transgenic UAS:nlsGFP reporter as well as the original pou3f3a and pou3f3b mutant lines. pou3f3a−/−; pou3f3bGal4ff/el502; UAS:nlsGFP embryos display similar opercular defects as pou3f3a−/−; pou3f3bel502/el502 mutants, indicating that the knock-in behaves as a null or severely hypomorphic allele (SI Appendix, Fig. S4 A and F). Sequential confocal imaging in controls revealed posterior-directed migration of pou3f3b-labeled mesenchymal cells in the hyoid arch, which contributed to both sox10:DsRed+ chondrocytes of the hyomandibular cartilage and RUNX2:mCherry+ osteoblasts of the opercle bone (Fig. 3G and SI Appendix, Fig. S4 D and E and Movie S1). In double mutants, comparable amounts of pou3f3b-labeled mesenchymal cells were present at 49 hpf, and yet outgrowth slowed by 72 hpf and completely stalled by 95 hpf, reflected by a significant decrease in the area of the pou3f3b-labeled dorsal hyoid arch as early as 72 hpf (Fig. 3 G and H). This failure of opercular outgrowth may be due in part to reduced proliferation, as there was a trend toward fewer bromodeoxyuridine-positive (BrdU+) pou3f3b-labeled cells at 72 hpf that became highly significant by 96 hpf (Fig. 3 I and J) (confirmed by pHH3 and PCNA staining; SI Appendix, Fig. S8). The failure of sustained opercular growth is likely due to defects in the mesenchyme and not the PEM. Expression of PEM markers shha, bmp7b, and fgf24 was unaltered in mutants at 48 hpf (Fig. 3K). Further, transplantation of wild-type neural crest precursor cells into pou3f3a+/−; pou3f3b−/− mutants fully rescued opercle bone formation, indicating that Pou3f3 function in arch mesenchyme is sufficient for development of the gill cover skeleton (Fig. 3L).
Requirement of the Dr1B Enhancer for Gill Cover Growth.
Given the essential roles for Pou3f3a/b in gill cover formation, we next set out to assess the specific requirements of the deeply conserved 1B enhancer. To do so, we used CRISPR/Cas9 to create a 1,025-bp deletion of the entire Dr1B sequence (pou3f3bci3010; Fig. 4A). Adult fish carrying one copy of this allele and one copy of the original el502 mutant allele developed normal bony gill covers (n = 8 of 8). We therefore further assessed the effect on a sensitized mutant background by comparing adult pou3f3a−/−; pou3f3bel502/ci3010 fish with pou3f3a−/−; pou3f3bel502/+ siblings that retained the enhancer. Compared with the mutants with the wild-type enhancer, fish lacking the 1B enhancer showed a significant reduction in the size of the gill cover opercle bone (Fig. 4 B and C). The 1B enhancer is thus required for the postlarval growth of the gill cover in zebrafish, although the fact that its deletion does not phenocopy the null allele suggests the existence of other complementary enhancers in the zebrafish genomic locus. Consistent with this hypothesis, at embryonic stages, arch pou3f3b expression is still detectable in homozygous pou3f3bci3010/ci3010 mutants (Fig. 4D). The lack of conservation of the 1B enhancer at the pou3f3a locus and its apparent absence from the pou3f3b locus in several teleost genomes also point to the presence of complementary arch mesenchyme enhancers for Pou3f3 genes (SI Appendix, Fig. S5).
Misexpression of Pou3f3b in the Posterior Arches Induces Ectopic Cartilages.
