Abstract
Background and objectives
Antimicrobial resistance is a growing global concern and has spurred increasing efforts to find alternative therapeutics. Bacteriophage therapy has seen near constant use in Eastern Europe since its discovery over a century ago. One promising approach is to use phages that not only reduce bacterial pathogen loads but also select for phage resistance mechanisms that trade-off with antibiotic resistance—so called ‘phage steering’.
Methodology
Recent work has shown that the phage OMKO1 can interact with efflux pumps and in so doing select for both phage resistance and antibiotic sensitivity of the pathogenic bacterium Pseudomonas aeruginosa. We tested the robustness of this approach to three different antibiotics in vitro (tetracycline, erythromycin and ciprofloxacin) and one in vivo (erythromycin).
Results
We show that in vitro OMKO1 can reduce antibiotic resistance of P. aeruginosa (Washington PAO1) even in the presence of antibiotics, an effect still detectable after ca.70 bacterial generations in continuous culture with phage. Our in vivo experiment showed that phage both increased the survival times of wax moth larvae (Galleria mellonella) and increased bacterial sensitivity to erythromycin. This increased antibiotic sensitivity occurred both in lines with and without the antibiotic.
Conclusions and implications
Our study supports a trade-off between antibiotic resistance and phage sensitivity. This trade-off was maintained over co-evolutionary time scales even under combined phage and antibiotic pressure. Similarly, OMKO1 maintained this trade-off in vivo, again under dual phage/antibiotic pressure. Our findings have implications for the future clinical use of steering in phage therapies.
Lay Summary: Given the rise of antibiotic-resistant bacterial infection, new approaches to treatment are urgently needed. Bacteriophages (phages) are bacterial viruses. The use of such viruses to treat infections has been in near-continuous use in several countries since the early 1900s. Recent developments have shown that these viruses are not only effective against routine infections but can also target antibiotic resistant bacteria in a novel, unexpected way. Similar to other lytic phages, these so-called ‘steering phages’ kill the majority of bacteria directly. However, steering phages also leave behind bacterial variants that resist the phages, but are now sensitive to antibiotics. Treatment combinations of these phages and antibiotics can now be used to greater effect than either one independently. We evaluated the impact of steering using phage OMKO1 and a panel of three antibiotics on Pseudomonas aeruginosa, an important pathogen in hospital settings and in people with cystic fibrosis. Our findings indicate that OMKO1, either alone or in combination with antibiotics, maintains antibiotic sensitivity both in vitro and in vivo, giving hope that phage steering will be an effective treatment option against antibiotic-resistant bacteria.
Keywords: antibiotic resistance, bacteriophage, combination therapy, phage steering
INTRODUCTION
Increasing levels of antimicrobial resistance have led to the exploration of alternative treatments and strategies [1, 2]. Lytic bacteriophage (phage) has demonstrated therapeutic potential, particularly for recalcitrant bacterial infections [3]. Phage therapy works in a similar way to conventional chemotherapies—bacterial densities are reduced to the extent that the immune system can clear the infection [4–7]. However, as with conventional therapies, treatment may fail if phages are not able to sufficiently reduce bacterial density, often attributable to bacteria evolving phage resistance during therapy. Many strategies have been proposed that go beyond simple phage monotherapy with the aim of preventing bacterial resistance. These capitalize on ecological and evolutionary events in the disease ecosystem and include multi-phage cocktails, evolutionarily trained ‘sur-mesure’ phage, engineered phage or combining phages with antibiotics [1].
One particularly promising approach is to anticipate resistance evolution to phages and use this as part of a therapeutic strategy, recently termed ‘phage steering’ [8]. This involves exploiting an evolutionary trade-off between resistance to the phage and either sensitivity to other antimicrobials (a ‘double bind’ [9]) or reduced bacterial virulence [2, 10]. Several phage resistance mechanisms have been identified that generate such trade-offs, one of the most promising being the reduction in efflux pump efficiency associated with phage resistance [3, 11]. This form of phage steering has the potential to prevent the emergence of antibiotic resistance or even revert a resistant bacterial infection to antibiotic sensitivity [8].
Several candidate phages have been identified for phage steering and are currently being used as compassionate therapies to great effect [2, 10]. However, despite progress in understanding the ecology and evolution of phage steering in vitro [3, 12–14] and in vivo [7, 15, 16], we are still in the early phases of a scientific understanding of the phenomenon as a therapeutic tool. There are two main challenges to employing phage steering as a predictable alternative to other therapies. First, using phages to steer pathogenic phenotypes relies on the development of resistance to the phage treatment and that, through a trade-off, resistance somehow compromises the bacterial population. Bacteria can evolve resistance to phage by mutation of receptors on the bacterial cell outer membrane, and, conversely, the evolution of bacterial resistance provides an opportunity for phage mutants able to use modified receptors to exploit otherwise resistant bacteria. Phages and bacteria readily co-evolve in vitro through reciprocal selection for infection and resistance phenotypes (for review, see [17]), respectively, and there is some evidence that bacterial pathogens can co-evolve with phages in vivo (e.g. [18]). Nevertheless, it is unclear how co-evolution may impact therapeutic outcomes. Specifically, it will be important to know the conditions under which co-evolution occurs and whether/how this could affect the evolution of the trade-offs required for phage steering to be effective.
Second, we need to assess how reliable and repeatable phage steering is in vivo. This is particularly important for phage–antibiotic combinations, as they are likely to become the most frequent form of phage therapy [19]. Previous in vitro studies have shown that combined phage–antibiotic introductions are more effective than either separately at reducing bacterial densities [20–23]. Moreover, in contrast to in vitro investigation, we know little about what determines therapeutic outcomes in vivo where disease ecosystem structure [24] and immune system dynamics [4, 6] can play important roles. In cases where the phage selects against antibiotic resistance, the expected outcome of combinations will likely depend on the strength of selection pressure and the rate of any co-evolution between the phages and their bacterial hosts. Recent modelling work [4] suggests that non-linear synergy between phages, antibiotics, and the innate-immune system may be required for therapeutic success.
