Abstract
Background
High-affinity nitrate transporter 2 (NRT2) genes have been implicated in nitrate absorption and remobilization under nitrogen (N) starvation stress in many plant species, yet little is known about this gene family respond to various stresses often occurs in the production of rapeseed (Brassica napus L.).
Results
This report details identification of 17 NRT2 gene family members in rapeseed, as well as, assessment of their expression profiles using RNA-seq analysis and qRT-PCR assays. In this study, all BnNRT2.1 members, BnNRT2.2a and BnNRT2.4a were specifically expressed in root tissues, while BnNRT2.7a and BnNRT2.7b were mainly expressed in aerial parts, including as the predominantly expressed NRT2 genes detected in seeds. This pattern of shoot NRT expression, along with homology to an Arabidopsis NRT expressed in seeds, strongly suggests that both BnNRT2.7 genes play roles in seed nitrate accumulation. Another rapeseed NRT, BnNRT2.5 s, exhibited intermediate expression, with transcripts detected in both shoot and root tissues. Functionality of BnNRT2s genes was further outlined by testing for adaptive responses in expression to exposure to a series of environmental stresses, including N, phosphorus (P) or potassium (K) deficiency, waterlogging and drought. In these tests, most NRT2 gene members were up-regulated by N starvation and restricted by the other stresses tested herein. In contrast to this overall trend, transcription of BnNRT2.1a was up-regulated under waterlogging and K deficiency stress, and BnNRT2.5 s was up-regulated in roots subjected to waterlogging. Furthermore, the mRNA levels of BnNRT2.7 s were enhanced under both waterlogging stress and P or K deficiency conditions. These results suggest that these three BnNRT2 genes might participate in crosstalk among different stress response pathways.
Conclusions
The results presented here outline a diverse set of NRT2 genes present in the rapeseed genome that collectively carry out specific functions throughout rapeseed development, while also responding not just to N deficiency, but also to several other stresses. Targeting of individual BnNRT2 members that coordinate rapeseed nitrate uptake and transport in response to cues from multiple stress response pathways could significantly expand the genetic resources available for improving rapeseed resistance to environmental stresses.
Keywords: High-affinity nitrate transporter, NRT2, Rapeseed, Various stresses
Background
Nitrogen (N) is an essential plant macronutrient that is often limited in availability, which leads to millions of metric tons of nitrogenous fertilizer being applied at significant cost to agricultural fields worldwide [1, 2]. Furthermore, excessive application of N fertilizer degrades the environment through pollution of water and air, where it also contributes to global warming. Therefore, reducing the requirements for N fertilizer through improvements in crop N use efficiency (NUE) is a worthwhile research objectives for crop breeders and plant scientists [3].
For most plant species, uptake, assimilation, and translocation processes are vital early steps for successful acquisition and utilization of N from soil sources. Nitrogen assimilation requires the reduction of nitrate (NO3−) to ammonium, followed by incorporation of ammonium into amino acids, which depends on the acquisition and transport of inorganic N sources and transfer of amino acids and proteins between multiple plant organs [4]. Since NO3− is the most abundant inorganic N in soils [1], NO3− uptake and translocation, therefore, play crucial roles in NUE.
The composition of N in the soil is affected by many factors, including mositure, temperature, wind, soil type and pH, which has led to large variations in nitrate concentrations among soil environments [3, 5]. In order to cope with the heterogeneity of NO3− in the soil, plants have evolved two NO3− absorption systems, namely a low-affinity transport system (LATS), and a high-affinity transport system (HATS), which are mediated by nitrate transporter 1 (NRT1) and nitrate transporter 2 (NRT2), respectively [6]. Overall, NRT2 activity supplants NRT1 activity as N availability diminishes. In one set of comparable results, expression in Xenopus oocytes has led to Km value estimates of 5.9 mM for the Arabidopsis LATS transporter AtNRT1.2 and 30 μM for the barley HATS transporter HvNRT2.1 [7, 8]. This nearly 200 fold variation in nitrate affinity suggests that HATS NRT2 may be key targets in efforts to improve NUE, especially under low N availability conditions.
Seven high-affinity nitrate transporters (NRT2.1-NRT2.7) have been identified in Arabidopsis thaliana (L.) [6, 9]. To date, AtNRT2.1, AtNRT2.2 and AtNRT2.4 have been implicated in root NO3− uptake [10, 11]. Interestingly, AtNRT2.1 plays a predominant role in NO3− absorption, while AtNRT2.2 is capable of compensating for lost NO3 uptake when AtNRT2.1 is incapacitated [12]. For its part, AtNRT2.4 not only acts in NO3− absorption, but also contributes to loading NO3− in shoot phloem vessels [11]. Another high-affinity nitrate transporter in Arabidopsis, AtNRT2.5, is up-regulated under N limiting conditions in both shoots and roots of mature plants, where it supports efficient uptake of nitrate, as well as, nitrate loading into the phloem during nitrate remobilization [13]. AtNRT2.7 has been reported to localize on the vacuolar membrane, it plays specifics roles in seed nitrate accumulation [14]. Neither NRT2.3 nor NRT2.6 have been implicated in NO3− uptake or translocation. Similar activities by homologues of Arabidopsis NRT2 genes in crop species have been partially confirmed, though efforts have been limited. Work with rice has revealed five NRT2 genes, including OsNRT2.3a, which was found to play a key role in root to shoot nitrate translocation under N limiting conditions [15].
Rapeseed (Brassica napus L.) is a globally important oil crop that is cultivated widely for production of vegetable oil, biofuels and industrial products, such as lubricants and surfactants [16, 17]. As an allotetraploid, the Brassica napus (genome AnAnCnCn) is a product of genome doubling in a hybridization between Brassica oleracea (L.) (genome CoCo) and Brassica rapa (L.) (genome ArAr) ~ 7500 years ago [18]. This suggests that the Brassica napus genome might harbor multiple homologs for individual NRT2 members in Arabidopsis [18]. Rapeseed requires substantial amounts of N, and, as a result, is extremely sensitive to changes in soil N availability [19]. Reducing the input of N fertilizer while maintaining yields of rapeseed will, therefore, likely entail improving the NUE of rapeseed. Furthermore, rapeseed is sensitive to various environmental stresses, such as macronutrient deficiencies, waterlogging and drought stresses, these will lead to the nutrients imbanlance, influence the N metabolism or other metabolic pathways, finally suppress the growth and yield of rapeseed [20–25]. It’s necessary to understand the adaptive patterns of NRT2 family genes in response to these stresses which commonly appeared in the production of rapeseed. Thus, the aim of this study is (i) to identify all NRT2s family members in the Brassica napus genome, (ii) analyze the genomic and transcriptomic characteristics of BnNRT2s family members, including exon-intron organization, conserved motifs, phylogeny, gene duplication, tissue specific expression, and their responsive profiles under various stresses, and (iii) determine core gene members contributing to NO3− absorption and transportation under N limiting conditions or other environmental stresses. Overall, this report reveals the characteristics of NRT2 genes in rapeseed and also provides candidate gene resources for improving NUE and reducing N fertilizer applications to rapeseed fields.
Results
Genome-wide identification and bioinformatic analysis of rapeseed NRT2 family genes
To identify NRT2 genes members in rapeseed, the amino acid sequences of AtNRT2s were used to perform a BLAST search of the Brassica Database (BRAD), which returned 17 genes identified as NRT2 family members in rapeseed, as listed in Table 1. These BnNRT2 genes were labelled according to their top Arabidopsis matches as BnNRT2.1 s (7 genes), BnNRT2.2 s (2 genes), BnNRT2.3 s (2 genes), BnNRT2.4 s (2 genes), BnNRT2.5 s (2 genes) and BnNRT2.7 s (2 genes). No homologues of AtNRT2.6 were identified in the rapeseed genome. According to the BRAD physical map, the identified BnNRT2 gene members mapped onto 9 chromosomes (chrA02, chrA06, chrA08, chrA09, chrA10; chrC02, chrC03, chrC08, chrC09), with BnNRT2 genes distributed almost equally in the A and C genomes (Fig. S1). For example, BnNRT2.5a and BnNRT2.5b mapped to chrC03 and chrA08, respectively, while BnNRT2.7a and BnNRT2.7b were respectively mapped to chrC02 and chrA02 (Fig. S1). This provide undeniable evidence for the presence of more NRT2s gene members in rapeseed than in Arabidopsis, which is due to genome multiplication in the hybridization between B.oleracea (genome CoCo) and B. rapa (genome ArAr). Gene duplication analysis in rapeseed indeed found that of 14 identified BnNRT2 duplicated pairs, 10 gene pairs were derived from whole-genome duplication, and 4 genes pairs were derived from transposed duplication (Table S1).
Table 1.