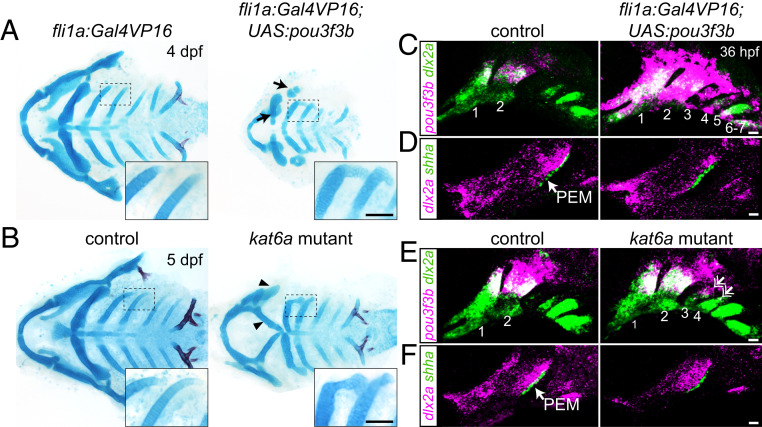
Next, we tested whether forced expression of Pou3f3b in the posterior arches was sufficient to induce the formation of ectopic gill cover-like structures in zebrafish. To do so, we used a fli1a:Gal4VP16 transgene to drive a UAS:pou3f3b transgene throughout postmigratory arch mesenchyme. At 4 dpf, Pou3f3b misexpression resulted in ectopic posterior-directed cartilaginous processes extending from the dorsal tips of the third and fourth arch-derived ceratobranchial cartilages, consistent with ectopic expression of pou3f3b observed in the third and fourth arches at 36 hpf (Fig. 5 A and C). The facial skeleton, in particular the jaws, was also hypoplastic, potentially reflecting that pou3f3b was also ectopically expressed in the normally Pou3f3-negative ventral jaw-forming region. We also observed similar ectopic cartilaginous processes in zebrafish kat6a (moz) mutants, in which Hox genes are down-regulated in the posterior gill-forming arches (19) and pou3f3b becomes ectopically expressed in the third and fourth arches (Fig. 5 B and E). Intriguingly, the central 200 bp of the 1B enhancer was the only sequence in the murine Pou3f3 locus bound by HOXA2 and its Pbx/Meis cofactors in chromatin immunoprecipitation-sequencing (ChIP-seq) assays of hyoid arch cells (20) (Fig. 2A and SI Appendix, Fig. S6). Hox transcription factors might therefore act through the 1B enhancer to prevent posterior arch expression of Pou3f3 orthologs in the bony fish clade. The two predicted Hox-binding motifs within this 200-bp HOXA2 peak are well conserved between cartilaginous and bony fish (SI Appendix, Fig. S6), although differences in the neighboring 5′ and 3′ sequences could affect transcription factor binding (21). Whereas posterior arch expression of pou3f3b was linked to ectopic skeletal elements, lethality by 7 dpf in both pou3f3b-misexpression and kat6a mutant embryos precluded us from assessing whether more extensive ectopic gill covers might form. We also noted no ectopic shha+ PEMs in gill arch epithelia of pou3f3b-misexpression or kat6a−/− embryos (Fig. 5 D and F) (19). Generation of the full multiple gill cover state seen in cartilaginous fishes might therefore require the establishment of PEMs in posterior arch epithelia in addition to expression of Pou3f3 orthologs in posterior arch mesenchyme.
Fig. 5.
Misexpression of pou3f3b in posterior arch mesenchyme induces ectopic posterior-directed cartilages. Cartilage (blue) and bone (red) staining reveals ectopic cartilaginous projections in the third and fourth arches (Insets) of fli1a:Gal4VP16; UAS:pou3f3b (A) and kat6a−/− (B) larvae. The hyoid skeleton was reduced in fli1a:Gal4VP16; UAS:pou3f3b embryos (arrows in A) and homeotically transformed in kat6a mutants (arrowheads in B). (C and E) In situ hybridization for pou3f3b (pink) relative to the pan-mesenchyme marker dlx2a (green) shows ectopic expression of pou3f3b in neural crest-derived mesenchyme (primarily arches 1 to 4) in fli1a:Gal4VP16; UAS:pou3f3b embryos and in the dorsal third and fourth arches (double arrows) of kat6a mutants. (D and F) In situ hybridizations show that the PEM marker shha remains confined to the hyoid arch (arrow) in fli1a:Gal4VP16; UAS:pou3f3b and kat6a−/− embryos. In situ images are maximum-intensity projections. (Scale bars: A and B, 50 μm; C–F, 20 μm.)
