Abstract
The current pandemic of novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) poses a significant global public health threat. While urgent regulatory measures in control of the rapid spread of this virus are essential, scientists around the world have quickly engaged in this battle by studying the molecular mechanisms and searching for effective therapeutic strategies against this deadly disease. At present, the exact mechanisms of programmed cell death upon SARS-CoV-2 infection remain to be elucidated, though there is increasing evidence suggesting that cell death pathways play a key role in SARS-CoV-2 infection. There are several types of programmed cell death, including apoptosis, pyroptosis, and necroptosis. These distinct programs are largely controlled by the proteins of the death domain (DD) superfamily, which play an important role in viral pathogenesis and host antiviral response. Many viruses have acquired the capability to subvert the program of cell death and evade the host immune response, mainly by virally encoded gene products that control cell signaling networks. In this mini-review, we will focus on SARS-CoV-2, and discuss the implication of restraining the DD-mediated signaling network to potentially suppress viral replication and reduce tissue damage.
Subject terms: Cell death, Infection, Inflammasome
Facts
There is an increasing evidence that cell death pathways play a key role in SARS-CoV-2 infection.
Cell death and inflammation pathways are largely controlled by the proteins of the DD superfamily.
The DD superfamily comprises the DD, death effector domain (DED), caspase activation recruitment domain (CARD), and pyrin domain (PYD) subfamilies.
Open questions
Restraining of SARS-CoV-2 viral replication and virus-induced tissue damage through modulation of DD network might pave the way toward new therapeutic approaches.
Understanding the different stoichiometry of DD, DED, PYD, and CARD macromolecular platforms might provide the link toward uncovering the cross talk between these pathways.
The role of positive and negative regulation of programmed cell death pathways by SARS-CoV-2 infection in the viral spread.
Targeting the DD superfamily proteins might play a pivotal role in the inflammation control during SARS-CoV-2 infection.
Introduction
During the end of 2019 and the beginning of 2020, multiple human cases of novel coronavirus infection were reported, caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2). Although some initial cases were linked to a local seafood market in Wuhan, its origin, intermediate hosts, and how it was transmitted to humans are still largely unknown. Coronaviruses (CoVs) are enveloped, nonsegmented, positive-sense single-stranded RNA viruses1,2. With genomes ranging in size from 26 to 32 kilobases, they are among the largest known viral RNA (vRNA) genomes. The virion has a nucleocapsid composed of genomic RNA and phosphorylated nucleocapsid (N) protein, which is buried inside phospholipid bilayers and covered by two different types of spike proteins: the spike glycoprotein trimmer (S) that can be found in all CoVs, and the hemagglutinin–esterase that exists in some CoVs. The membrane (M) protein (a type III transmembrane glycoprotein) and the envelope (E) protein are located among the S proteins in the virus envelope. SARS-CoV-2 as well as earlier reported SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) infections may remain asymptomatic in the early stage, until causing severe pneumonia, dyspnea, renal insufficiency, and even death3,4. Currently, no promising antiviral treatment is available. There are, however, numerous compounds with proven effectiveness against SARS-CoV and MERS-CoV, which have not yet been tested for the newly emerged SARS-CoV-2.
There are several types of regulated cell death5. Apoptosis is a form of regulated cell death that eliminates damaged and excessive cells to maintain tissue homeostasis. There are two key ways of apoptosis induction: the intrinsic and the extrinsic pathway6. The intrinsic pathway is initiated via mitochondrial outer membrane permeabilisation, which is followed by the release of several proapoptotic factors from the mitochondria, resulting in the initiation of the cell death cascade5. The extrinsic cell death pathway is triggered by the stimulation of the death receptors (DRs)7,8. Six DR family members have been characterized so far: TNF-R1, CD95 (FAS/APO-1), DR3, TRAIL-R1/DR4, TRAIL-R2/DR5, and DR69. All members of the DR family are characterized by the presence of the death domain (DD) playing the key role in the transduction of apoptotic and antiapoptotic signals7,10,11.
