Abstract
A recent outburst of the pandemic caused by a member of the coronaviridae family identified as SARS-CoV-2. The highly contagious nature of the virus allows it to spread rapidly worldwide and caused severe healthcare and economic distress. So far, no proper line of treatment or vaccines has been available against SARS-CoV-2. Since, the infected people rapidly increased, causing the saturation of healthcare systems with coronavirus disease (COVID-19) patients. As the virus spread to new locations it also acquired various mutations. Here, in this study, we focused on identifying mutations in one of the crucial complex of SARS-CoV-2, the Nsp10-Nsp16 2′-O-methyltransferase complex. This complex plays indispensable role in the post-transcriptional modifications of viral RNA by its capping. We analysed 208 sequences of Nsp10-Nsp16 reported from India and compared with first reported sequence from Wuhan, China. Our analysis revealed a single mutation in Nsp10 and five mutations in Nsp16 protein. We also show that these mutations are leading to alteration in the secondary structure of Nsp10-Nsp16. Further, the protein modelling studies revealed that the mutation of both Nsp10-Nsp16 impacts the protein dynamicity and stability. Altogether, this study provides novel insights into the variations observed in the proteins of SARS-CoV-2 that might have functional consequences.
Keywords: COVID-19, SARS-CoV-2, Mutations, Nsp10-Nsp16, Infectious diseases, India
Highlights
-
•
The non-structural protein of SARS-CoV-2, Nsp 10 and Nsp16 is mutated as it spreads to new locations.
-
•
SARS-CoV-2 harbours six mutations in Nsp 10-Nsp16 complex among Indian Isolates.
-
•
These mutation alters secondary structure of both Nsp 10 and Nsp16 and also affects protein dynamicity.
1. Introduction
The severe acquired respiratory syndrome coronavirus-2 (SARS-CoV-2) has caused a global pandemic with Coronavirus disease-19 (COVID-19) [1]. The COVID-19 causes respiratory distress with mild to severe symptoms and lethal to the patients with chronological illness or compromised immune system [[2], [3], [4]]. As of June 14 2020, approximately 8 million people are infected with SARS-CoV-2 and more than 0.4 million reported deaths worldwide. This pandemic started from Wuhan province, China and thought to emerge from wet seafood market area of Wuhan [5]. SARS-CoV-2 is highly contagious and within a span of two months it spread to almost all countries worldwide [6]. The up-to-date and real-time information on virus spread and deaths caused by COVID-19 are publicly accessible online by worldometers [7]. Various countries have already declared COVID-19, an emergency and initiated large scale preparations to tackle this pandemic that includes travel restrictions, social distancing, personal and community hygiene, lockdown to limit spread and many others [8].
SARS-CoV-2 has around 30 kb single-stranded, positive sense RNA genome [5]. Its genome encodes four structural, sixteen non-structural and nine accessory proteins [9]. The RNA is capped at 5′end and also has a 3′end poly-a tail that enables them to be translated as they infect host cells [10]. The 5′end of nascent mRNA is modified by a combination of enzymes that includes Nsp10-Nsp16 complex which mediated 2′-O-methylation of the coronavirus RNA [11]. Studies show that the Nsp10 stimulates the enzymatic activity of Nsp-16 upon binding; however, Nsp16 is the main enzymatic partner of the complex that is a m7GpppA-specific, S-Adenosyl methionine-dependent, 2′-O-MTase [12]. The Nsp10 possess an interacting surface through which it binds to Nsp16 and provides favourable conditions for the enzymatic reaction to take place [13]. The Nsp16 have a conserved four amino acid sequence ‘KDKE’ in its catalytic pocket that contributes to methyltransferase activity [14,15]. Basically, this complex modifies viral RNA that leads to close resemblance with the host or human cell RNAs. As a consequence of this modification the viral RNA remains undetected by host cell immune surveillance mechanisms allowing viral RNA to translate as well as to multiply. It has been observed that the abrogation of 2′-O-MTase activity of Nsp10-Nsp16 complex of SARS-CoV leads to significant reduction in viral replication and recovery of animal models also significantly increases [16]. Therefore, Nsp10-Nsp16 complex has emerged as an attractive SARS-Cov-2 target and researchers are trying to identify drugs that can specifically inhibit this complex enzymatic activity.