Molecular characterization of the NRT2 family proteins in rapeseed
| No. | Gene name | Gene ID | CDS (bp) |
Amino acids (aa) |
MY | PI | Subcellular localization |
|---|---|---|---|---|---|---|---|
| 1 | BnNRT2.1a | BnaC08g02430D | 1395 | 464 | 50,555.41 | 8.79 | PM |
| 2 | BnNRT2.1b | BnaAnng40490D | 1062 | 353 | 38,542.64 | 9.16 | PM |
| 3 | BnNRT2.1c | BnaAnng35750D | 1062 | 353 | 38,491.53 | 9.14 | PM |
| 4 | BnNRT2.1d | BnaA06g04560D | 1383 | 460 | 50,267.16 | 8.98 | PM |
| 5 | BnNRT2.1e | BnaA06g04570D | 1395 | 464 | 50,665.68 | 8.94 | PM |
| 6 | BnNRT2.1f | BnaA09g49050D | 1266 | 421 | 45,571.37 | 9.14 | PM |
| 7 | BnNRT2.1 g | BnaC08g43380D | 1392 | 463 | 50,319.18 | 8.86 | PM |
| 8 | BnNRT2.2a | BnaC08g43370D | 1509 | 502 | 54,398.46 | 8.91 | PM |
| 9 | BnNRT2.2b | BnaA09g49040D | 1521 | 506 | 54,827.93 | 9.02 | PM |
| 10 | BnNRT2.3a | BnaC09g54030D | 1443 | 480 | 52,018.08 | 8.49 | PM |
| 11 | BnNRT2.3b | BnaA10g13570D | 1443 | 480 | 52,013.20 | 9.01 | PM |
| 12 | BnNRT2.4a | BnaAnng28170D | 1182 | 393 | 43,761.98 | 8.96 | C (SP) |
| 13 | BnNRT2.4b | BnaC09g35990D | 1371 | 456 | 50,062.26 | 9.33 | M |
| 14 | BnNRT2.5a | BnaC03g56990D | 1497 | 498 | 54,128.70 | 9.11 | PM |
| 15 | BnNRT2.5b | BnaA08g24500D | 1500 | 499 | 54,174.72 | 9.01 | PM |
| 16 | BnNRT2.7a | BnaC02g05530D | 1509 | 502 | 53,454.55 | 7.61 | V |
| 17 | BnNRT2.7b | BnaA02g02200D | 1455 | 484 | 52,062.10 | 7.54 | V |
PM plasma membrane, C cytoplasmic, SP signal peptide, M Mitochondrial, V Vacuolar
The length of BnNRT2 proteins range from 353 (BnNRT2.1b and BnNRT2.1c) to 506 (BnNRT2.2b) amino acid residues (Table 1). The percent identity between BnNRT2s (Table S2) ranged from 26.9% (BnNRT2.1b and BnNRT2.7a) to 98.8% (BnNRT2.5a and BnNRT2.5b). The minimum PI value calculated was 7.54 for BnNRT2.7b, which means that all BnNRT2s are basic proteins. Subcellular localization predictions indicate that most BnNRT2s localize on plasma membranes, except for BnNRT2.4a, which was predicted as a cytoplasmic protein with a signal peptide, BnNRT2.4b, which was predicted as a mitochondrial protein, and both BnNRT2.7 s, which were predicted as vacuolar proteins (Table 1). This variety in predicted subcellular localization suggests that BnNRT2s, like AtNRT2s, perform different transporter functions. The results of transmembrane helix predictions also support this conclusion, with individual BnNRT2s harboring 8 to 10 transmembrane helices (Fig. S2).
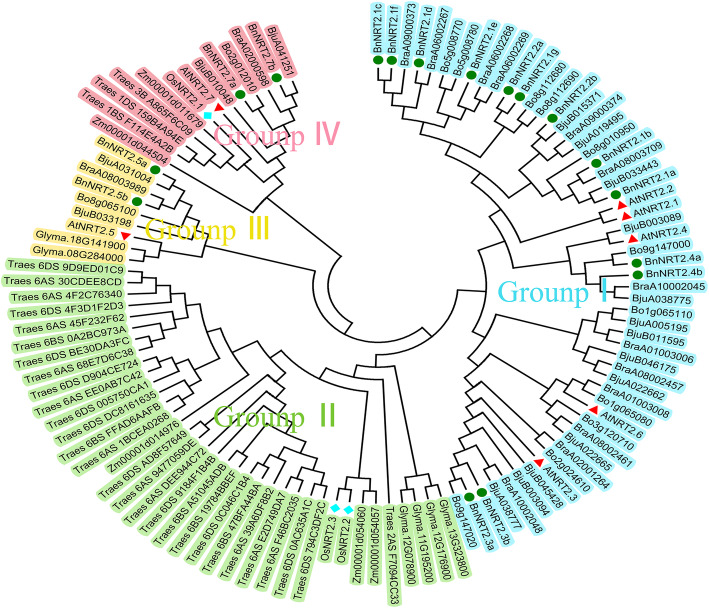
To understand the phylogenetic relationships between NRT2 proteins in rapeseed and other species, an unrooted phylogenetic tree of NRT2 family members was constructed. As shown in Fig. 1, the tree was divided into four groups named groups I-IV. Among these four groups, BnNRT2.1 s, BnNRT2.2 s, BnNRT2.3 s and BnNRT2.4 s were distributed in group I, while BnNRT2.5 s and BnNRT2.7 s were distributed in groups III and IV, respectively, which further indicates that functional divergence has occurred between BnNRT2.5 s and BnNRT2.7 s. Comparisons of the analyzed species revealed that rapeseed NRT2 proteins align closely with their homologs from A. thaliana, but not with NRT2s from O. sativa, a monocotyledon with NRT2 protein members that fell mainly into group II.
Fig. 1.
Phylogenetic relationships of NRT2 family proteins in diverse species. A total of 114 protein sequences were subjected to multiple sequence alignment in ClustalW prior to constructing a phylogenetic tree using the Neighbor-Joining method in MEGA 6.06. Different groups are labeled by different colors. The aligned NRT2 proteins originating from A.thaliana, B.napus and O.sativa are marked by triangles, circles and squares, respectively
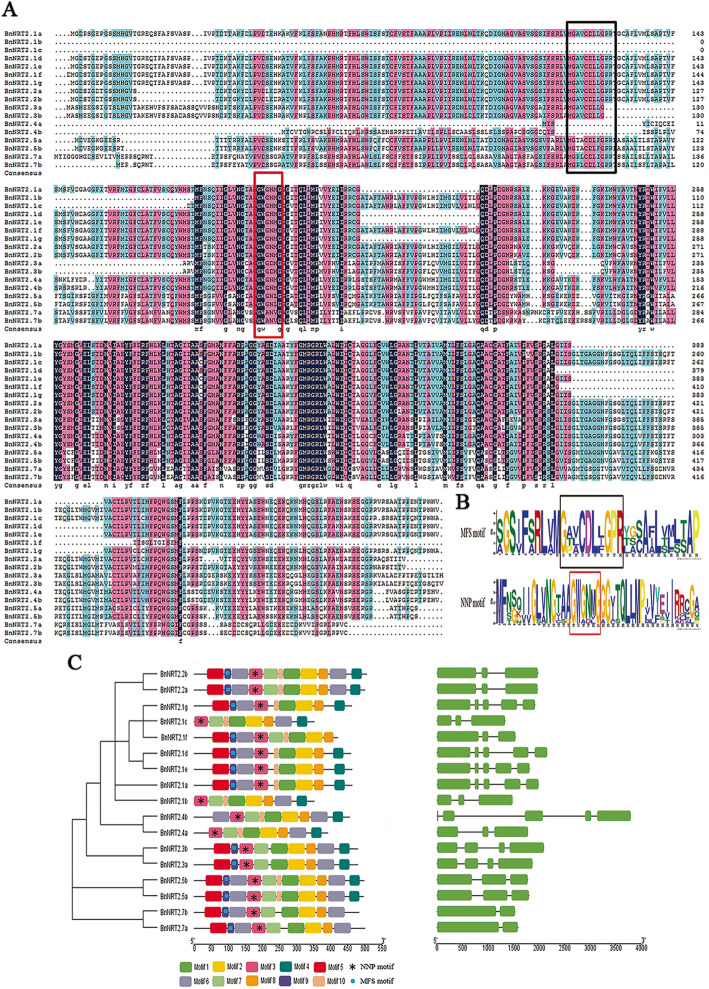
The multiple sequence alignment results of BnNRT2 amino acid sequences, as performed in DNAMAN, showed that NRT2 proteins are likely nitrate/nitrite porter family (NNP) members of the Major Facilitator Superfamily (MFS), with both MFS (G-x-x-x-D-x-x-G-x-R) and NNP (G-W/L-G-N-M/A-G) signatures identified in BnNRT2 sequences (Fig. 2a). Weblogo outputs revealed that nearly all of the BnNRT2 protein members have a highly conserved NNP motif (G-W-G/A-N/D-M/L/V-G). The MFS motif (G-A/T/P-V/L/A-C-D-L-L/I/F-G-P-R), in contrast, was not found in BnNRT2.1b, BnNRT2.1c, BnNRT2.4a and BnNRT2.4b (Fig. 2a, b). Further analysis of BnNRT2 amino acid sequences in the MEME web app with the maximum number of motifs set to 10 revealed that all of the identified BnNRT2 members contain the conserved domain labelled as “motif 1-motif 2-motif 8” (Fig. 2c).
Fig. 2.
Gene structures and conserved domains in the NRT2 family protein of rapeseed. a A multiple sequence alignment of BnNRT2 family proteins constructed using DNAMAN software. The black frame and red frame represent the domain of MFS signature sequences and NNP signature sequences, respectively. b Amino acid sequences of the motif conserved across rapeseed NRT2s proteins, which contain signature sequences of MFS and NNP domains. The bigger the font size, the more likely the amino acid is at that location across all BnNRT2s proteins; c Boxes with different colors represent different conserved motifs, with the MFS and NNP motifs being denoted by circles and asterisks in the boxes, and a gene structure map of BnNRT2s was correspondingly constructed in GSDS2.0. Green boxes indicate exons, and black lines represent introns
Gene structure analysis of BnNRT2s showed that BnNRT2.2 s, BnNRT2.3 s, BnNRT2.5 s and BnNRT2.7 s have the same number of introns and exons (Fig. 2c). Within contrast, BnNRT2.4a and BnNRT2.4b lack the first two exons (Fig. 2c), which might be indicative of functional divergence. What’s more, the gene structures of BnNRT2.1 members can be divided into two types: 1) those with four introns and five exons (BnNRT2.1a, BnNRT2.1d, BnNRT2.1e and BnNRT2.1 g); and 2) those with two introns and three exons (BnNRT2.1b, BnNRT2.1c and BnNRT2.1f) (Fig. 2c).
