Significance
All existing blood biomarkers of kidney function such as creatinine and cystatin C undergo renal clearance and are thus inversely correlated with estimated glomerular filtrate rate (eGFR). Using a proteomic approach, we highlight the renal release of testican-2 among individuals undergoing invasive catheterization and show that higher blood testican-2 levels are associated with higher eGFR and slower rate of subsequent eGFR decline in two large racially diverse cohorts. In conjunction with microscopy and single-cell RNA sequencing of human kidney samples, these studies advance the concept that a protein released by the podocyte can provide insight into the kidney’s health and prognosis. Cell-based studies also raise the possibility that this protein has functional effects on neighboring glomerular endothelial cells.
Keywords: testican-2, proteomics, chronic kidney disease
Abstract
In addition to their fundamental role in clearance, the kidneys release select molecules into the circulation, but whether any of these anabolic functions provides insight on kidney health is unknown. Using aptamer-based proteomics, we characterized arterial (A)-to-renal venous (V) gradients for >1,300 proteins in 22 individuals who underwent invasive sampling. Although most of the proteins that changed significantly decreased from A to V, consistent with renal clearance, several were found to increase, the most significant of which was testican-2. To assess the clinical implications of these physiologic findings, we examined proteomic data in the Jackson Heart Study (JHS), an African-American cohort (n = 1,928), with replication in the Framingham Heart Study (FHS), a White cohort (n = 1,621). In both populations, testican-2 had a strong, positive correlation with estimated glomerular filtration rate (eGFR). In addition, higher baseline testican-2 levels were associated with a lower rate of eGFR decline in models adjusted for age, gender, hypertension, type 2 diabetes, body mass index, baseline eGFR, and albuminuria. Glomerular expression of testican-2 in human kidneys was demonstrated by immunohistochemistry, immunofluorescence, and electron microscopy, while single-cell RNA sequencing of human kidneys showed expression of the cognate gene, SPOCK2, exclusively in podocytes. In vitro, testican-2 increased glomerular endothelial tube formation and motility, raising the possibility that its secretion has a functional role within the glomerulus. Taken together, our findings identify testican-2 as a podocyte-derived biomarker of kidney health and prognosis.
Emerging affinity-based proteomic technologies enable the measurement of hundreds to thousands of circulating proteins in human blood (1). These methods achieve specific protein capture using nucleotide-labeled antibodies or nucleic acid-based aptamers. The use of nucleotide reagents facilitates large-scale multiplexing, enhancing throughput relative to traditional immunoassays and mass spectrometry-based approaches. Recent studies have applied these methods to well-phenotyped clinical cohorts to identify markers of myocardial injury and cardiovascular risk (2–4), various cross-sectional health indicators (5), and a signature of inflammatory proteins that predicts end-stage kidney disease (ESKD) onset among individuals with type 1 and 2 diabetes (6). Thus, coupled to high-quality clinical samples, affinity-based proteomics is a promising tool to identify biomarkers and elucidate new biology.
Potential confounding by glomerular filtration rate (GFR) requires careful consideration in nephrology biomarker research. Although small molecules such as metabolites are more likely to be affected, some proteins are also subject to renal clearance. In an early application of aptamer-base proteomics, Gold et al. (7) measured 614 proteins in plasma obtained from 42 individuals with a range of chronic kidney disease (CKD) severity and found that 60 proteins differed significantly across CKD stage. Nine of the top 11 proteins were relatively small (<25 kDa) and had an inverse correlation with estimated GFR (eGFR), suggesting they may be markers of renal clearance—indeed, two of the top findings were cystatin C and β2-microglobulin, both of which are known to undergo glomerular filtration followed by tubular absorption and catabolism.
One approach to assess how the kidney modulates circulating biomarkers is to assess their renal arteriovenous gradients, i.e., to compare levels in the renal vein to the renal artery (or aorta) using samples acquired via invasive catheterization. This approach has been used to demonstrate the heterogeneous impact the kidneys have on the blood metabolome and to identify glycerol-3-phosphate as a kidney-derived metabolite that modulates systemic mineral metabolism (8, 9). Here, we extend this paradigm to include an aptamer-based analysis of the blood proteome. Using these physiologic insights as a springboard, we then examine the associations between circulating proteins and both cross-sectional and longitudinal measures of eGFR in two large community-based cohorts, the Jackson Heart Study (JHS) and the Framingham Heart Study (FHS). In conjunction with microscopy and single-cell RNA sequencing analysis of human kidney tissue, these studies advance the concept that a protein released by the kidney into circulation can provide insight into the organ’s health. Furthermore, the data provide a resource for the research community that outlines the relationship between kidney function and the circulating proteome.
Results
Arteriovenous Sampling Demonstrates the Heterogeneous Impact of Kidney Function on the Blood Proteome and Highlights the Renal Release of Testican-2.
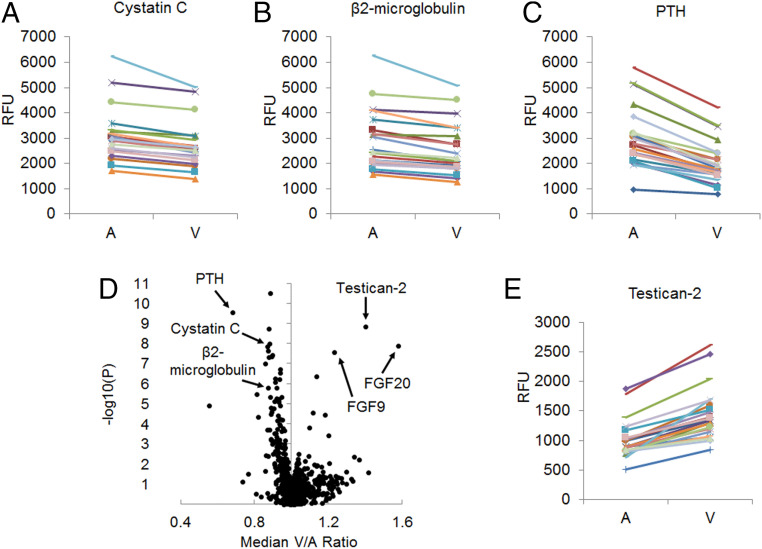
We collected paired arterial and renal venous plasma samples from 22 individuals undergoing cardiac catheterization (Table 1). The patients had a mean age of 69.5 ± 13.0 and mean eGFR of 66.6 ± 21.3 mL/min/1.73 m2. Aptamer-based profiling was used to assay 1,305 proteins, and as expected, the filtration marker cystatin C decreased from artery (A) to renal vein (V) in every individual, confirming appropriate blood sampling (Fig. 1A). Similar findings were observed for β2-microglobulin and parathyroid hormone (PTH) (Fig. 1 B and C), which are also known to undergo substantial renal clearance. Fig. 1D shows the median V/A ratio for all measured proteins (full results in SI Appendix, Table S1). Thirty-eight proteins had a statistically significant change from A to V at a Bonferroni adjusted threshold of P < 3.8 × 10−5. Of these, 32 proteins had a V/A < 1, consistent with net clearance by the kidney, whether via glomerular filtration, tubular secretion, and/or renal catabolism. Interestingly, six significant proteins had a V/A > 1, consistent with net release by the kidney. Testican-2 (V/A = 1.40, P = 1.5 × 10−9) had the most significant augmentation from A to V, increasing in all individuals (Fig. 1E), followed by fibroblast growth factor 20 (FGF20) (V/A = 1.58, P = 1.3 × 10−8), fibroblast growth factor 9 (FGF9) (V/A = 1.23, P = 2.8 × 10−8), insulin growth factor binding protein 7 (IGFBP-7) (V/A = 1.14, P = 4.3 × 10−7), urokinase plasminogen activator (uPA) (V/A = 1.12, P = 2.8 × 10−5), and Cathepsin V (V/A = 1.18, P = 3.5 × 10−5). Of note, renin also had V/A >1 as expected (V/A = 1.04, P = 0.019), although this did not reach Bonferroni significance, while an erythropoietin gradient was not observed (V/A = 1.02, P = 0.58).