The recent advances from the SARS-CoV-2 genomic sequencing from different countries indicate that the SARS-CoV2 is rapidly evolving by acquiring mutations in its genome [17]. It has been suggested that the new variants of SARS-CoV-2 might attain better adaptation at new geographical areas that can make it more potent than the virus that emerged from Wuhan, China. Such as, D614G mutant (Spike glycoprotein) has emerged as the predominant form of SARS-CoV-2 [18,19]. Moreover, as the virus spread to new locations including India, the genomic sequences of SARS-CoV-2 from Indian COVID-19 patients were also conducted and deposited in NCBI-virus repositories. In order to understand the variations in SARS-CoV-2, we initiated this study with the aim to identify new mutants from India. Here, we identified six mutation from Nsp10-Nsp16 complex and discussed their possible implication on its structure and function.
2. Material and methods
2.1. Sequence retrieval
The sequences of SARS-CoV-2 from Indian COVID-19 patients are deposited in NCBI-virus database which is publicly available. We downloaded Orf1ab protein sequence (7096 residues) from this database that harbours all non-structural proteins encoded by SARS-CoV-2 RNA genome (Nsp1-16). As of June 10 2020, there are 208 protein sequences of Orf1ab deposited form India. The list of Orf1ab protein accession number (208 sequences) used in this analyses are mentioned in Table 1. The protein accession number of Orf1ab (reference sequence) used in this analysis is YP_009724389 which was reported from Wuhan, China at the beginning of the pandemic [5]. We extracted Nsp10 and Nsp16 protein sequence from Orf1ab. Nsp10 is a 139 residues long protein that starts from 4254 till 4392 of Orf1ab and Nsp16 is a 298 residues long protein that starts from 6799 till 7096 of Orf1ab. Both Nsp10 and Nsp16 are proteolytically cleaved from Orf1ab polypeptide by the protease encoded in SARS-CoV-2 genome.
Table 1.
List showing the protein accession number of SARS-CoV-2 Orf1ab that harbours Nsp10 and Nsp16 sequences from India used in this study.
2.2. Polypeptide residues alignment
To identify the variation in the amino acid sequences of Nsp10 and Nsp16 isolated from India, we compared Indian sequences with the first reported sequence from Wuhan province, China also known as reference sequence [5]. The protein accession number of reference sequence used in this analysis is YP_009724389. For this analysis we used Clustal Omega tool [20]. This online program can perform alignment task with thousands of input polypeptide sequences and this program uses seeded guide trees and HMM profile-profile techniques to generate alignments. After the alignment the variation in amino acid residues from the reference sequence was carefully marked and noted for further analysis.
2.3. Secondary structure prediction
To understand the contribution of the mutant amino acid in the secondary structure of Nsp10 and Nsp16, we used secondary structure prediction tool [21]. This prediction tool provides the information about the possible variation in the localised secondary structures such as helix, sheet coiled coil and turns upon mutation.
2.4. Protein modelling to measure protein dynamicity
The three dimensional structures of protein attains the best possible native conformation. The mutation can lead to the variation in protein structure that can impact the dynamicity of the protein. To understand, if any, variation in protein dynamicity is contributed by the mutations identified in Nsp10 and Nsp16, we used protein stability and dynamicity predictor programme called DynaMut [22]. This program requires the solved structure of the target protein. Therefore, we upload the reported structure of Nsp10 -Nsp16 complex (RCSB PDB ID: 6W75) [23] to the DynaMut webserver. This tool provides the difference in free energy between wild type and mutant protein that represents the protein stability. This tool also provides information regarding the difference in vibrational entropy between wild type and mutant protein that demonstrates the protein rigidification and flexibility. Furthermore, DynaMut program can be used to visualise the intramolecular bonds contributed by the selected residues (wild type or mutant) in the three dimensional pockets where these amino acid resides. This information is crucial to understand the close-range variations contributed by the amino acid residues.
