Highlights
-
•
15 studies were included - 13 SARS-CoV-2 culture and 2 contact tracing studies.
-
•
Mild-to-moderate COVID-19 patients highly unlikely to be infectious after day 10.
-
•
Immunocompromised and severe-to-critical patients may be infectious for >10 days.
-
•
This evidence may be used to inform duration of isolation guidance.
Keywords: SARS-CoV-2, COVID-19, Isolation, Infectiousness, Review, Coronavirus, RNA, Virus culture, Contact tracing
Abstract
Objectives
To summarise the evidence on the duration of infectiousness of individuals in whom SARS-CoV-2 ribonucleic acid is detected.
Methods
A rapid review was undertaken in PubMed, Europe PubMed Central and EMBASE from 1 January 2020 to 26 August 2020.
Results
We identified 15 relevant studies, including 13 virus culture and 2 contact tracing studies. For 5 virus culture studies, the last day on which SARS-CoV-2 was isolated occurred within 10 days of symptom onset. For another 5 studies, SARS-CoV-2 was isolated beyond day 10 for approximately 3% of included patients. The remaining 3 virus culture studies included patients with severe or critical disease; SARS-CoV-2 was isolated up to day 32 in one study. Two studies identified immunocompromised patients from whom SARS-CoV-2 was isolated for up to 20 days. Both contact tracing studies, when close contacts were first exposed greater than 5 days after symptom onset in the index case, found no evidence of laboratory-confirmed onward transmission of SARS-CoV-2.
Conclusion
COVID-19 patients with mild-to-moderate illness are highly unlikely to be infectious beyond 10 days of symptoms. However, evidence from a limited number of studies indicates that patients with severe-to-critical illness or who are immunocompromised, may shed infectious virus for longer.
Introduction
Severe acute respiratory syndrome coronavirus, type 2 (SARS-CoV-2) is a highly infectious virus that is responsible for tens of millions of cases of coronavirus disease 2019 (COVID-19) worldwide.1 SARS-CoV-2 constitutes a significant public health concern due to its high basic reproduction rate (R0 of 4–5 in the unmitigated setting), the absence of immunity in the human population, the lack of effective treatment or vaccination approaches, the high SARS-CoV-2 viral loads detectable in respiratory samples, and the occurrence of transmission early in the disease course, often pre-symptomatically, or by those asymptomatic or pauci-symptomatic.2, 3, 4, 5, 6, 7 Due to the lack of effective antiviral therapy or a vaccine, the rapid diagnosis, contact tracing, and isolation of suspected COVID-19 cases are of critical importance in the control of this pandemic.3
Isolation is defined as separating those with symptoms of, or diagnosed with, COVID-19, from people who are not infected.8 This is distinct from quarantine, which is defined as separating and restricting the movements of people who were exposed or potentially exposed to COVID-19, as a precautionary measure to prevent transmission should they later become infected.8 In circumstances in which cases are confirmed, isolation should continue until a patient is no longer expected to be infectious. However, recommendations regarding the appropriate duration of isolation for those who test positive for SARS-CoV-2 ribonucleic acid (RNA) have evolved since the emergence of the virus in China in December 2019, and vary across jurisdictions, generally ranging from 7 days9 to 14 days.10
SARS-CoV-2 infection is primarily diagnosed based on detecting the presence of viral RNA by molecular testing, usually by reverse transcription polymerase chain reaction (RT-PCR) in a specimen from an individual's respiratory tract. However, detection of viral RNA does not necessarily mean that a person is infectious, i.e. that they are capable of transmitting the virus to another person.3 Several factors determine viral transmission risk: these include whether a virus is still replication-competent (or viable); the amount of replicative virus; the presence of symptoms, such as a cough which can spread infectious droplets; the individual's local mucosal immune response to the virus; and the behavioural and environmental factors associated with the infected individual and their contacts.11 Determination of the presence of viable virus may be accomplished by monitoring the ability of SARS-CoV-2 to replicate in laboratory-based cell culture. Such virus culture serves as a marker of potential viral infectivity.12 Therefore, for the purpose of determining the infectiousness of a patient, and the duration of isolation required to minimise the risk of onward transmission, virus culture is significantly more informative than simple detection of viral RNA which could reflect non-replicative virus.11 , 13 – 15 It is important to note that cells used to culture SARS-CoV-2 in the laboratory have been developed to be susceptible to, and enhance replication, of a wide range of viruses and therefore do not necessarily “mimic” the susceptibility of cells in a patient.16 In addition, virus culture is usually used to identify the causative agent of an infection, and not to infer the infectivity of the individual from whom the sample was collected.12 Therefore, the culture of SARS-CoV-2 derived from a patient's clinical sample does not necessarily prove that the patient is infectious at the time of sampling, but rather that they were potentially infectious due to the presence of viable virus. Evidence from epidemiological investigations can supplement the knowledge gained from virus culture studies by describing the transmission dynamics between index cases and their close contacts. Together, virus culture studies and epidemiological investigations (i.e., contact tracing studies) provide a more complete understanding of the duration of infectiousness.15
The aim of this rapid review was to summarise the evidence on the potential duration of patient infectiousness, as derived from virus culture and contact tracing studies, for those individuals in whom SARS-CoV-2 RNA is detected, and hence inform guidance on the duration of isolation.
Materials and methods
We conducted a rapid review to answer the following research question: What is the duration of infectiousness in those in whom SARS-CoV-2 RNA is detectable? This rapid review followed a standardised protocol,17 in keeping with Cochrane rapid review methodology guidance.18 We searched PubMed, Europe PubMed Central and EMBASE from 1 January 2020 to 26 August 2020. The search combined terms for COVID-19 and SARS-CoV-2 with terms for infectiousness, virus culture, and contact tracing. Reference lists of systematic reviews were also screened. No language restrictions were applied. Only articles including human subjects were included. The search strategy is provided in the supplementary material (Appendix 1).
All potentially eligible papers, including non-peer-reviewed pre-prints, were exported to Covidence19 and screened for relevance. Any study that addressed the research question and met the inclusion criteria was included (Table 1 ). Primary research studies presenting empirical data on duration of infectiousness of SARS-CoV-2 in human populations were included. Specifically, the included studies were laboratory studies where virus culture was performed, and contact tracing studies with case-contact pairs.
Table 1.
Inclusion and exclusion criteria.
| Inclusion Criteria | Exclusion criteria |
|---|---|
| Primary studies presenting empirical data on duration of infectiousness of SARS-CoV-2 in human populations, including: Contact tracing studies involving case-contact pairs Laboratory studies where virus culture was performed. |
|
For each included study, data on the study design, participant demographics and relevant outcomes were extracted. For virus culture studies, outcomes included the days post symptom onset on which virus culture was attempted, the days on which SARS-CoV-2 culture was positive, and the viral load of the original samples (where reported). Where graphical distributions were presented to model the probability of a positive culture on each day post symptom onset, data from curves were extracted using WebPlotDigitzer, version 4.3.20 In order to present the data specifically for the time period following symptom onset (as opposed to pre-symptomatic culture positivity), and to allow for visual comparison of curves using common axes, extracted data derived from individual studies were plotted on a common chart. Plotting of data was performed using RStudio version 1.1.442.