Table 1.
Clinical characteristics of the human renal arteriovenous sampling cohort
| n = 22 | |
| Age, y | 70 ± 13 |
| Women, % | 32% |
| Race/ethnicity | 21 Caucasian, 1 Hispanic |
| Hypertension, % | 86% |
| Type 2 diabetes, % | 32% |
| Coronary artery disease, % | 64% |
| Congestive heart failure, % | 27% |
| Serum creatinine, mg/dL | 1.05 ± 0.30 |
| eGFR, mL/min/1.73 m2 | 66.6 ± 21.3 |
Values are mean ± SD or percentage.
Fig. 1.
Proteomic profiling of arteriovenous samples highlights kidney release of testican-2. Paired arterial [A] and renal venous [V] levels for (A) Cystatin C, (B) β2-microglobulin, and (C) PTH demonstrating reduction from A to V in all individuals (n = 22). (D) Volcano plot showing median V/A ratio for all 1,317 proteins detected by aptamer with P values plotted on the y axis; two-sided paired t tests. (E) Paired A and V levels for testican-2, demonstrating increase from A to V in all individuals (n = 22). RFU, relative fluorescent units.
Testican-2 Levels Are Positively Associated with eGFR in Two Large Community-Based Cohorts.
The high throughput of aptamer-based proteomics permits examination of large study populations. We first examined plasma proteomics data obtained from the JHS (n = 1,928), a cohort of African-American participants, a population that is disproportionately affected by CKD (10). To enhance generalizability across racially and regionally diverse groups, we validated findings in FHS, a White cohort of Northern European descent (n = 1,621). Study characteristics of these two cohorts are shown in Table 2. The mean eGFR was 93 ± 23 mL/min/1.73 m2 in JHS, while the mean eGFR was 89 ± 20 mL/min/1.73 m2 in FHS. Albuminuria, defined as a spot urine albumin-to-creatinine ratio ≥30 mg/g in JHS or greater than or equal to trace on urine dipstick in FHS, was present in 14% and 24% of study participants with available measurements, respectively. All JHS participants were African-American, including 14% with a high-risk Apolipoprotein L1 (APOL1) genotype, whereas all FHS participants were White. Mean body mass index (BMI) (32 versus 26 kg/m2) and the prevalence of diabetes (23% versus 8%) and hypertension (57% versus 35%) were higher in JHS than FHS.
Table 2.
Clinical characteristics of the JHS and FHS study cohorts
| JHS | FHS | |||
| Total sample (n = 1,928) | Subset with eGFR slope (n = 1,371) | Total sample (n = 1,621) | Subset with eGFR slope (n = 1,332) | |
| Age, y | 56 ± 13 | 55 ± 12 | 55 ± 10 | 54 ± 10 |
| Women, % | 61% | 61% | 53% | 54% |
| Race/ethnicity | 100% AA | 100% AA | 100% CW | 100% CW |
| eGFR, mL/min/1.73 m2 | 93 ± 23 | 95 ± 21 | 89 ± 20 | 90 ± 20 |
| Albuminuria, % | 14%* | 11%† | 24%‡ | 23%§ |
| BMI, kg/m2 | 32 ± 7.3 | 32 ± 7.2 | 27.5 ± 5.0 | 27.5 ± 5.0 |
| Diabetes, % | 23% | 20% | 8% | 6% |
| Hypertension, % | 57% | 54% | 35% | 33% |
| Current smoker, % | 13% | 11% | 19% | 18% |
| High risk APOL1 genotype, % | 14% | 13% | — | — |
| eGFR decline, mL/min/1.73 m2 per y | −1.2 ± 1.9 | −1.0 ± 2.7 | ||
Values are mean ± SD or percentage. AA, African-American; CW, Caucasian White. Albuminuria in JHS defined as microalbumin/creatinine ratio ≥30 mg/g. Albuminuria in FHS defined as greater than or equal to trace on urine dipstick.
n = 1,033.
n = 673.
n = 1,556.
n = 1,284.
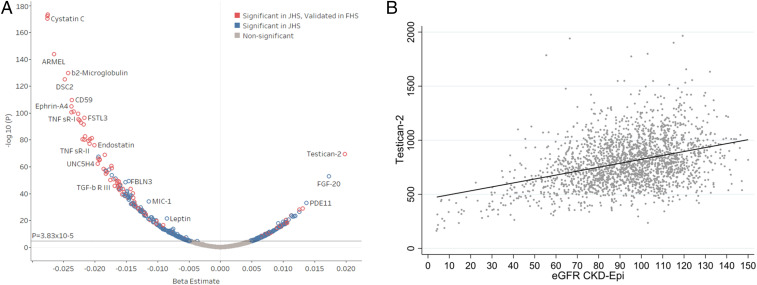
First, we examined the cross-sectional associations between proteins and baseline eGFR in linear regression models adjusting for age and gender in both cohorts. JHS was further adjusted for APOL1 risk status. Fig. 2A shows the 465 proteins significantly associated with baseline eGFR in JHS (Bonferroni threshold, P < 3.8 × 10−5). We replicated 126 protein associations with eGFR in FHS as an independent cohort (repeat Bonferroni threshold, P < 1.1 × 10−4; Fig. 2A, red circles). Of the 126 validated protein associations, the majority of proteins were inversely associated with eGFR. We confirmed previously identified markers inversely associated with eGFR including cystatin C (JHS: β = −0.028, P = 6.6 × 10−171; FHS: β = −0.014, P = 5.6 × 10−31) and β2-microglobulin (JHS: β = −0.024, P = 1.7 × 10−130; FHS: β = −0.020, P = 2.7 × 10−24). However, the majority of protein associations have not been described in the context of kidney function, and notably 33 proteins were positively associated with eGFR (i.e., lower eGFR associated with lower levels circulating proteins). Testican-2 had the single strongest positive association with baseline eGFR in JHS (Fig. 2B; β = 0.020, P = 3.6 × 10−70), with replication in FHS (β = 0.010, P = 3.8 × 10−13). Notably, two of the other proteins found to undergo net release by the kidney (V/A > 1) in the arteriovenous cohort, FGF20 and FGF9, also had positive cross-sectional associations with eGFR: FGF20 (JHS: β = 0.017, P = 1.3 × 10−53; FHS: β = 0.003, P = 4.9 × 10−2), FGF9 (JHS: β = 0.010, P = 6.4 × 10−19; FHS: β = 0.004, P = 4.3 × 10−3).