3. Results
3.1. Identification of Nsp10 and Nsp16 mutation present among indian SARS-CoV-2 isolates
To identify the variations in the polypeptide sequence of Nsp10 and Nsp16, we compared the Indian SARS-Cov-2 sequences with the reference sequence reported from Wuhan, China. We used Clustal omega tool to perform the multiple alignments. In this analysis we used 208 reported sequences of Nsp10 and Nsp16 from India. Our data demonstrate the presence of single mutation in Nsp10 at 20th position where the wild type alanine residue is substituted by valine in the mutant protein (Fig. 1A). Similarly, we identified five mutations in Nsp16 namely at 7, 111, 116, 236 and 285 positions (Fig. 1B). The two mutations are localised towards the end of N and C-terminus of Nsp16 (P7S and N285D). Other three mutations identified are L111F, A116S and P236L (Fig. 1B).
Fig. 1.
Mutational analysis of Nsp10 and Nsp16. The amino acid sequences of 208 Indian SARS-CoV-2 were compared with first reported sequence from Wuhan, China. The mutations are marked in the schematic diagram of Nsp10 (A) and Nsp16 (B).
3.2. Mutations in Nsp10 and Nsp16 alter the secondary structure of the protein
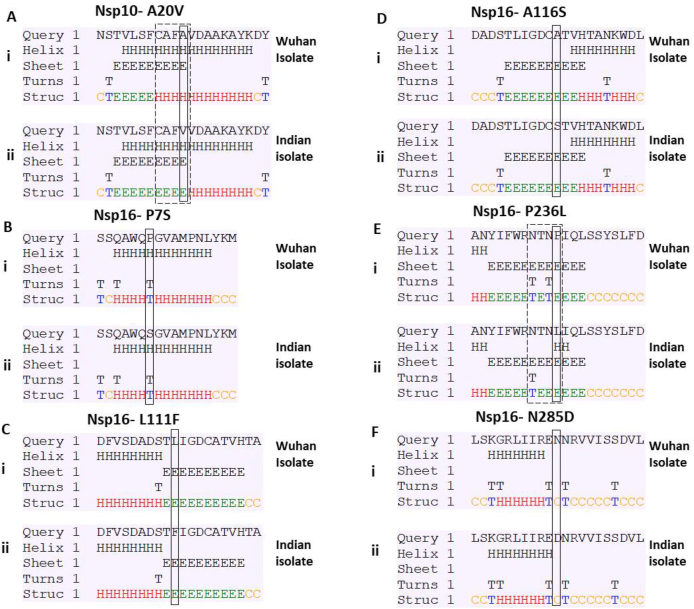
Next, to further understand the possible impact of the identified mutations on protein structure, we studied secondary structure of the protein. The secondary structure of protein relies on the amino acid sequence present in the polypeptide chain. Since, the mutations alter the amino acid sequence; thereby, they can alter the secondary structure of the protein. We used secondary structure prediction tool to identify, if any, variations in secondary structure are contributed by the mutant residues. Our analysis revealed that Nsp10-A20V mutation was leading to alteration in secondary structure (Fig. 2A, compare panel i and ii). The helical structures present from 17 to 20 residues in wild type sequence are replaced by beta sheet (Fig. 2A) in the mutant. Similar analysis with Nsp16 mutations revealed that only P236L (Fig. 2E, compare panel i and ii) was leading to alterations in secondary structure; however, rest of the four mutations (Fig. 2B, C, D and F) does not cause any variation. Nsp16-P236L mutation leads to loss in a turn structure as shown in Fig. 4E. Altogether, we observed the change in secondary structure of both Nsp10 and Nsp16 due to the mutations.