For contact tracing studies, outcomes included the number of days post symptom onset in the infector at which infection in contacts was observed to have occurred, and the secondary attack rate. As studies were predominantly case series or case reports in design, a de novo tool, adapted from related tools, was used for quality appraisal.17 Screening, data extraction, and quality appraisal were undertaken by single reviewers, however studies excluded at full-text screening stage, extracted data, and quality appraisal judgments were double-checked for accuracy by another reviewer.18 Given the inconsistent presentation of data across the studies, and the heterogeneity of methods used to culture SARS-CoV-2, a quantitative synthesis of results could not be reliably performed. However, a narrative synthesis, including graphical representation of findings, is presented.
Results
Search results
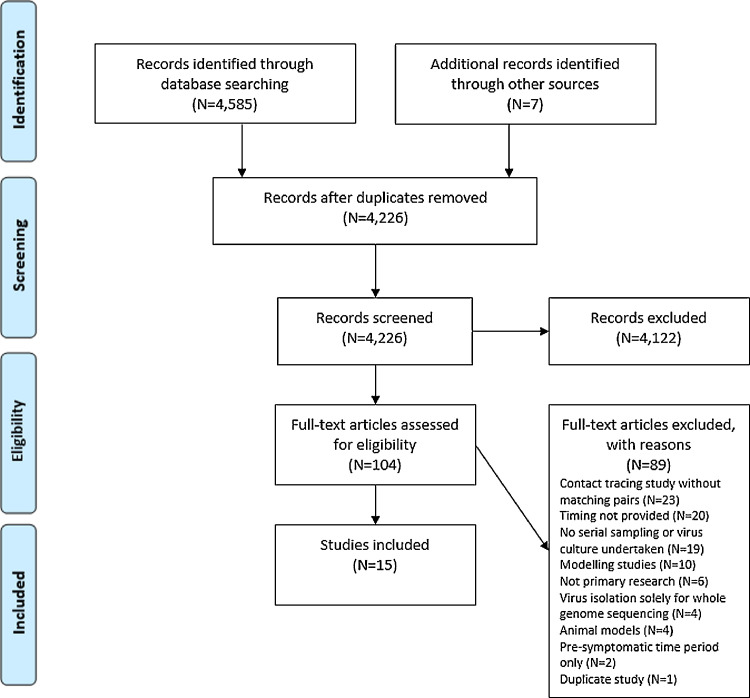
Of 4226 studies screened after duplicate removal, 15 were included (Fig. 1 ).14 , 21 – 34 In addition, unpublished data linked to 1 of the included studies28 were also included.35 Thirteen of these studies attempted to culture SARS-CoV-2 (Appendix 2),14 , 21 – 23 , 25 – 33 and 2 studies conducted contact tracing of case-contact pairs (Appendix 3).24 , 34 All 15 studies reported results in relation to time since symptom onset. No studies were found that reported duration of infectiousness in asymptomatic individuals (i.e. patients that never develop symptoms).
Fig. 1.
PRISMA diagram indicating the flow of inclusion of studies for review.
Population characteristics of included studies
Two studies each were conducted in the United States (US),21 , 28 Taiwan,24 , 30 the United Kingdom (UK)31 , 34 and Germany;25 , 33 while 1 study each was conducted in Switzerland,29 Australia,22 Canada,23 Spain,26 South Korea,27 Hong Kong,14 and the Netherlands.32
Of the 13 SARS-CoV-2 virus culture studies, 11 were case series14 , 21 – 23 , 26 – 29 , 31 – 33 and 2 were case reports.25 , 30 The contact tracing studies were both described as prospective, case-ascertained studies.24 , 34 Across all 13 virus culture studies, SARS-CoV-2 culture was attempted from samples collected from at least 808 COVID-19 patients, with SARS-CoV-2 isolated in at least 206 of these patients. A total of at least 1652 samples had SARS-CoV-2 culture attempted and at least 413 of these were positive. Five studies did not clearly provide information on the number of samples and/or patients included.22 , 23 , 26 , 30 , 33
Twelve studies included COVID-19 patients only after onset of symptoms.14 , 22 , 23 , 25 – 30 , 32 – 34 Three studies additionally included patients before they become symptomatic (i.e., pre-symptomatic).21 , 24 , 31 The earliest SARS-CoV-2 virus culture attempt was 13 days before symptom onset,31 whereas the latest SARS-CoV-2 culture attempt was 67 days after symptom onset.14 The contact tracing study by Cheng et al. included contacts who had their first day of exposure to the index case between day 4 before and up to day 26 after symptom onset in the index case. Contacts were subsequently followed for 14 days after the last exposure to the index case.24 The contact tracing study by Lopez Bernal et al. included household contacts who were exposed to the index case at any time between symptom onset and 14 days thereafter.34 Follow-up of the contacts was limited to 14 days from onset of symptoms in the index case.34
Asymptomatic patients were included in 3 studies; however, no information was provided from these studies that could be used to inform the potential duration of infectiousness of asymptomatic patients. Results from serial sampling and attempted virus culture in asymptomatic patients were not reported,21 , 31 and in 1 of the contact tracing studies it was reported that no onward transmission occurred from the 9 asymptomatic patients included, despite 91 close contacts being identified for these patients.24
Virus culture study findings
Days post symptom onset on which SARS-CoV-2 could be cultured
Fig. 2 presents, for each individual study, the first and last days, post symptom onset, on which SARS-CoV-2 culture was attempted, and the first and last days on which SARS-CoV-2 culture was successful. These values are also listed in Table 2 alongside additional details of the studies.
Fig. 2.
Days since symptom onset at which SARS-CoV-2 virus culture was attempted (blue, patterned) and successful virus culturing (grey) took place in each study, displayed in descending order of last day of successful virus culturing.
Table 2.