In the present study, we investigated the outcomes of phage steering on antibiotic resistance in the pathogenic bacterium Pseudomonas aeruginosa (strain PAO1) both in vitro and in vivo. Specifically, we tested the sustainability of the steering effect over co-evolutionary time scales (e.g. [25]). Briefly, in experimental microcosms, PAO1 bacteria were continuously cultured with phage OMKO1 either in the absence or presence of one of each of three antibiotics (tetracycline, erythromycin and ciprofloxacin). After 10 serial transfers, we determined levels of antibiotic resistance in the co-evolved bacteria. In a next step, we evaluated the efficacy of both single or combined introductions of phage and the antibiotic erythromycin in an in vivo system by using Galleria mellonella (wax moth) larvae as hosts for the bacteria. We monitored wax moth survival and determined levels of antibiotic resistance in the surviving bacteria. We found clear evidence of phage impact on bacterial hosts and a phage steering effect of increased bacterial sensitivity for all three antibiotics tested in vitro. Importantly, phages reduced bacterial antibiotic resistance levels when simultaneously given with erythromycin in vivo. Our results suggest that steering phages are effective either when used as stand-alone therapies or when used in combination with antibiotics.
MATERIALS AND METHODS
Bacterial culture and phage preparation
Pseudomonas aeruginosa (Washington PAO1) was grown at 37°C (constant shaking at 200 rpm) in King’s B (KB) medium [26]. For phage amplification, 10 µl of purified OMKO1 phage [3] was added to mid-log growing PAO1, then incubated overnight (18 h) with intermittent shaking (1 min every 30 min) at 37°C (Bülher compact shaker, model KS-15 Control). 10% vol/vol of chloroform was added, the mixture vortexed and centrifuged (1 min 10 000 RCF) to separate the phases. The supernatant containing phages was pipetted into a sterile 2 ml Eppendorf and stored at 4°C. For the complete phage preparation protocol, see [25].
Experiment 1: Short-term in vitro selection for phage resistance
In this experiment, we generated phage-resistant bacteria and tested for a correlated decrease in antibiotic resistance, previously described in [3]. PAO1 bacteria from a −80°C freezer stock was grown on KB agar plates overnight at 37°C. Three single colonies were isolated with a sterile loop, each inoculated individually into 6 ml of KB medium and incubated for 4 h shaking (200 rpm) at 37°C. After 4 h, 10 µL of OMKO1 (ca. 1 × 106) were added, and the three phage-bacteria replicate cultures incubated with intermittent shaking (1 min every 30 min) at 37°C for 18 h. The surviving bacteria were streaked onto a KB agar plate and incubated at 37°C overnight. Phage resistance was confirmed by cross streaking the bacteria against the phage [27].
To assess the level of antibiotic resistance of phage-resistant bacteria, the minimum inhibitory concentration (MIC) [28] was determined for three antibiotics with different modes of action (tetracycline, erythromycin and ciprofloxacin). Briefly, phage-resistant bacteria were re-streaked onto KB agar plates overnight, then grown in liquid KB overnight to ensure phage-free cultures. Optical densities of these cultures were measured at a wavelength of 600 nm (OD600) and then re-suspended to a uniform OD600 of 1.0. In the same way, we prepared ancestral (phage-susceptible) bacterial cultures. In 96-well microtitre plates, a 2-fold dilution series of each antibiotic was prepared, starting at 800 µg/ml and finishing at 6.25 µg/ml for tetracycline and erythromycin, and 10 to 0.156 µg/ml for ciprofloxacin. Bacteria were added to each well to a final OD600 of 0.05 and incubated at 37°C for 18 h. Bacteria were considered as having grown if the OD600 was above 0.1 after 18 h. The MIC was determined as the lowest antibiotic concentration at which no observable bacterial growth occurred. Three individual colonies were isolated from each replicate culture and three from the ancestor, and their MIC determined for three ‘technical’ replicates. The mean over the technical replicates was used for further analysis.
Experiment 2: In vitro bacterial (co-)evolution
We tested longer term (10 serial transfers over 20 days) bacterial evolutionary responses to antibiotics and phage. Bacteria were exposed to a single antibiotic (tetracycline, erythromycin or ciprofloxacin), in the presence of the phage OMKO1 or in the presence of both antibiotic and phage. In a control treatment, neither antibiotic nor phages were added. The experiment was conducted in 2-ml microcosms of KB medium in 24-well microtitre plates, incubated at 37°C with intermittent 200 rpm orbital agitation for 1 min every 30 min (Bülher compact shaker, model KS-15 Control). For each of the four treatments, 4 replicate lines were initiated in separate microcosms, for a total of 32 lines across all treatments (4 control + 4 phage + 3 × 4 antibiotic-only + 3 × 4 phage plus antibiotic). Each microcosm was inoculated with 20 μl of PAO1 (ca. 5 × 106 cells) from an overnight population initiated from a single ancestral P. aeruginosa PAO1 colony. For treatments with phage, 5 μl of the ancestral phage stock (ca. 5 × 106 phage particles) were added together with the inoculated bacteria, producing a 1:1 ratio of bacteria and phage. For treatments with antibiotics, we used half the MIC at the start and again at each serial transfer (200 μg/μl tetracycline, 100 μg/μl erythromycin and 0.5 μg/μl ciprofloxacin). For each microcosm, a 20-μl sample of culture (containing both bacteria and phage) was transferred to a new microcosm with fresh KB medium every 48 h, for a total of 10 transfers (ca. 70 bacterial generations for controls). At each transfer, a 50-μl sample was stored in 50% glycerol at −80°C for subsequent analysis. At the end of the 10 transfers, we determined the antibiotic resistance of the evolved bacteria (MIC, as described above) for 3 arbitrarily chosen colonies from each replicate line. MIC measurements were taken for three technical replicates per colony and averaged for statistical analysis.
