Abstract
Covid-19 is an infectious respiratory disease due to a coronavirus named SARS-CoV-2. A critical step of the infection cycle is the binding of the virus spike S protein to the cellular ACE-2 receptor. This interaction involves a receptor binding domain (RBD) located at the center of the S trimer, whereas the lateral N-terminal domain (NTD) displays a flat ganglioside binding site that enables the virus to bind to lipid rafts of the plasma membrane, where the ACE-2 receptor resides. S protein binding to lipid rafts can be blocked by hydroxychloroquine, which binds to gangliosides, and by azithromycin, which binds to the NTD. Based on these data, we identified the NTD of SARS-CoV-2 as a promising target for both therapeutic and vaccine strategies, a notion later supported by the discovery, in convalescent Covid-19 patients, of a neutralizing antibody (4A8) that selectively binds to the NTD. The 4A8 epitope overlaps the ganglioside binding domain, denying any access of the virus to lipid rafts when the antibody is bound to the S protein. Thus, our data explain why antibody binding to the tip of the NTD results in SARS-CoV-2 neutralization. The high level of conservation of the ganglioside binding domain of SARS-CoV-2 (100% identity in 584 of 600 isolates analyzed worldwide) offers unique opportunities for innovative vaccine/therapeutic strategies.
Keywords: Coronavirus, Vaccine, SARS-CoV-2, Ganglioside, Azithromycin, Hydroxychloroquine, Lipid raft
Abbreviations: ACE-2, angiotensin converting enzyme-2; COVID-19, coronavirus disease 19; NTD, N-terminal domain; RBD, receptor binding domain; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, SARS-coronavirus-2
1. Introduction
Covid-19 is a threatening infectious pandemic disease due to the novel coronavirus SARS-Cov-2 [1]. Considerable research efforts are currently engaged to find both prophylactic and therapeutic solutions to fight the virus. As other members of the coronavirus family, SARS-Cov-2 infects host target cells via a specific interaction with angiotensin converting enzyme 2 (ACE-2), a membrane bound protein with an ubiquitous expression in various tissues including the lung, heart, vessels, gut, kidney, and brain [2]. ACE-2 is recognized by the trimeric spike (S) viral protein which belongs to the viral envelope and protrudes outwards with a typical corona-like appearance [3,4].The receptor binding domain (RBD), which is located at the center of the spike trimer interacts with a single ACE-2 receptor. This domain has received most attention from researchers and laboratories worldwide for therapeutic and vaccine strategies [3,[5], [6], [7]].
Despite the lack of a clear-cut consensual therapy for Covid-19 patients, several repositioned drugs, especially anti-malarial (hydroxychloroquine) and antibiotics (azithromycin), are currently used in several countries [8,9]. Retrospective studies cumulating thousands of Covid-19 patients in Belgium, France, Italy, and the USA gave remarkably converging evidences that hydroxychloroquine (often used in combination with azithromycin), significantly reduced mortality rates, especially when administered at early infection stages [[10], [11], [12], [13]]. In vitro, hydroxychloroquine and azithromycin act synergistically to reduce SARS-Cov-2 infection [14], giving a rationale for the combination therapy used in Covid-19 patients. Since none of these drugs is primarily an antiviral compound, the mechanistic explanation of such synergy is puzzling. Recently, we used in silico methods to study the mode of action of hydroxychloroquine and azithromycin at the molecular level. We first identified a ganglioside binding domain at the tip of the N-terminal domain (NTD) of the spike protein, which may allow a functional interaction of the virus with lipid rafts of the plasma membrane independently of the RBD [15]. Then we showed that hydroxychloroquine has a remarkable affinity for sialic acids which are key structural determinants of glycoproteins and gangliosides [15]. Therefore, hydroxychloroquine can bind to lipid raft gangliosides and neutralize virus binding and infection [15]. In a subsequent study we demonstrated that the ganglioside binding domain is also recognized by azithromycin [16]. Overall, our data showed that hydroxychloroquine and azithromycin could block SARS-CoV-2 binding to lipid raft gangliosides through two distinct synergistic mechanisms: hydroxychloroquine binds to gangliosides, azithromycin binds to the ganglioside binding domain of the NTD. Based on these data we suggested to focus on the NTD - rather than the RBD - to define innovative therapies and vaccine strategies against Covid-19 [16]. The importance of lipid rafts in SARS-CoV-2 infection [[17], [18], [19]] is consistent with the established role of these microdomains in the infection cycle of related coronaviruses [20,21]. It may also explain how SARS-CoV-2 infects ganglioside-rich cell types such as epithelial intestinal [22] and brain cells [17].