Details of numbers of samples and patients for SARS-CoV-2 cultivation (positive cultures/attempted) and days on which culturing attempted and successful.
| Numbers of samples and patients (culture positive/attempted) and associated sample incubation duration |
Days since symptom onset on which virus cultivation attempted |
Days since symptom onset on which virus culture positive |
|||||
|---|---|---|---|---|---|---|---|
| N culture positive samples/ N samples where culture attempted | N patients culture positive / N patients in whose samples culture was attempted | Duration (days) for which cultured cells incubated | First day attempted | Last day attempted (or further information) | Minimum | Maximum | |
| Studies including symptomatic period only | |||||||
| Basile | 56/234 (23.9%) | NR/195 | 5 | 0 | 29 | 0 | 18 |
| Bullard | 26/90 (28.9%) | NR | 4 | 0 | 21 | 0 | 7 |
| Decker | 2/2 (100%) | 1/1 (100.0%) | NR | Only attempted on days 18 and 21. | 18 | 21 | |
| Folgueira | 49/106 (46.2%) Mild: 18/50 Severe: 31/56 | NR/105 Mild: NR/50 Severe: NR/55 | 5 | 1 | At least day 32 | 1 | 10 (mild) 32 (severe) |
| Jeong | 3/9 (33.3%) (respiratory samples) | 2/5 (40.0%) | 4 | 8 | 30 | 11 | 15 |
| Kujawski | 13/17 (76.5%) respiratory samples) | 9/9 (100.0%) | NR | 0 | 8 | 0 | 8 |
| CDC unpublished (Midgley et al., Kujawski group) | Unclear from Figure | Unclear/14† | NR | 0 | 30 | 0 | 8 |
| L'Huillier | 12/23 (52.2%) | 12/23 (52.5%) | 6 | 0 | 5 | 1 | 5 |
| Liu | NR/NR | 1/1 (100%) | NR | NR | >18 days (exact days not reported) | NR | 18 |
| Perera | 16/68 (23.5%) | 16/35 (45.7%) | 6 days (72 h followed by an additional 72 h) | 0 | 67 | 0 | 7 |
| van Kampen | 62/690 (9.0%) | 23/129 (17.8%) | 7 days | 0 | 39 | 0 | 20 |
| Wölfel | 9/34 (26.5%) respiratory (43 samples in total)* | NR/9 | 6 days | 3 | 13 | 3 | 8 |
| Studies including pre-symptomatic and symptomatic period | |||||||
| Arons | 32/55 (58%) | 31/43 (72.1%) | NR | −7 | 13 | −6 | 13 (where all symptoms considered) 9 (fever, cough, shortness of breath only) |
| Singanayagam | 133/324 (41%) | 111/253 (44%) | Up to 14 days | −13 | 60 | −13 | 12 |
NR: Not reported.
Note that these figures are estimated from the number of markers displayed on Fig. 1d in Wölfel et al.; it is possible that the numbers are underestimated due to potential overlay of markers.
CDC unpublished data by Midgley et al. includes all patients included in Kujawski et al. plus 5 additional patients.
For 5 of the 13 studies (with culture attempted in at least 76 patients), the last day on which SARS-CoV-2 could be cultured occurred within the first 10 days since onset of symptoms.14 , 23 , 28 , 29 , 33 However, 1 of these studies, the sole study including only children, did not appear to attempt virus culture beyond day 5.29 For the most part, the clinical severity of patients was mild-to-moderate in these 5 studies, though Perera et al. noted that 3 of the 35 patients in the study had been critically ill, while the remainder had mild disease or had been asymptomatic.14
In 2 studies, (with culture attempted in 296 patients) the last day on which SARS-CoV-2 culture was reported occurred between 10 and 14 days post onset of symptoms.21 , 31 Singanayagam (with 133 SARS-CoV-2 culture positive samples from 253 patients) obtained only 2 culture positive samples between days 10 and 14 (1 on each of days 11 and 12). The authors also noted that more than half of the samples had been collected at least 8 days from symptom onset, and that 21% of these samples yielded SARS-CoV-2 in culture, the majority of which (25 of 27) was positive between 8 and 10 days after symptom onset. None of these late culture-positive samples included patients who were immunosuppressed or severely ill.31 In Arons et al., 1 of 53 samples (for which the date of symptom onset was known) from 43 patients yielded SARS-CoV-2 on day 13; SARS-CoV-2 was not observed in other samples collected after day 10, though no virus culture was attempted beyond day 13.21 There was also some uncertainty with regards to symptom onset in this study, with the longest time between the onset of typical COVID-19 symptoms (i.e., fever, cough or shortness of breath) and detection of SARS-CoV-2 culture positive recorded at 9 days, but 13 days after onset of less typical COVID-19 symptoms (i.e., chills, malaise, increased confusion, rhinorrhoea, nasal congestion, sore throat, myalgia, dizziness, headache, nausea or diarrhoea).
The remaining 6 studies, comprising 2 case reports and 4 case series (with culture attempted in 436 patients), reported successful SARS-CoV-2 culture between days 15 and 32 post symptom onset.22 , 25 – 27 , 30 , 32 Three of the 4 case series included samples from over 100 patients.22 , 26 , 32 Two of these studies included patients with severe disease.26 , 32 Folgueira et al. (N = 105 patients) presented the maximum day on which SARS-CoV-2 was cultured for patients with mild symptoms (max day: 10) and severe symptoms (max day: 32).26 Van Kampen et al. (N = 129 patients) reported SARS-CoV-2 culture results up until day 20. This study notably included only severely or critically ill patients, and included a large proportion of patients (23%) who were immunocompromised.32 The third large case series, Basile et al. (N = 195), reported that the latest day on which a SARS-CoV-2 was cultured was day 10 for all but 1 patient (maximum day: 18).22 The fourth case series, Jeong et al. (N = 9) reported positive cultures for 2 patients; the patient with SARS-CoV-2 cultured in saliva on day 15 was noted to be critically ill at the time of sampling.27 Both of the case reports which identified SARS-CoV-2 in culture beyond day 14 were of hospitalised patients with mild disease,25 , 30 although 1 of the case reports described an immunocompromised patient who had recently had a heart transplantation.25
Viral load and duration of infectiousness
The assays used to detect SARS-CoV-2 RNA could be regarded as semi-quantitative, using the cycle threshold (Ct) value as a surrogate marker of the SARS-CoV-2 viral load. For each study, those samples collected on the maximum day on which SARS-CoV-2 was isolated in culture are outlined in Appendix 2. Of note, lower Ct values indicate a higher viral load.
Of the 6 included studies that examined the relationship between viral load and culture of SARS-CoV-2, all 6 found an inverse correlation.14 , 22 , 23 , 31 – 33 Singanayagam et al. (N = 324 samples) estimated that the probability of successfully culturing SARS-CoV-2 from samples with a Ct value greater than 35 was 8.3% (95% CI: 2.8%–18.4%).31 Basile et al. (N = 234 samples) concluded that any clinical sample with a Ct value of ≥37 was not indicative of replicative (or potentially transmissible) virus.22 Bullard et al. (N = 90 samples) estimated that for every 1 unit increase in Ct value, the odds of culturing SARS-CoV-2 decreased by 32%.23 While Folgueira et al. reported culturing SARS-CoV-2 on day 32 post symptom onset in a patient with severe COVID-19, they observed a very low viral load, with a Ct value of 39.1.26 Hence it is probable (assuming the Ct value accurately reflects the amount of virus in the patient's respiratory tract) albeit not impossible, that the patient was not highly infectious at this particular time.