Fig. 2.
Testican-2 has a strong, positive cross-sectional association with eGFR. (A) Volcano plot showing multivariable-adjusted protein associations with baseline eGFR in the JHS and FHS. The blue circles represent Bonferroni significant associations (P < 3.8 × 10−5) in JHS. The red circles represent proteins found to be significant in both JHS (P < 3.8 × 10−5) and FHS (P < 1.1 × 10−4). Models adjusted for age, sex, BMI, diabetes, hypertension, and APOL1 risk status (JHS only). (B) Scatter plot showing cross-sectional relationship between baseline eGFR and testican-2 levels in JHS.
In contrast to the strong associations with eGFR, testican-2 showed no association with albuminuria in JHS (β = −0.06, P = 0.42) and a nominal association with albuminuria in FHS (β = 0.11, P = 0.047) in logistic regression models adjusted for age, sex, and baseline eGFR. We performed secondary analyses evaluating protein associations with baseline eGFR in a multivariable-adjusted model adjusting for albuminuria in the smaller subgroups with available albuminuria data in both cohorts. Testican-2 remained the top positive association in JHS (n = 1,033; β = 0.018, P = 4.1 × 10−28) and was highly significant in FHS (n = 1,556; β = 0.013, P = 3.4 × 10−16). Results for all 126 proteins significantly associated with baseline eGFR in JHS and FHS, including additional adjustment for albuminuria, are shown in SI Appendix, Table S2.
Higher Baseline Testican-2 Levels Are Associated with Lower Rate of eGFR Decline and Lower Risk of Incident CKD.
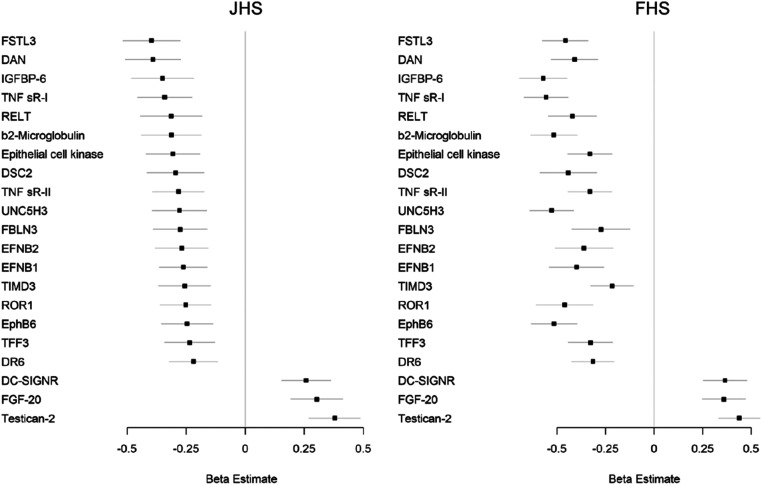
Follow-up data on longitudinal change in eGFR were available in n = 1,371 and n = 1,332 individuals who had undergone proteomic profiling in JHS and FHS, respectively. Baseline characteristics of these individuals were similar to the overall profiled cohorts (Table 2). Based on a mean follow-up of 8.0 ± 0.9 y, the mean eGFR decline in JHS was −1.2 mL/min/1.73 m2. In JHS, we found 26 proteins significantly associated with eGFR slope in linear regression models adjusted for age, sex, plate, BMI, diabetes, hypertension, APOL1 risk status, and baseline eGFR (Bonferroni threshold, P < 3.8 × 10−5). The top association was testican-2, with higher levels associated with less eGFR loss over time (β = 0.38, P = 1.7 × 10−11).
For replication, we examined the association between proteins identified in JHS with eGFR slope in FHS, where mean eGFR decline over a mean 7.0 ± 0.9 y was −1.0 mL/min/1.73 m2. As shown in Fig. 3 and SI Appendix, Table S3, 21 proteins associated with eGFR slope in JHS remained significant in FHS in linear regression models adjusted for age, sex, BMI, diabetes, hypertension, and baseline eGFR, with the same direction of association (Bonferroni threshold, P < 1.9 × 10−3). Testican-2 was again highly significantly associated with slower rate of eGFR decline (β = 0.44, P = 1.5 × 10−15). FGF20 and dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGNR) were the only other proteins associated with slower rate of eGFR decline in both JHS and FHS (negative β value). As shown in SI Appendix, Table S4, testican-2 remained significantly associated with eGFR slope in both JHS (β = 0.38, P = 4.8 × 10−6) and FHS (β = 0.45, P = 1.1 × 10−15) following additional adjustment for albuminuria in the subsets of individuals who had urine albumin measurements.
Fig. 3.
Testican-2 is associated with eGFR decline in both the JHS and FHS. Proteins associated with eGFR decline in multivariable-adjusted analyses in JHS (Left, P < 3.8 × 10−5) and replicated in FHS (Right, P < 1.9 × 10−3). Models adjusted for age, sex, BMI, diabetes, hypertension, APOL1 risk status (JHS only), and baseline eGFR.
Of the 18 proteins associated with greater eGFR decline in JHS and FHS, almost all were strongly inversely associated with eGFR at baseline (SI Appendix, Table S3). In our renal arteriovenous analysis, 4 of these 18 proteins had V/A <1 at the Bonferroni threshold (insulin-like growth factor-binding protein 6 [IGFBP-6], β2-microglobulin, deadenylating nuclease [DAN], and trefoil factor 3 [TFF3]) and an additional 5 had V/A <1 at P < 0.05 (tumor necrosis factor receptor superfamily member 19L [RELT], desmocollin-2 [DSC2], tumor necrosis factor receptor superfamily member 1 [TNF sR-1], follistatin-related protein 3 [FSTL3], and ephrin B1 [EFNB1]). Nine of these 18 proteins had no significant change from A to V in the renal arteriovenous analysis (ephrin type-B receptor 6 [EphB6], epithelial cell kinase, ephrin B2 [EFNB2], netrin receptor [UNC5H3], tyrosine-protein kinase transmembrane receptor [ROR1], tumor necrosis factor receptor superfamily member 6 [DR6], tumor necrosis factor receptor superfamily member 1B [TNF sR-II], fibulin 3 [FBLN3], and hepatitis A virus cellular receptor 2 [TIMD3]).
Finally, we tested the association of baseline levels of testican-2 with the risk of incident CKD, defined as eGFR decline to <60 mL/min per 1.73 m2. Among individuals with eGFR ≥60 mL/min/1.73 m2 at baseline, increased testican-2 levels were associated with reduced risk of incident CKD in both cohorts after adjusting for age, sex, BMI, diabetes, hypertension, APOL1 status (in JHS), and baseline eGFR (JHS: odds ratio [OR], 0.70; 95% CI, 0.55–0.90; P = 0.005; n = 1,199; FHS: hazard ratio [HR], 0.83; 95% CI, 0.69–1.00; P = 0.046; n = 1,387). In the smaller subsets with available measurements, the direction of association between baseline testican-2 and incident CKD remained consistent in both cohorts and was significant in FHS following additional adjustment for albuminuria (JHS OR, 0.73; P = 0.15; n = 649; FHS HR, 0.80; P = 0.024; n = 1,336).