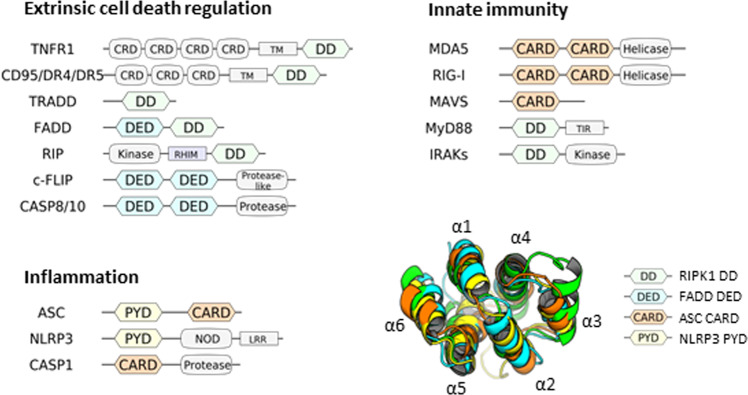
The DD family is a subfamily of the so-called DD superfamily9,12. The DD superfamily shares a highly conservative structural fold that includes six to seven α-helices (Fig. 1). The DD superfamily comprises the DD, death effector domain (DED), caspase activation recruitment domain (CARD), and pyrin domain (PYD) subfamilies (Fig. 1). The members of this superfamily are capable of assembling into oligomeric structures that are formed via homotypic interactions between DD superfamily members13. In this way, the regulation of programmed cell death, inflammation, and host defense against intracellular pathogens is largely controlled by the proteins from the DD superfamily12. In particular, the key regulatory step involves the assembly of macromolecular complexes, including death-inducing signaling complex (DISC), necrosome, ripoptosome, faddosome, myddosome, and PIDDosome that serve as platforms for the initiation of a particular signaling pathway14–22. The individual function and sequence of proteins from the DD superfamily likely diverged in the course of evolution to provide multiple strategies for host defense and programmed cell death activation. However, at the same time, the cross talk between these pathways through the proteins of DD superfamily has gained more attention recently. It turned out that the key players of the extrinsic apoptosis pathway, such as Fas-associated protein with DD (FADD), cellular FLICE inhibitory protein (c-FLIP), caspase-8, and RIPK1 play the major role in the regulation of different programs of cell death, including apoptosis, necroptosis, and pyroptosis, as well as innate immune responses12,17,23–26. Thus, targeting the DD superfamily proteins of the extrinsic pathway might play a pivotal role in the inflammation control during the viral infection and restore the innate immune response.
Fig. 1. Domain organization of the major DD superfamily members controlling inflammation, cell death, and the innate immune response.
DD, CARD, DED, and PYD are shown in green, blue, yellow, and orange, respectively. The superimposed representative structures of DDs of RIPK1 DD (PDB identifier 6AC5), FADD DED (PDB identifier 1A1W), ASC CARD (PDB identifier 5GPQ), and NLRP3 PYD (PDB identifier 3QF2) are shown. N- and C-terminal regions were truncated to improve visual perception.
Host cells eliminate virally infected cells via cell death pathways, including apoptosis and necroptosis, which aborts virus infection27. On the other hand, some viruses take advantage of inducing cell death as a way to release and disseminate progeny viruses or as a strategy to evade the immune system28. Activation of the necroptosis response in host cells, in turn can lead to antiviral inflammation29. In this review, we will mainly focus on the restraining of SARS-CoV-2 viral replication and virus-induced tissue damage through modulation of the signaling pathways mediated by DD superfamily members, and possible treatment therapies.
Targeting extrinsic cell death as a strategy to combat SARS-CoV-2 replication at an early stage of the infection
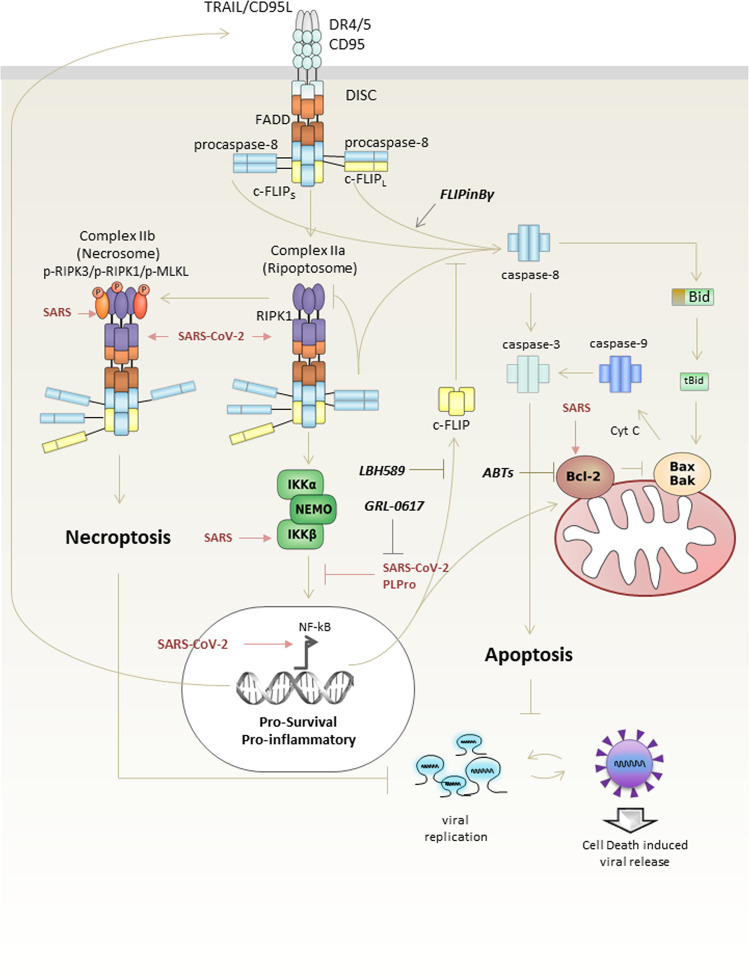
The extrinsic cell death pathway is triggered via DRs9. Activation of CD95 or TRAIL-R1/2 with cognate ligands or agonistic antibodies results in the formation of the DISC at the cellular membrane20. DISC consists of oligomerized receptors, the adaptor protein FADD, initiator zymogens procaspase-8 and -10, and the c-FLIP (Fig. 2). All interactions at the DISC are based on the intricate network of DD and DED interactions. DDs provide the basis for FADD and DR interactions, while DEDs are essential for the formation of the so-called DED filaments that de facto present the platform for procaspase-8 dimerization and activation30–32. Procaspase-8 activation at the DED filaments is inhibited by c-FLIP proteins33. One long and two short c-FLIP isoforms, named Long (L), Short (S), and Raji (R), i.e., c-FLIPL, c-FLIPS, and c-FLIPR, were reported34. c-FLIPL at the DISC can act both in a proapoptotic as well as in an antiapoptotic manner35,36. The proapoptotic function of c-FLIPL is mediated by the formation of catalytically active procaspase-8/c-FLIPL heterodimers37,38. Short c-FLIP isoforms, c-FLIPS and c-FLIPR, act solely in