Summary estimates, including probability of culturing SARS-CoV-2
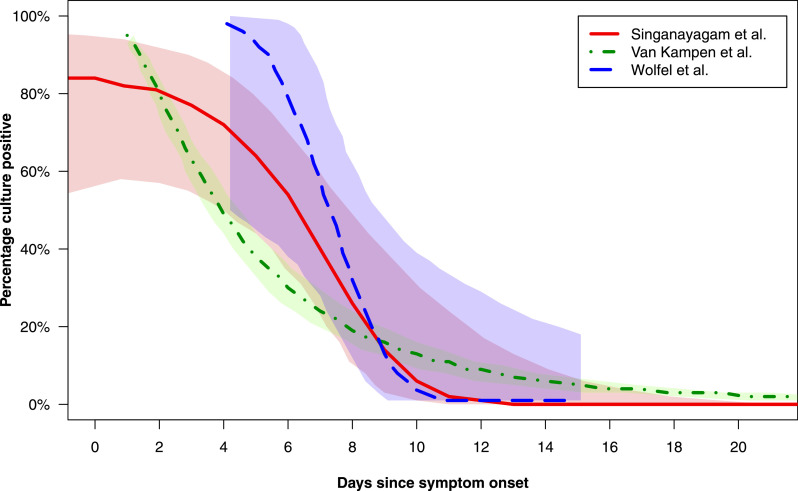
Three studies presented graphical estimates of the probability of culturing SARS-CoV-2 versus the number of days since symptom onset.31 – 33 These probability distributions are reproduced in Fig. 3 in an amended form (data for days 0 to 20 presented only) with overlay of curves to allow visual comparison of the probability results. Van Kampen et al.32 and Wölfel et al.33 both presented probit (dose-response) distributions for this outcome and estimated the day post symptom onset on which the probability of culturing SARS-CoV-2 culture fell below 5%. These results were as follows: van Kampen et al., 15.2 days (95% CI 13.4–17.2);32 Wölfel et al., 9.8 days (95% CI 8.5–21.8).33 Singanayagam et al.31 presented the results of a mixed effects logistic regression analysis (to account for the clustering of samples within patients); estimates of the percentage of samples with replicative SARS-CoV-2 were presented for each of days 7 to 15 post symptom onset, and are reproduced in Table 3 . At 10 days post symptom onset, the probability of a sample with replicative SARS-CoV-2 was 6% (95% CI 0.9–31.2); however, these data carry a high degree of uncertainty, as shown by the wide confidence intervals.
Fig. 3.
Probability curves presented in 3 studies (Singanayagam et al., van Kampen et al., Wölfel et al.) which attempted to model the probability of successful virus culture versus duration of days of symptoms.31, 32, 33Curves are presented with adaptation from the original presentation in the respective manuscripts in order to provide information for the first 20 days post symptom onset and to permit visual comparison of curves via overlay on a single plot. Lines (solid, dashed or dotted) depict estimated probability of positive virus culture versus days post symptom onset, while shaded areas depict 95% confidence intervals around these estimates.
Table 3.
Estimated percentage of samples with infectious SARS-CoV-2 for days 7–15 after symptom onset (adapted from Singanayagam et al.).
| Day post symptom onset | Estimated% culture positive (95% CI) | Observed number of samples culture positive | Observed number of samples tested | |
|---|---|---|---|---|
| 7 | 40.1 | (22.8 - 60.4) | 10 | 14 |
| 8 | 25.8 | (11.0 - 49.4) | 9 | 33 |
| 9 | 13.7 | (3.7 - 39.6) | 10 | 34 |
| 10 | 6.0 | (0.9 - 31.2) | 6 | 23 |
| 11 | 2.2 | (0.2 - 23.9) | 1 | 6 |
| 12 | 0.7 | (0.0 - 17.9) | 1 | 3 |
| 13 | 0.2 | (0.0 - 13.1) | 0 | 4 |
| 14 | 0.03 | (0.0 - 9.4) | 0 | 2 |
| 15 | 0.006 | (0.0 - 6.7) | 0 | 2 |
| Total: 37 | Total: 121 | |||
While unpublished, Midgley et al. additionally presented a Kaplan-Meier analysis of time to a negative culture SARS-CoV-2 following illness onset.35 This analysis represents an expanded analysis of the results reported by Kujawski et al.,35 including a greater number of patients from the same setting (n = 14 versus n = 12 (9 of whom had samples collected for virus culture)), though the total number of samples tested is not known. Midgley et al. reported that the probability of successful SARS-CoV-2 culture fell from 50% at day 4 after illness onset, to 20% at day 8, and approached 0% after day 9.
Analysis and interpretation of virus culture study findings
Data for maximum day that SARS-CoV-2 was cultured
Several jurisdictions suggest a period of isolation for COVID-19 patients of a minimum of 10 days since the onset of symptoms.11 , 15 , 36 As such, day 10 represents a relevant reference timepoint when considering the results of the SARS-CoV-2 culture studies included within this review.
While 8 of the 13 studies successfully cultured SARS-CoV-2 on days beyond day 10 post symptom onset, these results require contextualisation. Important aspects to consider include the number of samples and patients in whom this outcome occurred with respect to the overall number of samples and patients tested, and the patient populations included.
In total, these 8 studies included at least 337 SARS-CoV-2 culture-positive samples obtained from 732 patients.22 , 25 – 27 , 30 – 32 In 4 of the 8 studies, only 1 patient was SARS-CoV-2 culture-positive beyond day 10,21 , 22 , 25 , 30 and a further study identified only 2 samples (of 133 positive samples) yielding SARS-CoV-2 culture beyond day 10.31 These 5 studies included at least 223 SARS-CoV-2 culture positive samples from 493 patients in whom culture was attempted (Table 2).21 , 22 , 25 , 30 , 31 They reported a maximum of 6 patients who were SARS-CoV-2 culture-positive beyond day 10 (assuming conservatively that the 2 positive samples beyond day 10 in Singanayagam et al. arose from 2 unique patients).31
While these 5 studies in total reported a maximum of 6 patients from whom SARS-CoV-2 was cultured beyond day 10,21 , 22 , 25 , 30 , 31 the total number of patients with positive samples is unknown (Table 2). However, amongst these 5 studies, the proportion of patients from whom SARS-CoV-2 was cultured ranged from 44%31 to 100%25 , 30 (the latter representing case reports) (Table 2). Applying a worst-case scenario by adopting the lower proportion, i.e., assuming a positivity rate amongst patients of 44%, this results in an estimated minimum of 217 patients across these 5 studies. As such, 6 patients amongst 217 patients (maximum proportion of approximately 3%) are estimated to have had samples containing replicative SARS-CoV-2 in vitro beyond day 10 post symptom onset.
Within the remaining 3 studies (with culture attempted in 239 patients), wherein the latest day of successful culture exceeded day 10, it is less clear how many patients contributed to the samples resulting in this outcome.26 , 27 , 32 Folgueira et al.26 and Van Kampen et al.32 noted that patients from whom SARS-CoV-2 could be isolated were severely or critically ill at the time of sampling. The remaining study, Jeong et al., included 2 patients with this outcome, 1 of whom was critically ill at the time of sampling (day 15), and the other of whom was severely ill, but recovering at the time of sampling (day 11).27 As such, patients with severe and critical illness may represent a relevant subgroup within the data.