Fig. 2.
Prediction of secondary structure of Nsp10 and Nsp16. The amino acid sequences near the mutation site were uploaded on CFSSP web tool that predict secondary structure. Each panel (A–F) shows the secondary structure of the wild type and mutated input sequences. The panel (i) represents the wild type or Wuhan sequence while panel (ii) represents the mutated Indian Sequence. The mutation site is highlighted in the rectangular box. The variation observed in the secondary structure between Wuhan and Indian sequence is highlighted by dashed box.
Fig. 4.
Analysis of intramolecular interactions contributed by wild type and mutant residues. (A–F) represent the mutations of Nsp 10 and Nsp16 as shown in respective panels. Each panel has wild type and mutant residues highlighted in light green colour. The intramolecular interactions made by mutant and wild type residues are also highlighted. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. The mutation contributes to alteration in protein dynamics and stability of Nsp10 and Nsp16
To substantiate our data, we analysed the conformational stability of the protein by using DynaMut webserver program. First, we analysed the differences in the free energy (ΔΔG) between wild type and the mutant protein. The positive ΔΔG value corresponds to protein stabilisation, and negative value represents protein destabilisation as shown in Table 2. The analysis with Nsp10-A20V mutant revealed the value of ΔΔG (kcal/mol) as −0.129 that indicates protein destabilisation. Similarly, we also analysed the ΔΔG values of Nsp16 mutants. Our data demonstrate the mutation at two position leads to destabilisation in protein structure namely at Nsp16-P7S and Nsp16-N285D with negative ΔΔG (kcal/mol) values of −0.533 and −0.077, respectively. However, the mutation at other three positions leads to stabilisation in protein structure namely at Nsp16-L111F, Nsp16-A116S, and Nsp10-P236L with positive ΔΔG (kcal/mol) values of 0.691, 0.023 and 0.656, respectively.
Table 2.
The table shows the data from protein modelling studies of wild type and mutant Nsp10 and Nsp16. DynaMut webserver was used for this analysis. The values of ‘difference in free energy (ΔΔG)’ and ‘difference in vibrational entropy (ΔΔSVib)’ between wild-type and mutant Nsp10 and Nsp16 proteins are shown.
| S. No | Protein | PDB file used for analysis | Wild type residue | Residue position | Mutant residue | DynaMut predicted ΔΔG (kcal/mol) | DynaMut predicted ΔΔSVib ENCoM (kcal.mol-1.K-1) |
|---|---|---|---|---|---|---|---|
| 1 | Nsp10 | 6w75.pdb | A | 20 | V | −0.129 | −0.038 |
| 2 | Nsp16 | 6w75.pdb | P | 7 | S | −0.533 | 0.05 |
| 3 | Nsp16 | 6w75.pdb | N | 285 | D | −0.077 | 0.151 |
| 4 | Nsp16 | 6w75.pdb | L | 111 | F | 0.691 | −0.245 |
| 5 | Nsp16 | 6w75.pdb | A | 116 | S | 0.023 | −0.108 |
| 6 | Nsp16 | 6w75.pdb | P | 236 | L | 0.656 | −0.34 |
In order to understand how the alteration in stabilisation/destabilisation of protein affects dynamicity of the Nsp10 and Nsp16, we also calculated the difference in vibrational entropy (ΔΔSVib) between the wild-type and mutant protein. The vibrational entropy provides the variation in protein structure in terms of increase or decrease in molecular flexibility as shown in Table 2. The analysis with Nsp10-A20V mutant revealed the ΔΔSVib (kcal.mol−1. K−1) value as −0.038, which represents decrease in molecular flexibility. We did similar analysis with Nsp16 mutants. Our data demonstrate the mutation at two position leads to increase in molecular flexibility namely at Nsp16-P7S and Nsp16-N285D with positive ΔΔSVib (kcal.mol−1. K−1) values of 0.050 and 0.151, respectively. However, the mutation at other three position leads to decrease in molecular flexibility namely at Nsp16-L111F, Nsp16-A116S, and Nsp10-P236L with negative ΔΔSVib (kcal.mol−1. K−1) values of −0.245, −0.108 and −0.340, respectively. The variation in vibrational entropy can also be visualised as shown in Fig. 3. The blue colour represents rigidification in protein structure, and red colour represents gain in flexibility. The data show that Nsp10-A20V (Fig. 3A) mainly contributes to rigidity in protein structure. The data with Nsp16 mutants show that Nsp16-P7S and Nsp16-N285D mainly contributes to gain in flexibility (Figure B and F). However, Nsp16-L111F, Nsp16-A116S, and Nsp10-P236L causes increase in rigidity of protein structure (Fig. 3C, D and E). Altogether, our protein modelling data strongly suggests that the mutations identified in this study alter dynamic structure and stability of Nsp10 and Nsp16.