an antiapoptotic manner34. Upon CD95 or TRAIL-R1/2 stimulation, procaspase-8 might be activated in other complexes, in particular, in the complex IIa or ripoptosome17,18 (Fig. 2). This complex is formed upon deubiquitinylation of another key protein of the DD networks—RIPK1, and similar to DISC comprises DED proteins: FADD, procaspase-8/10, and c-FLIP. Upon stimulation of the DRs and inhibition of caspase-8, RIPK1 serves as a core for the formation of the complex IIb or necrosome promoting necroptosis39,40 (Fig. 2). This macromolecular platform comprises RIPK1, RIPK3, FADD, procaspase-8/10, and c-FLIP. Strikingly, stimulation of CD95 or TRAIL-R1/2 can also lead to the induction of non cell death pathways, including the induction of transcription factor NF-kB24,41,42.
Fig. 2. Induction of cell death via DR activation and SARS/SARS-CoV-2 infection.
The key signaling platforms involved in programmed cell death through CD95 and DR4/5, as well as their cross talk are shown. The proteins that are regulated by SARS and SARS-CoV-2 infections are denoted in purple. Small-molecule targets are indicated with black arrows.
Coronaviridae cause various diseases, including bronchitis, gastroenteritis, hepatitis, and systemic diseases on humans, bats, rodents, animals, and birds. Both host and viral factors affect coronavirus virulence and the disease severity in animals, which has allowed to study the key mechanisms of virus influence on cell death networks. For instance, it has been shown that infection of cell lines with canine coronavirus type II (CCoV-II) controls both intrinsic and extrinsic apoptosis to promote the replication of the virus43,44. Strikingly, one of the key mechanisms of virus replication control involves the inhibition of extrinsic apoptosis at early stages of infection via c-FLIP proteins44. c-FLIP expression is suppressed by the forkhead transcription factor FOXO3A45,46. The level of FOXO3A mRNA was found to be increased 12 h after CCoV-II infection, which might lead to triggering extrinsic apoptosis44. Moreover, CCoV-II infection is characterized by elevated levels of TRAILR 12 h post infection, suggesting an important role for the extrinsic apoptosis pathway at the later stages of canine coronavirus infection44.
Similar mechanisms might be involved in SARS-CoV-2 infections. Inducing extrinsic apoptosis at the early stage of infection might impair virus replication, and therefore this response might be counteracted by the virus through c-FLIP upregulation. SARS-CoV-2 virus can take advantage of this c-FLIP-mediated cell death delay for its own replication. Indeed, the most recent studies have demonstrated transcriptional changes for the key regulatory proteins and miRNAs of the DR pathways upon SARS-CoV-2 infection47–49. In particular, the upregulation of miRNA-155 was reported, which along with other targets represses FOXO3A that is a key suppressor of c-FLIP expression50. Remarkably, the c-FLIP expression level was found to be enhanced in several SARS-CoV-2-infected cell lines, together with key mediators of the DR signaling networks, such as A20 and DR547. Elevated c-FLIP expression was observed at very early time points after SARS-CoV-2 infection, as well as in postmortal lung biopsies of COVID-19 patients. In line with these data, it is logical to assume that blocking extrinsic apoptosis via upregulation of c-FLIP proteins is one of the ways for SARS-CoV-2 to sustain its replication.
Besides c-FLIP protein, other targets of SARS-CoV-2 in the DR network are currently being uncovered. The upregulation of DR5 and A20 mRNAs, which is mentioned above, probably plays a key role at the later stages of infection by triggering cell death (Fig. 2). Likewise, using proteomic analysis, it was shown that SARS-CoV-2 directly interacts with RIPK1, one of the major components of DR complexes, e.g., complex IIa/ripoptosome or IIb/necrosome2. However, the role of this interaction in the viral life cycle remains to be elucidated. It might control apoptosis or necroptosis induction at the complex IIa or IIb, respectively, at the later stages of the viral response. Accordingly, SARS-CoV infection was able to target RIPK3, another key component of complex IIb, facilitating necroptotic cell, which might be similar for SARS-CoV-251.