Analysis of putative cis-acting regulatory elements (CREs) in the promotor regions of BnNRT2 family genes
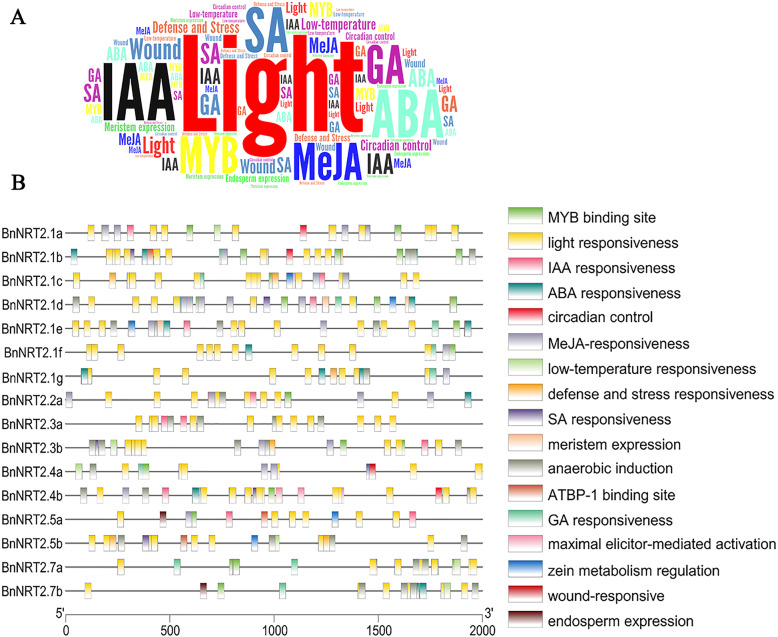
Transcriptional expression is regulated in large part through binding of transcription factors (TFs) to cis-acting regulatory elements (CREs). To gain insight into potential regulation of BnNRT2s, we performed CREs analysis on the 2000 bp region upstream of the start codon for each BnNRT2. As indicated in Fig. 3a, the most abundant CREs in all of the analyzed BnNRT2 promotor regions were light responsive elements, followed by several hormone-related elements and MYB binding sites. To be specific, the CREs of rapeseed NRT2 genes can be divided into four categories, namely MYB binding sites, abiotic stress responsive elements, hormone-related elements, growth and development regulative elements (Fig. 3b).
Fig. 3.
Identification of putative cis-acting regulatory elements (CREs) in the promoter regions of BnNRT2 family genes. a The abundance of CREs in the promotor regions (2000 bp genomic sequences upstream the start codon) of BnNRT2s were determined in the WordArt program. Font size increases with the abundance of CREs. b Genomic distribution of CREs in promotor regions of BnNRT2s. Boxes filled with different colors represent different CREs
MYB proteins are multifunctional TFs that are involved in many plant processes [26]. MYB binding sites were found in all BnNRT2 promoter regions, except those upstream of BnNRT2.1 g, BnNRT2.3a and BnNRT2.5b (Fig. 3b). Light responsive elements were found in abundance among the promoter regions of all the BnNRT2 gene members, while low-temperature responsive elements were also found in the promoter regions of BnNRT2.1a, BnNRT2.3b, BnNRT2.4a, BnNRT2.5b and BnNRT2.7 s (Fig. 3b). The broader category of abiotic stress responsive elements, which refers to light responsiveness, low-temperature responsiveness, defense and stress responsiveness, anaerobic induction and wound-responsive, was represented extensively throughout BnNRT2 promoter regions (Fig. 3b). In addition, hormone-related elements were well represented among BnNRT2 promoter regions, with ABA, MeJA, SA, GA, and IAA responsive elements widely identified in the promotor regions of rapeseed NRT2 genes (Fig. 3b). In contrast, growth and development regulatory elements were more sparsely distributed among BnNRT2 promoter regions. The identified growth and development CREs included circadian control related elements occurring in the promotor regions of BnNRT2.1a, BnNRT2.1b and BnNRT2.4b, meristem expression related elements occurring in the promotor regions of BnNRT2.1b, BnNRT2.1d and BnNRT2.1e, and endosperm expression related elements occurring only in the promotor regions of BnNRT2.5a and BnNRT2.7b (Fig. 3b).
Tissue specific expression of rapeseed NRT2 family genes
To explore the tissue specific expression profiles of BnNRT2 genes, samples for RNA-seq and qRT-PCR analyses were collected from various tissues throughout the rapeseed lifecycle. Specifically, RNA-seq was performed with pistil, stamen, ovule, stem, leaf, root, silique, and pericarp samples, while qRT-PCR tests were run on flower, stem, junior leaf, senior leaf, root, seed and pericarp samples from adult plants. Early expression was also tested for BnNRT2.5a, and BnNRT2.5a in qRT-PCR tests performed with root and leaf samples at seedling stage of rapeseed.
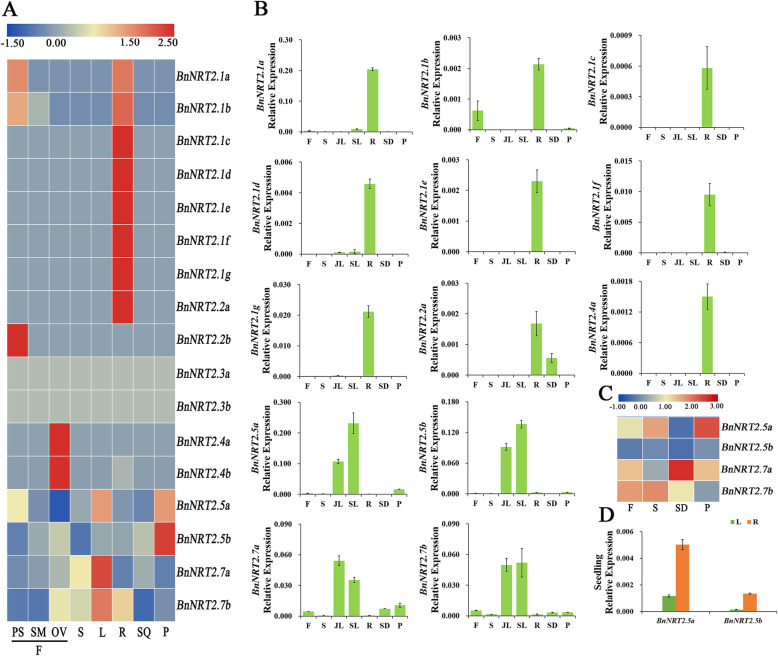
Combined results from RNA-seq and qRT-PCR analyses revealed that in adult plants, BnNRT2.1a-g, BnNRT2.2a and BnNRT2.4a were mainly expressed in the root, with BnNRT2.1a being the most abundant root BnNRT2, and BnNRT2.1b transcripts abundant in both roots and flowers, especially pistils (Fig. 4a). Further observation of BnNRT2 expression in qRT-PCR assays (Fig. 4b, c) largely confirmed the results from RNA-seq analysis (Fig. 4a). Notably, all BnNRT2.1 genes were predominantly expressed in roots in both assays. Meanwhile, among shoot expressed BnNRT2s, BnNRT2.5 and BnNRT2.7 family members were primarily expressed in leaf tissue (Fig. 4b), though they were also obviously detectable in flowers, stems and pericarps, as well as, in seeds for BnNRT2.7 family members (Fig. 4c). Further qRT-PCR assays of BnNTR2.5 tissue specific expression profiles conducted with hydroponically reared seedlings also returned detectable expression for both BnNRT2.5a and BnNRT2.5b, though shoot expression was much lower for these genes in seedlings than in adult plants to the point that expression was higher in roots than in shoots (Fig. 4d).
Fig. 4.
Tissue specific expression profiles of BnNRT2 family genes as determined in RNA-seq and qRT-PCR assays. a Heat maps of relative expression from RNA-seq analysis of BnNRT2 expression in eight different tissues. PS: pistil, SM: stamen, OV: ovule, S: stem, L: leaf, R: root, SQ: silique, P: pericarp. b-d Summary of tissue specific expression of BnNRT2 genes determined in qRT-PCR assays. b Expression of BnNRT2s of different tissues in adult plants at different growth stages. Each tissue of adult plants were collected from field plots supplemented with N supplied as 180 kg/hm2 urea, in detail, junior leaf (JL), senior leaf (SL) and root (R) were collected at vegetative growth stage (namely over-wintering stage), flower (F) and stem (S) were collected at flowering stage, then seed (SD) and pericarp (P) were collected at reproductive growth stage. c The expression of BnNRT2.5 s in seedling tissues (21-days-old rapeseed seedlings). Rapeseed seedlings were reared in hydroponics cultures supplied with normal N treatment (9.5 mM N), L: leaf, R: root. d Expressions of BnNRT2.5 s and BnNRT2.7 s in the flower (F), stem (S), seed (SD) and pericarp (P) though qRT-PCR assays were further delineated in heat maps
BnNRT2 family genes respond to N deprivation in the field
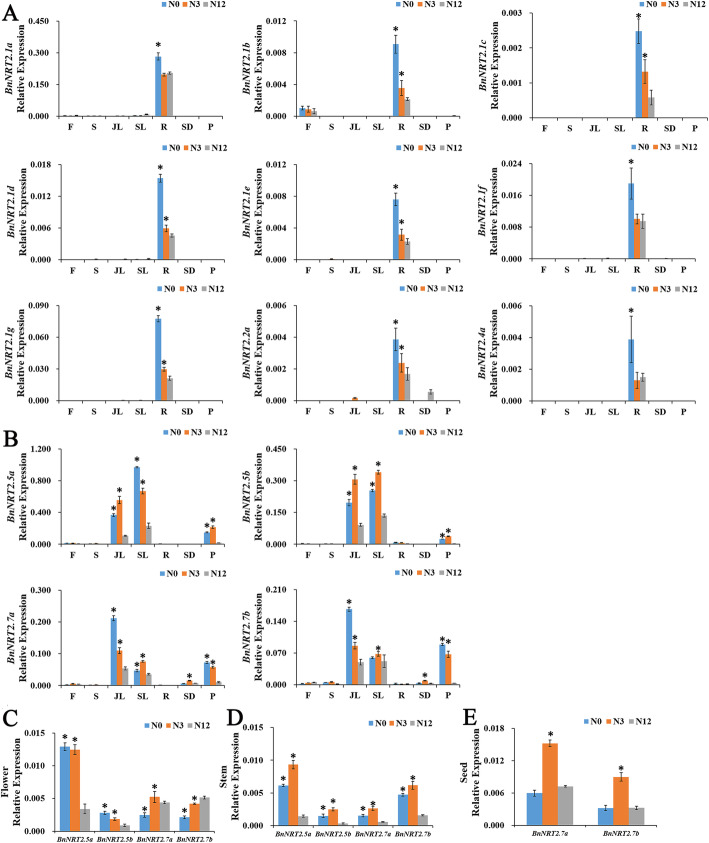
To investigate the possible roles of NRT2s in rapeseed responses to N deprivation, the expression of BnNRT2 family genes was assayed in various rapeseed tissues from plants grown in field plots producing significant N effects on biomass among urea application treatments (Fig. S3A, B). The results show dramatic up-regulation of BnNRT2.1 s, BnNRT2.2a and BnNRT2.4a in roots (Fig. 5a), and the expressions of BnNRT2.5 s and BnNRT2.7 s were significantly up-regulated in leaves, stems and pericarps of rapeseed plants responding to N deprivation (Fig. 5b). In addition, the abundance of BnNRT2.5 transcripts also significantly increased in flowers of rapeseed plants responding to N deprivation (Fig. 5c). The responses of BnNRT2.7 s to N deprivation ran counter to the responses observed for other BnNRT2 members, with expression peaking not in plants grown in the lowest N supply treatment plots, but rather in plants reared in intermediate or high N supply plots (Fig. 5c, d, e).