Experiment 3: Host mortality and antibiotic resistance evolution in the wax moth infection model
We tested the combined effects of phage and antibiotic on bacteria-induced wax moth mortality and bacterial antibiotic resistance evolution. For the wax moth mortality assay, we used a modified protocol based on [29]. Four experimental treatments were established. Late-instar larvae were injected with a total of 20 µl PBS containing ancestral PAO1 bacteria (5 × 102 CFU), supplemented with (i) phage only (5 × 103 PFU), (ii) the antibiotic erythromycin only [concentration: 100 µg/ml suspended in phosphate-buffered saline (PBS)], (iii) both phage and antibiotic or (iv) neither phage nor antibiotic (=positive control). This fully factorial experiment was replicated four times and within each replicate run we used 6 larvae per treatment, giving a total of 96 larva. In an additional negative control treatment, 18 larvae were injected with 20 µl PBS, without bacteria. Larvae were kept at 37°C for 48 hours following injection and the survival of the treated larvae was checked at 6-h intervals for the first 24 h, then at 12-h intervals. Larvae were considered dead when three successive stimulations along their ventral surface produced no response. All dead larvae were stored intact in 25% glycerol at −80°C at the end of the experiment at 48 h.
Bacterial antibiotic resistance was determined from 4 wax moth larvae in each of two arbitrarily selected replicate lines for each treatment (therefore, 8 wax moths analysed per treatment). Each individual larva was homogenized at 48 h using pestle sticks (Bel-Art instruments). The resulting homogenates were then serially diluted and plated onto Pseudomonas Isolation Agar (PIA, DifCO). The MIC was determined for three arbitrarily selected single colonies per wax moth analysed, and the mean recorded as described for Experiment 2. To determine if phage resistance had evolved in treatments (i) and (iii), the 3 single colonies used to determine the MIC (48 colonies total) were grown overnight at 37°C in KB and then washed twice in PBS and cross-streaked [27] against the ancestral OMKO1 phage (1E8 PFU/ml). In treatment (i), 21 of the 24 colonies showed total resistance and the remaining 3 showed intermediate resistance. In treatment (iii), 20 of the 24 colonies showed total resistance and the remaining 4 showed intermediate resistance, while the ancestral PAO1 was fully sensitive. Our research on wax moths was conducted at the University of Montpellier, France, and adhered to national and institutional regulations concerning research on animals.
Statistical analysis
For the initial screen of phage-resistant PAO1 (Experiment 1), MIC data of bacterial resistance to the three antibiotics were analysed by a t-test (Welch two-sample test), comparing evolved phage-resistant isolates with phage-susceptible ancestral bacteria. Analysis of Variance (R 3.01 base package, supplemented with the ‘car’ and ‘lsmeans’ packages using the Tukey adjustment) was used for the MIC data from Experiment 2 (in vitro evolution). A fully factorial model of phage and antibiotic treatments was fitted, with replicate selection line nested in the interaction term.
To analyse variation in larval mortality associated with bacteria in Experiment 3, we fitted a parametric survival model with time to death as the response variable, using the Weibull distribution function (R packages ‘survival’ and ‘survminer’). A Weibull distribution function was chosen for having the lowest AIC score in comparison to models with other functions (e.g. lognormal, exponential). Right censoring was used for cases of larvae surviving the full 48 h post-injection. Phage and antibiotic treatments (and their interaction) were fitted as explanatory factors in a factorial model, as above. The identity of experimental replicate run was removed from an initial full model, since it explained a non-significant portion of the variance (P > 0.1). Its removal did not bias our results for the remaining effects in the model. We further analysed variation in MIC among bacteria isolated from dead larvae, with phage and antibiotic treatments (and their interaction) as explanatory factors. MIC data in all analyses were square-root transformed to meet model assumptions. These analyses were carried out in JMP 14 (SAS).
RESULTS
Experiment 1: Short-term selection for phage resistance leads to reduced MIC in vitro
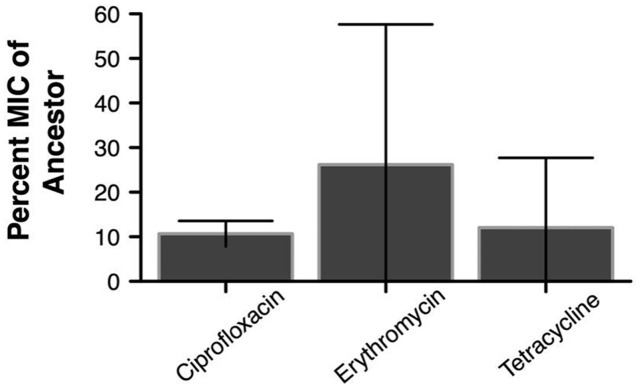
As shown previously [3, 11], PAO1 bacteria that had become resistant to phage OMKO1 after 1 day of exposure were found to have significantly lower antibiotic resistance (Fig. 1 and Supplementary Fig. S1). For all three antibiotics tested, the group of phage-resistant bacteria isolates had >70% lower MICs than the phage-susceptible ancestral bacteria (Welch t-tests with df adjusted for unequal variances: ciprofloxacin: t2.04 d.f = 14.9, P = 0.004; erythromycin: t3.2 d.f = 8.5, P = 0.013 and tetracycline: t3.92 d.f = 16.63, P = 0.004).