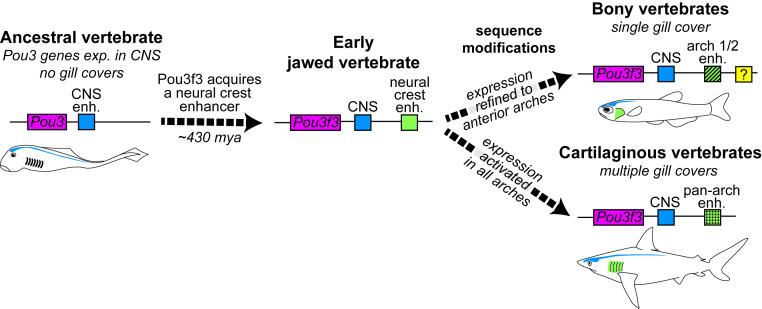
We propose that in an ancestral gnathostome, cooption of the neural gene Pou3f3 into neural crest-derived mesenchymal cells of the arches, through acquisition of the 1B enhancer, conferred a capacity for mesenchymal growth and skeletal differentiation that led to the formation of a gill cover (Fig. 6). Recent discoveries of a stem gnathostome (22) and a stem chondrichthyan (23) with prominent hyoid arch bony opercula are consistent with a scenario in which the last common ancestor of extant jawed vertebrates possessed a single, hyoid arch-derived gill cover. The four posterior gill covers of cartilaginous fishes would therefore have evolved after the two lineages diverged (24), perhaps coincident with sequence changes in the 1B enhancer permitting robust Pou3f3 expression in the posterior arches. Additional enhancers reinforcing arch expression likely also evolved during vertebrate diversification, at least in the teleost fish lineage. Sequence modifications in otherwise highly conserved enhancers have been linked to other more recent evolutionary transitions, e.g., the loss of limbs in snakes (Shh ZRS enhancer) (25) and the fin-to-limb transformation in tetrapods (Gli3 intronic enhancer) (26). It remains to be determined whether other opercular novelties are associated with modifications to the 1B sequence or Pou3f3 activity more generally, such as the precocious opercular outgrowth with external gill development observed in bichir (27) or the modification of the opercular apparatus to seal off the neck when ancestral tetrapods migrated onto land (1, 28).
Fig. 6.
Model of Pou3f3 regulation in the evolution of vertebrate gill covers. Pou3 genes are ancestrally expressed in the CNS (blue element). In early jawed vertebrates, emergence of a new enhancer in the Pou3f3 locus (green element) allowed cooption of its expression into arch mesenchyme. Subsequent sequence changes in this enhancer then reinforced anterior arch expression in bony vertebrates and pan-arch expression in cartilaginous vertebrates. Additional enhancers (e.g. yellow element) likely evolved to reinforce arch expression, at least in the teleost lineage. Intersection of these patterns with PEM signaling centers produces the single- versus multiple gill cover patterns observed in the two gnathostome lineages today.
Materials and Methods
The Institutional Animal Care and Use Committees of the University of Southern California (USC) (no. 10885, 20540), Cincinnati Children’s Hospital Medical Center (no. 2018-0076), the University of Colorado at Boulder (no. 2392), and the Marine Biological Laboratory (no. 17-31, 18-32) approved all of the animal procedures carried out in this study. Mice were housed in accordance with NIH guidelines.
Husbandry and Specimen Collection.
Zebrafish (Danio rerio) were reared in embryo medium (29) at 28.5 °C and staged according to ref. 30. Mixed-background wild-type mouse embryos were collected from timed-pregnant females at 9.5 and 11.5 d post coitus. Adult sea lamprey (P. marinus) were maintained and embryos were raised and collected as described (31), following Tahara staging (32). Skate embryos (L. erinacea) were reared in a flow-through seawater system at 15 °C at the Marine Biological Laboratory in Woods Hole, MA, and fixed as described (33). Holocephalan embryos (C. milii) were collected from the field and fixed as described (3).
Zebrafish Lines and Genotyping.
Existing mutant and transgenic lines used in this study include kat6a/mozb719 (19), trps^j1271aGt (referred to as trps1:GFP) (34), Tg(fli1a:Gal4VP16)el360 (35), Tg(fli1a:eGFP)y1 (36), TgBAC(hand2:eGFP)pd24Tg (37), Tg(RUNX2:mCherry) (gift of Dr. Shannon Fisher, Boston University, Boston, MA), Tg(sox10:GFPCAAX) (38), and Tg(sox10:DsRedExpress)el10 (39). Three mutant lines (pou3f3ael489, pou3f3bel502, and pou3f3bci3010), a targeted knock-in line (pou3f3bGal4ff-el795), and two transgenic lines [Tg(UAS:pou3f3b; α-crystallin:Cerulean)el578 and Tg(UAS:nlsGFP; α-crystallin:Cerulean)el609] were generated as part of this study (see "Generation of Zebrafish Mutant and Transgenic Lines"). Transgenic lines were maintained by selectively raising larvae expressing fluorescent marker proteins. To identify carriers of the mutant alleles, the caudal fin was biopsied under tricaine anesthesia (Western Chemicals) at 14 or 90 dpf, and the tissue was digested with proteinase K and genotyped by PCR/restriction fragment-length polymorphism assays using GoTaq DNA polymerase (Promega). The primers used to genotype the pou3f3a and pou3f3b mutant and Dr1B enhancer deletion lines were pou3f3a-F2 (5′-ACCACGCATACTTTTCCAGC-3′) and pou3f3a-R2 (5′-CTCCTTGCATGAAGTCGCTC-3′); pou3f3b-Fout (5′-TCGATAGTGCACTCGGACTC-3′) and pou3f3b-Rout (5′-CCAGGCTGCGAGTATATGAGA-3′); and Dr1B-60F (5′-CCCCTCCAGGCAAATATTAGAT-3′) and Dr1B-58R (5′-TCAAAGCAAGCTCAACCCTG-3′). The reaction was run at 95 °C for 3 min, followed by 35 cycles of 95 °C for 15 s, 56 or 58 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. The 311- and 473-bp products for pou3f3a and pou3f3b were digested with HinfI at 37 °C and ApaI at 25 °C, respectively, which cut the wild-type alleles only. The Dr1B primers amplify a 542-bp product in carriers of the deletion.