Probability estimate data
Two of the included studies (with culture attempted in 262 patients) which largely included patients with mild-to-moderate disease,31 , 33 and additional unpublished estimates associated with 1 of the included studies,28 , 35 estimated that the probability of culturing SARS-CoV-2 falls below 6% at day 10 (Fig. 3). A third study (with culture attempted in 129 patients) which modelled the probability of virus culturing versus time, suggested that the probability of a SARS-CoV-2 positive culture would fall below 5% at day 1532 (Fig. 3). However, this latter study uniquely included only hospitalised patients with severe or critical illness.
Singanayagam et al., the virus culture study in our review with the greatest number of patients (N = 253 patients in whom culture was attempted), and which predominantly included patients with mild-to-moderate disease, reported numerical estimates of the probability of a sample producing a SARS-CoV-2 positive culture at each day from day 7 onwards (Table 3).31 These data carry a high degree of uncertainty, as shown by wide confidence intervals, but showed a steep decline in the probability of culturing SARS-CoV-2 over time. For example, the estimated percentage of samples culture-positive on day 8 stood at 25.8% (95% CI 11.0–49.3) while the corresponding figure for day 10 was 6.0% (95% CI 0.9–31.2).
Contact tracing study findings
Two of the 15 included studies were contact tracing studies.24 , 34 Cheng et al.24 undertook a contact tracing study of 100 confirmed COVID-19 cases (ranging from asymptomatic to critical disease) in Taiwan. Amongst the 2761 close contacts of the 100 COVID-19 cases, there were 22 paired index-secondary cases. This study found that the overall secondary clinical attack rate was 0.7% (22 cases from 2761 contacts; 95% CI, 0.4%−1.0%). The attack rate was higher amongst the 1818 close contacts whose exposure to index cases started within 5 days of index case symptom onset (22 cases from 1818 contacts; 1.0%; 95% CI, 0.6%−1.6%) compared with those who were exposed later (0 cases from 852 contacts; 0%; 95% CI, 0%−0.4%). The 299 close contacts with exclusively pre-symptomatic exposure (up to 4 days before symptom onset in the index case) were also found to be at risk of infection (2 cases from 299 contacts; 0.7%; 95% CI, 0.2%−2.4%). The authors of this study concluded that there is a relatively short period of infectiousness of SARS-CoV-2, with higher transmissibility from 4 days before and up until 5 days after symptom onset. Based on the lack of onward transmission when close contacts were first exposed more than 5 days after symptom onset in the index case, the authors deduce that there is a lower transmission risk at the later stage of the disease.24
Lopez Bernal et al.34 undertook a contact tracing study of 269 lab-confirmed COVID-19 cases in the UK, who lived with at least 1 other person. Local health protection teams contacted household contacts of these patients daily for 14 days after onset of symptoms in the index case. In total, 472 household contacts were identified. Of these 472 contacts, 65 (13.8%) had a laboratory-confirmed diagnosis of COVID-19 using RT-PCR testing, and another 96 (20.3%) had probable COVID-19 based on the onset of symptoms of fever, anosmia (i.e., loss of smell) or respiratory symptoms. The remaining 311 contacts were classified as non-cases as they did not experience any symptoms suggestive of COVID-19 within 14 days of symptom onset in the index case. The household secondary attack rate was found to be 37% (95% CI, 31%−43%) including both laboratory-confirmed and probable secondary cases.
Of the 161 laboratory-confirmed or probable secondary cases, 41 had a point source exposure (i.e., a maximum exposure window of 1 day) to an index case and data available to allow analysis of timing of exposure. Laboratory-confirmed and probable secondary cases considered together (N = 41) were exposed a mean of 2.37 days (standard deviation (SD) 3.36) and a median of 1 day (interquartile range (IQR) 0–4) after symptom onset in the index case; exposure ranged from 0 days to the maximum follow up of 14 days. As identification of contacts was limited to 14 days from onset of symptoms in the index case, no information was provided regarding secondary cases in household contacts outside this exposure window. Similarly, of the contacts identified in the 14-day window, follow-up was truncated at 14 days from symptom onset in the index case, so no information was provided regarding symptoms that may have arisen after this follow-up period.
Restricting to lab-confirmed secondary cases only (N = 12), exposure occurred a mean of 1.33 days (SD 1.61) and a median of 1 day (IQR 0–1.25) after symptom onset in the index case, ranging from 0 to 5 days. In contrast, non-cases were exposed a mean of 2.71 days (SD 2.74) and a median of 2 days (IQR 0–5) after symptom onset in the index case, ranging from 0 to 9 days. No analysis was undertaken in this study to determine whether these time differences were statistically significant.34
Methodological quality of included studies
There are some important methodological limitations associated with all of the included studies. All 13 SARS-CoV-2 culturing studies were retrospective in nature and were either case series14 , 21 – 23 , 26, 27, 28, 29 , 31 – 33 or case reports,25 , 30 where patients do not appear to have been selected or sampled systematically. However, the contact tracing study by Cheng et al. was generally well-conducted due to its prospective nature, the systematic pairing of index cases and close contacts over time, and the follow up of contacts for 14 days after last exposure to the index case to assess for symptom onset.24 Hence, the majority of evidence in this review comes from studies of low quality design. Importantly, it would appear that most studies did not specifically set out to identify the temporal relationship between time post symptom onset and presence of infectious virus, but rather reported culture positivity based on samples collected for other purposes.23 , 26 – 29 , 31 , 33 Thereby, some uncertainty remains regarding the true duration of patient infectiousness, as the upper limit of outliers may not have been fully clarified. Recall bias is another issue which may have important implications on outcomes across all studies, as patients may not have been able to accurately determine when symptoms began, particularly if they were quite mild and non-specific.37
There are some concerns regarding the inconsistent approaches used for cell culture amongst studies, and the interpretation of findings by the study authors. If, as noted in some studies,14 , 29 a second passage of the sample is required to observe a cytopathic effect (CPE), and thereby to confirm culture positivity, this suggests a very low replicative viral load in the original sample and may reflect that the individual from whom the sample was collected is unlikely to have been infectious at the time of sampling.38 Furthermore, given that longer periods for a CPE to develop are indicative of a lower viral replicative titre in the original sample,16 where studies incubate the cell lines for prolonged periods, and only observe a CPE towards the end of this period, this may also lead to overestimation of the clinical significance of the findings.31
A de novo tool was also used to quality appraise case reports and case series.39 This tool identified additional concerns regarding the limited reporting of demographic information,22 , 23 , 30 , 31 , 33 the unclear criteria for selection of cases,14 , 23 , 26 , 27 , 32 , 33 the non-consecutive inclusion of cases,14 , 22 , 23 , 26 , 27 , 32 , 33 uncertainty regarding the appropriateness of statistical analysis14 , 27 , 31 – 33 and the inconsistent use of RT-PCR testing for diagnosis.34 Four of the studies included in this review are published as pre-prints, so have not yet been formally peer-reviewed, raising additional concerns about overall quality and the potential for results to change prior to formal publication.22 , 26 , 32 , 34
Discussion
Thirteen SARS-CoV-2 virus culture studies of various quality and size, and 2 large contact tracing studies, were included in this rapid review. The evidence to date from virus culture studies would appear to suggest that COVID-19 patients with mild-to-moderate illness are highly unlikely to be infectious beyond 10 days from symptom onset. Evidence from large contact tracing studies24 , 34 appears to support this finding. However, evidence from a limited number of studies indicates that patients with severe-to-critical illness, and or those who are immunocompromised, may be infectious for a prolonged period, possibly for 20 days or more.25 , 26 , 32 Virus culture studies suggested a steady decline in the probability of culturing SARS-CoV-2 in the first 10 days following symptom onset; the largest included study estimated the probability as falling from 40% on day 7 to 26% on day 8, 14% on day 9 and 6% by day 10. However, these results were subject to considerable uncertainty, and the clinical significance of these findings is unknown given potentially prolonged incubation of cultured cells.31 Notably, there are important methodological limitations associated with all of the included studies. The number of studies for consideration remains few in number and sample sizes are often small; hence it is essential that the findings from these studies are interpreted in light of these caveats. Furthermore, virus culture studies determine if a sample contains replicative competent SARS-CoV-2; while a positive virus culture indicates potential infectiousness of the individual from whom the sample was collected, risk of transmission (i.e., clinical infectivity) is also influenced by the viral load, local immune response to SARS-CoV-2, clinical symptoms and environmental factors, and behaviour of the infected individual and their contacts.