Genome-Wide Association Study Corroborates Aptamer Specificity for Testican-2.
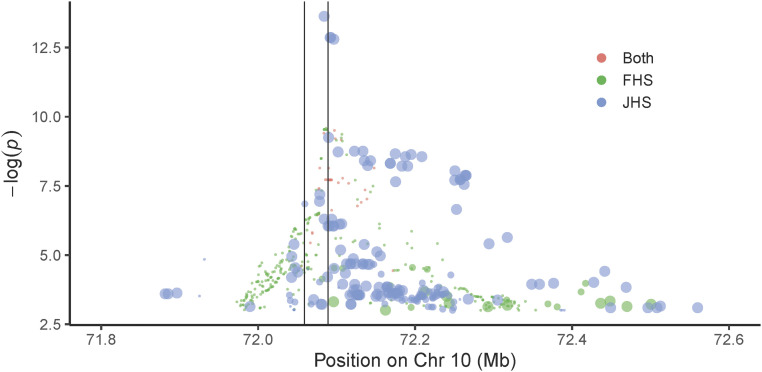
Genome-wide association study (GWAS) of aptamer-derived proteomics datasets can support aptamer specificity, i.e., when significant associations are identified within the gene locus that encodes the protein of interest (11). Importantly, we found that variants in SPOCK2, the cognate gene encoding testican-2, were significantly associated with circulating testican-2 levels in both JHS and FHS. The specific variant associations and allele frequencies differed between cohorts (SI Appendix, Table S5). In JHS, the variant with the strongest association was rs116264251 (effect, −0.32; P = 2.3 × 10−14), which has an alternate allele frequency of 0.17 in Africans, but only 0.0002 in Europeans (gnomAD) (12). The variant with the strongest association in FHS was rs1245547 (effect, −0.25; P = 2.3 × 10−10), which was common in both European and African samples (alternate allele frequencies of 0.45 and 0.88, respectively, in gnomAD) (12). The two cohorts had 35 variants in common, with the strongest variants clustered at the transcription start site and upstream of the SPOCK2 gene (Fig. 4). A summary of orthogonal data supporting aptamer specificity for all proteins significantly associated with baseline eGFR or eGFR decline in both JHS and FHS is shown in SI Appendix, Table S6.
Fig. 4.
SPOCK2 variants are associated with circulating testican-2 levels in JHS and FHS. Modified zoom locus plot where red circles represent variants observed in both JHS and FHS; green circles represent variants observed in FHS only, and blue circles represent variants observed in JHS only. The size of the blue circles indicates the alternate allele frequency in Europeans; the largest circles indicate European MAF < 0.001, the medium circles indicate 0.001 < MAF < 0.05, and small circles indicate MAF ≥ 0.05. These data demonstrate that many JHS variants are not observed in European cohorts. The vertical lines are drawn to indicate the SPOCK2 locus, with transcription running from Right to Left.
Microscopy and Single-Cell RNA Sequencing Demonstrate Podocyte-Specific Expression of Testican-2.
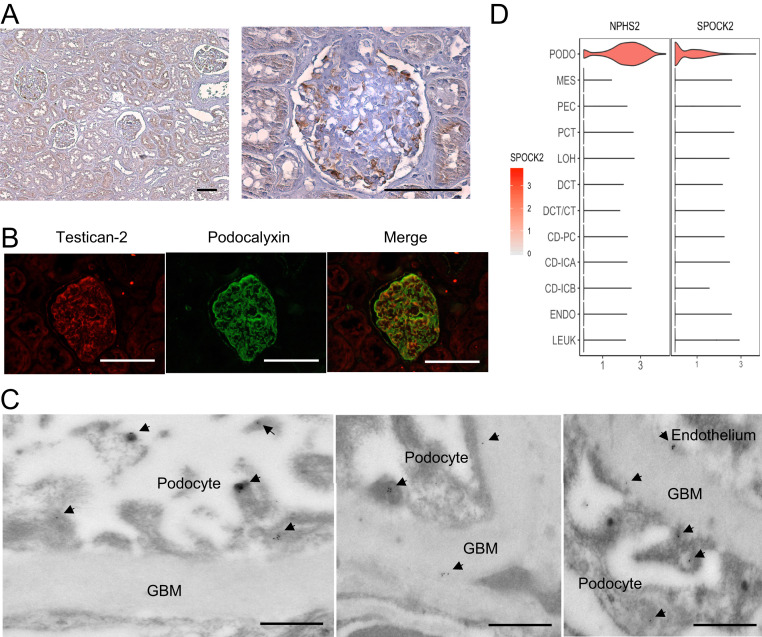
To localize kidney testican-2 expression, we assessed human kidney tissue from individuals with normal eGFR by immunohistochemistry, demonstrating staining in both glomeruli and tubular epithelium (Fig. 5A), consistent with results from the Human Protein Atlas (SI Appendix, Fig. S1). By immunofluorescence, testican-2 expression was strongest in glomeruli, colocalizing with the podocyte protein podocalyxin (Fig. 5B). Furthermore, using immunogold labeling and transmission electron microscopy, we found that testican-2 is expressed in podocyte foot processes, with testican-2 also visualized within the glomerular basement membrane (Fig. 5C). Finally, to better parse cell specific SPOCK2 gene expression in the human kidney, we mapped its expression to a single nucleus RNA-sequencing (snRNA-seq) dataset obtained from three healthy human kidney samples (13,860 cells, with an average of 2,505 detected genes per cell) (13). Strong SPOCK2 expression was detected only in podocytes, without significant expression in any other kidney cell type (Fig. 5D).
Fig. 5.
Testican-2 is expressed in human podocytes. (A) Testican-2 expression in human kidney tissue assessed by immunohistochemistry. (Scale bar: 100 μm.) Representative image from one of two independent samples. (B) Representative photographs of immunofluorescence for testican-2 (Left), podocalyxin (Center), and merge (Right) of human kidney tissue. (Scale bar: 100 μm.) (C) Representative transmission electron micrographs of human kidney tissue with immunogold labeling of testican-2. (Scale bar: 500 nm.) The arrowheads point to gold-labeled testican-2 in podocyte foot processes, glomerular basement membrane (GBM), and at the GBM–endothelial interface. (D) Violin plot demonstrating SPOCK2 expression across all kidney cell types. The podocyte-specific gene NPHS2 serves as a positive control. There is negligible expression in other cell clusters. snRNA-seq data are from ref. 13. CD-ICA, collecting duct, intercalated cells type A; CD-ICB, collecting duct, intercalated cells type B; CD-PC, collecting duct-principal cells; DCT, distal convoluted tubule; DCY/CT, distal convoluted tubule/connecting tubular cells; ENDO, endothelium; LEUK, leukocytes; LOH, loop of Henle; MES, mesangial cells; PCT, proximal convoluted tubule; PEC, parietal epithelial cells; PODO, podocytes.