Generation of Zebrafish Mutant and Transgenic Lines.
pou3f3a and pou3f3b mutant lines were created with TALENs targeting the following sequences: pou3f3a-L (5′-TCTCTCATCAGCCTCGCTCG-3′) and pou3f3a-R (5′-TCCCGGCTGCATGCCACCAC-3′); and pou3f3b-L (5′-TGCACTCTGGGACTGCGCTG-3′) and pou3f3b-R (5′-TCTGGTGGGGACCTAAATGT-3′). TALEN constructs were assembled using a PCR-based platform (40), digested with StuI, and used as templates for RNA synthesis with the mMessage mMachine T7 Ultra kit (Ambion/Life Technologies). Pairs of TALEN RNAs were injected into one-cell-stage embryos at 100 ng/μL, and mosaic germline founders were identified by screening their offspring for frame-shift alleles using PCR and restriction digest assays followed by Sanger sequencing. The pou3f3ael489 (c.71_78del; p.Ser24Trpfs*41) and pou3f3bel502 (c.408_409del; p.Arg136Serfs*16) mutations occur upstream of the conserved POU and homeobox domains. No pou3f3a+/−; pou3f3b−/− or double mutants were recovered as adults, although single pou3f3a and pou3f3b mutants and pou3f3a−/−; pou3f3b+/− fish were fully viable and fertile.
To delete the Dr1B enhancer, we injected Cas9 RNA (100 ng/µL) with guide RNAs (gRNAs) targeting flanking sequences 5′-GGGCTCCCCAACATCTGCAC-3′ and 5′-GTGTGGGGAGCAATACTC-3′ (100 ng/µL) into embryos from an outcross of pou3f3ael489/+; pou3f3bel502/+ double heterozygotes to wild-type Tubingen. We screened for founders that passed on the predicted deletion and were heterozygous for pou3f3ael489 and wild-type for the pou3f3bel502 allele. The pou3f3bci3010 allele (NC_007117.6:g.14845039_14846063del) is a 1,025-bp deletion that removes the ∼610-bp enhancer plus ∼400 bp of flanking sequences. No subviability was noted in adult F2 pou3f3a−/−; pou3f3bel502/ci3010 fish.
The pou3f3bGal4ff-el795 targeted knock-in line was made using a CRISPR/Cas9-based protocol (41). Briefly, three gRNAs targeting sequences upstream of the pou3f3b translational start site (5′-AAACATATTCATAAGGTTAA-3′, 5′-GGTTAACGGAATGGCCACAG-3′, and 5′-AGCAAAGAGAAAGTATCTGC-3′) were coinjected at 100 ng/µL into Tg(UAS:nls:GFP) embryos together with Cas9 RNA (100 ng/µL), a circular hsp70l:Gal4ff:pA construct, and a fourth gRNA targeting a bait sequence within the construct, which linearized the donor DNA in vivo (41). Germline founders that recapitulated the embryonic expression pattern of pou3f3b were identified by crossing back to Tg(UAS:nls:GFP) upon reaching maturity.
The UAS:Pou3f3b; α-crystallin:Cerulean and UAS:nlsGFP; α-crystallin:Cerulean constructs were created with the Gateway Tol2kit (42) by combining pMEs containing the pou3f3b or nlsGFP coding sequence with p5E-UAS, p3E-polyA and pDestTol2AB2 in an LR reaction. The stable lines Tg(UAS:pou3f3b; α-crystallin:Cerulean)el578 and Tg(UAS:nlsGFP; α-crystallin:Cerulean)el609 were established by injecting each construct together with transposase RNA (30 ng/µL each) into wild-type embryos and selecting for fish whose offspring expressed Cerulean in the lens.