An important step toward the design of an efficient Covid-19 vaccine is the demonstration that neutralizing anti-SARS-CoV-2 antibodies can be found in convalescent patients. Due to the prominent role of ACE-2, most scientists believed that the epitopes recognized by those neutralizing antibodies should be part of the RBD. This proved to be the case for some of these neutralizing antibodies, but not for all. Indeed, two recent studies have characterized monoclonal neutralizing antibodies (nAb) that recognize the tip of the NTD and do not interfere at all with the ACE-2-spike interaction [23,24]. This unexpected result led the authors of one of these studies to state: “it is unclear how binding to the tip of the NTD results in neutralization of SRAS-CoV-2“ [24]. Our molecular dynamics studies shed, ahead of this publication, some light on this issue. Indeed, we concluded that “the NTD region of the SARS-CoV-2 spike protein, which is recognized by both gangliosides and azithromycin, should be considered as a target for neutralizing antibodies in vaccine strategies” [16].
2. Structural relationship between the 4A8 epitope and the ganglioside binding domain of SARS-CoV-2
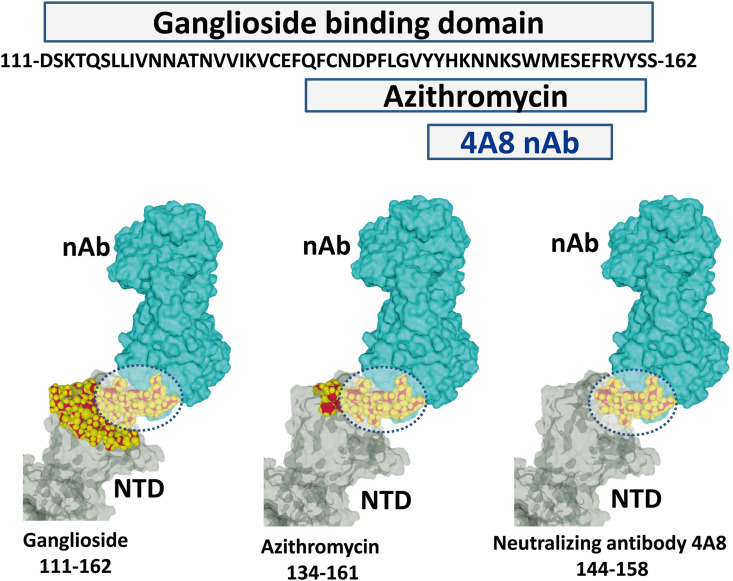
The ganglioside binding domain of the SARS-CoV-2 S protein consists in a flat electropositive surface at the tip of the NTD. This region encompasses amino acid residues 111 to 162 (Fig. 1 ). This domain includes a shorter motif (residues 134–161) that is recognized by azithromycin, a sugar-containing macrolide antibiotic which has some ganglioside mimicry properties [16]. The 4A8 antibody that has been characterized in convalescent Covid-19 patients recognizes a discontinuous epitope of the NTD [23] which includes a significant part of the ganglioside binding domain (amino acid residues 144–158). Thus, as illustrated in Fig. 1, the 4A8 antibody may prevent any access of the spike protein to lipid raft gangliosides through a competitive inhibition mechanism.
Fig. 1.