Figure 1.
Percent (± 95% CI) of MIC of phage-resistant bacteria tested for three antibiotics relative to ancestral phage-susceptible bacteria. PAO1 bacteria resistant to OMKO1 were obtained after 24 h of in vitro growth in the presence of the phage. MIC levels were determined for each of three antibiotics; all three were significantly reduced from the initial ancestor level (t-test, Welch two-sample test)
Experiment 2: Long-term exposure to phage prevents the emergence of antibiotic resistance
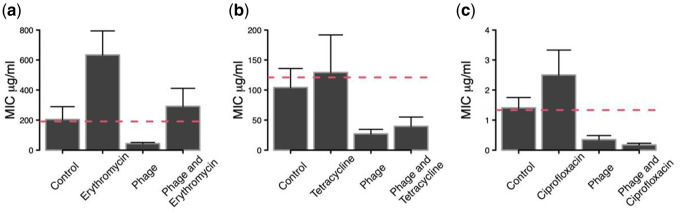
Visual inspection indicated that bacteria maintained substantial densities after the second serial transfer (ca. >1.0E8; J. Gurney, pers. obs.; see also [30] and [31]). Analyses of bacterial resistance (MIC) after 10 transfers revealed a general negative effect of phage treatment on MIC (significant main effects; Table 1). Bacteria that had been co-cultured with phage showed >60% reductions in resistance to all three antibiotics compared to bacteria that had not been exposed to phage (Fig. 2). In contrast, although mean MICs for antibiotic-alone treatments were always greater than controls, there was no clear signal when considering all treatments that included antibiotics (Table 1; Fig. 2). Erythromycin significantly increased resistance (main effect of antibiotic treatment), tetracycline had no significant effect, while the effect of ciprofloxacin depended on the presence of phage (significant antibiotic × phage interaction; Table 1).
Table 1.
Analyses of variance of antibiotic resistance (MIC) for bacteria isolated from selection lines undergoing treatments with and without exposure to phage OMKO1 or each of three antibiotics (erythromycin, tetracycline and ciprofloxacin)
| Source | df | Erythromycin |
Tetracycline |
Ciprofloxacin |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MS | F | P | MS | F | P | MS | F | P | ||
| Phage | 1 | 679.2 | 42.4 | <0.0001 | 277.2 | 66.8 | <0.0001 | 9.03 | 164.2 | <0.0001 |
| AB | 1 | 1315.4 | 82.1 | <0.0001 | 9.4 | 2.3 | 0.1586 | 0.12 | 2.3 | 0.1514 |
| Phage × AB | 1 | 4.1 | 0.3 | 0.6230 | <0.1 | <0.1 | 0.9329 | 0.83 | 15.2 | 0.0021 |
| Selection line (phage, AB) | 32 | 192.3 | 0.7 | 0.7182 | 4.1 | 0.6 | 0.8077 | 0.05 | 0.9 | 0.5810 |
| Residual | 54 | 22.1 | 6.7 | 0.06 | ||||||
All treatment effects were tested against the selection line effect. Significant effects in bold.
Figure 2.
Levels of antibiotic resistance [MIC ± 95% CI] of bacteria evolved in the presence of an antibiotic only, phage only or both phage and antibiotic combined. (a) Erythromycin, (b) tetracycline and (c) ciprofloxacin. Bacteria were isolated after 10 serial transfers (20 days) of treatment and their MICs determined. Bacteria from antibiotic treatments were only tested against the same antibiotic; bacteria from phage-only treatments were tested against all three antibiotics. Phage OMKO1 either reduced the emergence of resistance or sensitized the bacteria to the antibiotic. Dashed red line is the mean MIC for the ancestor
Detailed pairwise treatment comparisons of MIC values showed two patterns of phage action. First, adding phage together with the antibiotic reduced bacterial resistance relative to antibiotic-alone treatments (all P < 0.002; Fig. 2a–c). In at least one case (ciprofloxacin), phage and antibiotic clearly had non-additive effects (significant phage × antibiotic interaction, Table 1), such that the MIC in the combined treatment was more similar to that of the phage-alone treatment (rather than the average of the two single treatments). Second, all three phage-alone treatments and two combined phage-antibiotic treatments reduced MIC levels below that of bacteria from untreated controls (all P < 0.05; Fig. 2). Only for erythromycin did we observe some level of antibiotic resistance evolution in the combined treatment, but this was not significant (P > 0.05) (Fig 2a). Taken together, these patterns indicate that phages not only prevented antibiotic resistance evolution, but also increased susceptibility of the bacteria to ciprofloxacin and tetracycline (Fig. 2b and c).
Experiment 3: Phage reduces bacteria-related larval mortality
Bacterial injection reduced larval survival relative to the PBS-injected negative control group (1 dead larva out of 18, data not shown) (Fig. 3). In the absence of phage or antibiotic, bacterial infection was ∼100% lethal within 24 h (a single larva survived to 48 h with erythromycin). Even with anti-bacterial treatments (phage and/or antibiotic) >40% of the larvae died within 48 h.
Figure 3.