To test the function of putative Pou3f3 enhancers, sequences downstream of pou3f3a or pou3f3b that were enriched for open chromatin in fli1a:EGFP+; sox10:DsRed+ versus fli1a:EGFP−; sox10:DsRed− cells (Dr4, Dr1, Dr1A, and Dr1B) were PCR-amplified from genomic DNA and cloned into pDONRp4p1r to generate p5E Gateway vectors. gBlocks were generated for the human, spotted gar, little skate, and elephant fish 1B homologs (Integrated DNA Technologies; SI Appendix, Table S1) and used to make p5E vectors. All p5Es were combined with pME-E1B:GFP, p3E:polyA, and pDestTol2AB2 to generate reporter constructs that use the minimal E1B promoter to drive GFP expression and carry the α-crystallin:Cerulean marker. These were microinjected with transposase as described above. Two independent stable alleles per construct were analyzed in the F1 and/or F2 generation.
In Situ Hybridization.
Published zebrafish probes include dlx2a (43), pou3f3a, pou3f3b-3′UTR (9), runx2b (44), shha (45), and sox9a (46). Complementary DNAs (cDNAs) for an exon-only pou3f3b probe, bmp7b, fgf24, lamprey Pou3a, and mouse Pou3f3 were PCR-amplified by Herculase II Fusion DNA Polymerase (Agilent) (SI Appendix, Table S2) and cloned into pCR-Blunt II-TOPO or pJet1.2 (Thermo Fisher Scientific). Fragments of skate and holocephalan Pou3f3 were PCR-amplified from total embryonic cDNA using REDTaq DNA polymerase (Millipore Sigma) and cloned into the pGemT-easy vector system (Promega). Lamprey Pou3b and Pou3c cDNA sequences were purchased as fragments cloned into pUC57 (Synbio Technologies) (SI Appendix, Table S3), and RNA probe templates were PCR-amplified with M13 primers. In all other cases, plasmids were linearized, and antisense probes incorporating digoxigenin- or dinitrophenol-labeled nucleotides were synthesized with Sp6 or T7 polymerase.
Embryos were collected at the designated stages and fixed overnight in 4% paraformaldehyde (PFA), passed through a methanol gradient, and stored at −80 °C until use. In situ hybridizations were performed as previously described for zebrafish (47), mouse (48) (modified to include maleic acid buffers), skate, elephant fish (49, 50), and lamprey (51). A minimum of three mutant or transgenic embryos was compared with a similar number of control siblings, and representative images are presented.
Staining and Cell Proliferation Assays.
The cranial musculature was labeled by incubating PFA-fixed sox10:GFPCAAX+; pou3f3a; pou3f3b mutant and control larvae with Alexa633-conjugated phalloidin (1:100 in phosphate-buffered saline [PBS]) following permeabilization in five 60-min washes of 1% Triton X-100. Alcian Blue and Alizarin Red staining to reveal cartilage and bone structure was performed as previously described for larvae (52) and juvenile/adult fish (53). For immunostaining, PFA-fixed embryos or larvae were passed through a methanol gradient and permeabilized with cold acetone at −20 °C for 7 to 13 min prior to immunostaining with anti-Pou3f3 (1:200; no. ab180094; Abcam), anti-pHH3 (1:500; no. 06-570; Millipore Sigma), or anti-PCNA (1:150; no. 13-3900; Thermo Fisher) and anti-GFP (1:200; no. ab13970; Abcam) primary antibodies and Alexa568- and Alexa488-conjugated secondary antibodies.
To acquire a snapshot of proliferating cells in the growing operculum, 72 and 96 hpf larvae were incubated with 4.5 mg/mL BrdU in 15% dimethyl sulfoxide for 10 min and then fixed in 4% PFA for 3 h at room temperature, passed through a methanol gradient, and stored at −80 °C. Larvae were subsequently rehydrated, genotyped, digested with proteinase K (20 µg/mL) at 25 °C for 5 min, postfixed in 4% PFA for 20 min, permeabilized in cold acetone for 13 min, and then treated with 2 N HCl for 1 h at 25 °C before immunostaining with anti-GFP (1:300) and anti-BrdU (1:200; no. NB500-169; Novus Biologicals) primary antibodies and Alexa568- and Alexa488-conjugated secondary antibodies in blocking solution containing 0.5% Triton X-100.
Transplants.