The 4A8 epitope overlaps the ganglioside and azithromycin binding domains of the NTD in the SARS-CoV-2 spike protein. A part of the 4A8 epitope lies within the 144–158 amino acid sequence. This region covers most of the azithromycin binding domain (amino acids 134–161) and a significant part of the ganglioside binding domain (amino acids 111–162). The ganglioside binding domain has a flat surface that can simultaneously accommodate two gangliosides [16]. The SARS-CoV-2 spike protein bound to the 4A8 neutralizing antibody (nAb) was obtained from pdb file # 7c2l [23].
As previously reported, the ganglioside binding domain of the spike protein is very well conserved among SARS-CoV-2 isolates worldwide [15,16]. Nevertheless, the study of a larger sample of SARS-CoV-2 sequences performed in late September 2020 allowed to identify minor amino acid variations of the motif. Over 600 sequences retrieved from the NCBI database [25], 584 were identical to the reference virus. However the remaining 16 sequences (2.7%) displayed a single amino acid change at the position highlighted in Fig. 2 . Interestingly, these substitutions seem to be concentrated in the azithromycin and the 4A8 antibody binding sites, so that they may confer some level of resistance of the virus to these antiviral agents. However, none of these mutations is expected to affect the binding of the NTD to gangliosides because the amino acid substitutions concern amino acid residues that could still be involved in glycan/ganglioside recognition [[26], [27], [28]]: basic residues (arginine), aromatic (histidine, phenylalanine, tyrosine) and hydroxylated (serine). Most importantly, the QFN triad (Q-134/F-135/N-137) that we identified as the most promising target for vaccine strategies [16] is fully conserved in 100% of the 600 isolates analyzed in the present study (Fig. 2). In contrast, The RBD is the most variable part of the coronavirus genome [1], which is a considerable pitfall for vaccine strategies [29].
Fig. 2.
Amino acid sequence alignments of the ganglioside binding domain of the SARS-CoV-2 spike protein. Among 600 sequences of the ganglioside binding domain (fragment 111–162) retrieved from the NCBI server [25], 584 were identical to the reference sequence (6VSB_A). The 16 remaining sequences (each with a single mutation) are listed below the reference with amino acid changes highlighted. The degree of conservation observed in each column is symbolized by an asterisk (∗) when the residue is fully conserved, a colon () for distinct residues with strongly similar properties (scoring > 0.5 in the Gonnet PAM 250 matrix), and a period (.) for distinct residues with weakly similar properties (scoring ≤ 0.5 in the Gonnet PAM 250 matrix [16,17].
3. Molecular mechanism of 4A8 neutralization: key role of lipid rafts
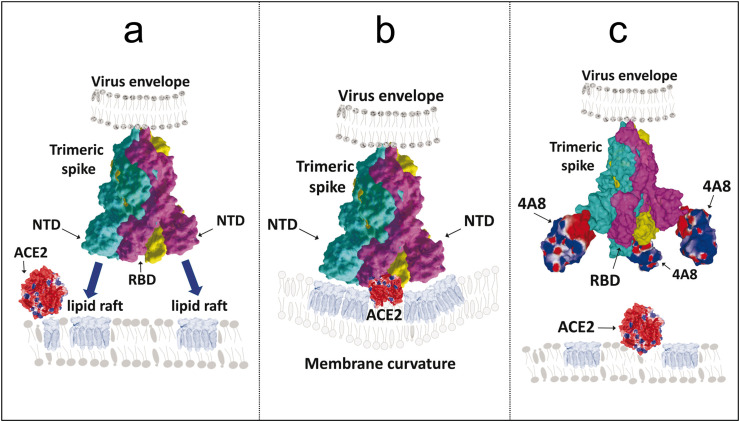
The potential mechanism of 4A8 antibody neutralization is illustrated in Fig. 3 . The trimeric structure of the spike protein is compatible with the simultaneous binding of each virus spike to three distinct gangliosides, which may belong to separate lipid rafts (Fig. 3a). It is likely that such a multivalent binding process could have an important effect on local membrane curvature which in turn could facilitate lipid raft coalescence (Fig. 3b) [28] leading to the recruitment of the ACE-2 receptor, which is also a raft-associated protein [30]. In this context, the initial concentration of virus particles within lipid rafts increases the probability of finding a functional ACE-2 receptor [30], as previously shown for HIV-1 fusion coreceptors [31,32]. In any case, the 4A8 antibody, when bound to the NTD, can prevent any access of the spike protein to lipid rafts and thus to ACE-2 (Fig. 3c). Given that each spike trimer can bind three 4A8 antibodies [23], but only one anti-RBD antibody [33], it might be easier to generate an efficient immune neutralization by targeting the NTD instead of the RBD. The only pitfall would be the selection of resistant viruses with selected mutations in the ganglioside binding domain. For this reason, we suggest focusing on the most conserved part of tip of the NTD, especially the one that contains the QFN triad previously characterized as the core of the azithromycin binding site [16].