Testican-2 Promotes Glomerular Endothelial Cell Tube Formation and Migration In Vitro.
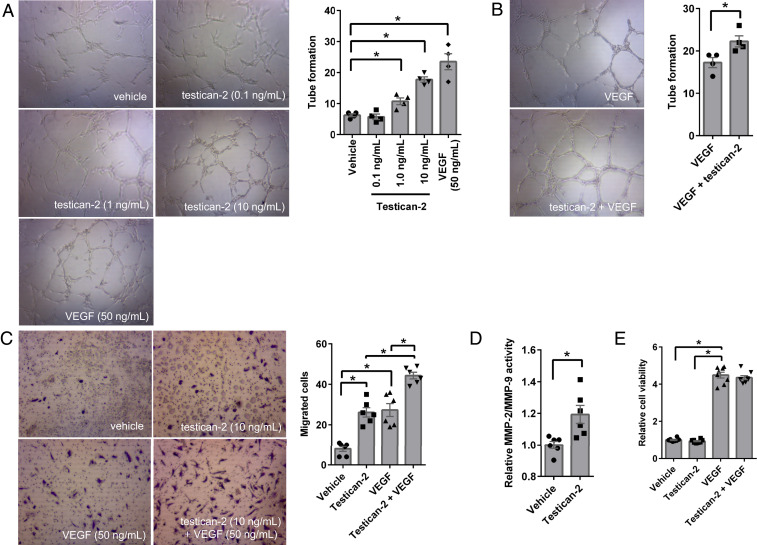
Testican-2 function in the kidney has not been studied. It does not appear to be expressed in adult mice, introducing an obstacle to functional investigation in vivo (14). Because it is a secreted protein, with electron microscopy demonstrating transit from podocyte foot processes through the glomerular basement membrane, we tested the effect of testican-2 on primary human glomerular endothelial cells (HGECs). These endothelial cells line the vascular side of the glomerular basement membrane, opposite the podocytes that line the urinary side. In an in vitro angiogenesis assay, testican-2 increased HGEC tube formation (Fig. 6A), and its effect was additive with vascular endothelial growth factor (VEGF) (Fig. 6B). Furthermore, testican-2 increased HGEC migration across extracellular matrix (Fig. 6C). Prior reports have shown that testican isoforms, including testican-2, are able to modulate matrix metalloproteinases (MMPs) (15). We found that testican-2 increases MMP-2/MMP-9 activity in HGEC cultured media, outlining a potential mechanism for its impact on HGEC motility (Fig. 6D). Unlike VEGF, testican-2 had no impact on HGEC proliferation (Fig. 6E).
Fig. 6.
Testican-2 increases glomerular endothelial cell tube formation and migration. Tube formation analysis of primary human glomerular endothelial cells (HGECs) treated with (A) vehicle, testican-2 (0.1, 1, and 10 ng/mL), or VEGF (50 ng/mL), or (B) VEGF (50 ng/mL) with or without testican-2 (10 ng/mL) for 8 h. Representative images at 10× magnification; graphs show results quantitated for n = 4. (C) Cell migration analysis of HGECs treated with vehicle, testican-2 (10 ng/mL), VEGF (50 ng/mL), or VEGF (50 ng/mL) plus testican-2 (10 ng/mL) for 22 h. Representative images at 10× magnification, graph shows results quantitated for n = 6. (D) MMP-2/MMP-9 activity in cultured supernatant from HGECs treated with vehicle or testican-2 (10 ng/mL) for 24 h; n = 6. (E) Cell proliferation of HGECs treated with vehicle, testican-2 (10 ng/mL), VEGF (50 ng/mL), or VEGF (50 ng/mL) plus testican-2 (10 ng/mL) for 72 h; n = 8. The bar graphs show means ± SEM. *P < 0.05, unpaired t test (A, B, and D) or ANOVA with Tukey’s multiple-comparisons test (C and E).
Discussion
We examined the relationship between kidney function and the blood proteome using a combined physiologic and epidemiologic approach. We find that dozens of proteins undergo renal clearance and/or are inversely correlated with GFR, including cystatin C, β2-microglobulin, and PTH, which serve as illustrative “positive controls.” Cystatin C is widely expressed and is a clinically established biomarker of glomerular filtration that provides prognostic information about the risk of progressive kidney disease and death (16). β2-microglobulin is a component of MHC class I molecules and at the high concentrations achieved in patients with ESKD can deposit in joint spaces, causing dialysis-related amyloidosis (17). PTH is a hormone released by the parathyroid gland, and elevated levels in kidney disease cause adverse bone remodeling (18). Thus, proteins that accumulate with kidney dysfunction can inform both prognosis and disease pathophysiology.
Our data highlight numerous protein elevations that warrant further study as potential markers and mediators of kidney disease and its complications, including proteins significantly associated with both reduced eGFR at baseline and greater eGFR decline over time in JHS and FHS. Several of these proteins demonstrate a significant reduction from A to V in our renal arteriovenous analysis, suggesting that increased levels are at least in part attributable to reduced renal clearance (e.g., β2-microglobulin). By contrast, others demonstrate no change from A to V, suggesting other mechanisms of protein elevation, including increased production. Regardless of why they are elevated, these proteins have the potential to impact a range of biologic pathways. Notably, our results highlight four inflammatory markers (TNF sR-1, TNF sR-II, RELT, and DR6) recently linked to the progression of kidney disease in both targeted assessment of these proteins as well as discovery proteomic analyses in studies of diabetic kidney disease progression (6). In addition, our findings include two ligands for Eph-related tyrosine kinases (EFNB1 and EFNB2) and an Eph receptor (EphB6). Eph/ephrin signaling has broad effects and has been implicated in maintenance of the barrier function of the glomerular slit diaphragm (19) and attenuation of renal fibrosis in diabetic nephropathy in mice (20).
In addition to its fundamental role in clearance, the kidney has several established anabolic functions. It produces metabolites such as 1,25-hydroxyvitamin D, arginine, and tyrosine—findings that we have highlighted with metabolomic analysis of the arteriovenous samples utilized for proteomics herein (8, 21). It also produces proteins such as renin and erythropoietin, which regulate blood pressure and red blood cell production, respectively. Our current results raise interest in other potential kidney-derived factors, particularly testican-2. First, testican-2 levels increased from artery to renal vein in all 22 individuals that underwent invasive sampling. Second, testican-2 levels have a strong, positive correlation with eGFR. This stands in contrast to all existing blood biomarkers of kidney function such as creatinine, blood urea nitrogen, and cystatin C, which undergo renal clearance and are thus inversely correlated with eGFR. Third, higher testican-2 levels are associated with slower eGFR decline over time and lower risk of incident CKD, even after adjusting for baseline eGFR and demographic factors, a finding that was significant in two large community-based cohorts that span >2,600 racially and geographically diverse individuals. Taken together, these findings nominate testican-2 as a marker of kidney health that provides risk information independent from currently available metrics.