Fig. 3.
The visual representation of protein dynamicity. Each panel (A–F) demonstrates the individual mutations identified from Indian SARS-CoV-2 isolates. The gain in molecular flexibility is represented by red colour and the increase in rigidity is represented by red colour. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. The mutant residues contribute to alteration in intramolecular interactions in Nsp 10 and Nsp16
Based on our previous data (Fig. 2, Fig. 3) that show the impact of mutations on secondary structure and protein stability motivated us to closely analyse the pocket of the protein where these residues are located. In order to visualise the location of the wild type and mutant residues in the Nsp10 and Nsp16, we used DynaMut webserver. The data show the variations in the intramolecular bonding patterns when the wild type residues are replaced by the mutant (Fig. 4). The detailed analysis revealed that Nsp10-A20V is located in the loop region of the protein structure. Similarly, Nsp16-P7S residue is located in the interface of the loop and beta-sheet (Fig. 4B) and Nsp16-L111F resides in the beta-sheet (Fig. 4C). We observed Nsp16- A116S location in the short helical region (Fig. 4D). Nsp16-P236L and Nsp16-N285D are located in the loop region (Fig. 4E and F). The intramolecular interactions contributed by each residue are highlighted in respective panels and drastic variations are clearly visible in panel C, D and F (Fig. 4). Altogether, our data strongly suggest the alteration in intramolecular interactions in the mutant Nsp10 and Nsp16.
4. Discussion
The COVID-19 caused by SARS-CoV-2 is rapidly spreading to new locations worldwide. The viruses have the tendency to mutate to adapt to new climatic and geographical areas. Various studies show that RdRp, N-protein, Spike glycoprotein are three hotspot proteins that are mutated as this virus spread out from Wuhan, China [17]. Here, we identified six mutations in Nsp10-Nsp16 complex from India. The interface of Nsp10 and Nsp16 is comprised of various residues including L244 and M247 positions of Nsp16 [24]. Our analysis shows a mutation at P236L (Fig. 1B) which is in close vicinity of the crucial L244 and M247 residues. Since, the proline is a bulky group and it affects larger area of the pocket where it resides. We propose that the mutation Nsp16-P236L might affect the interaction between Nsp10 and Nsp16.
The Nsp10-Nsp16 complex is involved in the 2′-O-methylation of the coronavirus RNA [11] and it has become one of the most promising candidates for drug targeting. This complex modifies viral RNA so that it closely resembles host cell mRNA that facilitates the viral protein synthesis and also evasion from host cell immune system. Earlier study with SARS-CoV has shown that inhibiting 2′-O-methyltansferase activity reduces the rate of replication and also alleviates respiratory distress in model organisms [16]. A recent study was conducted to screen a library of thousands of compound with an aim to identify drugs that can efficiently bind and inhibit the 2′-O-methyltansferase activity of Nsp10-Nsp16 complex (doi.org/10.26434/chemrxiv.12279671. v1). The study identified various promising candidates and top two (Velpatasvir and JFD00244) of them bind to a pocket which resides mutant residues identified in our study. The Velpatasvir and JFD00244 require aspartate residue at 114 positions of Nsp16 for its interaction. Our study shows that two mutations in Nsp16 at L111F and A116S which is in close vicinity of the crucial aspartate residue. A recent study reported the interaction of Nsp10-Nsp16 complex with Sinefungin, a panMTase inhibitor [24]. This study also shows the efficient interaction of Sinefungin requires aspartate residue at 114 position [24]. Therefore, these data strongly suggest that this part of the Nsp16 protein (residues 110 to 120) might be a target for drug interactions and the virus is mutating in that region to evade drug interactions.