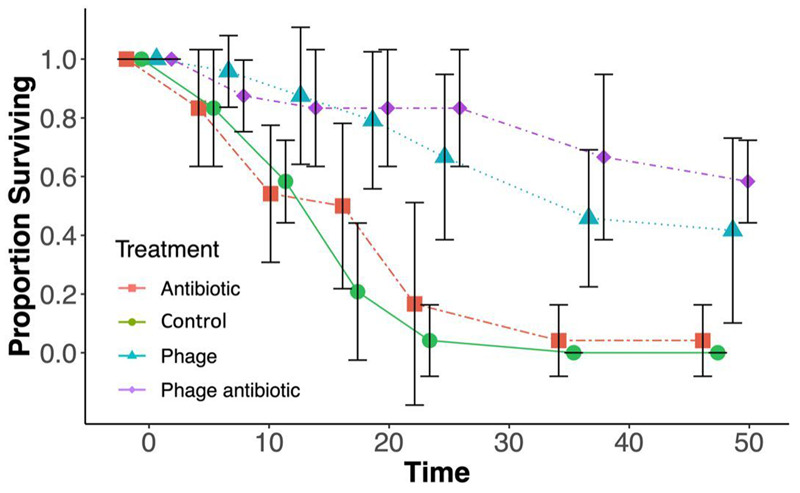
Mean (±95% CI) proportion of surviving wax moth larvae under the four different treatments. Larvae were infected with a lethal dose of PAO1 and observed for 48 h. All three treatment lines provided a greater mean survival time. Both phage alone (blue triangles) and phage plus erythromycin (purple diamonds) produced an additive survival rate when compared with erythromycin alone (red squares), representative markers are staggered to the left or right of the control for ease of reading. Mean survival of 24 replicates
The addition of the phage OMKO1 and of the antibiotic each significantly increased larval survival (phage: Wald χ2 = 58.8, P < 0.0001 and antibiotic: χ2 = 5.1, P = 0.0242; Fig. 3). However, while the phage treatment prolonged survival by ca. 20 h and left >50% of the larvae alive at the end of the 48-h assay, the positive effect of the antibiotic was only marginal (4 h longer life; 96% mortality by 48 h) and its significance depended on the distribution function chosen (e.g. P > 0.2 for a lognormal survival function). There was no significant interaction between antibiotic and phage treatment (χ2 < 1.0, P > 0.8), indicating that the two anti-microbial agents acted additively. Thus, the combined injection of phage and antibiotic was most successful at prolonging larval survival, even though it was not significantly different from the phage-alone treatment (pairwise contrast P > 0.2).
Experiment 3: Phage maintains selection against antibiotic resistance in vivo
Prior to MIC analysis, single colonies were cross-streaked against the ancestral OMKO1 and found to be resistant [27]. Factorial ANOVA showed a highly significant overall effect of phage treatment on the MIC of evolved bacteria (F1,28 = 132.7, P < 0.0001), whereas the main effect of the antibiotic treatment and the antibiotic × phage interaction were small and neither were significant (F1,28 = 0.44, P > 0.5 and F1,28 = 0.48, P > 0.4, respectively). While the antibiotic alone led to only a slight increase in MIC, the action of the phage strongly reduced the MIC of the surviving bacteria (Fig. 4). Thus, like in the in vitro experiment, the presence of phage increased susceptibility of the bacteria to the antibiotic, resulting in lower MICs than in the untreated control lines.
Figure 4.
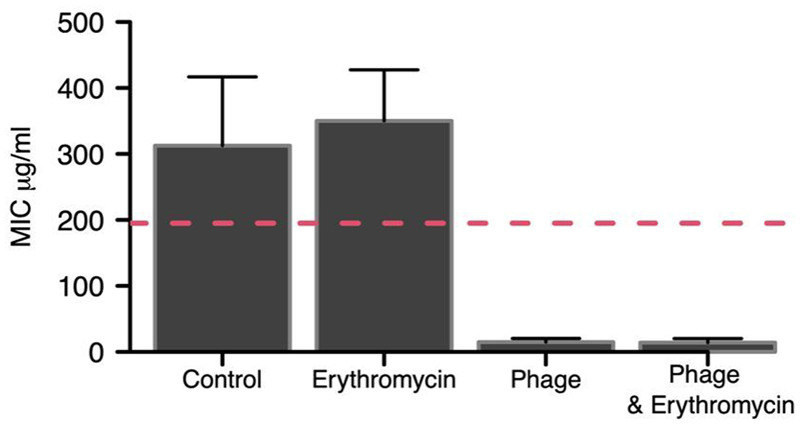
Mean MIC (±95% CI) of recovered PAO1 bacteria from wax moth infection assay. Bacteria were recovered from 8 wax moth larvae per treatment. Both the phage and phage + antibiotic (erythromycin) lines produced clear reductions in the MIC level post-infection. The reason for the control line MIC above the ancestor is unknown, but could be associated with increased expression of the efflux system responsible for observed antibiotic resistance [54]. Dashed red line is the mean MIC for the ancestor
DISCUSSION
We present evidence of a phage-associated reduction in antibiotic resistance in bacterial populations of P. aeruginosa, both in vitro and in vivo. In line with previous work [3], the addition of phage OMKO1 reduced antibiotic resistance in the majority of treatment conditions (by up to 90%; Fig. 1) and impeded the emergence of resistance in those it did not reduce (Fig. 2). In vivo assays largely confirmed the in vitro experiments. Treatment with OMKO1 led to both reductions in antibiotic resistance as measured by MIC and, when combined with certain antibiotics, improved larval survival compared to either antimicrobial used separately. Below, we discuss the precedent for phage steering, how our results advance our understanding of phage steering, study limitations and future research directions.