Fig. 5.
Responses of BnNRT2 expression profiles to alterations in N supply as determined in qRT-PCR assays. a Responses of root specific BnNRT2 genes (BnNRT2.1 s, BnNRT2.2a and BnNRT2.4a). b Responses of shoot specific BnNRT2 genes (BnNRT2.5 s and BnNRT2.7 s). c Responses of BnNRT2.5 s and BnNRT2.7 s in flower. d Responses of BnNRT2.5 s and BnNRT2.7 s in stem. e Responses of BnNRT2.7 s in seed. For these assays, rapeseed was cultivated in the field plots supplied with 180, 45 or 0 kg/hm2 N. The rapeseed plants grown in 180 kg/hm2 N-supplied plots were regarded as the control plots at each sampling time. Asterisks mark significant differences in expression between control plots and plots supplied 45 or 0 kg/hm2 of urea (P < 0.05)
Expression profiles of BnNRT2 family genes in response to macronutrient deficiency in hydroponics
In this study, the expression of BnNRT2 genes was assayed for responses to nutrient imbalances in hydroponics. To this end, rapeseed seedlings were subjected to N, phosphorus (P) and potassium (K) deficiency treatments in hydroponics cultures in order to test for responses in the expression BnNRT2 genes to deficiencies in any of these three macronutrients. As shown in Fig. 6, most BnNRT2.1 s (with exception of BnNRT2.1a and BnNRT2.1e) and BnNRT2.2a were significantly up-regulated in roots subjected to N deficiency, but dramatically down-regulated by P or K deficiency. Interestingly, BnNRT2.1a appeared to be specifically up-regulated in roots in response to K deficiency, while BnNRT2.1e was significantly down-regulated in roots in response to P and K deficiency, but not N deficiency (Fig. 6). Moreover, BnNRT2.4 and BnNRT2.5 members were specifically and remarkably up-regulated by N deficiency in roots (Fig. 6). In contrast to root specific responses, leaf expressions of BnNRT2.7 s was significantly up-regulated by each of the tested macronutrient deficiencies (Fig. 6), suggesting that these genes might be involved in crosstalk of N, P, and K homeostasis signaling pathways.
Fig. 6.
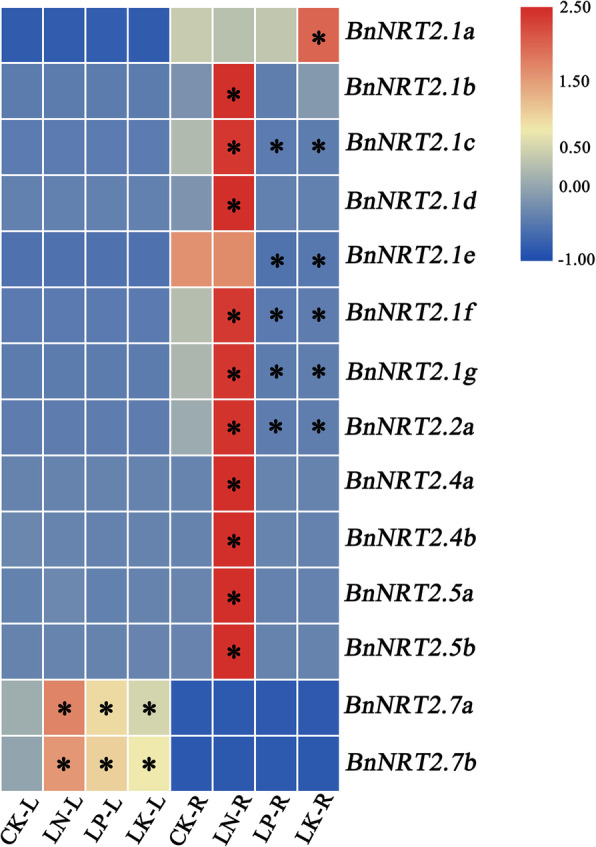
Heat maps of BnNRT2 family genes expression profiles responding to macronutrient deficiency simulated in hydroponics. For this assay, rapeseed seedlings were cultivated in hydroponics cultures containing low N, low P or low K nutrient solution. Seedlings reared under full nutrient conditions were regarded as control samples, and P = 0.05 was used as the threshold to identify significant differences in expression between different conditions. Significant treatments are marked with asterisks. CK: full nutrient condition, LN: N deficiency, LP: P deficiency, LK: K deficiency, L: leaf, R: root
Responses of BnNRT2 gene expression to drought or waterlogging stress
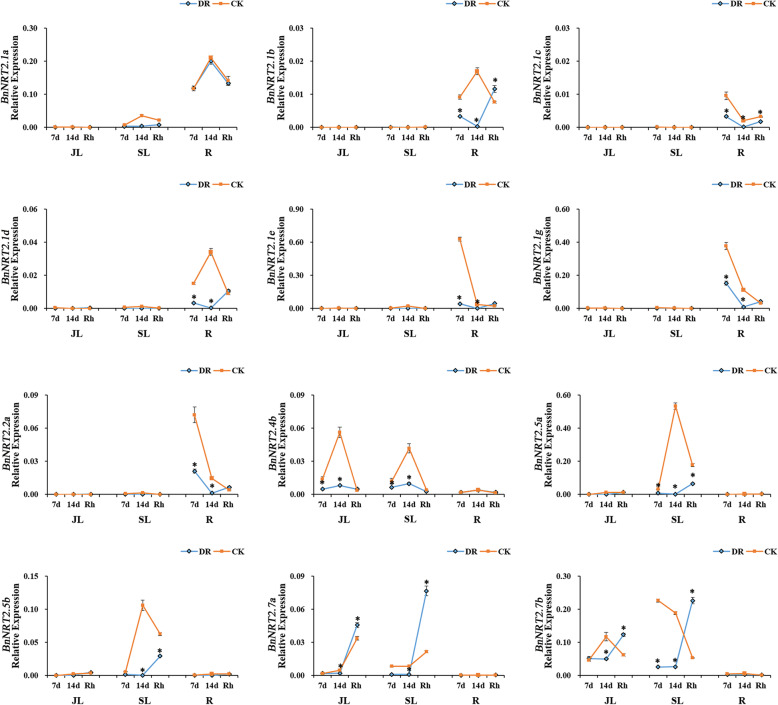
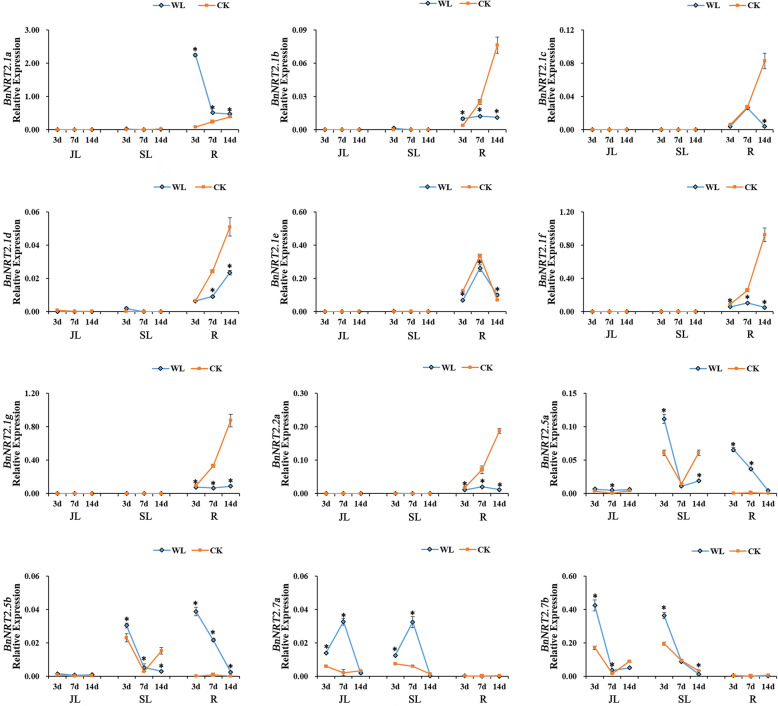
Drought and waterlogging have extremely serious impacts on agricultural production, with severe impacts on rapeseed plants often occurring vegetative growth. In this study, drought and waterlogging stresses were simulated in pot cultures in order to investigate the expression patterns of BnNRT2s genes in response to these stresses. Under drought stress conditions, the transcript abundance of all BnNRT2 genes, except BnNRT2.1a, dramatically decreased relative to expression in control pots over the course of 14 days of drought stress, and then restored to control levels upon rehydration (Fig. 7). Expression of BnNRT2.1a in drought treated pots was maintained at the levels observed in control pots (Fig. 7). At the other extreme, under waterlogging stress conditions, BnNRT2.1a and BnNRT2.5 members were significantly up-regulated in the root, while BnNRT2.7a expression was significantly up-regulated in leaves (Fig. 8). At the same time, root expression of BnNRT2.1e in remained high in waterlogged pots and did not vary relative to expression in control pots, while, the transcript abundances of BnNRT2.1f, BnNRT2.1 g and BnNRT2.2a were remarkably down-regulated in roots responding to waterlogging stress (Fig. 8).