The in vivo function of testican-2 is unknown. Encoded by the SPOCK2 gene, it is a 424-amino acid protein predicted to form part of the extracellular matrix, with an N-terminal signal peptide and C-terminal glycosaminoglycan binding sites. Testican-2 was originally cloned from a human fetal brain cDNA library, but subsequent gene expression databases demonstrate widespread expression, including in the human kidney (14). Both immunohistochemistry and immunofluorescence of human kidney tissue are consistent with glomerular expression, and at single-cell resolution, SPOCK2 gene expression is confined to podocytes in adult human kidney samples. Previously studied in the context of neurons and neurite outgrowth, the role of glomerular testican-2 is unknown (22). Our electron micrographs demonstrate testican-2 protein in both podocyte foot processes and the glomerular basement membrane, a specialized, proteoglycan-rich extracellular matrix that lies between podocytes and endothelial cells (23). Interestingly, testican-2 increases glomerular endothelial cell tube formation and migration and increases MMP-2/MMP-9 activity in vitro. However, unlike VEGF, an established podocyte-derived angiogenic factor (24), testican-2 does not increase glomerular endothelial cell proliferation. The utility of testican-2 as a biomarker may be unrelated to these potential functional effects; for example, it could simply reflect an association with podocyte number or synthetic function. This requires further analysis. Similarly, whether testican-2 appearance in blood is solely attributable to protein passage across the glomerular basement membrane and endothelium, or also by shedding into Bowman’s space with subsequent tubular reabsorption, requires further study.
Although we emphasize testican-2 because of its strong association with eGFR and eGFR slope, our results raise interest in other potential kidney-derived factors as well. Prior work has outlined a critical role for FGF20 and FGF9 in kidney development, demonstrating that they are necessary and sufficient to maintain kidney progenitor stemness. Deletion of both results in kidney agenesis in mice, deletion of FGF20 has been identified in a human kindred with kidney agenesis, and FGF9 is widely used as a reagent for the differentiation of pluripotent stem cells to kidney organoids (25–27). The finding that both are higher in the renal vein than arterial circulation and that their circulating levels are positively associated with eGFR further enhances interest in these proteins, for example to understand whether they play a functional role in cell growth or differentiation in the adult kidney. DC-SIGNR, a transmembrane receptor involved in the innate immune systems, also warrants further consideration (28)—although it does not change across the renal arteriovenous circulation, like testican-2 its levels are positively associated with baseline eGFR and higher baseline levels are associated with slower eGFR decline over time.
Proteomic discovery requires validation. In addition to seeking replication of findings in an independent cohort, this involves confirmation of assay specificity. A key strength of our study is the use of GWAS to show that single-nucleotide polymorphisms (SNPs) within the SPOCK2 locus are significantly associated with aptamer-based measurement of testican-2 levels. We found a strong association between genetic variants in and near the SPOCK2 gene with circulating levels of testican-2 across both racially distinct populations. Notably, a large number of variants associated with testican-2 levels in each cohort were different. This is likely explained by the finding that JHS variants were monomorphic or much less common in White populations (12), limiting the power to detect an association in Whites. Additionally, since linkage is higher among Whites, there may be more variants associated with testican-2 aptamer levels due to linkage with causal variants in FHS, possibly explaining variants observed in FHS only (29). While our results support aptamer specificity for testican-2, they also highlight the potential role for racial genetic heterogeneity in determining circulating protein levels.
Our study has several limitations. Although large and well-characterized, the study cohorts we examined had a relatively small number of individuals with prevalent CKD, precluding examination of protein levels at very low eGFR and precluding analysis of CKD progression to hard outcomes like ESKD or the requirement for kidney transplantation. Instead, we focused on eGFR slope and incident CKD among individuals largely with normal kidney function at baseline. In addition, proteinuria in JHS and FHS was mild, and in FHS was limited to urinary dipstick. This limited our ability to assess the relationship between testican-2 and proteinuria. Furthermore, because we performed proteomics at a single time point in both cohorts, we do not have information on biomarker trajectory for testican-2 or other proteins of interest highlighted in our analyses. Finally, we note that aptamer-based measurements provide relative, not absolute quantitation. Thus, future studies will require use of a more targeted, quantitative assay for testican-2 to fully characterize the protein’s performance as a biomarker of kidney health and prognosis.
In summary, aptamer-based proteomics provides insight into the kidney’s broad and heterogeneous impact on the circulating proteome. Complete results on renal arteriovenous protein gradients, as well as significant protein associations with eGFR and eGFR slope in both JHS and FHS, are shared as a resource for the research community. Protein associations common across JHS and FHS may represent shared biology across African-American and White populations. Here, we highlight specific interest in testican-2, a kidney-derived molecule that is positively associated with eGFR and longitudinal eGFR stability—confidence in the aptamer-based testican-2 data are buttressed by both single-cell and population-based genomic analyses. More work is required to develop a targeted assay for testican-2; to test the association between blood testican-2 levels and hard renal outcomes, particularly among higher risk individuals with hypertension, diabetes, and abnormal kidney function; and to extend our initial mechanistic inquiry of testican-2 in the kidney.
Materials and Methods
Arterial and Renal Venous Plasma Sampling.
Details on this protocol have been published (8, 9). For the current study, we include data from 22 individuals who had proteomic profiling performed. In brief, we recruited consecutive patients referred to the Massachusetts General Hospital Cardiac Catheterization Laboratory for elective right and left heart catheterization. During the procedure, plasma was sampled from catheters placed in the ostium of a renal vein and the abdominal aorta. All patients were fasting, and all samples were obtained prior to coronary artery catheterization. The Modification of Diet in Renal Disease equation was used to calculate eGFR for all study participants (30).
Cohort Study Populations.
The JHS is a longitudinal population-based cohort study prospectively investigating the determinants of cardiovascular disease (CVD) among African Americans living in the Jackson, Mississippi, metropolitan area. Recruitment, sampling, and data collection methods have been described (31). Among 5,306 individuals enrolled at baseline (2000–2004), proteomic profiling was performed on 2,147 samples, from which 1,928 participants were included in the current study. The first batch of JHS participants (n = 315) was sampled across participants in a nested case-control study for incident coronary heart disease (CHD), while the remaining participants (n = 1,613) were selected randomly across the entire cohort. Incident CHD was defined as incident myocardial infarction, fatal CHD, or coronary artery bypass graft or stent (32). Creatinine data were collected at baseline (examination 1) and at examination 3. The APOL1 high-risk status was defined as having two copies of G1 and/or G2 risk alleles, and the low-risk status was defined as having 0 or 1 risk alleles. Albuminuria was defined as a urinary albumin-to-creatinine ratio of ≥30 (mg/g) (33).
The FHS Offspring cohort was formed in 1971 with the enrollment of 5,124 individuals in a community-based longitudinal cohort study (34). Of the over 3,529 individuals in the cohort at examination 5 (1991–1995), 1,913 had proteomic profiling performed. The first batch of participants (n = 821) was sampled from an incident CVD case-cohort study, whereas the second batch of participants (n = 1092) was sampled randomly across the remaining participants in the entire cohort. Incident CVD cases were selected on the basis of CVD events or diagnoses occurring after examination 5. In FHS, proteinuria was defined as a value of trace or higher on urine dipstick assay.