In its simplest form, phage steering can be achieved when antibiotic resistance and phage resistance are based on the same virus receptor, especially direct lytic phage attachment to proteins of an efflux pump. For example, in a separate in vitro study, Burmeister et al. [32] showed how phage steering can occur through interactions between phage U136B and the outermost TolC protein of Escherichia coli efflux pumps. The mechanism of phage steering in our study is less clear, as there is increasing evidence that phage OMKO1 may interact with multiple surface factors of P. aeruginosa via binding to Type-IV pili, a portion of the lipopolysaccharide structure (LPS) and the OprM protein of the MexXY efflux pumps (P. Turner, unpublished data). One possibility is that phage-OMKO1 steering can be impacted by evolution of phage resistance that is caused by truncated LPS, which can make bacteria more permeable to antibiotics [33]. Alternatively, retraction of the Type-IV pilus may permit phage OMKO1 to target OprM protein of efflux pumps, analogous to phage ϕ6 attachment to Type-IV pili of Pseudomonas syringae bacteria, where pilus retraction facilitates virus-particle interaction with the cell surface [34, 35]. These possibilities are the subjects of ongoing study and are reminders that receptor binding in viruses can be challenging to elucidate. Moreover, trade-offs in favour of the patient cannot be expected to be universal: the evolution of phage resistance may also increase bacterial virulence or antibiotic resistance [36]. Therefore, steering bacterial phenotypes will require extensive testing of the proposed phage [36] (for review, see [37]). Altogether, we emphasize that phage steering could be achieved via one or more alternative mechanisms responsible for antibiotic resistance.
The main findings of our work relate to how phage treatment influences bacterial MIC. The PAO1 isolate used in this study was determined to be clinically resistant to ciprofloxacin by both the CLSI and BSAC breakpoints [38, 39]. Neither erythromycin nor tetracycline is routinely used clinically for P. aeruginosa treatment and therefore do not have established clinical resistance levels (CLSI and BSCA do not set breakpoints). Nevertheless, the ancestor MICs for both erythromycin and tetracycline reported here were in line with previous studies [40, 41], and post-phage treatment MICs for both were significantly reduced (Fig. 1). All three antibiotics are known to be effluxed by OprM-MexXY systems [40], which appear to be targeted by phage OMKO1.
The long-term (20-day) experiment yielded a number of interesting and promising results. OMKO1 was able to significantly reduce resistance or impede the emergence of resistance in all three antibiotic treatments, even in the presence of the antibiotic (Fig. 2). This demonstrates that OMKO1 might be used as an adjuvant in antibiotic treatments by selecting for antibiotic sensitivity and contributing along with the immune system to bacterial clearance [4]. Such treatments have been undertaken as expanded access compassionate care cases and are currently being analysed.
As a proof of principle for such treatments and to test the interaction of phages, antibiotics, and an innate-immune system, we performed a simple infection assay using wax moth larvae. The larvae were injected with a lethal quantity of bacteria with or without erythromycin and/or phage, and then observed for 48 h. The mean survival time increased for all three treatment conditions compared to bacteria injected alone (Fig. 3). Fitting a parametric survival model however failed to show a synergistic effect of phage and antibiotic, and rather gave evidence for an additive response. Our results nevertheless support the basic findings of other studies showing improved treatment outcomes with phage for a range of in vivo systems ranging from Caenorhabditis elegans [15] to mammals including humans [7, 11, 42–45]. Our work is the first to our knowledge to show a reduction in resistance in any in vivo system. This result, taken together with the in vitro experiments, suggests that phage steering can occur in vivo and could be evolutionary stable during the course of treatment.
Although we have shown that OMKO1 can reduce bacterial numbers and maintain antibiotic sensitivity even in the presence of the antibiotic, unanswered questions remain. First, we did not measure bacterial growth rates during the evolution experiment and an alternative hypothesis to explain our observation of increased sensitivity is lower bacterial growthrates, and not lower efflux pump activity [46]. Although previous work on this same bacterial isolate–phage interaction indicated that resistance resulted in lowered efflux pump activity [3]—the putative explanation for lowered MIC—further work is necessary to assess a possible contribution of lowered growth rates. Second, we did not determine the extent to which phage maintains selection on the target, in this case the oprM-mexXY system. This could be investigated by examining efflux pump genetics during a similar long-term experiment, where both phage and bacteria potentially (co)evolve and both are independently allowed to evolve in response to the antibiotic [47]. During this evolution experiment when both phages and antibiotics are present, selection leading to antibiotic sensitivity by the phage is in opposition to selection for resistance imposed by the antibiotic. According to our observations, selection by the phage OMKO1 generally dominates antibiotic effects. A possible, untested explanation for why erythromycin resistance was selected in vitro, whereas enhanced sensitivity emerged in vivo is antibiotic-sensitive variants in the latter not encountering sufficient drug concentrations (see, e.g. Ref. [48]). Alternatively, the bacteria in the in vivo experiment may not have had sufficient standing variation for an antibiotic resistance mutation to emerge. Further study is necessary to evaluate these hypotheses. Finally, introducing other bacterial species that capture the multispecies nature of many infections may also change how resistance develops [49]. Co-evolution under different environmental pressures such as other bacterial species may shift dynamics between ‘arms race’ and ‘fluctuating’ with implications for phage resistance [50, 51]. Shifting co-evolutionary dynamics may change selection from the receptor target to other resistance mechanisms such as CRISPR-Cas [49]. Theoretical work however indicates that selection will often result in receptor modification [52].
The use of wax moths as in vivo experimental systems has two main drawbacks. First, the immune system is limited to an innate response. Although an innate response can be key to successful phage therapy [4, 6], the influence of the adaptive response [as would be the case in human subjects and other animal models such as mice] is currently unknown. Second, the bacteria causing the infection and its treatment were introduced simultaneously, which [possibly excluding prophylaxis] is clearly irrelevant to real treatments. This simultaneous administration likely means that the bacteria had little time to grow and adapt to the host or to form structures such as biofilms that may limit the ability of phage to infect [53]. Physical limitations of the wax moth model mean that it is difficult establish an infection and then apply the treatment, as death can be rapid and multiple injections are likely to produce non-infection-related mortality. Future study should investigate different delays between infection and the introduction of antibiotics and phages in murine models, where immune responses are more similar to human in situ contexts.