Fig. 7.
qRT-PCR assay expression profiles of BnNRT2 family genes responding to drought stress. For this assay, rapeseed seedlings at the five leaf growth stage were subjected to drought stress for 14 day before rehydration. The tissues for qRT-PCR assays were harvested on the 7th and 14th days of drought stress, as well as, on the 7th day after rehydration. The blue line connects observations in the drought stress treatment (DR) and the orange line connects observations in the control treatment (CK). Rh: rehydration. Asterisks mark significant differences in expression between control and drought stress treatments (P < 0.05). JL: junior leaf, SL: senior leaf, R: root
Fig. 8.
qRT-PCR assay expression profiles of BnNRT2 family genes responding to waterlogging stress. For this assay, rapeseed seedlings at the five leaf growth stage were subjected to waterlogging stress for 14 days. Tissues for qRT-PCR assays were harvested on the 3rd, 7th and 14th days of waterlogging stress. The blue line connects observations in the waterlogging stress treatment (WL), and the orange line connects observations in the control treatment (CK). Asterisks mark significant differences in expression between control and waterlogging stress treatments (P < 0.05). JL: junior leaf, SL: senior leaf, R: root
Discussion
NRT2 proteins are common in bacteria, animals and plants. Though NRT2 gene family members from a number of species have been described in detail, information on nitrate transporters in rapeseed, especially respond to various stresses remains limited. In this study, 17 putative NRT2 genes were identified from the genome of rapeseed, which means that individual Arabidopsis NRT2 genes (7 genes) may have multiple homologs in rapeseed. Indeed, that was observed in this study, but with the less expected outcome that a total of seven BnNRT2.1 genes and two BnNRT2.2 genes were identified based on their respective closest matches in Arabidopsis being the highly similar pair of NRT2 genes, AtNRT2.1 and AtNRT2.2 (Fig. 1 and Table 1). The extent of BnNRT2.1 genes in rapeseed suggests that this complex gene family might be the product of allopolyploidy or duplication events in the rapeseed evolutionary history [18], and our results indeed found that most gene duplications of the NRT2 gene family resulted from whole-genome duplication (Table S1). In addition, results from the macronutrient deficiency experiment reported herein clearly indicate that duplication of BnNRT2.1 genes has led to divergence of the specific roles played by members of this family in responses to varied macronutrient stress conditions. Conserved domain analysis indicated that all 17 of the putatively identified NRT2 proteins harbor the signature sequence (G-W-G/A-N/D-M/L/V-G) of NNP family proteins (Fig. 2), which are also found in other NRT2 transporters, such as the NarK protein in E. coli and NRT2 in Heb. cylindrosporum [9, 27].
Cis-acting regulatory element analysis of BnNRT2 promoter regions revealed a great abundance of light responsive element (Fig. 3), which implies that BnNRT2 gene expression is closely associated with photosynthesis and might be diurnally regulated. Previous studies with Arabidopsis have elucidated up-regulation of AtNRT2.1 in daylight and down-regulation during the night, which can be inhibited by sucrose [28, 29]. Taking our results together with past reports supports the conclusion that, as light and sucrose regulated mediators of NO3− uptake and transport, rapeseed NRTs are key members of systems coordinating carbon and N metabolism. Furthermore, the duplication of BnNRT2.1 genes and variation in their responses to N, P and K deficiency treatments, suggests that NRT genes are involved in modulating multiple nutrient response pathways.
Several previous studies have suggested that limited external NO3− supplies can promote lateral root elongation through regulation of meristematic activity in mature root tips, where, in rapeseed, cells in the meristematic region are packed more densely than in the elongation zone [30]. In Arabidopsis, AtNRT2.1 and AtNRT2.2, two nitrate transporter genes mainly expressed in the root, play key roles in the promotion of lateral root elongation [31, 32]. Recent research has also demonstrated that knock-down of CsNRT2.1 in cucumber strongly reduces root length and lateral root numbers under low NO3−conditions [33]. In the present study, BnNRT2.1 s were predominantly expressed in roots, and most were up-regulated during N deficiency stress (Figs. 5 and 6). Plus, lateral root lengths and root tips also increased in response to N deficiency (Fig. S3C, D). Therefore, we conclude that BnNRT2.1 family members are likely involves in the regulation of the lateral root elongation under N limited conditions.
More recently, EXO70A3 was identified as a downstream target of PIN4, an auxin efflux carrier, in adaptive drought responses that lead to the production of a deeper root system architecture (RSA) [34]. Analogously, a deeper RSA induced by N deficiency might be modulated by EXO70A3, with NRT2.1 genes possibly involve downstream of auxin pathway related RSA modulation, which is supported by the fact that in CREs analysis, IAA responsive elements were found in the promoter sequences of BnNRT2.1 s (Fig. 3).
In Arabidopsis, Orsel [35] have outlined expression profiles of NRT2 family members, with AtNRT2.1, AtNRT2.4, AtNRT2.5 and AtNRT2.6 being preferentially expressed in roots, AtNRT2.7 specifically expressed in aerial parts, AtNRT2.3 expression intermediate between the previous two profiles, and AtNRT2.2 undetectable in either roots or shoots. Similarly, in the rapeseed expression profiles observed herein, BnNRT2.1 s and BnNRT2.4 s were mainly expressed in the root, while BnNRT2.7 s were detected only in aerial parts, including leaves, seeds, flowers and stems (Fig. 4b, c). On the other hand, in contrast to the observations of Orsel [35], BnNRT2.2a transcripts were mainly detected in roots, and no BnNRT2.3 transcripts were detected in either roots or shoots. We did observe that BnNRT2.5 s were like BnNRT2.7 s and expressed mainly in the shoots of adult plants (Fig. 4b, c). However, in the seedlings reared in hydroponics cultures, BnNRT2.5 s were mainly expressed in the root (Fig. 4d). This could be an artefact of the hydroponics system, or it might suggest that BnNRT2.5 s expression in roots changes as the plant matures. Finally, BnNRT2.7 s were the only BnNRT2s detected in seeds (Fig. 4b), which is consistent with Chopin’s [14] finding that AtNRT2.7 plays a specific role in nitrate accumulation in the seed.
To date, a number of NRT2.1-homologous genes have been cloned and characterized in higher plant species including Arabidopsis thaliana [10, 36], Glycine max [37], Triticum aestivum [38], Oryza sativa [39], Zea mays [40, 41], Cucumber [33], and Chinese cabbage [42], many of which have been demonstrated to be major HATS-type genes involved in root NO3− uptake under N limitation. In accordance with previous reports, the mRNA levels of BnNRT2.1 s in the roots observed in this study were up-regulated with declines in N supply (Fig. 5). What’s more, recent research has identified the gene BnaA06g04560D (BnNRT2.1d) as a key factor in NUE through genomic and transcriptomic analysis experiments involving two Brassica napus genotypes contrasting in NUE [43]. Additionally, BnNRT2.5 expression profiles vary between seedlings and adult plants, though expression increased with decreases in external N supply (Figs. 5 and 6). Coincidentally, previous research with NRT2.1, NRT2.2, NRT2.4 and NRT2.5 mutants has indicated that AtNRT2.5, which is expressed in the epidermis and the cortex of lateral roots in young seedlings and the minor veins of mature leaves in adult plants, works in combination with these other NRT2s to modulate efficient uptake of nitrate, as well as, to participate in nitrate loading into the phloem during nitrate remobilization under N-starvation conditions [13]. BnNRT2.5 s might perform similar functions in rapeseed as AtNRT2.5 does in Arabidopsis, which could be a worthwhile subject of future investigations.
The absorption of N directly impacts crop yield, and, therefore, maintaining appropriate N fertilizer supplies is an important component of managing crop yields. However, numerous stresses, such as waterlogging, drought and nutrients deficiencies, may reduce crop yields. As expected, in the present study, it was found that waterlogging, drought, P deficiency and K deficiency suppress the expression of rapeseed NRT2 gene members (Figs. 6, 7 and 8). Specifically, BnNRT2.1a was up-regulated under waterlogging stress and K deficiency conditions, BnNRT2.7 members were up-regulated under waterlogging stress and P or K deficiencies conditions (Figs. 6 and 8), and BnNRT2.5 members were up-regulated in roots under waterlogging stress (Fig. 8). This is consistent with the previous findings of some Pi, Fe transporters [44, 45], which were also regulated by different nutrient stresses, and it is well-documented that the cross-talk exist among ion signals when plant respond to different nutrient stresses [45–47]. The enhanced expressions of BnNRT2s genes under P or K deficiencies probably because the N uptake or metabolism would be influenced by these deficiencies, we speculated that these NRT2 genes could sense the status of external P or K level. However, the synergistic regulation mechanisms of different nutrients homeostasis need to be further investigated. Additionally, the enhanced expression of BnNRT2s might contribute to efficient NO3− uptake and transport of rapeseed under waterlogging stresses. Thus, it is reasonable to infer that BnNRT2.1a, BnNRT2.5 s and BnNRT2.7 s might play key roles in modulating N uptake and transport in rapeseed plants experiencing multiple stresses.