Fig. 3.
Why antibody binding to the tip of the NTD blocks SARS-Cov-2 infection. (a) In the trimeric spike of SARS-Cov-2 envelope the RBD is located at the center of the spike, whereas the three NTD (one in each protein subunit) are positioned laterally [16]. The ganglioside binding domain is a large flat surface located at the tip of each NTD, allowing a multiple engagement of the whole spike to several lipid rafts. (b) the NTD binding to gangliosides induces the coalescence of lipid rafts together with a local modulation of membrane curvature that may facilitate the recruitment of the ACE-2 receptor. (c) Anti-NTD antibodies neutralize SARS-Cov-2 by blocking the initial step of the infection cycle, i.e. virus-ganglioside interactions in lipid rafts. Preventing virus binding to lipid rafts is thus likely to render ACE-2 inaccessible. Note that the complementary-determining region (CDR) of the 4A8 antibody has a negative surface potential (colored in red). Since the tip of the NTD has a positive surface potential, it fits with both the CDR of the 4A8 antibody and the negative charges of lipid raft gangliosides. In all panels, the trimeric structure of the SARS-CoV-2 spike is represented in surface rendition with subunits A, B and C colored in cyan, purple, and yellow. The surface potential of the A48 antibody is indicated in conventional colors (positive areas in blue, negative in red, apolar in white). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions and perspectives
Gangliosides play a pivotal role in virus infection. First, they provide a negatively charged flat surface which attracts the electropositive tip of virus envelope glycoproteins [27]. Second, they exert membrane chaperone properties that activate the viral glycoproteins through conformational modifications [27,28]. Third, they facilitate the recruitment of protein virus receptors which are generally associated with lipid rafts [[31], [32], [33], [34]]. Fourth, they are closely associated with cholesterol which controls the fusion process [34,35]. A universal mechanism of virus fusion with host cell membranes, that considers all these key features has been described [27]. It applies to several pathogenic viruses including HIV-1 [28,31,32].
The initial interaction between virus envelope glycoproteins such as SARS-Cov-2 S protein and cell surface gangliosides can be blocked by several distinct mechanisms: i) hydroxychloroquine and potentially other antimalarial drugs have a unexpected affinity for gangliosides [15]; ii) azithromycin and potentially other macrolide antibiotics recognize the core of the ganglioside binding domain [16]; neutralizing antibodies directed against the tip of the NTD of the S glycoprotein prevent any access of the virus to lipid rafts, independently of RBD-ACE-2 interactions. All these strategies, which received clinical validation in several countries [[10], [11], [12], [13],23,24], converge to disrupt virus-raft interactions through competitive mechanisms [15,16].