The delayed apoptosis of host cells is of advantage for the replication of coronavirus and aggravating the infection at its early stages. Therefore, inducing extrinsic apoptosis and caspase-8 activation via targeting c-FLIP or caspase-8 at the beginning of the infection seems to be an important strategy to inhibit virus replication, and limit the number of viruses. In this regard, recent reports on small molecules activating caspase-8 via targeting caspase-8/c-FLIPL heterodimers, or small molecules activating caspase-8 at the initial steps after death ligand administration might be of particular interest52 (Fig. 2). In light of this hypothesis, the application of histone deacetylase inhibitors, such as LBH589, that have been accepted as effective agents against c-FLIP, presents an additional promising strategy53 (Fig. 2). Broad-spectrum antivirals, for instance, dsRNA-activated caspase oligomerizer, which selectively induces apoptosis in virus-containing host cells, also can be evaluated for their effectiveness against SARS-CoV-254.
As mentioned above, coronaviruses also control the intrinsic apoptosis pathway. SARS Orf7a was shown to interact with antiapoptotic members of the Bcl-2 family, which might facilitate apoptosis at the later infection stages55. Bcl-xL was reported to play a role in suppression of proapoptotic activities of SARS in T cells56. The putative role of similar regulation in SARS-CoV-2 infection is yet to be studied. Moreover, the small-molecule ABT-263 targeting Bcl-2/Bcl-XL was reported to have antiviral properties and facilitate apoptosis induction upon infection, with several DNA and RNA viruses, including MERS-CoV57. It makes targeting Bcl-2 proteins another promising strategy to sensitize cells infected with SARS-CoV-2 to apoptosis.
The NF-κB pathway is considered as a major antiapoptotic pathway24,47. Activation of NF-κB leads to the upregulation of the most important apoptosis inhibitors, such as c-FLIP, antiapoptotic Bcl-2 family members, and XIAPs. According to the analysis of mRNA expression, several key components of the NF-κB pathway were shown to be upregulated upon SARS-CoV-2 infection47–49 (Fig. 2). Along with other NF-κB targets, increased expression of inhibitors of the apoptotic pathway, such as c-FLIP and XIAPs, was detected in this analysis. Moreover, according to recent proteomic analyses, multiple protein products of genes from the NF-κB pathway were shown to interact with SARS-CoV-2 proteins2. Hence, selective targeting of the NF-κB machinery in combination with inducing apoptosis might pave the way toward development of specific strategies for down modulating the replication of SARS-CoV-2. In this regard, it has to be mentioned that both SARS-CoV and MERS-CoV were reported to have multiple mechanisms to regulate the NF-κB induction by facilitating its activation or, on the opposite, suppressing it58. These different ways of influencing the NF-κB pathway might play a role at different stages of the infection.
Hyperactivation of the NF-κB pathway is one classical feature of viral infections, and might be important for the early stages of SARS-CoV-2 infection to block apoptosis. The induction of NF-κB activity is reported to be mediated by SARS-CoV 3a and 7a, and nsp1 proteins58. Several small-molecule inhibitors developed to inhibit the NF-κB activation, such as proteasome inhibitors, represent another valuable strategy to target the antiapoptotic proinflammatory response induced by SARS-CoV-2 and induce apoptosis59.
The suppression of NF-κB activity seems to be a crucial step for viral infection in promoting cell death. Among several mechanisms of NF-κB suppression by SARS-CoV is the direct interaction of the SARS-CoV M protein with IKKβ, leading to suppression of the NF-κB activation60. The proteolytic activity of SARS-CoV papain-like protease (PLpro) protease was also found to repress the NF-κB activation by removing the K48-linked ubiquitin chains from IκBα61,62. The latter is a crucial step in the activation of NF-κB leading to the degradation of IκBα and nuclear translocation of NF-κB. Interestingly, SARS-CoV-2 PLpro was reported to be less active in deubiquitinating critical components of the NF-κB pathway compared to SARS PLpro, but more active in suppressing type I interferon responses63. Small-molecule inhibitors of PLpro were reported to be active against both SARS and SARS-CoV-2, including the compound GRL-061761,63. Their potential for clinical use is currently being evaluated. In addition, it has to be mentioned that SARS-CoV-2 infection may regulate extrinsic cell death and NF-κB pathways through interferon regulatory factors (IRFs), and therefore small-molecule inhibitors of PLpro are of high interest as an additional point of interference with extrinsic cell death and NF-κB pathways.
Taken together, dual regulation of cell death and NF-κB activity can play important role in regulation of the proinflammatory response, controlling the programmed cell death at different time intervals of viral replication. In this regard, it is also striking that MERS-CoV has been reported to induce apoptosis in primary T lymphocytes3. Moreover, there is the first evidence of involvement of the CD95 pathway in SARS-CoV-2-infected lymphoid cells, which would suppress the immune system of SARS-CoV-2-infected patients. In this case, the opposite strategy of restraining apoptosis would be particularly helpful in the battle between the host immune system and SARS-CoV-2.