Study Approval for Human Studies.
The human study protocols were approved by the institutional review boards of Massachusetts General Hospital, Boston University Medical Center, Beth Israel Deaconess Medical Center, the University of Mississippi Medical Center, Jackson State University, and Tougaloo College and adhered to the Declaration of Helsinki. All participants provided written informed consent.
Proteomic Profiling.
The single-stranded DNA aptamer-based SOMAscan proteomics platform was applied to FHS and JHS cohort plasma samples that were stored at −80 °C (35). FHS samples were collected between 1991 and 1995 and had undergone two prior freeze–thaw cycles. JHS samples were collected between 2000 and 2004 and had never been previously thawed. FHS samples were profiled in two batches: the first batch, using Version 1.1, contained 1,129 aptamers, and the second batch, using Version 1.3, contained 1,305 aptamers. The JHS cohort samples were assayed in three batches with Version 1.3. Aptamers that were unique to FHS SOMAscan, Version 1.1, were not included in our analyses, so only 1,062 aptamers in FHS Batch 1 were included in FHS analyses. FHS median intra-assay coefficient of variation (CV) was <4% and median interassay CV was <7% across batches. The average JHS median intra-assay and interassay CVs were 2.7% and 5.2%, respectively, across all plates and batches.
Protein Associations with Baseline eGFR.
To reduce skewness of the data, protein measures were log-transformed, standardized within batch, and rank-normalized over the entire sample. For FHS, the rank-normalized values were regressed onto plate. The transformed proteins were used for all subsequent analyses. Linear regression was used to model the association between proteins (dependent variable) and eGFR (independent variable). The eGFR was calculated by the Chronic Kidney Disease Epidemiology Collaboration equation, using serum creatinine values at visit 1 for JHS and serum creatinine at examination 5 for FHS (36). In JHS, covariates used to adjust the analyses included age, sex, APOL1 risk status, and proteomic assay plate. Because albuminuria data were available for a smaller sample size in JHS (n = 673), we adjusted for it in secondary models. In FHS, covariates used to adjust the analysis included age and sex. As in JHS, proteinuria adjustment was added in secondary models.
Protein Associations with eGFR Decline and Incident CKD.
As only two values for serum creatinine were available in JHS (examination 1 and examination 3), eGFR decline was defined as the eGFR slope between examination 1 and examination 3 and calculated using the equation eGFR decline = (eGFRexam3 – eGFRexam1)/(timeyears from examination 1 to examination 3). Mean follow-up time in JHS was 8.0 ± 0.9 y. Linear regression was used to model the association between eGFR decline (dependent variable) and proteins (independent variable). Covariates used to adjust the analyses included age, sex, plate, and eGFRexam1, hypertension, diabetes, BMI, and APOL1 risk status. To best replicate the model for eGFR decline used in JHS for analyses in the FHS, eGFR decline in FHS was defined as the eGFR slope between examination 5 and examination 7 and was calculated using the equation eGFR decline = (eGFRexam7 – eGFRexam5)/(timeyears from examination 5 to examination 7). Mean follow-up time in the FHS analysis was 7.0 ± 0.9 y. Linear regression was used to model the association between eGFR decline (dependent variable) and proteins (independent variable). Covariates used to adjust the analyses included age, sex, and eGFRexam5, hypertension, diabetes, and BMI. Logistic regression models were used to evaluate association of testican-2 with incident CKD in JHS. Given availability of two follow-up time points in FHS, proportional hazards regression was used to evaluate testican-2 association with incident CKD. Incident CKD was defined as eGFR ≤60 mL/min/1.73 m2 occurring after baseline examination after exclusion of prevalent CKD cases (n = 141 in JHS and n = 137 in FHS). In both JHS and FHS, the multivariable models were additionally adjusted for albuminuria in secondary analyses.
Genome-Wide Association Analyses.
GWAS in JHS was run using build GRCh38 of the human genome and FHS was run using build GRCh37. In order to compare results between cohorts, genomic positions for each RefSNP ID from FHS were annotated in Build 38 using package biomaRt (R) to access the Ensembl database (access date 11/13/19) limited to variants within a 1-MB region around SPOCK2. Full details are provided in SI Appendix.
Immunohistochemistry and Immunofluorescence.
Human kidney samples were acquired from macroscopically normal portions of kidney tissue obtained from patients with renal tumors with normal eGFR. The kidney samples were fixed with 4% paraformaldehyde overnight, embedded in paraffin, and sectioned onto slides. Primary antibodies were used as follows: rabbit anti-testican-2 (Sigma); goat anti-podocalyxin (R&D Systems) (both diluted 1:200, incubated overnight). After washing the tissue, slides were incubated for 1 h at room temperature in the dark with the secondary antibody (donkey anti-rabbit IgG-conjugated Alexa Fluor 568 and donkey anti-goat IgG-conjugated Alexa Fluor 488, diluted 1:200). For immunohistochemistry, DAB kit (Thermo Scientific) was used according to the manufacturer’s instruction.
Electron Microscopy.
Small pieces of human kidney cortex tissue (1 mm × 1 mm) were prepared as previously described (37, 38). Ultrathin (70 nm) sections were cut using a Reichert Ultracut E ultramicrotome and collected onto formvar-coated grids (EMS). Sections were incubated for 1 h at room temperature on drops of primary antibody (rabbit polyclonal anti-testican-2; Sigma; HPA044605), diluted 1:50 or 1:100 in DAKO antibody diluent; some sections were pretreated with SDS prior to incubating in primary antibody. Specimens were rinsed several times in PBS, incubated on drops of a secondary gold conjugate for 1 h at room temperature (goat anti-rabbit IgG, 10 nm; Ted Pella; #15726), then rinsed several times with distilled deionized water and contrast-stained using 2.0% aqueous uranyl acetate. Grids were examined at 80 kV in a JEOL 1011 transmission electron microscope equipped with an AMT digital camera and proprietary image capture software (Advanced Microscopy Techniques).
Cell Culture Experiments.
Primary HGECs (Cell Systems) were cultured in ATCC Vascular Cell Basal Medium (ATCC PCS-100-30) plus Endothelial Cell Growth Kit-BBE (ATCC PCS-100-040) at 37 °C, 5% CO2. Cells in passages 4 to 10 were used in all experiments. In vitro angiogenesis was examined using the Angiogenesis Assay kit (ECM625; EMD Millipore), HGEC migration was examined using the Corning BioCoat Matrigel Invasion Chambers (catalog #354480), MMP-2/MMP-9 activity in the cultured media of HGECs was examined using the Fluorogenic InnoZyme Gelatinase (MMP-2/MMP-9) Activity Assay kit (CBA003; EMD Millipore), and HGEC proliferation/viability was examined using the Resazurin Cell Viability kit (PK-CA707-30025; PromoCell). Complete details are provided in SI Appendix.