CONCLUSIONS AND FUTURE APPLICATIONS
Our study examined whether phage OMKO1 could steer antibiotic resistance successfully during in vitro experimental evolution and in an animal model, without regard to the spectrum of phage-resistance and drug-resistance mutations underlying phenotypic changes in bacterial populations. These experiments considered the underlying genetics as unopened ‘black boxes’, analogous to treatment situations such as chemotherapy administered against cancer and antibiotic therapy targeting bacterial infections, where the mutational responses to therapy are typically neither elucidated nor monitored. These established therapies assume the mutational spectra for drug resistance can vary but should generally not impede a successful treatment outcome in the patient. Measuring at the site of an infection is generally impractical; thus, these therapies presume the beneficial phenotypic responses will prevail over unfavourable clinical outcomes, based on in vitro or in vivo experimental evidence. Similarly, the widespread use of phage therapy may move forward by relying on confidence that the treatment can be delivered safely and effectively, while ignoring many of the underlying mechanistic details. Our ongoing work examines the generalities for P. aeruginosa resistance to phage OMKO1, by mapping the mutational spectra of fitness and phenotypic effects for these mutations in PAO1 and for strains isolated in the clinic.
In summary, as expected, phage OMKO1 was able to achieve a reduction in MIC for all three antibiotics (Fig. 1). Our results support the action of a trade-off between the reduction of MIC and phage resistance (Fig. 2), suggesting that the phage was able to maintain selection against resistance over long-term, co-evolutionary time scales even in the presence of antibiotics. This effect was confirmed in vivo, where the phage improved survival and reduced antibiotic resistance (Figs. 3 and 4).
Supplementary data
Supplementary data is available at EMPH online.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers and James Bull for their insightful comments.
Funding
B.K.C. and P.E.T. thank the Project High Hopes Foundation for generous support. M.E.H. thanks the McDonnell Foundation for funding (Studying Complex Systems research award 220020294).
Conflict of interest: P.E.T. discloses a financial interest in Felix Biotechnology Inc., a company that seeks to commercially develop phages for use as therapeutics.
REFERENCES
- 1. Hochberg ME. An ecosystem framework for understanding and treating disease. Evol Med Public Health 2018;2018:270–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kortright KE, Chan BK, Koff JL, Turner PE. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019;25:219–32. [DOI] [PubMed] [Google Scholar]
- 3. Chan BK, Sistrom M, Wertz JE. et al. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 2016;6:26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodriguez-Gonzalez RA, Leung C-Y, Chan BK, et al. Quantitative models of phage-antibiotics combination therapy. mSystems. 2020;5(1):e00756-19. doi: 10.1128/mSystems.00756-19 [DOI] [PMC free article] [PubMed]
- 5. Leung CYJ, Weitz JS. Modeling the synergistic elimination of bacteria by phage and the innate immune system. J Theor Biol 2017;429:241–52. [DOI] [PubMed] [Google Scholar]
- 6. Roach DR, Leung CY, Henry M. et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 2017;22:38–47.e4. [DOI] [PubMed] [Google Scholar]
- 7. Levin BR, Bull JJ. Phage therapy revisited: the population biology of a bacterial infection and its treatment with bacteriophage and antibiotics. Am Nat 1996;147:881–98. [Google Scholar]
- 8. Gurney J, Brown SP, Kaltz O, Hochberg ME. Steering phages to combat bacterial pathogens. Trends Microbiol 2019;28:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gatenby RA, Brown J, Vincent T. Lessons from applied ecology: cancer control using an evolutionary double bind. Cancer Res 2009;69:7499–502. [DOI] [PubMed] [Google Scholar]
- 10. León M, Bastías R. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol 2015;6:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan BK, Turner PE, Narayan D. et al. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Medi Public Health 2018;2018:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ricci V, Piddock L. Exploiting the role of TolC in pathogenicity: identification of a bacteriophage for eradication of Salmonella Serovars from poultry. Appl Environ Microbiol 2010;76:1704–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chart H, Row B, Threlfall EJ, Ward LR. Conversion of Salmonella enteritidis phage type 4 to phage type 7 involves loss of lipopolysaccharide with concomitant loss of virulence. FEMS Microbiol Lett 1989;60:37–40. [DOI] [PubMed] [Google Scholar]
- 14. Brockhurst MA, Buckling A, Rainey PB. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc Biol Sci 2005;272:1385–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Javier S, James R. Phage-resistance of Salmonella enterica serovar Enteritidis and pathogenesis in Caenorhabditis elegans is mediated by the lipopolysaccharide. Electron J Biotechnol 2007;10:2007. [Google Scholar]
- 16. Smith HW, Huggins MB. Successful treatment of experimental Escherichia coli infections in mice using phage: its general superiority over antibiotics. J Gen Microbiol 1982;128:307–18. [DOI] [PubMed] [Google Scholar]
- 17. Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 2014;38:916–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Sordi L, Lourenço M, Debarbieux L. “I will survive”: a tale of bacteriophage-bacteria coevolution in the gut. Gut Microbes 2019;10:92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torres-Barcelo C, Hochberg ME. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol 2016;24:249–256. [DOI] [PubMed] [Google Scholar]