An important feature of the ganglioside binding motif is that it forms a flat platform at the tip of the NTD of the spike protein. Indeed, this large smooth surface forms a perfect landing pad for the spike trimer (Fig. 3), especially as its electronegative surface potential due to the negative charges of sialic acids fits with the electropositive potential of the NTD. Interestingly, it has been emphasized that such a flat glycan binding domain conflicts with the evolutionary host tropism/adaptation and survival pattern observed for all other coronaviruses, described by the “Canyon Hypothesis” [36]. According to this model, mutations near or at the glycan binding domain are hidden so that the virus escapes detection by host glycan binding immune receptors [37]. Hence it might not be the case for SARS-CoV-2, which, together with other sequence peculiarities, raises question concerning the proximal origin of this virus and its improved functional interaction with lipid rafts [36]. Although this puzzle warrants further discussion, we suggest using ganglioside-NTD interactions as molecular targets for therapeutic and vaccine strategies against Covid-19. In this context, a particular focus on the conserved QFN triad of the NTD [16] deserves careful consideration.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare no conflict of interest.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.C Walls A., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seyedpour S., Khodaei B., Loghman A.H., Seyedpour N., Kisomi M.F., Balibegloo M., Nezamabadi S.S., Gholami B., Saghazadeh A., Rezaei N. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: a systematic review of in vitro and in vivo studies. J. Cell. Physiol. 2020 Sep 9 doi: 10.1002/jcp.30032. [DOI] [PubMed] [Google Scholar]
- 6.Datta P.K., Liu F., Fischer T., Rappaport J., Qin X. SARS-CoV-2 pandemic and research gaps: understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy. Theranostics. 2020;10:7448–7464. doi: 10.7150/thno.48076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Optimization of the production process and characterization of the yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1), a SARS vaccine candidate. J. Pharm. Sci. 2017;106:1961–1970. doi: 10.1016/j.xphs.2017.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha B.K., Bonnier A., Chong W. Antimalarials as antivirals for COVID-19: believe it or not! Am. J. Med. Sci. 2020;S0002–9629:30371–30372. doi: 10.1016/j.amjms.2020.08.019.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth A., Prathapan P. Azithromycin: the first broad-spectrum Therapeutic. Eur. J. Med. Chem. 2020;207:112739. doi: 10.1016/j.ejmech.2020.112739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catteaua L., Dauby N., Montourcy M., Bottieau E., Hautekiet J., Goetghebeur E., van Ierssel S., Duysburgh E., Van Oyen H., Wyndham-Thomas C., Van Beckhoven D. Low-dose hydroxychloroquine therapy and mortality in hospitalised patients with COVID-19: a nationwide observational study of 8075 participants. Int. J. Antimicrob. Agents. 2020;56:106144. doi: 10.1016/j.ijantimicag.2020.106144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagier J.-C., Million M., Gautret P., Colson P., Cortaredona S., Giraud-Gatineau A., Honoré S., Gaubert J.-Y., Fournier P.-E., Tissot-Dupont H., Chabrière E., Stein A., Deharo J.-C., Fenollar F., Rolain J.-M., Obadia Y., Jacquier A., La Scola B., Brouqui P., Drancourt M., Parola P., Raoult D. Outcomes of 3,737 COVID-19 patients treated with hydroxychloroquine/azithromycin and other regimens in Marseille, France: a retrospective analysis. Trav. Med. Infect. Dis. 2020;36:101791. doi: 10.1016/j.tmaid.2020.101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Castelnuovo A., et al. Use of hydroxychloroquine in hospitalised COVID-19 patients is associated with reduced mortality: findings from the observational multicentre Italian CORIST study. Eur. J. Intern. Med. 2020;S0953–6205:30335–30336. doi: 10.1016/j.ejim.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arshad S., Kilgore P., Chaudhry Z.S., Jacobsen G., Wang 4 D.D., Huitsing K., Brar I., Alangaden G.J., Ramesh M.S., McKinnon J.E., O’Neill W., Zervos M., Ford H. COVID-19 Task Force, Treatment with hydroxychloroquine, azithromycin, and combination in patients hospitalized with COVID-19. Int. J. Infect. Dis. 2020;97:396–403. doi: 10.1016/j.ijid.2020.