Conclusions
In conclusion, 17 NRT2 genes distributed on nine chromosomes were identified in the rapeseed genome. BnNRT2 genes appear to play important roles during rapeseed growth and development, as indicated by their spatial and temporal expression patterns, especially under low N conditions. Notably, the expression of several BnNRT2 increased in response to various stresses, indicating that these NRT2 genes contribute to stress adaption through regulation of NO3− uptake and transport. Taken together, this study included a genome-wide analysis of the rapeseed NRT2 gene family and provides valuable information for understanding the possible functions of these genes, which could prove valuable in breeding efforts aiming to produce rapeseed plants capable yielding well in stressful environments.
Methods
Identification of NRT2 genes in rapeseed
Sixteen previously reported NRT2 protein sequences, including 7 from A.thaliana, 3 from Oryza sativa (L.), 3 from Glycine max (L.), and 3 from Zea mays (L.) [39, 48, 49], were used to perform BLAST searches of the gene database for A.thalianas, B.napus, G.max, O.sativa, Z.mays, T.aestivum, B.rape, B.oleracea and B.juncea. Then, the 16 starting protein sequences were aligned to construct an HMM profile in HMMER (v3.1), which was used in a domain searches of the BLAST hits using an E-value threshold of 0.001. Rapeseed proteins that were returned in the BLAST search, and which contained NRT2 domains defined in the HMMER search, were finally labelled as rapeseed NRT2 proteins. The gene databases queried in this study include The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/) for A. thaliana genes and gene products; Genoscope (http://www.genoscope.cns.fr/brassicanapus/) for B.napus [18]; the Brassica Database (BRAD) (http://brassicadb.org/brad/) for B.rape, B.oleracea and B.juncea [50–52]; URGI (https://wheat-urgi.versailles.inra.fr/) for T.aestivum [53]; the Maize Genetics and Genomics Database (meizeGDB) (https://maizegdb.org/) for Z.mays; and Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html) for the other species [54]. The NRT2 genes of rapeseed in this study were named as follows: Species abbreviation (Bn) + name of gene homologs in A. thaliana (such as NRT2.1) + a lowercase letter to distinguish among homologs in rapeseed for individual A.thaliana NRT2 genes. For example, BnNRT2.5a and BnNRT2.5b are two different rapeseed homologs of AtNRT2.5 from Arabidopsis.
Bioinformatics analyses of NRT2 genes in rapeseed
The genomic locations of rapeseed NRT2 genes were determined in BLASTn searches using the nucleotide sequences in Genoscope (http://www.genoscope.cns.fr/blat-server/cgi-bin/colza/webBlat). The resulting coordinates were plotted in a genomic location map using MapInspect v. 2010 (http://www.softsea.com/review/MapInspect.html). The gene duplication modes were identified among rapeseed NRT2 genes using DupGen_finder (https://github.com/qiao-xin/DupGen_finder) in B. napus [55]. The number of amino acids and theoretical pI were analyzed in the online Expasy software ProParam (https://web.expasy.org/cgi-bin/protparam/protparam) [56]. TargetP v 1.1 (http://www.cbs.dtu.dk/services/TargetP/) [57], WOLF PSORT (http://www.genscript.com/wolf-psort.html) [58] and ProtComp v. 9.0 (http://linux1.softberry.com/) were used to predict the subcellular localization of NRT2 proteins in rapeseed. Prediction of transmembrane helices in NRT2 proteins was performed via TMHMM 2.0 web application (http://www.cbs.dtu.dk/services/TMHMM/).
All the NRT2 protein sequences of A.thalianas, B.napus, G.max, O.sativa, Z.mays, T.aestivum, B.rape, B.oleracea and B.juncea were compiled in a FASTA file that was then subjected to multiple sequence alignment in ClustalW, which was then used to construct phylogenetic trees using the Neighbor-Joining method in MEGA 6.06, with the bootstrap value set at 1000, and the other parameters set to default values [59].
Exon-Intron structure information was obtained by importing each cDNA sequence (CDS) and genomic sequence of NRT2 gene family members into the Gene Structure Display Server (GSDS 2.0) (http://gsds.cbi.pku.edu.cn/) [60]. Conserved motifs/domains were then deciphered by subjecting translated rapeseed NRT2 protein sequences to the Multiple Expectation maximization for Motif Elicitation (MEME v5.1.0) algorithm of the MEME suite of analysis tools (http://meme-suite.org/tools/meme) [61].
Promoter sequences of rapeseed NRT2 genes were analyzed by downloading the 2000 bp region upstream from the start codon (ATG) for each gene from the Brassica Database (BRAD) (http://brassicadb.org/brad/), and then submitting these promoter regions for inspection at plantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [62]. Putative cis-acting regulatory elements were then prepared for graphical views using TBtools v0.6.
Plant materials and treatments
A Chinese grown rapeseed cultivar zhongshuang 11 (B. napus cv. ZS11), which bred by Oil Crops Research Institute of Chinese Academy of Agricultural Sciences, was the source of plant material applied in this study. Tissue specific expression analysis of BnNRT2s was performed in eight tissues by RNA-seq analysis, and then confirmed by rearing rapeseed plants in the field with N supplied as 180 kg/hm2 urea and harvesting seven tissues at several developmental stages for qRT-PCR assays, in detail, junior leaf, senior leaf and root were collected at vegetative growth stage (namely over-wintering stage), flower and stem were collected at flowering stage, then seed and pericarp were collected at reproductive growth stage. Three independent biological replicates were included for tissue specific analysis.
To investigate possible responses of BnNRT2s to nutrient deficiency, 7-day-old seedlings were exposed to low nitrogen (LN), low phosphorus (LP), and low potassium (LK) conditions for 14 days, in which time nutrient deficiency symptoms became evident. In more detail, nutrient deficiency conditions were simulated in hydroponics cultures with the base solution being 1/2 strength modified Hoagland nutrient solution, containing 2.5 mM Ca (NO3)2, 2.5 mM KNO3, 1 mM NH4NO3, 0.25 mM K2SO4, 0.5 mM KH2PO4, 1 mM MgSO4, 0.08 mM EDTA-Fe, 0.02 mM H3BO4, 4.5 μM MnCl2, 0.3 μM ZnSO4, 0.16 μM CuSO4, and 0.16 μM (NH4)6Mo7O24. Control solution contained 9.5 mM N, 0.5 mM P and 3.5 mM K. The nutrient deficiency treatment solutions were the same as the control solution, except LN contained 0.475 mM N (5% of control N), LP contained 10 μM P (2% control P), and LK contained 10 μM K (0.29% of control K). Plants were grown in a 24 °C/20 °C day/night temperature regime with a 14-h/10-h light/dark photoperiod. Each treatment had four independent biological replicates. Nutrient (N, P, K)-deficient rapeseed leaves and roots were separately sampled for further RNA extraction and qRT-PCR analysis.
Additionally, BnNRT2 responses to N availability were performed in a field experiment conducted in Huanggang city of Hubei province. Experimental plots were planted with the rapeseed genotype ZS11 and subjected to routine management practices. Nitrogen was supplied to plots as urea (46% N) at the rates of 0, 45, or 180 kg/hm2. Each treatment had three independent biological replicates. Seven tissues at several developmental stages under different N rates were sampled for qRT-PCR assays, in detail, junior leaf, senior leaf and root were collected at vegetative growth stage (namely over-wintering stage), flower and stem were collected at flowering stage, then seed and pericarp were collected at reproductive growth stage.
Moreover, drought and waterlogging stress were also tested in pot cultures set up in accordance with previous research [63]. For drought stress observations weighed pots were maintained with water content ranging between 25 to 35% for a drought treatment, and between 65 to 75% in control pots. Rapeseed seedlings at the five leaf growth stage were subjected to drought stress for 14 day before rehydration. Rapeseed junior leaf, senior leaf and root were separately sampled after 7 days, 14 days drought treatment and rehydration for 7 days. Rapeseed seedlings at the five leaf growth stage also were subjected to waterlogging stress for 14 days. Waterlogging stress, meanwhile, was conducted by keeping the water surface 1 ~ 2 cm above the soil surface, with the control pots being the same as those used in the observations of drought stress. Tissues (junior leaf, senior leaf and root) for qRT-PCR assays were harvested on the 3rd, 7th and 14th days of waterlogging stress. Each treatment in this pot experiment had three independent biological replicates.
RNA-seq analysis and quantitative real time RT-PCR (qRT-PCR)
For tissue specific analysis of rapeseed NRT2 family genes by RNA-seq technique, the detail of generation of RNA-Seq data had been described in detail previously [64, 65], and the publically RNA-seq dataset of various tissues collected from different rapeseed growth stages were deposited in a public repository (SRA accession: PRJNA474576, 65] and applied in this study, the Fragments per kilobase of exon per million fragments mapped (FPKM) was used to calculate gene expression levels, and a heat map of gene expression profiles (log2) of BnaNRT2 genes was generated using pheatmap package in R software after data normalization (https://cran.r-project.org/web/packages/pheatmap/).