- 20. Torres-Barceló C, Arias-Sánchez FI, Vasse M. et al. A window of opportunity to control the bacterial pathogen Pseudomonas aeruginosa combining antibiotics and phages. PLoS One 2014;9:e106628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Escobar-Páramo P, Gougat-Barbera C, Hochberg ME. Evolutionary dynamics of separate and combined exposure of Pseudomonas fluorescens SBW25 to antibiotics and bacteriophage. Evolu Appl 2012;5:583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang QG, Buckling A. Phages limit the evolution of bacterial antibiotic resistance in experimental microcosms. Evol Appl 2012;5:575–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaudhry WN, Concepcion-Acevedo J, Park T. et al. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 2017;12:e0168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lourenço M, Chaffringeon L, Lamy-Besnier Q, et al. The spatial heterogeneity of the gut limits bacteriophage predation leading to the coexistence of antagonist populations of bacteria and their viruses. bioRxiv 2019. doi: 10.1101/810705:810705. [DOI] [Google Scholar]
- 25. Betts A, Kaltz O, Hochberg ME. Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proc Natl Acad Sci U S A 2014;111:11109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 1954;44:301–307. [PubMed] [Google Scholar]
- 27. Brockhurst MA, Morgan AD, Fenton A, Buckling A. Experimental coevolution with bacteria and phage: the Pseudomonas fluorescens—Φ2 model system. Infect Genet Evol 2007;7:547–552. [DOI] [PubMed] [Google Scholar]
- 28. Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 2001;48:5–16. [DOI] [PubMed] [Google Scholar]
- 29. Harrison F, Browning LE, Vos M, Buckling A. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol 2006;4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres-Barceló C, Gurney J, Gougat-Barberá C. et al. Transient negative effects of antibiotics on phages do not jeopardise the advantages of combination therapies. FEMS Microbiol Ecol 2018;94(8):10.1093/femsec/fiy107. doi:10.1093/femsec/fiy107 [DOI] [PubMed] [Google Scholar]
- 31. Betts A, Gray C, Zelek M. et al. High parasite diversity accelerates host adaptation and diversification. Science 2018;360:907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burmeister AR, Bender RG, Fortier A, et al. Two lytic bacteriophages that depend on the Escherichia coli multi-drug efflux gene tolC and differentially affect bacterial growth and selection. BioRxiv 2018. doi: 10.1101/397695:397695. [DOI]
- 33. Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 2009;1794:808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Daugelavičius R, Cvirkaitč V, Gaidelytč Ara. et al. Penetration of enveloped double-stranded RNA bacteriophages phi13 and phi6 into Pseudomonas syringae cells. J Virol 2005;79:5017–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dennehy JJ, Friedenberg NA, Yang YW, Turner PE. Virus population extinction via ecological traps. Ecol Lett 2007;10:230–40. [DOI] [PubMed] [Google Scholar]
- 36. Burmeister AR, Fortier A, Roush C, Lessing AJ. et al. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc Natl Acad Sci 2020;117:11207–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Torres-Barceló C. The disparate effects of bacteriophages on antibiotic-resistant bacteria. Emerg Microbes Infect 2018;7:1–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Testing TECoAS. 2019. Clinical Breakpoints (Bacterial v9.0 and Fungal v9.0). http://www.eucast.org/clinical_breakpoints. (12 April 2019, date last accessed).
- 39. Programme BRRS. 2019. http://www.bsacsurv.org/ (12 April 2019, date last accessed).
- 40. Morita Y, Tomida J, Kawamura Y. Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol 2014;4:422–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tseng JT, Bryan LE. Mechanisms of R factor R931 and chromosomal tetracycline resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1973;3:638–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Capparelli R, Nocerino N, Iannaccone M. et al. Bacteriophage therapy of Salmonella enterica: a fresh appraisal of bacteriophage therapy. J Infect Dis 2010;201:52–61. [DOI] [PubMed] [Google Scholar]
- 43. Capparelli R, Parlato M, Borriello G. et al. Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob Agents Chemother 2007;51:2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Law N, Logan C, Yung G. et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 2019;47(4):665-668 doi:10.1007/s15010-019-01319-0. [DOI] [PubMed] [Google Scholar]
- 45. Schooley RT, Biswas B, Gill JJ. et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii Infection. Antimicrob Agents Chemother 2017;61:e00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wright RCT, Friman V-P, Smith MCM, Brockhurst MA. Resistance evolution against phage combinations depends on the timing and order of exposure. mBio 2019;10:e01652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brockhurst MA, Koskella B. Experimental coevolution of species interactions. Trends Ecol Evol 2013;28:367–375. [DOI] [PubMed] [Google Scholar]
- 48. Hill L, Veli N, Coote PJ. Evaluation of Galleria mellonella larvae for measuring the efficacy and pharmacokinetics of antibiotic therapies against Pseudomonas aeruginosa infection. Int J Antimicrob Agents 2014;43:254–61. [DOI] [PubMed] [Google Scholar]
- 49. Alseth EO, Pursey E, Luján AM, et al. Bacterial biodiversity drives the evolution of CRISPR-based phage resistance in Pseudomonas aeruginosa bioRxiv 2019. doi: 10.1101/586115:586115. [DOI] [PMC free article] [PubMed]
- 50. Hall AR, Scanlan PD, Morgan AD, Buckling A. Host–parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett 2011;14:635–642. [DOI] [PubMed] [Google Scholar]
- 51. Gurney J, Aldakak L, Betts A. et al. Network structure and local adaptation in co-evolving bacteria–phage interactions. Mol Ecol 2017;26:1764–1777. [DOI] [PubMed] [Google Scholar]
- 52. Gurney J, Pleska M, Levin BR. Why put up with immunity when there is resistance: an excursion into the population and evolutionary dynamics of restriction-modification and CRISPR-Cas. Philos Trans R Soc Lond B Biol Sci 2019;374:20180096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chan BK, Abedon ST. Bacteriophages and their enzymes in biofilm control. Curr Pharm Des 2014;21:85–99. [DOI] [PubMed] [Google Scholar]
- 54. Cornforth DM, Dees JL, Ibberson CB. et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci USA 2018;115:E5125–E5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.