Unilateral cell transplants were performed as described (54), with the contralateral side serving as an internal control. In brief, naïve ectoderm from fli1a:EGFP donor embryos was transplanted into the neural crest fate-mapped domain of pou3f3a; pou3f3b mutant hosts at 6 hpf. At 5 dpf, transplanted larvae were live-stained with 140 µM Alizarin red for 30 min prior to imaging, and host genotype was confirmed by tail biopsy.
Imaging.
Colorimetric in situs and adult skeletons were imaged using LAS software with a Leica S8APO or M165FC stereomicroscope and larval skeletons with a Leica DM2500 compound microscope. Adult pou3f3b fish were imaged with an AxioCam MRm on a Zeiss StereoDiscovery microscope immediately following euthanasia. Transgenic or fluorescently stained samples were imaged with a Zeiss LSM800 or a Nikon A1R inverted confocal and are shown as single optical sections or maximum-intensity projections, as indicated. Brightness and contrast were modified evenly across samples using Adobe Photoshop CS6 or CC2019. Time-lapse imaging of control pou3f3bGal4ff; UAS:nlsGFP; sox10:DsRed fish was performed with a 20× objective beginning at 56 hpf as previously described (54), with a 61.2-µm z-stack imaged every 18 min.
ATAC-Seq.
fli1a:EGFP; sox10:DsRed double-positive embryos were selected under a fluorescent-dissecting microscope at 36 hpf, dechorionated, and dissociated in batches of 25 as previously described (55). Cells were sorted based on GFP and DsRed expression on a MoFlo Astrios instrument (Beckman-Coulter). Approximately 15,000 double-positive cells and 20,000 double-negative cells were collected into PBS with 5% fetal bovine serum and used to construct ATAC-seq libraries using a low-input μATAC-seq protocol (modified from ref. 56). Briefly, cells were centrifuged at 700 × g, and the supernatant was carefully removed using a 200-μl pipette tip with a broad opening. Next, 20 μL lysis buffer [10 mM Tris⋅HCl (pH 7.4), 5 mM MgCl2, 10% dimethylformamide (DMF), 0.2% Nonidet P-40] was added to the cell pellet and mixed by pipetting up and down 5 to 10 times. A 30-μL aliquot of reaction buffer (10 mM Tris⋅HCl [pH 7.4], 5 mM MgCl2, 10% DMF) containing homemade Tn5 transposase (57) was then immediately added to the lysis, and the samples were incubated for 20 min at 37 °C. DNA fragments were extracted with the Qiagen MinElute kit and amplified by PCR for five cycles using NEBNext Master Mix and indexed primers. Additional cycles were determined by performing qPCR on 1 of 10 of the amplified libraries. Following amplification, the libraries were cleaned using AMPURE beads to preserve only those fragments longer than 100 bp. The libraries were then pooled together before sequencing on an Illumina NextSEq 550, generating 75-bp paired-end reads that were aligned to GRCz10 with Bowtie2 and ENCODE settings. To identify potential regulatory sequences, we uploaded the bigwig data files to the University of California, Santa Cruz (UCSC) genome browser and visually scanned the intergenic sequences between pou3f3a/b and their respective nearest neighbors for peaks that were highly enriched in fli1a:EGFP; sox10:DsRed double-positive (i.e., arch neural crest cells) (55) versus double-negative cells from the rest of the body. Sequences that retained conservation with other teleosts or more deeply with other vertebrates were given priority for transgenic analysis. Potential Hox-binding motifs in the 1B sequence were predicted using the JASPAR core database (58).
Data Analysis.
The numbers of BrdU+GFP+, PCNA+GFP+, or pHH3+/GFP+ cells within the operculum were determined using the Spot Colocalization MATLAB extension in Imaris (Bitplane). Genotypes (n > 6 each) were compared using unpaired two-tailed t tests (assuming normal distributions), with P < 0.05 deemed significant. Area of the dorsal hyoid arch was calculated using the polygon tool in ImageJ (NIH) and analyzed by two-way repeated-measures ANOVA (α = 0.05) followed by Bonferroni’s test for multiple comparisons. Area of the adult opercle bone was also measured in ImageJ and analyzed by one-way ANOVA with Tukey’s multiple-comparisons test. Standard length, from snout to the base of the caudal fin, was measured as described (59). Statistical analyses were performed using GraphPad Prism Version 8.