Targeting DD-mediated networks at the later stages of SARS-CoV-2 infection
Cytokine storm that occurs at the later stages of SARS-CoV-2 infection is reported to be one of the major reasons of high lethality due to the SARS-CoV-2 virus64. It is characterized by the release of the proinflammatory cytokines, such as TNF-α, IL6, and others. Another form of programmed cell death, termed pyroptosis, significantly contributes to the maturation of proinflammatory cytokines, such as IL-1β65. The maturation of IL-1β occurs upon assembly of the inflammasome, which serves as an initiator macromolecular platform in the pyroptosis pathway66 (Fig. 3). The inflammasome comprises pattern recognition receptors, including PYD-containing 3 (NLRP3), adaptor protein apoptosis-associated speck-like protein containing a CARD (ASC), and inflammatory procaspases, such as procaspase-165. The assembly of the inflammasome is based on interactions between CARDs of procaspase-1 and ASC adaptor protein12. Inflammasome assembly leads to the autocatalytic activation of procaspase-1, and subsequent processing of pro-IL-1β and pro-IL-18 into their mature forms. A hyperproduction of proinflammatory cytokines leads to the cytokine storm that has been well-documented for SARS, and is considered to play a major role in SARS-CoV-2 infection as well64 (Fig. 3). In particular, for SARS-CoV infection, ORF and E proteins were reported to strongly facilitate NLRP3 inflammasome activation and similar mechanisms might play a role in SARS-CoV-2 infection67,68.
Fig. 3. Influence of SARS, MERS, and SARS-Cov-2 infection on the DD network.
The key signaling platforms comprising DD superfamily members are depicted, as well as their cross talk. The proteins that are regulated by SARS, MERS, and SARS-Cov-2 infection are denoted in purple.
Furthermore, it might be suggested that reprogramming of pyroptosis through targeting the proteins of the extrinsic cell death pathway to induce different forms of programmed cell death may lead to a less pathogenic phenotype of SARS-CoV-2. This strategy can be achieved due to the tight cross talk between pyroptosis and apoptosis mediated by the members of extrinsic cell death pathways25,26,69. In particular, in recent studies it was shown that inflammasome adaptor ASC recruits procaspase-8 through a PYD/DED interaction32. Moreover, it was shown that catalytically inactive caspase-8 has a scaffold function in pyroptosis induction by promoting formation of ASC specks25,26. However, there are also studies indicating that the recruitment of procaspase-8 to the inflammasome results in apoptosis induction70,71. This proapoptotic activity is dependent on caspase-8 and negatively regulated by elevated c-FLIP levels. This further underlines the importance of compounds reducing c-FLIP levels, as promising agents against SARS-CoV-2 along with the compounds activating caspase-852,53. Interestingly, c-FLIP has been recently shown to protect macrophages from LPS-induced pyroptosis via inhibition of complex II formation; thus, playing a dual role in pyroptosis and apoptosis regulation72. Accordingly, the strategy of increasing caspase-8 activity should certainly take into account the overall c-FLIP levels in the cell and their influence on caspase-8 activation.
vRNA is sensed by the innate immune system and triggers an antiviral response. Several members of Toll-like receptors (TLR) are involved in the recognition of viral dsRNA and innate immune system activation73,74. The major adaptor protein of TLR signaling is MyD88 that comprises DD. Thereby, MyD88 recruits the DD proteins IRAK4 and IRAK1 to form the oligomeric macromolecular complex termed Myddosome75. Myddosome allows proximity-induced phosphorylation of IRAKs, and their activation to trigger innate immunity pathway activation via TNF-receptor-associated factor 3 (TRAF3) and TRAF676. The Myddosome pathway triggers the activation of a number of genes controlling the immune response (Fig. 3). Accordingly, it is not surprising that SARS-CoV-2 apparently developed multiple strategies to evade the Myddosome pathway (Fig. 3). In particular, SARS-CoV-2 was reported to target TBK1 and TBKBP1, key proteins involved in the activation of interferon-induced gene expression2. Importantly, similar mechanisms have been reported for MERS and SARS infections (Fig. 3). Moreover, SARS-CoV PLPro was reported to catalyze deubiquitination of TRAF3 and TRAF6, inhibiting the TLR7 innate immunity pathway77. This pathway is also the one leading to the suppression of IRF3 activation by MERS-CoV and SARS-CoV27. As mentioned above, small-molecule inhibitors of PLPro demonstrated potent activity against SARS-CoV and SARS-CoV-2 infection61,63, likely through complex activity including suppression of viral replication and restoration of the IRF3 pathway63.
Alternative to the Myddosome pathway, TLR3 and TLR4 receptors can engage TRADD via interactions with the TRIF domain78. This, in turn, results in the recruitment of RIPK1, likely through the formation of a DD platform that is capable of FADD and subsequent caspase-8/c-FLIP recruitment. This complex is reported to regulate inflammatory and cell death responses, and can be negatively modulated by the cellular inhibitor of apoptosis protein cIAP79. Interestingly, major proteins of RIPK1-dependent cell death activation were shown to be targeted by CoV proteins; however, there were no reports on the direct regulation of caspase-8-dependent cell death activation. This suggests that the modulation of caspase-8 activity in the complex formed through TLR3/TLR4 activation might also present a promising strategy for antiviral effects leading to apoptosis activation. Likewise, c-FLIP downregulation with the chemotherapeutic agent paclitaxel is reported to enhance the apoptotic cell death through TLR3 activation, which might imply a promising strategy80. Taken together, this further underlines the importance of developing compounds specifically increasing caspase-8 activity, also for the latter stages of the SARS-CoV-2 infection.