Supplementary Material
Acknowledgments
Electron microscopy was performed in the Microscopy Core of the Massachusetts General Hospital Center for Systems Biology/Program in Membrane Biology. This work was supported by Grants R01NR017399 and U01DK106981 (E.P.R.), R01HL133870 (J.G.W. and R.E.G.), and R01HL132320 (T.J.W., R.S.V., and R.E.G.). The JHS is supported and conducted in collaboration with Jackson State University (Grant HHSN268201800013I), Tougaloo College (Grant HHSN268201800014I), the Mississippi State Department of Health (Grant HHSN268201800015I), and the University of Mississippi Medical Center (Grants HHSN268201800010I, HHSN268201800011I, and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute on Minority Health and Health Disparities. The FHS is supported by Grants NO1HC25195, HHSN268201500001I, and 75N92019D00031 from the NHLBI. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the NHLBI; the NIH; or the US Department of Health and Human Services.
Footnotes
Competing interest statement: D.J.F. and M.R.P. are equity holders in Apolo1bio, receive research support from Vertex, and are inventors on patents related to APOL1 diagnostics and therapeutics.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009606117/-/DCSupplemental.
Data and Materials Availability.
FHS proteomic data have been deposited in the databases of Genotypes and Phenotypes (dbGAP) (https://www.ncbi.nlm.nih.gov/gap/) (accession no. pht006013.v3.p12). JHS proteomics data have been deposited to the JHS Data Coordinating Center and will also be deposited in dbGAP; until posted in dbGAP, all JHS data are available from the JHS Data Coordinating Center on request (JHSccdc@umc.edu). All other results and analytic methods are available within the manuscript or from the authors on request. Noncommercial study materials will be made available for the purposes of reproducing the results or replication of the procedure, as respective institutional review board and material transfer agreements permit.
References
- 1.Smith J. G., Gerszten R. E., Emerging affinity-based proteomic technologies for large-scale plasma profiling in cardiovascular disease. Circulation 135, 1651–1664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ngo D. et al., Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation 134, 270–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mosley J. D. et al., Probing the virtual proteome to identify novel disease biomarkers. Circulation 138, 2469–2481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganz P. et al., Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA 315, 2532–2541 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Williams S. A. et al., Plasma protein patterns as comprehensive indicators of health. Nat. Med. 25, 1851–1857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niewczas M. A. et al., A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat. Med. 25, 805–813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gold L. et al., Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5, e15004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee E. P. et al., A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 24, 1330–1338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simic P. et al., Glycerol-3-phosphate is an FGF23 regulator derived from the injured kidney. J. Clin. Invest. 130, 1513–1526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.U.S. Renal Data System , USRDS 2016 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States, (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, 2016). [Google Scholar]
- 11.Benson M. D. et al., Genetic architecture of the cardiovascular risk proteome. Circulation 137, 1158–1172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karczewski K. J. et al. ; Genome Aggregation Database Consortium , The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson P. C. et al., The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. U.S.A. 116, 19619–19625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vannahme C. et al., Molecular cloning of testican-2: Defining a novel calcium-binding proteoglycan family expressed in brain. J. Neurochem. 73, 12–20 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Nakada M., Miyamori H., Yamashita J., Sato H., Testican 2 abrogates inhibition of membrane-type matrix metalloproteinases by other testican family proteins. Cancer Res. 63, 3364–3369 (2003). [PubMed] [Google Scholar]
- 16.Shlipak M. G. et al.; CKD Prognosis Consortium , Cystatin C versus creatinine in determining risk based on kidney function. N. Engl. J. Med. 369, 932–943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrell J., Bastani B., Beta 2-microglobulin amyloidosis in chronic dialysis patients: A case report and review of the literature. J. Am. Soc. Nephrol. 8, 509–514 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Drüeke T. B., Massy Z. A., Changing bone patterns with progression of chronic kidney disease. Kidney Int. 89, 289–302 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Fukusumi Y. et al., Nephrin-binding ephrin-B1 at the slit diaphragm controls podocyte function through the JNK pathway. J. Am. Soc. Nephrol. 29, 1462–1474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y. et al., Activation of EphA1-Epha receptor axis attenuates diabetic nephropathy in mice. Biochem. Biophys. Res. Commun. 486, 693–699 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Rhee E. P. et al., Metabolomics of chronic kidney disease progression: A case-control analysis in the chronic renal insufficiency cohort study. Am. J. Nephrol. 43, 366–374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartmann U., Maurer P., Proteoglycans in the nervous system—the quest for functional roles in vivo. Matrix Biol. 20, 23–35 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Miner J. H., The glomerular basement membrane. Exp. Cell Res. 318, 973–978 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eremina V., Quaggin S. E., The role of VEGF-A in glomerular development and function. Curr. Opin. Nephrol. Hypertens. 13, 9–15 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Barak H. et al., FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev. Cell 22, 1191–1207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takasato M. et al., Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Morizane R. et al., Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppel E. A., van Gisbergen K. P., Geijtenbeek T. B., van Kooyk Y., Distinct functions of DC-SIGN and its homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition and immune regulation. Cell. Microbiol. 7, 157–165 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Shifman S., Kuypers J., Kokoris M., Yakir B., Darvasi A., Linkage disequilibrium patterns of the human genome across populations. Hum. Mol. Genet. 12, 771–776 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Levey A. S. et al.; Modification of Diet in Renal Disease Study Group , A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Ann. Intern. Med. 130, 461–470 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Taylor H. A. et al., Toward resolution of cardiovascular health disparities in African Americans: Design and methods of the Jackson Heart Study. Ethn. Dis. 15, S6-4-S6-17 (2005). [PubMed] [Google Scholar]
- 32.Keku E. et al., Cardiovascular disease event classification in the Jackson Heart Study: Methods and procedures. Ethn. Dis. 15, S6-62-S6-70 (2005). [PubMed] [Google Scholar]
- 33.Young B. A. et al., Risk factors for rapid kidney function decline among African Americans: The Jackson Heart Study (JHS). Am. J. Kidney Dis. 68, 229–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmood S. S., Levy D., Vasan R. S., Wang T. J., The Framingham Heart Study and the epidemiology of cardiovascular disease: A historical perspective. Lancet 383, 999–1008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Candia J. et al., Assessment of variability in the SOMAscan assay. Sci. Rep. 7, 14248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levey A. S. et al.; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) , A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 150, 604–612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice W. L. et al., High resolution helium ion scanning microscopy of the rat kidney. PLoS One 8, e57051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vedovelli L. et al., Altered V-ATPase expression in renal intercalated cells isolated from B1 subunit-deficient mice by fluorescence-activated cell sorting. Am. J. Physiol. Renal Physiol. 304, F522–F532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FHS proteomic data have been deposited in the databases of Genotypes and Phenotypes (dbGAP) (https://www.ncbi.nlm.nih.gov/gap/) (accession no. pht006013.v3.p12). JHS proteomics data have been deposited to the JHS Data Coordinating Center and will also be deposited in dbGAP; until posted in dbGAP, all JHS data are available from the JHS Data Coordinating Center on request (JHSccdc@umc.edu). All other results and analytic methods are available within the manuscript or from the authors on request. Noncommercial study materials will be made available for the purposes of reproducing the results or replication of the procedure, as respective institutional review board and material transfer agreements permit.