One of the notable findings from 2 of the included studies was that patients who were found to be SARS-CoV-2 culture-positive for a prolonged period were immunosuppressed.25 , 32 In particular, 1 of the case reports describes a mild clinical course of COVID-19 in a heart transplant recipient on immunosuppressant therapies (mycophenolate mofetil, ciclosporin and prednisone) from whom SARS-CoV-2 was cultured 21 days after symptom onset.25 Although the evidence pertaining to prolonged infectiousness in immunosuppressed populations is very limited, these findings do raise important research and policy questions. Immunosuppressant therapies have been observed to delay the clearance of SARS-CoV-2 viral RNA in previous studies.40 , 41 However, some of these agents are also being investigated as treatments for severe and critical COVID-19 to modulate the hyperinflammatory response (e.g., tocilizumab).42 , 43 Little is known regarding the presentation and course of disease in patients who are immunocompromised; it is unknown whether all patients who are in an immunocompromised state necessarily present with severe COVID-19 disease, or how important the degree of immunosuppression is for clearance of the virus.43 More evidence is required to understand the relationship between immunosuppression and the illness severity and duration of infectiousness of SARS-CoV-2. However, in these individuals it is very likely that any molecular investigations would indicate a significant SARS-CoV-2 viral load.
Only 1 study provided information on SARS-CoV-2 culture specifically in children.29 However, as virus culture was only attempted once per patient, to a maximum of 5 days post symptom onset, this study provides insufficient information to inform policy decisions surrounding isolation requirements specifically for children testing positive for SARS-CoV-2 RNA. Children have generally been under-represented in COVID-19 studies to-date, although this may be a function of testing practices which initially typically prioritised those with more severe symptoms, healthcare workers and those residing in long term care settings. Given reports of milder symptoms in children, during the earlier stages of the pandemic they were less likely to be tested and diagnosed.44 As children return to school en masse, increased testing and cases may occur in these settings, thus more data may emerge regarding the potential duration of infectiousness in this population.
The two included contact tracing studies both provide epidemiological evidence to support the findings from the virus culture studies.24 , 34 Notably, there were substantial differences in the observed secondary attack rates between the 2 contact tracing studies (0.7% vs. 37%). These differences are likely to be explained by the different exposure settings at play; the study by Lopez Bernal et al. focused on transmission amongst household contacts,34 a setting in which SARS-CoV-2 is known to be highly transmissible,45 whereas Cheng et al. included all possible modes of transmission.24 Differences in case or contact definitions and general approaches within the different public health systems may also explain this substantial disparity.24 , 34 Despite these differences, both studies reported that when close contacts were first exposed greater than 5 days after symptom onset in the index case, there was no evidence of laboratory-confirmed onward transmission of SARS-CoV-2.
The findings from our rapid review are largely in agreement with 4 previous reviews conducted in this general area.3 , 46 – 48 All of these reviews concluded that infectiousness generally declines 7–10 days after symptom onset, and point to uncommon outlier cases where this duration is exceeded. Another common finding across 3 of these reviews was the prolonged duration of SARS-CoV-2 RNA detection, sometimes for 2–3 months after onset of symptoms, along with cases of repeat SARS-CoV-2 RNA detection after a patient has clinically recovered.3 , 46 , 48 Hence, patients are unlikely to be infectious for the entire duration of viral RNA detection as the presence of viral RNA may not represent transmissible or replication-competent virus.3 Our rapid review is the first to attempt to quantify the proportion of COVID-19 patients that are potentially infectious beyond day 10 post symptom onset, and draws on more recent evidence, including both virus culture and contacting tracing studies. Hence, our rapid review adds greater clarity to this complex issue.
Evidence from our rapid review has directly informed the decision to reduce the duration of isolation in individuals who test positive for SARS-CoV-2 RNA in Ireland, from 14 to 10 days (with the additional clinical requirement of being fever-free for 5 days).49 , 50 Though guidance still varies on this topic globally, several public health agencies have updated their recommendations in recent times in light of the growing evidence of this rapid reduction in infectiousness post symptom onset, and a greater understanding of the prolonged shedding of non-transmissible SARS-CoV-2 RNA.11 , 15 , 36 , 51
The World Health Organization (WHO) currently recommends that a COVID-19 patient (regardless of symptom severity) can end isolation, without requiring re-testing, 10 days after symptom onset and at least 3 additional days without symptoms.11 Previously, WHO recommendations published in January 2020 stated that COVID-19 patients could only be released from isolation if they were clinically recovered and received 2 negative RT-PCR results on sequential samples taken at least 24 h apart.11 In contrast, the European Centre for Disease Prevention and Control (ECDC) issued a recommendation in April 2020, in the context of widespread community transmission, that COVID-19 patients in the community with mild or asymptomatic illness can end isolation 8 days after the onset of symptoms.51 This recommendation is conditional on resolution of fever and clinical improvement of other symptoms for at least 3 days.51 Previous recommendations issued by ECDC in March 2020 advised that mild cases of COVID-19 could be discharged from hospital, if clinically appropriate, with a further 14 days of isolation post-discharge.52 The Centers for Disease Control and Prevention (CDC) in the United States (US) recommended in July 2020 that isolation can generally be discontinued (in patients with mild-to-moderate illness) 10 days after symptom onset and resolution of fever for at least 24 h (without the use of fever-reducing medications) and with improvement of other symptoms.53 This may be contrasted with the previously recommended period of isolation of 14 days. However, the CDC also now recommends that isolation may be required for up to 20 days after symptom onset in a limited number of severe cases, and recommend that for patients who are severely immunocompromised, a test-based strategy (using RT-PCR) may be considered to discontinue isolation, in consultation with infectious diseases experts.15 For all other patients, the CDC no longer recommend a test-based strategy to discontinue isolation.15 The CDC also points to the lack of equivalent data from children and infants and the need for more data regarding immunocompromised individuals. As new evidence emerges for these subgroups of interest, recommendations regarding the duration of isolation in these populations may change.