5. Conclusions
We report mutation in SARS-CoV-2 Nsp10-Nsp16 complex which is a 2′-O-methyltransferase required for viral RNA capping. The mutations reported here might help this virus to evade the pharmacological agents that target Nsp10-Nsp16 complex activity. Further, our study emphasizes the importance of variation studies to be conducted on emerging SARS-CoV-2 variants. It seems that this virus is mutating fast to attain novel functional property that will enable them to survive and adapt better.
CRediT authorship contribution statement
Gajendra Kumar Azad: Conceptualization, Supervision, Methodology, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of competing interest
Authors declare no conflict of interests.
Acknowledgements
We would like to acknowledge Patna University, Patna, Bihar, India for providing infrastructural support for this study.
References
- 1.Zhou P., Yang X. Lou, Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R. Di, Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabi F.A., Al Zoubi M.S., Al-Nasser A.D., Kasasbeh G.A., Salameh D.M. 2020. Sars-cov-2 and Coronavirus Disease 2019: what We Know So Far. Pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worldometers . 2020. COVID-19 Coronavirus Pandemic, Worldometers. [Google Scholar]
- 8.Güner R., Hasanoğlu İ., Aktaş F. Covid-19: prevention and control measures in community. Turk. J. Med. Sci. 2020 doi: 10.3906/sag-2004-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khailany R.A., Safdar M., Ozaslan M. 2020. Genomic Characterization of a Novel SARS-CoV-2, Gene Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020 doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Encinar J.A., Menendez J.A. Potential drugs targeting early innate immune evasion of SARS-coronavirus 2 via 2’-O-methylation of viral RNA. Viruses. 2020 doi: 10.3390/v12050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-o-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lécine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus Nsp 10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014 doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antivir. Res. 2014 doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugari A., Betzi S., Decroly E., Bonnaud E., Hermant A., Guillemot J.C., Debarnot C., Borg J.P., Bouvet M., Canard B., Morelli X., Lécine P. Molecular mapping of the RNA cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menachery V.D., Yount B.L., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-O-methyltransferase activity. J. Virol. 2014 doi: 10.1128/jvi.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020 doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korber B., Fischer W., Gnanakaran S.G., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E., Bhattacharya T., Parker M.D., Partridge D.G., Evans C.M., de Silva T., LaBranche C.C., Montefiori D.C., S.C.-19 G. Group . BioRxiv; 2020. Spike Mutation Pipeline Reveals the Emergence of a More Transmissible Form of SARS-CoV-2. [DOI] [Google Scholar]
- 19.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashok Kumar T. Wide Spectr; 2013. CFSSP: Chou and Fasman Secondary Structure Prediction Server. [DOI] [Google Scholar]
- 22.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemus M.R., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Jaroszewski L., Godzik A., Joachimiak A., Satchell K.J. BioRxiv; 2020. The Crystal Structure of Nsp10-Nsp16 Heterodimer from SARS CoV-2 in Complex with S-Adenosylmethionine. [DOI] [Google Scholar]
- 24.Krafcikova P., Silhan J., Nencka R., Boura E. BioRxiv; 2020. Structural Analysis of the SARS-CoV-2 Methyltransferase Complex Involved in Coronaviral RNA Cap Creation. [DOI] [PMC free article] [PubMed] [Google Scholar]