The DD, DED, and CARD interaction network also plays an important role in the intracellular dsRNA recognition pathway, which is mediated by RIG-I and MDA-5. Both RIG-I and MDA-5 comprise N-terminal CARDs that recruit MAVS through its CARD leading to the formation of macromolecular complex at the mitochondria. This complex also comprises DD- and DED-containing TRADD/RIPK1/caspase-8/FADD proteins leading to a macromolecular platform, which mediates the activation of IRF3 and NF-kB81. The MAVS/caspase-8 platform was shown to link RNA virus innate antiviral signaling to apoptosis82. The involvement of this pathway has been shown for SARS-CoV and MERS-CoV27, which allows to suggest an important role in SARS-CoV-2 infection as well. In line with this hypothesis, recent proteomics studies have demonstrated that MAVS might be indirectly regulated by SARS-CoV-2 through interactions with TOMM702. The latter is a critical adaptor linking MAVS to the TBK/IRF3 complex at the mitochondria. Moreover, the role of DED proteins in the regulation of the innate immunity pathway suggests that targeting caspase-8 and c-FLIP represents another potential antiviral strategy. In this regard, an important role belongs to uncovering the distinct roles of procaspase-8 and c-FLIP in the activation of the intracellular dsRNA recognition pathway.
The development of therapeutic approaches based on the targeting the DD superfamily network is very important for combatting SARS-CoV-2 infection. At the early stages of infection, as discussed above, inducing apoptosis of infected cells via increasing caspase-8 activity, using small molecules or triggering apoptosis via TRAIL receptor agonists might be beneficial. In this regard, it might be hypothesized that a combinations of drugs can be derived from the clinical trials of TRAIL receptor agonists in cancer cells (Supplementary Table 1). Targeting the DD superfamily network plays a pivotal role in inflammation control during the later stages of SARS-CoV-2 infection. In this regard, a key role should belong to anti-inflammatory modulators, such as the above mentioned small-molecule inhibitors of PLpro and TNFα antagonists. The latter are in clinical trials performed by several groups (Table 1). In addition, the above discussed mechanisms of cross talk between apoptosis, pyroptosis, and inflammation via DD proteins have to be specially considered in future studies for the development of therapeutic approaches against SARS-CoV-2 infection.
Table 1.
Clinical trials of TNFα inhibitors in COVID-19.
| TNFα inhibitors | Number of patients | Combination | Clinical phase | Reference |
|---|---|---|---|---|
| Infliximab (Remicade, Kineret) | 17 | Premedication with Tylenol or diphenhydramine and prednisone | II | NCT04425538 |
| Emapalumab (Gamifant) | 54 | None | II/III | NCT04324021 |
| XPRO1595 | 366 | None | II/III | NCT04370236 |
Taken together, DD superfamily members play a pivotal role in the formation of macromolecular platforms that initiate multiple signaling pathways from cell death to the inflammatory response. Moreover, the different stoichiometry of DD, DED, PYD, and CARD-containing proteins that provide the major core of these macromolecular platforms largely defines the cross talk between these pathways. Hence, targeting the key DD superfamily members presents a very important strategy in the development of treatment therapies for SARS-CoV-2 infections.
Supplementary information
Acknowledgements
We acknowledge Phillip Schwartz Stiftung, Wilhelm Sander-Stiftung (2017.008.02), Center of dynamic systems (CDS), funded by the EU-program ERDF (European Regional Development Fund), DFG (LA 2386), Russian Foundation for Basic Research (19-54-45015) and Russian State Budget Project (АААА-А17-117092070032-4) for supporting our work. Open Access funding enabled and organized by Projekt DEAL.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by Ivano Amelio
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41420-020-00331-w) contains supplementary material, which is available to authorized users.