For verify the tissue specific expression and further analyze the differential expression profiles of rapeseed NRT2 family genes under various stresses, the total RNA of each harvested samples were extracted using the HiPure Plant RNA Mini Kit according to manufacturer instructions (Magen, Guangzhou, China). Then, samples were treated with RNase-free DNaseI (Invitrogen, Grand Island, NY,USA) before being included as templates for cDNA synthesis using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Tokyo, Japan). Following cDNA synthesis, the relative expression of rapeseed NRT2 gene family members was detected by qRT-PCR using the SYBR® Premix Ex Taq™ II kit (Tli RNaseH Plus) (TaKaRa, Tokyo, Japan) and a CFX96™ Real-Time PCR Detection System (BIO-RAD,USA). The PCR program was run as follows: 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 30 s. The PCR reaction volume was 20 μL in total, which included 2 μL diluted cDNA, 10 μL SYBR Premix Ex Taq II (Tli RNaseH Plus)(2×), 0.6 μL primers and 6.8 μL RNAfree water (TaKaRa, Tokyo, Japan). The Cq value of candidate genes were normalized relative to the Cq value of reference gene act-7, and relative expression was calculated using the 2 -ΔCq method. The specific primers for rapeseed NRT2 genes (Table S3) and the reference gene act-7 were designed in Primer-NCBI (https://www.ncbi.nlm.nih.gov/tools/primerblast/index.cgi?LINK_LOC=BlastHome), and confirmed by Melt curve analysis, with the reaction program as follows: 95 °C for 10 s, 65 °C to 95 °C for 5 s (+ 0.5 °C per cycle).
Statistical analyses
All means and standard errors of data in the present study were calculated in Microsoft Excel 2010. All the comparisons between control and stress treatment in this study were performed using analysis of variance (ANOVA) method in SPSS statistics 25 with P values < 0.05 considered statistically significant.
Supplementary information
Additional file 1: Figure S1. Physical map of NRT2 family genes in the genome of rapeseed (Brassica napus L.)
Additional file 2: Figure S2. Membrane-spanning regions predicted for NRT2 family proteins in rapeseed (Brassica napus L.).
Additional file 3: Figure S3. Phenotypic and physiological changes of rapeseed plants grown under different N supply treatments. (A) Phenotypes and biomasses of rapeseed plants grown under different N supply treatments in the field plots supplied with 180, 45 or 0 kg/hm2 N. (B) Biomasses of rapeseed plants grown under different N supply treatments in the field plots supplied with 180, 45 or 0 kg/hm2 N. (C) Phenotypes of rapeseed roots grown under different N supply treatments (CK: 9.5 mM N, LN: 0.475 mM N) in hydroponics. (D) Lateral root length and root tips of rapeseed plants grown under different N supply treatments (CK: 9.5 mM N, LN: 0.475 mM N) in hydroponics.
Additional file 4: Table S1. The duplicated rapeseed NRT2 gene pairs identified in this study.
Additional file 5: Table S2. The comparison of identity between the BnNRT2 protein members.
Additional file 6: Table S3. Specific primers of rapeseed NRT2 genes used in qRT-PCR assays.
Acknowledgements
We greatly thank the group of Prof. Huawei in Oil Crops Research Institute of Chinese Academy of Agricultural Sciences, Wuhan, Hubei province, P.R. China for the providers of RNA-seq data of rapeseed for analysis.
Abbreviations
- N
nitrogen
- NUE
nitrogen use efficiency
- NO3−
nitrate
- LATS
low-affinity transport system
- HATS
high-affinity transport system
- NRT1
nitrate transporter 1
- NRT2
nitrate transporter 2
- NNP
nitrate/nitrite porter family
- MFS
major facilitator superfamily
- CREs
cis-acting regulatory elements
- TFs
transcription factors
- P
phosphorus
- K
potassium
- FPKM
Fragments per kilobase of exon per million fragments mapped
Authors’ contributions
TJF performed the experiments, analyzed the data and wrote the manuscript. WTC and CLY conducted the bioinformatic analysis. HPP and LYS preformed experiments in hydroponics. GCM and HXJ performed the pot experiments. XLH and SXJ preformed field trial. QL and LX designed and supervised research, interpreted the data and revised the manuscript. All authors have read and approved the manuscript.
Funding
This work was funded by grants from the National Key Research and Development Program (2016YFD0100202–5) to perform research design and most of the experiments. The National Project of Modern Agricultural Industry Technology System in China (CARS-22-G-05) funded us in bioinformatics and data analysis. The Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI) funded us to cultivate experimental materials in the field.
Availability of data and materials
The Arabidopsis, Wheat and Maize NRT2 protein sequences were collected from TAIR (http://www.arabidopsis.org), URGI (https://wheat-urgi.versailles.inra.fr/) and meizeGDB (https://maizegdb.org/) database. The Rice and Soybean NRT2 protein sequences were collected from Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html). And the NRT2 protein sequences of B.rape, B.oleracea and B.juncea were collected from BRAD (http://brassicadb.org/brad/). The genome and protein sequences of B.napus were downloaded from Genoscope (http://www.genoscope.cns.fr/brassicanapus/). The RNA sequencing data used in this study were deposited in the NCBI SRA database (SRA accession: PRJNA474576). All data generated or analyzed during this study were included in this published article and its supplementary information files. The materials are available upon request by contacting the corresponding author.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xing Liao, Email: liaox@oilcrops.cn.
Lu Qin, Email: qinlu-123@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02648-1.
References
- 1.Konishi M, Yanagisawa S. Emergence of a new step towards understanding the molecular mechanisms underlying nitrate-regulated gene expression. J Exp Bot. 2014;65(19):5589–5600. doi: 10.1093/jxb/eru267. [DOI] [PubMed] [Google Scholar]
- 2.Good AG, Shrawat AK, Muench DG. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 2004;9(12):597–605. doi: 10.1016/j.tplants.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Hirel B, Le Gouis J, Ney B, Gallais A. The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot. 2007;58(9):2369–2387. doi: 10.1093/jxb/erm097. [DOI] [PubMed] [Google Scholar]
- 4.Meyer C, Stitt M. Nitrate reduction and signalling. In: Lea PJ, Morot-Gaudry JF, editors. Plant Nitrogen. Berlin: Springer-Verlag; 2001. pp. 37–59. [Google Scholar]
- 5.Jackson LE, Stivers LJ. Root distribution of lettuce under commercial production: implications for crop uptake of nitrogen. Biol Agric Hortic. 1993;9(3):273–293. doi: 10.1080/01448765.1993.9754639. [DOI] [Google Scholar]
- 6.Wang YY, Hsu PK, Tsay YF. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012;17(8):458–467. doi: 10.1016/j.tplants.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Tong Y, Zhou JJ, Li Z, Miller AJ. A two-component high-affinity nitrate uptake system in barley. Plant J. 2005;41(3):442–450. doi: 10.1111/j.1365-313X.2004.02310.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang NC, Liu KH, Lo HJ, Tsay YF. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell. 1999;11(8):1381–1392. doi: 10.1105/tpc.11.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forde BG. Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta. 2000;1465(1–2):219–235. doi: 10.1016/S0005-2736(00)00140-1. [DOI] [PubMed] [Google Scholar]
- 10.Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, et al. An arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 2001;489(2–3):220–224. doi: 10.1016/S0014-5793(01)02096-8. [DOI] [PubMed] [Google Scholar]
- 11.Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell. 2012;24(1):245–258. doi: 10.1105/tpc.111.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass AD. Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant physio. 2007;143(1):425–433. doi: 10.1104/pp.106.091223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lezhneva L, Kiba T, Feria-Bourrellier AB, Lafouge F, Boutet-Mercey S, Zoufan P, et al. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014;80(2):230–241. doi: 10.1111/tpj.12626. [DOI] [PubMed] [Google Scholar]
- 14.Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, et al. The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell. 2007;19(5):1590–1602. doi: 10.1105/tpc.107.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Fan XR, Li Q, Feng HM, Miller AJ, Shen QR, et al. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 2012;160(4):2052–2063. doi: 10.1104/pp.112.204461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allender CJ, King GJ. Origins of the amphiploid species Brassica napus L investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 2010;10:54. doi: 10.1186/1471-2229-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackshaw R, Johnson E, Gan Y, May W, Mcandrew D, Barthet V, et al. Alternative oilseed crops for biodiesel feedstock on the Canadian prairies. Can J Plant Sci. 2011;91(5):889–896. doi: 10.4141/cjps2011-002. [DOI] [Google Scholar]
- 18.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science (New York, NY) 2014;345(6199):950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 19.Rathke GW, Christen O, Diepenbrock W. Effects of nitrogen source and rate on productivity and quality of winter oilseed rape (Brassica napus L.) grown in different crop rotations. Field Crop Res. 2005;94(2–3):103–113. doi: 10.1016/j.fcr.2004.11.010. [DOI] [Google Scholar]
- 20.Zou J, Lu JW, Li YS, Li XK. Regional evaluation of winter rapeseed response to K fertilization, K use efficiency, and critical level of soil K in the Yangtze river valley. Agr Sci China. 2011;6:911–920. doi: 10.1016/S1671-2927(11)60076-X. [DOI] [Google Scholar]
- 21.Wang Y, Liu T, Li XK, Ren T, Cong RH, Lu JW. Nutrient deficiency limits population development, yield formation, and nutrient uptake of direct sown winter oilseed rape. J Integr Agr. 2015;14(004):670–680. doi: 10.1016/S2095-3119(14)60798-X. [DOI] [Google Scholar]