Supplementary Material
Acknowledgments
We are grateful to Megan Matsutani, Jennifer DeKoeyer Crump, and Sandhya Paudel for fish care, Jeffrey Boyd at the USC Stem Cell Flow Cytometry Core Facility for fluorescence-activated cell sorting, and the USC Norris Medical Library Bioinformatics Service for sequence analysis. Funding was provided by National Institute of Dental and Craniofacial Research Grants R35 DE027550 (to J.G.C.), R00 DE026239 (to L.B.), and R21 DE025940A (to D.M.M.); National Institute on Deafness and Other Communication Disorders Grant R01 DC015829 (to Neil Segil, supporting H.V.Y.); NSF Grant IOS 1744837 (to D.M.M.); the A. P. Giannini Foundation (L.B.); startup funds from the Cincinnati Children’s Research Foundation (to L.B.); Scientific Grant Agency of Slovak Republic Grant VEGA 1/0415/17 (to D.J.); Royal Society University Research Fellowship UF130182 and Isaac Newton Trust Award 14.23z (to J.A.G.); and a Biotechnology and Biological Sciences Research Council Doctoral Training Partnership studentship (C.H.). The bioinformatics software and computing resources were funded by the USC Office of Research, the USC Norris Medical Library, the Cincinnati Children’s Research Foundation, and the Hearing Health Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011531117/-/DCSupplemental.
Data Availability.
The ATAC-seq data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE140636) (62).
References
- 1.Graham A., Richardson J., Developmental and evolutionary origins of the pharyngeal apparatus. Evodevo 3, 24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson J., Shono T., Okabe M., Graham A., The presence of an embryonic opercular flap in amniotes. Proc. Biol. Sci. 279, 224–229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillis J. A. et al., Holocephalan embryos provide evidence for gill arch appendage reduction and opercular evolution in cartilaginous fishes. Proc. Natl. Acad. Sci. U.S.A. 108, 1507–1512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wall N. A., Hogan B. L., Expression of bone morphogenetic protein-4 (BMP-4), bone morphogenetic protein-7 (BMP-7), fibroblast growth factor-8 (FGF-8) and sonic hedgehog (SHH) during branchial arch development in the chick. Mech. Dev. 53, 383–392 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Gillis J. A., Dahn R. D., Shubin N. H., Shared developmental mechanisms pattern the vertebrate gill arch and paired fin skeletons. Proc. Natl. Acad. Sci. U.S.A. 106, 5720–5724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draper B. W., Stock D. W., Kimmel C. B., Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development 130, 4639–4654 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Knight R. D., Javidan Y., Nelson S., Zhang T., Schilling T., Skeletal and pigment cell defects in the lockjaw mutant reveal multiple roles for zebrafish tfap2a in neural crest development. Dev. Dyn. 229, 87–98 (2004). [DOI] [PubMed] [Google Scholar]
- 8.McEvilly R. J., de Diaz M. O., Schonemann M. D., Hooshmand F., Rosenfeld M. G., Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295, 1528–1532 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Askary A. et al., Genome-wide analysis of facial skeletal regionalization in zebrafish. Development 144, 2994–3005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong J. et al., Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development 135, 2905–2916 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Square T. et al., A gene expression map of the larval Xenopus laevis head reveals developmental changes underlying the evolution of new skeletal elements. Dev. Biol. 397, 293–304 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lee A. P., Kerk S. Y., Tan Y. Y., Brenner S., Venkatesh B., Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol. Biol. Evol. 28, 1205–1215 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Attanasio C. et al., Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342, 1241006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minoux M. et al., Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science 355, eaal2913 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Prescott S. L. et al., Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163, 68–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilderman A., VanOudenhove J., Kron J., Noonan J. P., Cotney J., High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep. 23, 1581–1597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brazeau M. D., Friedman M., The origin and early phylogenetic history of jawed vertebrates. Nature 520, 490–497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crump J. G., Swartz M. E., Eberhart J. K., Kimmel C. B., Moz-dependent Hox expression controls segment-specific fate maps of skeletal precursors in the face. Development 133, 2661–2669 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Miller C. T., Maves L., Kimmel C. B., Moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development 131, 2443–2461 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Donaldson I. J. et al., Genome-wide occupancy links Hoxa2 to Wnt-β-catenin signaling in mouse embryonic development. Nucleic Acids Res. 40, 3990–4001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yáñez-Cuna J. O., Dinh H. Q., Kvon E. Z., Shlyueva D., Stark A., Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 22, 2018–2030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu M. et al., A Silurian placoderm with osteichthyan-like marginal jaw bones. Nature 502, 188–193 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Dearden R. P., Stockey C., Brazeau M. D., The pharynx of the stem-chondrichthyan Ptomacanthus and the early evolution of the gnathostome gill skeleton. Nat. Commun. 10, 2050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coates M. I. et al., An early chondrichthyan and the evolutionary assembly of a shark body plan. Proc. Biol. Sci. 285, 20172418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvon E. Z. et al., Progressive loss of function in a limb enhancer during snake evolution. Cell 167, 633–642.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onimaru K. et al., A shift in anterior-posterior positional information underlies the fin-to-limb evolution. eLife 4, e07048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stundl J. et al., Bichir external gills arise via heterochronic shift that accelerates hyoid arch development. eLife 8, e43531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daeschler E. B., Shubin N. H., Jenkins F. A. Jr., A Devonian tetrapod-like fish and the evolution of the tetrapod body plan. Nature 440, 757–763 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Westerfield M., The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio), (University of Oregon Press, Eugene, OR, 2007). [Google Scholar]