References
- 1.Chang CK, et al. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature583, 459–468 (2020). [DOI] [PMC free article] [PubMed]
- 3.Chu H, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T Lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krammer PH, Arnold R, Lavrik IN. Life and death in peripheral T cells. Nat. Rev. Immunol. 2007;7:532–542. doi: 10.1038/nri2115. [DOI] [PubMed] [Google Scholar]
- 7.Lavrik I, Golks A, Krammer PH. Death receptor signaling. J. Cell Sci. 2005;118:265–267. doi: 10.1242/jcs.01610. [DOI] [PubMed] [Google Scholar]
- 8.Seyrek K, Lavrik IN. Modulation of CD95-mediated signaling by post-translational modifications: towards understanding CD95 signaling networks. Apoptosis. 2019;24:385–394. doi: 10.1007/s10495-019-01540-0. [DOI] [PubMed] [Google Scholar]
- 9.Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat. Immunol. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- 10.Lavrik IN. Systems biology of death receptor networks: live and let die. Cell Death Dis. 2014;5:e1259. doi: 10.1038/cddis.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strasser A. The physiological relevance of death receptor-mediated apoptosis. Nat. Rev. Mol. Cell Biol. 2014;15:633. doi: 10.1038/nrm3875. [DOI] [PubMed] [Google Scholar]
- 12.Park HH, et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park HH, Wu H. Crystallization and preliminary X-ray crystallographic studies of the oligomeric death-domain complex between PIDD and RAIDD. Acta Crystallogr Sect. F Struct. Biol. Cryst. Commun. 2007;63:229–232. doi: 10.1107/S1744309107007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Sci. Signal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linkermann A, Green DR. Necroptosis. N. Engl. J. Med. 2014;370:455–465. doi: 10.1056/NEJMra1310050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feoktistova M, et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenev T, et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Hughes MA, et al. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol. Cell. 2009;35:265–279. doi: 10.1016/j.molcel.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Muzio M, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death–inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/S0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 21.Vanden Berghe T, Hassannia B, Vandenabeele P. An outline of necrosome triggers. Cell Mol. Life Sci. 2016;73:2137–2152. doi: 10.1007/s00018-016-2189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sprick MR, et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 2000;12:599–609. doi: 10.1016/S1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 23.Weinlich R, Oberst A, Beere HM, Green DR. Necroptosis in development, inflammation and disease. Nat. Rev. Mol. Cell Biol. 2017;18:127–136. doi: 10.1038/nrm.2016.149. [DOI] [PubMed] [Google Scholar]
- 24.Henry CM, Martin SJ. Caspase-8 acts in a non-enzymatic role as a scaffold for assembly of a pro-inflammatory “FADDosome” complex upon TRAIL stimulation. Mol. Cell. 2017;65:715–729. doi: 10.1016/j.molcel.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Newton K, et al. Activity of caspase-8 determines plasticity between cell death pathways. Nature. 2019;575:679–682. doi: 10.1038/s41586-019-1752-8. [DOI] [PubMed] [Google Scholar]
- 26.Fritsch M, et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature. 2019;575:683–687. doi: 10.1038/s41586-019-1770-6. [DOI] [PubMed] [Google Scholar]
- 27.Fung TS, Liu DX. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 28.Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nailwal H, Chan FK. Necroptosis in anti-viral inflammation. Cell Death Differ. 2019;26:4–13. doi: 10.1038/s41418-018-0172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickens LS, et al. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol. Cell. 2012;47:291–305. doi: 10.1016/j.molcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schleich K, et al. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol. Cell. 2012;47:306–319. doi: 10.1016/j.molcel.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Fu TM, et al. Cryo-EM structure of caspase-8 Tandem DED filament reveals assembly and regulation mechanisms of the death-inducing signaling complex. Mol. Cell. 2016;64:236–250. doi: 10.1016/j.molcel.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozturk S, Schleich K, Lavrik IN. Cellular FLICE-like inhibitory proteins (c-FLIPs): fine-tuners of life and death decisions. Exp. Cell Res. 2012;318:1324–1331. doi: 10.1016/j.yexcr.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 34.Hughes MA, et al. Co-operative and hierarchical binding of c-FLIP and caspase-8: a unified model defines how c-FLIP isoforms differentially control cell fate. Mol. Cell. 2016;61:834–849. doi: 10.1016/j.molcel.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fricker N, et al. Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL. J. Cell Biol. 2010;190:377–389. doi: 10.1083/jcb.201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang DW, et al. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu JW, Jeffrey PD, Shi Y. Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl Acad. Sci. USA. 2009;106:8169–8174. doi: 10.1073/pnas.0812453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Micheau O, et al. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 2002;277:45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- 39.Zamaraev AV, Kopeina GS, Zhivotovsky B, Lavrik IN. Cell death controlling complexes and their potential therapeutic role. Cell Mol. Life Sci. 2015;72:505–517. doi: 10.1007/s00018-014-1757-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton K, et al. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature. 2019;574:428–431. doi: 10.1038/s41586-019-1548-x. [DOI] [PubMed] [Google Scholar]
- 41.Neumann L, et al. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol. Syst. Biol. 2010;6:352. doi: 10.1038/msb.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartwig T, et al. The TRAIL-induced cancer secretome promotes a tumor-supportive immune microenvironment via CCR2. Mol. Cell. 2017;65:730–742. doi: 10.1016/j.molcel.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggieri A, et al. Canine coronavirus induces apoptosis in cultured cells. Vet. Microbiol. 2007;121:64–72. doi: 10.1016/j.vetmic.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marfe G, et al. Involvement of FOXO transcription factors, TRAIL-FasL/Fas, and sirtuin proteins family in canine coronavirus type II-induced apoptosis. PLoS ONE. 2011;6:e27313. doi: 10.1371/journal.pone.0027313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Fulda S. Targeting c-FLICE-like inhibitory protein (CFLAR) in cancer. Expert Opin. Ther. Targets. 2013;17:195–201. doi: 10.1517/14728222.2013.736499. [DOI] [PubMed] [Google Scholar]
- 46.Shirley S, Micheau O. Targeting c-FLIP in cancer. Cancer Lett. 2013;332:141–150. doi: 10.1016/j.canlet.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emanuel, W. et al. Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention. Preprint at 10.1101/2020.05.05.079194 (2020).