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreani J., Le Bideau M., Duflot I., Jardot P., Rolland C., Boxberger M., Wurtz N., Rolain J.-M., Colson P., La Scola B., Raoult D. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb. Pathog. 2020;145:104228. doi: 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55:105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fantini J., Chahinian H., Yahi N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: what molecular dynamics studies of virus-host interactions reveal. Int. J. Antimicrob. Agents. 2020;56:106020. doi: 10.1016/j.ijantimicag.2020.106020. [DOI] [PubMed] [Google Scholar]
- 17.Fenrich M., Mrdenovic S., Balog M., Tomic S., Zjalic M., Roncevic A., Mandic D., Debeljak Z., Heffer M. SARS-CoV-2 dissemination through peripheral nerves explains multiple organ injury. Front. Cell. Neurosci. 2020;14:229. doi: 10.3389/fncel.2020.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirone L., Del Gatto A., Di Gaetano S., Saviano M., Capasso D., Zaccaro L., Pedone E. A multi-targeting approach to fight SARS-CoV-2 attachment. Front. Mol. Biosci. 2020;7:186. doi: 10.3389/fmolb.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radenkovic D., Chawla S., Pirro M., Sahebkar A., Banach M. Cholesterol in Relation to COVID-19: should we care about it? J. Clin. Med. 2020;9:1909. doi: 10.3390/jcm9061909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glende J., Schwegmann-Wessels C., Al-Falah M., Pfefferle S., Qu X., Deng H., Drosten C., Naim H.Y., Herrler G G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381:215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y., Liu D.X., Tam J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008;369:44–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engin A.B., Engin E.D., Engin A. Dual function of sialic acid in gastrointestinal SARS-CoV-2 infection. Environ. Toxicol. Pharmacol. 2020;79:103436. doi: 10.1016/j.etap.2020.103436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi X., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 25.NCBI 2020. https://www.ncbi.nlm.nih.gov/protein 25 September.
- 26.Yahi N., Fantini J. Deciphering the glycolipid code of Alzheimer’s and Parkinson’s amyloid proteins allowed the creation of a universal ganglioside-binding peptide. PloS One. 2014;9 doi: 10.1371/journal.pone.0104751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fantini J., Yahi N. Elsevier Academic Press; San Francisco: 2015. Brain Lipids in Synaptic Function and Neurological Disease. Clues to Innovative Therapeutic Strategies for Brain Disorders. [Google Scholar]
- 28.Fantini J., Garmy N., Mahfoud R., Yahi N. Lipid rafts: structure, function and role in HIV, Alzheimer’s and prion diseases. Expet Rev. Mol. Med. 2002;4:1–22. doi: 10.1017/S1462399402005392. [DOI] [PubMed] [Google Scholar]
- 29.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glende J., et al. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin- converting enzyme 2. Virology. 2008;381:215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fantini J., Hammache D., Piéroni G., Yahi N. Role of glycosphingolipid microdomains in CD4-dependent HIV-1 fusion. Glycoconj. J. 2000;17:199–204. doi: 10.1023/a:1026537122903. [DOI] [PubMed] [Google Scholar]
- 32.Hammache D., Yahi N., Piéroni G., Ariasi F., Tamalet C., Fantini J. Sequential interaction of CD4 and HIV-1 gp120 with a reconstituted membrane patch of ganglioside GM3: implications for the role of glycolipids as potential HIV-1 fusion cofactors. Biochem. Biophys. Res. Commun. 1998;246:117–122. doi: 10.1006/bbrc.1998.8531. [DOI] [PubMed] [Google Scholar]
- 33.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1101/2020.02.11.944462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawat S.S., Viard M., Gallo S.A., Blumenthal R., Puri A. Sphingolipids, cholesterol, and HIV-1: a paradigm in viral fusion. Glycoconj. J. 2006;23:189–197. doi: 10.1007/s10719-006-7924-4. [DOI] [PubMed] [Google Scholar]
- 35.Fantini J., Epand R.M., Barrantes F.J. Cholesterol-recognition motifs in membrane proteins. Adv. Exp. Med. Biol. 2019;1135:3–25. doi: 10.1007/978-3-030-14265-0_1. [DOI] [PubMed] [Google Scholar]
- 36.Seyran M., et al. Questions concerning the proximal origin of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.26478. Sep. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]