It is important to note that the majority of evidence in this review is derived from studies of low quality design, where determining the duration of infectiousness may not have been the primary objective and SARS-CoV-2 culturing methods were not consistent. Furthermore, the inconsistent reporting of data precluded any meaningful quantitative synthesis. Notably, virus culture studies are inherently fraught with challenges,54 but the requirement for biosafety level 3 (BSL-3) facilities is a particular barrier to research. Further work is required to develop standardised methods to optimise culturing of SARS-CoV-2.
Future research should ideally be prospective in design, following a large cohort of COVID-19 patients with a broad spectrum of disease and inclusive of subgroups of interest (such as children, asymptomatic and immunosuppressed patients). Patients should be regularly and systematically sampled and SARS-CoV-2 virus culturing attempted using standardised methods, until virus can no longer be successfully isolated.47 The time between sample inoculation and the detection of viral growth should be reported for all studies, as this is a reflection of the viral load of infectious virus in the original sample. The findings of such virus culture studies should be supplemented with epidemiological evidence of onward transmission at different time points, as derived from well-conducted, large and prospective contact tracing studies. A core outcome set should be measured and reported, stratified by different subgroups, thus enabling better synthesis by systematic reviewers to better facilitate interpretation and decision-making by policy-makers.55
Conclusion
The evidence to date suggests that COVID-19 patients with mild-to-moderate illness are highly unlikely to be infectious beyond 10 days from symptom onset. However, evidence from a very limited number of studies indicates that patients with severe-to-critical illness or those who are immunocompromised, may shed infectious virus for longer than 10 days. Future research is required to confirm these findings and to provide information on the duration of infectiousness in subgroups such as children, and asymptomatic and immunosuppressed patients.
Acknowledgments
Acknowledgments
The authors would like to thank Executive Assistant Debra Spillane (HIQA) and Information Specialist Andrew Simpson (RCSI), and acknowledge the support of the Health Technology Assessment, the Health Information and Standards, and the Communications & Stakeholder Engagement Directorates at HIQA.
Funding Statement
This research was funded in part by the Health Research Board under grant no. HRB-CICER-2016-1871.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.10.009.
Appendix. Supplementary materials
References
- 1.John Hopkins University & Medicine. COVID-19 Dashboard 2020 [updated 5 Sep 2020; cited 2020 5 Sep]. Available from: https://coronavirus.jhu.edu/map.html.
- 2.Tu Y.-.F., Chien C.-.S., Yarmishyn A.A., Lin Y.-.Y., Luo Y.-.H., Lin Y.-.T. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci. 2020;21(7):2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infection. 2020;81(3):357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabi F.A., Al Zoubi M.S., Kasasbeh G.A., Salameh D.M., Al-Nasser A.D. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9(3):231. doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey M., Griffin J., McAloon C.G., Byrne A.W., Madden J.M., McEvoy D. medRxiv; 2020. Pre-symptomatic transmission of SARS-CoV-2 infection: a secondary analysis using published data. 20094870, 05.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O Murchu E., Byrne P., Walsh K.A., Carty P.G., Maire C., De Gascun C.F. Immune response following infection with SARS-CoV-2 and other coronaviruses: a rapid review. Rev Med Virol. 2020 doi: 10.1002/rmv.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2):taaa021. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. Quarantine and isolation 2017 [updated 29 Sep 2017; cited 2020 27 Aug]. Available from: https://www.cdc.gov/quarantine/index.html.
- 9.French Ministry of National Education Youth and Sport. [Suspicion or confirmation of a covid-19 case: what to do] 2020 [updated 1 September 2020; cited 2020 19 September]. Available from: https://www.education.gouv.fr/suspicion-ou-confirmation-de-cas-covid-19-ce-qu-il-faut-faire-305730.
- 10.Government of Canada. Coronavirus disease (COVID-19): symptoms and treatment 2020 2020 [updated 14 Aug 2020; cited 2020 19 Sep]. Available from: https://www.canada.ca/en/public-health/services/diseases/2019-novel-coronavirus-infection/symptoms.html#s.
- 11.World Health Organization. Criteria for releasing COVID-19 patients from isolation 2020 [updated 17 Jun 2020; cited 2020 27 Aug]. Available from: https://www.who.int/publications/i/item/criteria-for-releasing-covid-19-patients-from-isolation.
- 12.Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20(1):49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Centre for Disease Control and Prevention. COVID-19 situation update worldwide, as of 26 August 2020 2020 [updated 26 Aug 2020; cited 2020 27 Aug]. Available from: https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases.
- 14.Perera R., Tso E., Tsang O.T.Y., Tsang D.N.C., Fung K., Leung Y.W.Y. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26(11) doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. Duration of isolation and precautions for adults with COVID-19 2020 [updated 16 Aug 2020; cited 2020 27 Aug]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html.
- 16.Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Health Information and Quality Authority. Protocol for evidence synthesis support - COVID-19. https://www.hiqa.ie/sites/default/files/2020-04/Protocol-for-evidence-synthesis-support_1-4-COVID-19.pdf: HIQA, 2020.
- 18.Garritty C., Gartlehner G., Kamel C., King V.J., Nussbaumer-Streit B., Stevens A. Interim guidance from the Cochrane rapid reviews methods group. Cochrane Rapid Rev. 2020 doi: 10.1016/j.jclinepi.2020.10.007. , March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veritas Health Innovation. Covidence systematic review software Melbourne, Australia. 2020 [Available from: www.covidence.org.
- 20.Rohtagi A. WebPlotDigitizer 4.3 2020 [updated 7 July 2020; cited 2020 22 Sep]. Available from: https://automeris.io/WebPlotDigitizer.