- 49.Sun, J. et al. Comparative transcriptome analysis reveals the intensive early-stage responses of host cells to SARS-CoV-2 infection. Preprint at 10.1101/2020.04.30.071274 (2020). [DOI] [PMC free article] [PubMed]
- 50.Yu DD, et al. Role of miR-155 in drug resistance of breast cancer. Tumour Biol. 2015;36:1395–1401. doi: 10.1007/s13277-015-3263-z. [DOI] [PubMed] [Google Scholar]
- 51.Yue Y, et al. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hillert, L. K. et al. Dissecting DISC regulation via pharmacological targeting of caspase-8/c-FLIPL heterodimer. Cell Death Differ.27, 2117–2130 (2020). [DOI] [PMC free article] [PubMed]
- 53.Scuto A, et al. The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph- acute lymphoblastic leukemia cells. Blood. 2008;111:5093–5100. doi: 10.1182/blood-2007-10-117762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rider TH, et al. Broad-spectrum antiviral therapeutics. PloS ONE. 2011;6:e22572. doi: 10.1371/journal.pone.0022572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan YX, et al. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 2007;81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, et al. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochemical J. 2005;392:135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bulanova, D. et al. Antiviral properties of chemical inhibitors of cellular anti-apoptotic Bcl-2 Proteins. Viruses9, 271 (2017). [DOI] [PMC free article] [PubMed]
- 58.Lim, Y. X., Ng, Y. L., Tam, J. P. & Liu, D. X. Human coronaviruses: a review of virus-host interactions. Diseases4, 26 (2016). [DOI] [PMC free article] [PubMed]
- 59.Miller SC, et al. Identification of known drugs that act as inhibitors of NF-kappaB signaling and their mechanism of action. Biochem. Pharm. 2010;79:1272–1280. doi: 10.1016/j.bcp.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fang X, et al. The membrane protein of SARS-CoV suppresses NF-kappaB activation. J. Med Virol. 2007;79:1431–1439. doi: 10.1002/jmv.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ratia K, Kilianski A, Baez-Santos YM, Baker SC, Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bekes M, et al. Recognition of Lys48-Linked di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol. Cell. 2016;62:572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin, D. et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 10.1038/s41586-020-2601-5 (2020). [DOI] [PMC free article] [PubMed]
- 64.Shi Y, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kepp O, Galluzzi L, Zitvogel L, Kroemer G. Pyroptosis - a cell death modality of its kind? Eur. J. Immunol. 2010;40:627–630. doi: 10.1002/eji.200940160. [DOI] [PubMed] [Google Scholar]
- 66.Sagulenko V, Vitak N, Vajjhala PR, Vince JE, Stacey KJ. Caspase-1 Is an apical caspase leading to caspase-3 cleavage in the AIM2 inflammasome response, independent of caspase-8. J. Mol. Biol. 2018;430:238–247. doi: 10.1016/j.jmb.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 67.Siu KL, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nieto-Torres JL, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tummers, B. et al. Caspase-8-dependent inflammatory responses are controlled by its adaptor, fadd, and necroptosis. Immunity52, 994–1006 (2020). [DOI] [PMC free article] [PubMed]
- 70.Vajjhala PR, et al. The inflammasome adaptor ASC induces procaspase-8 death effector domain filaments. J. Biol. Chem. 2015;290:29217–29230. doi: 10.1074/jbc.M115.687731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sagulenko V, et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013;20:1149–1160. doi: 10.1038/cdd.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muendlein HI, et al. cFLIPL protects macrophages from LPS-induced pyroptosis via inhibition of complex II formation. Science. 2020;367:1379–1384. doi: 10.1126/science.aay3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 75.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li, S. W. et al. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 17, 678 (2016). [DOI] [PMC free article] [PubMed]
- 78.Ermolaeva MA, et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat. Immunol. 2008;9:1037–1046. doi: 10.1038/ni.1638. [DOI] [PubMed] [Google Scholar]
- 79.Estornes Y, et al. dsRNA induces apoptosis through an atypical death complex associating TLR3 to caspase-8. Cell Death Differ. 2012;19:1482–1494. doi: 10.1038/cdd.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alkurdi L, et al. Release of c-FLIP brake selectively sensitizes human cancer cells to TLR3-mediated apoptosis. Cell Death Dis. 2018;9:874. doi: 10.1038/s41419-018-0850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Michallet MC, et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28:651–661. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 82.El Maadidi S, et al. A novel mitochondrial MAVS/Caspase-8 platform links RNA virus-induced innate antiviral signaling to Bax/Bak-independent apoptosis. J. Immunol. 2014;192:1171–1183. doi: 10.4049/jimmunol.1300842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.