- 22.Ploschuk RA, Miralles DJ, Colmer TD, Ploschuk EL, Striker GG. Waterlogging of winter crops at early and late stages: impacts on leaf physiology, growth and yield. Front Plant Sci. 2018;9:1863. doi: 10.3389/fpls.2018.01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu CM, Zhang SJ, Han PP, Hu XJ, Xie LH, Li YS, et al. Soil enzyme activity in soils subjected to flooding and the effect on nitrogen and phosphorus uptake by oilseed rape. Front Plant Sci. 2019;10:368. doi: 10.3389/fpls.2019.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XK, Lu GY, Long WH, Zou XL, Li F, Nishio T. Recent progress in drought and salt tolerance studies in Brassica crops. Breed Sci. 2014;64(1):60–73. doi: 10.1270/jsbbs.64.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Jiao J, Zhou MJ, Jin ZY, Yu YJ, Liang MX. Physiological and transcriptional responses of industrial rapeseed (Brassica napus) seedlings to drought and salinity stress. Int J Mol Sci. 2019;20(22):5604. doi: 10.3390/ijms20225604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41(5):577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- 27.Jargeat P, Rekangalt D, Verner MC, Gay G, Debaud JC, Marmeisse R, et al. Characterisation and expression analysis of a nitrate transporter and nitrite reductase genes, two members of a gene cluster for nitrate assimilation from the symbiotic basidiomycete Hebeloma cylindrosporum. Curr Genet. 2003;43(3):199–205. doi: 10.1007/s00294-003-0387-2. [DOI] [PubMed] [Google Scholar]
- 28.Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, et al. Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J. 1999;18(5):509–519. doi: 10.1046/j.1365-313X.1999.00480.x. [DOI] [PubMed] [Google Scholar]
- 29.Vidal EA, Gutiérrez RA. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol. 2008;11(5):521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Qin L, Walk TC, Han PP, Chen LY, Zhang S, Li YS, et al. Adaption of roots to nitrogen deficiency revealed by 3D quantification and proteomic analysis. Plant Physiol. 2019;179(1):329–347. doi: 10.1104/pp.18.00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Forde BG. Regulation of Arabidopsis root development by nitrate availability. J Exp Bot. 2000;51(342):51–59. doi: 10.1093/jxb/51.342.51. [DOI] [PubMed] [Google Scholar]
- 32.Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, et al. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006;140(3):909–921. doi: 10.1104/pp.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Li JQ, Yan Y, Liu WQ, Zhang WN, Gao LH, Tian YQ. Knock-down of CsNRT2.1, a cucumber nitrate transporter, reduces nitrate uptake, root length, and lateral root number at low external nitrate concentration. Front. Plant Sci. 2018;9:722. doi: 10.3389/fpls.2018.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogura T, Goeschl C, Filiault D, Mirea M, Slovak R, Wolhrab B, et al. Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell. 2019;178(2):400–412. doi: 10.1016/j.cell.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 35.Orsel M, Krapp A, Daniel-Vedele F. Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 2002;129(2):886–896. doi: 10.1104/pp.005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, et al. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 2006;142(3):1304–1317. doi: 10.1104/pp.106.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amarasinghe BH, de Bruxelles GL, Braddon M, Onyeocha I, Forde BG, Udvardi MK. Regulation of GmNRT2 expression and nitrate transport activity in roots of soybean (Glycine max) Planta. 1998;206(1):44–52. doi: 10.1007/s004250050372. [DOI] [PubMed] [Google Scholar]
- 38.Zhao XQ, Li YJ, Liu JZ, Li B, Liu QY, Tong YP, et al. Isolation and expression analysis of a high-affinity nitrate transporter TaNRT2.3 from roots of wheat. Acta Bot Sin. 2004;46(3):347–54.
- 39.Cai C, Wang JY, Zhu YG, Shen QR, Li B, Tong YP, et al. Gene structure and expression of the high-affinity nitrate transport system in rice roots. J Integr Plant Biol. 2008;50(4):443–451. doi: 10.1111/j.1744-7909.2008.00642.x. [DOI] [PubMed] [Google Scholar]
- 40.Plett D, Toubia J, Garnett T, Tester M, Kaiser BN, Baumann U. Dichotomy in the NRT gene families of dicots and grass species. PLoS One. 2010;5(12):e15289. doi: 10.1371/journal.pone.0015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamboni A, Astolfi S, Zuchi S, Pii Y, Guardini K, Tononi P, et al. Nitrate induction triggers different transcriptional changes in a high and a low nitrogen use efficiency maize inbred line. J Integr Plant Biol. 2014;56(11):1080–1094. doi: 10.1111/jipb.12214. [DOI] [PubMed] [Google Scholar]
- 42.Liu TK, Dai W, Sun FF, Yang XD, Xiong AS, Hou XL. Cloning and characterization of the nitrate transporter gene BraNRT2.1 in non-heading Chinese cabbage. Acta Physiol Plant. 2014;36(4):815–823. doi: 10.1007/s11738-013-1460-1. [DOI] [Google Scholar]
- 43.Li Q, Ding G, Yang N, White PJ, Ye X, Cai H, et al. Comparative genome and transcriptome analysis unravels key factors of nitrogen use efficiency in Brassica napus L. Plant Cell Environ. 2020;43(3):712–731. doi: 10.1111/pce.13689. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Wang X, Zhang H, Wang SL, Ye XS, Shi L, et al. Molecular identification of the phosphate transporter family 1 (PHT1) genes and their expression profiles in response to phosphorus deprivation and other abiotic stresses in Brassica napus. PLoS One. 2019;14(7):e0220374. doi: 10.1371/journal.pone.0220374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang YH, Garvin DF, Kochian LV. Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 2002;130(3):1361–1370. doi: 10.1104/pp.008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin L, Guo YX, Chen LY, Liang RK, Gu M, Xu GH, et al. Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS One. 2012;7(10):e47726. doi: 10.1371/journal.pone.0047726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maillard A, Etienne P, Diquélou S, Trouverie J, Billard V, Yvin JC, et al. Nutrient deficiencies modify the ionomic composition of plant tissues: a focus on cross-talk between molybdenum and other nutrients in Brassica napus. J Exp Bot. 2016;67(19):5631–5641. doi: 10.1093/jxb/erw322. [DOI] [PubMed] [Google Scholar]
- 48.von Wittgenstein NJ, Le CH, Hawkins BJ, Ehlting J. Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol Biol. 2014;14:11. doi: 10.1186/1471-2148-14-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto M, Vidmar JJ, Glass AD. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol. 2003;44(3):304–317. doi: 10.1093/pcp/pcg036. [DOI] [PubMed] [Google Scholar]
- 50.Wang XW, Wang HZ, Wang J, Sun RF, Wu J, Liu SY, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 51.Yang JH, Liu DY, Wang XW, Ji CM, Cheng F, Liu BN, et al. Author correction: the genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet. 2018;50(11):1616. doi: 10.1038/s41588-018-0227-4. [DOI] [PubMed] [Google Scholar]
- 52.Liu SY, Liu YM, Yang XH, Tong CB, Edwards D, Parkin IA, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alaux M, Rogers J, Letellier T, Flores R, Alfama F, Pommier C, et al. Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018;19(1):111. doi: 10.1186/s13059-018-1491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiao X, Li QH, Yin H, Qi KJ, Li LT, Wang RZ, et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019;20(1):38. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 57.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 58.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- 60.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics (Oxford, England) 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia LJ, Yang LQ, Sun NL, Li J, Fang YJ, Wang YP. Physiological and antioxidant enzyme gene expression analysis reveals the improved tolerance to drought stress of the somatic hybrid offspring of Brassica napus and Sinapis alba at vegetative stage. Acta Physiol Plant. 2016;38(4):88. doi: 10.1007/s11738-016-2111-0. [DOI] [Google Scholar]
- 64.Sun FM, Fan GY, Hu Q, Zhou YM, Guan M, Tong CB, et al. The high-quality genome of Brassica napus cultivar 'ZS11' reveals the introgression history in semi-winter morphotype. Plant J. 2017;92(3):452–468. doi: 10.1111/tpj.13669. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Dong CX, Hu M, Bai ZT, Tong CB, Zuo R, et al. Identification of flower-specific promoters through comparative transcriptome analysis in Brassica napus. Int J Mol Sci. 2019;20(23):5949. doi: 10.3390/ijms20235949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Physical map of NRT2 family genes in the genome of rapeseed (Brassica napus L.)
Additional file 2: Figure S2. Membrane-spanning regions predicted for NRT2 family proteins in rapeseed (Brassica napus L.).
Additional file 3: Figure S3. Phenotypic and physiological changes of rapeseed plants grown under different N supply treatments. (A) Phenotypes and biomasses of rapeseed plants grown under different N supply treatments in the field plots supplied with 180, 45 or 0 kg/hm2 N. (B) Biomasses of rapeseed plants grown under different N supply treatments in the field plots supplied with 180, 45 or 0 kg/hm2 N. (C) Phenotypes of rapeseed roots grown under different N supply treatments (CK: 9.5 mM N, LN: 0.475 mM N) in hydroponics. (D) Lateral root length and root tips of rapeseed plants grown under different N supply treatments (CK: 9.5 mM N, LN: 0.475 mM N) in hydroponics.
Additional file 4: Table S1. The duplicated rapeseed NRT2 gene pairs identified in this study.
Additional file 5: Table S2. The comparison of identity between the BnNRT2 protein members.
Additional file 6: Table S3. Specific primers of rapeseed NRT2 genes used in qRT-PCR assays.
Data Availability Statement
The Arabidopsis, Wheat and Maize NRT2 protein sequences were collected from TAIR (http://www.arabidopsis.org), URGI (https://wheat-urgi.versailles.inra.fr/) and meizeGDB (https://maizegdb.org/) database. The Rice and Soybean NRT2 protein sequences were collected from Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html). And the NRT2 protein sequences of B.rape, B.oleracea and B.juncea were collected from BRAD (http://brassicadb.org/brad/). The genome and protein sequences of B.napus were downloaded from Genoscope (http://www.genoscope.cns.fr/brassicanapus/). The RNA sequencing data used in this study were deposited in the NCBI SRA database (SRA accession: PRJNA474576). All data generated or analyzed during this study were included in this published article and its supplementary information files. The materials are available upon request by contacting the corresponding author.