- 21.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basile K., McPhie K., Carter I., Alderson S., Rahman H., Donovan L. medRxiv; 2020. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. 20153981, 07.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. : ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng H.Y., Jian S.W., Liu D.P., Ng T.C., Huang W.T., Lin H.H. Contact tracing assessment of COVID-19 transmission dynamics in taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med. 2020;180(9):1156–1163. doi: 10.1001/jamainternmed.2020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decker A., Welzel M., Laubner K., Grundmann S., Kochs G., Panning M. Prolonged SARS-CoV-2 shedding and mild course of COVID-19 in a patient after recent heart transplantation. Am J Transplant. 2020;00:1–7. doi: 10.1111/ajt.16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folgueira M.D., Luczkowiak J., Lasala F., Perez-Rivilla A., Delgado R. medRxiv; 2020. Persistent SARS-CoV-2 replication in severe COVID-19. 20127837, 06.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong H.W., Kim S.M., Kim H.S., Kim Y.I., Kim J.H., Cho J.Y. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.07.020. : S1198-743X(20)30427-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kujawski S.A., Wong K.K., Collins J.P., Epstein L., Killerby M.E., Midgley C.M. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.L'Huillier A.G., Torriani G., Pigny F., Kaiser L., Eckerle I. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg Infect Dis. 2020;26(10):2494–2497. doi: 10.3201/eid2610.202403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W.-.D., Chang S.-.Y., Wang J.-.T., Tsai M.-.J., Hung C.-.C., Hsu C.-.L. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England. Eurosurveillance. 2020;25(32) doi: 10.2807/1560-7917.ES.2020.25.32.2001483. January to May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N. Vol. 2020. medRxiv; 2020. (Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants). 20125310, 06.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 34.Lopez Bernal J., Panagiotopoulos N., Byers C., Garcia Vilaplana T., Boddington N.L., Zhang X. Vol. 2020. medRxiv; 2020. (Transmission dynamics of COVID-19 in household and community settings in the United Kingdom). 20177188, 08.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Midgley Claire. 2020. [Personal Communication] CDC Unpublished data of 14 COVID-19 patients. Available from: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html. [Google Scholar]
- 36.UK Government. Guidance for stepdown of infection control precautions and discharging COVID-19 patients 2020 [updated 30 Jul 2020. Available from: https://www.gov.uk/government/publications/covid-19-guidance-for-stepdown-of-infection-control-precautions-within-hospitals-and-discharging-covid-19-patients-from-hospital-to-home-settings/guidance-for-stepdown-of-infection-control-precautions-and-discharging-covid-19-patients.
- 37.G-u Kim, M-J Kim, Ra S.H., Lee J., Bae S., Jung J. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin Microbiol Infection. 2020;26(7):948.e1–948.e3. doi: 10.1016/j.cmi.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todd D., Connor T., Creelan J.L., Borghmans B., Calvert V.M., McNulty M. Effect of multiple cell culture passages on the biological behaviour of chicken anaemia virus. Avian Pathol. 1998;27(1):74–79. doi: 10.1080/03079459808419277. [DOI] [PubMed] [Google Scholar]
- 39.Health Information and Quality Authority. Protocol for evidence synthesis support - COVID-19 2020 [updated 19 Aug; cited 2020 27 Aug]. Available from: https://www.hiqa.ie/reports-and-publications/health-technology-assessment/protocol-evidence-synthesis-support-covid-19#:∼:text=This%20protocol%20outlines%20HIQA's%20process,their%20response%20to%20COVID%2D19.
- 40.Ling Y., Xu S.-.B., Lin Y.-.X., Tian D., Zhu Z.-.Q., Dai F.-.H. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antwi-Amoabeng D., Kanji Z., Ford B., Beutler B.D., Riddle M.S., Siddiqui F. Clinical outcomes in COVID-19 patients treated with tocilizumab: an individual patient data systematic review. J. Med. Virol. 2020;92:2516–2522. doi: 10.1002/jmv.26038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thng Z.X., De Smet M.D., Lee C.S., Gupta V., Smith J.R., McCluskey P.J. COVID-19 and immunosuppression: a review of current clinical experiences and implications for ophthalmology patients taking immunosuppressive drugs. Br J Ophthalmol. 2020 doi: 10.1136/bjophthalmol-2020-316586. bjophthalmol-2020-316586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludvigsson J.F. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020;109(6):1088–1095. doi: 10.1111/apa.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jing Q.-.L., Liu M.-.J., Zhang Z.-.B., Fang L-Q, Yuan J., Zhang A.-.R. Household secondary attack rate of COVID-19 and associated determinants in Guangzhou, China: a retrospective cohort study. Lancet Infectious Dis. 2020;20(10):1141–1150. doi: 10.1016/S1473-3099(20)30471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. Vol. 2020. medRxiv; 2020. (SARS-CoV-2, SARS-CoV-1 and MERS-CoV viral load dynamics, duration of viral shedding and infectiousness: a living systematic review and meta-analysis). 20162107, 07.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jefferson T., Spencer E., Brassey J., Heneghan C. Vol. 2020. medRxiv; 2020. (Viral cultures for COVID-19 infectivity assessment. Systematic review). 20167932, 08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park M., Pawliuk C., Nguyen T., Griffitt A., Dix-Cooper L., Fourik N. Determining the period of communicability of SARS-CoV-2: a rapid review of the literature. Vol. 2020. medRxiv; 2020. 20163873, 07.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Health Service Executive. Self-isolation: managing coronavirus at home 2020 [updated 14 Sep 2020; cited 2020 20 Sep]. Available from: https://www2.hse.ie/conditions/coronavirus/managing-coronavirus-at-home/self-isolation.html.
- 50.Health Information and Quality Authority. HIQA publishes an evidence summary on the duration of infectiousness of COVID-19 patients 2020 [updated 15 Sep 2020; cited 2020 20 Sep]. Available from: https://www.hiqa.ie/hiqa-news-updates/hiqa-publishes-evidence-summary-duration-infectiousness-covid-19-patients.
- 51.European Centre for Disease Control and Prevention. Guidance for discharge and ending isolation in the context of widespread community transmission of COVID-19 – first update 2020 [updated 8 April 2020; cited 2020 1 Sep]. Available from: https://www.ecdc.europa.eu/en/publications-data/covid-19-guidance-discharge-and-ending-isolation.
- 52.European Centre for Disease Control and Prevention. Discharge criteria for confirmed COVID-19 cases – When is it safe to discharge COVID-19 cases from the hospital or end home isolation? 2020 [updated 10 Mar 2020; cited 2020 20 Sep]. Available from: https://www.ecdc.europa.eu/en/publications-data/novel-coronavirus-sars-cov-2-discharge-criteria-confirmed-covid-19-cases.
- 53.Centers for Disease Control and Prevention. Discontinuation of Isolation for Persons with COVID-19 Not in Healthcare Settings 2020 [updated 20 Jul 2020; cited 2020 2 Sep]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-in-home-patients.html.
- 54.Pyrc K., Sims A.C., Dijkman R., Jebbink M., Long C., Deming D. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J Virol. 2010;84(21):11255–11263. doi: 10.1128/JVI.00947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clarke M., Williamson P.R. Core outcome sets and systematic reviews. Syst Rev. 2016;5:11. doi: 10.1186/s13643-016-0188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.