- 30.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Nikitina N., Bronner-Fraser M., Sauka-Spengler T., Culturing lamprey embryos. Cold Spring Harb. Protoc. 2009, pdb.prot5122 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Tahara Y., Normal stages of development in the Lamprey, Lampetra-Reissneri, (Dybowski). Zool. Sci. 5, 109–118 (1988). [Google Scholar]
- 33.Gillis J. A., Modrell M. S., Baker C. V., Developmental evidence for serial homology of the vertebrate jaw and gill arch skeleton. Nat. Commun. 4, 1436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talbot J. C., Johnson S. L., Kimmel C. B., hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507–2517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P. et al., Fox proteins are modular competency factors for facial cartilage and tooth specification. Development 145, dev165498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson N. D., Weinstein B. M., In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Kikuchi K. et al., Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397–404 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Askary A. et al., Iroquois proteins promote skeletal joint formation by maintaining chondrocytes in an immature state. Dev. Cell 35, 358–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das A., Crump J. G., Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet. 8, e1002710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanjana N. E. et al., A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171–192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura Y., Hisano Y., Kawahara A., Higashijima S., Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 4, 6545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwan K. M. et al., The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Akimenko M. A., Ekker M., Wegner J., Lin W., Westerfield M., Combinatorial expression of three zebrafish genes related to distal-less: Part of a homeobox gene code for the head. J. Neurosci. 14, 3475–3486 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul S. et al., Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development 143, 2066–2076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balczerski B. et al., Analysis of sphingosine-1-phosphate signaling mutants reveals endodermal requirements for the growth but not dorsoventral patterning of jaw skeletal precursors. Dev. Biol. 362, 230–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Y. L. et al., A zebrafish sox9 gene required for cartilage morphogenesis. Development 129, 5065–5079 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Barske L. et al., Essential role of Nr2f nuclear receptors in patterning the vertebrate upper jaw. Dev. Cell 44, 337–347.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogan B., Manipulating the Mouse Embryo: A Laboratory Manual, (Cold Spring Harbor Laboratory Press, Plainview, NY, ed. 2, 1994). [Google Scholar]
- 49.Gillis J. A. et al., Electrosensory ampullary organs are derived from lateral line placodes in cartilaginous fishes. Development 139, 3142–3146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Neill P., McCole R. B., Baker C. V., A molecular analysis of neurogenic placode and cranial sensory ganglion development in the shark, Scyliorhinus canicula. Dev. Biol. 304, 156–181 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerny R. et al., Evidence for the prepattern/cooption model of vertebrate jaw evolution. Proc. Natl. Acad. Sci. U.S.A. 107, 17262–17267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker M. B., Kimmel C. B., A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23–28 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Ullmann B., “Juvenile no acid bone & cartilage stain” in ZFIN Protocol Wiki, (Zebrafish Information Network [ZFIN], University of Oregon, 2011), https://wiki.zfin.org/display/prot. [Google Scholar]
- 54.Crump J. G., Swartz M. E., Kimmel C. B., An integrin-dependent role of pouch endoderm in hyoid cartilage development. PLoS Biol. 2, E244 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barske L. et al., Competition between jagged-notch and Endothelin1 signaling selectively restricts cartilage formation in the zebrafish upper face. PLoS Genet. 12, e1005967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picelli S. et al., Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fornes O. et al., JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parichy D. M., Elizondo M. R., Mills M. G., Gordon T. N., Engeszer R. E., Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Dev. Dyn. 238, 2975–3015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayor C. et al., VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Brudno M. et al.; NISC Comparative Sequencing Program , LAGAN and multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13, 721–731 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barske L., et al. , Evolution of vertebrate gill covers through shifts in an ancient gnathostome Pou3f3 enhancer. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140636. Deposited 18 November 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ATAC-seq data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE140636) (62).