Abstract
Objectives
In this review, we discuss current knowledge about the genetics and epigenetics of vestibular schwannoma (VS) in relation to hearing loss. A multi-step and sequential genetic algorithm suitable for the identification of Neurofibromatosis Type 2 (NF2) constitutional and somatic mutations is discussed.
Data Sources, Study Selection
A review was performed of the English literature from 1990 to 2019 using PubMed regarding genetics and epigenetics of vestibular schwannoma and NF2.
Conclusion
NF2 is a genetic disorder characterized by NF2 mutations that affect the function of a tumor suppressor called merlin. In particular, individuals with NF2 develop bilateral VS that can lead to hearing loss and even deafness. Recent advances in genetic and epigenetic studies have improved our understanding of the genotype-phenotype relationships that affect hearing in NF2 patients. Specific constitutional NF2 mutations including particular truncating, deletion, and missense mutations, have been associated with poorer hearing outcomes and more severe clinical manifestations. Epigenetic events, such as DNA methylation and histone modifications, also contribute to the development and progression of hearing loss in NF2 patients. Furthermore, the accumulation of multiple NF2 and non-NF2 genetic and epigenetic abnormalities at the level of the tumor may contribute to worse hearing outcomes. Understanding genetic and epigenetic signatures in individual NF2 patients and particularly in each VS will allow us to develop novel gene therapies and precision medicine algorithms to preserve hearing in NF2 individuals.
Introduction
Neurofibromatosis Type 2 (NF2) is a genetic disorder caused by loss-of-function mutations in the NF2 gene (MIM 607379) that code for the tumor suppressor, merlin.(1) Individuals with NF2 are predisposed to developing a variety of benign nervous system tumors and distinct clinical manifestations. Recent advances in genetic and epigenetic testing have allowed researchers to better characterize the genotype-phenotype relationships that occur in NF2 patients, especially in relation to hearing loss (HL).(2) HL serves as a significant source of morbidity for NF2 patients, with essentially all patients eventually progressing to profound deafness bilaterally.(3) By better characterizing the full range of mutations underlying NF2, and understanding how genetic and epigenetic signatures can affect the clinical presentations of NF2 patients, we can diagnose at-risk patients earlier and identify and test new therapies that can reverse pathologic mutations and epigenetic events to restore normal merlin function, preserve hearing, reduce tumor burden, and improve survival in NF2. This review focuses on genetics and epigenetics of NF2 as it pertains to hearing loss.
Clinical Presentation & Epidemiology
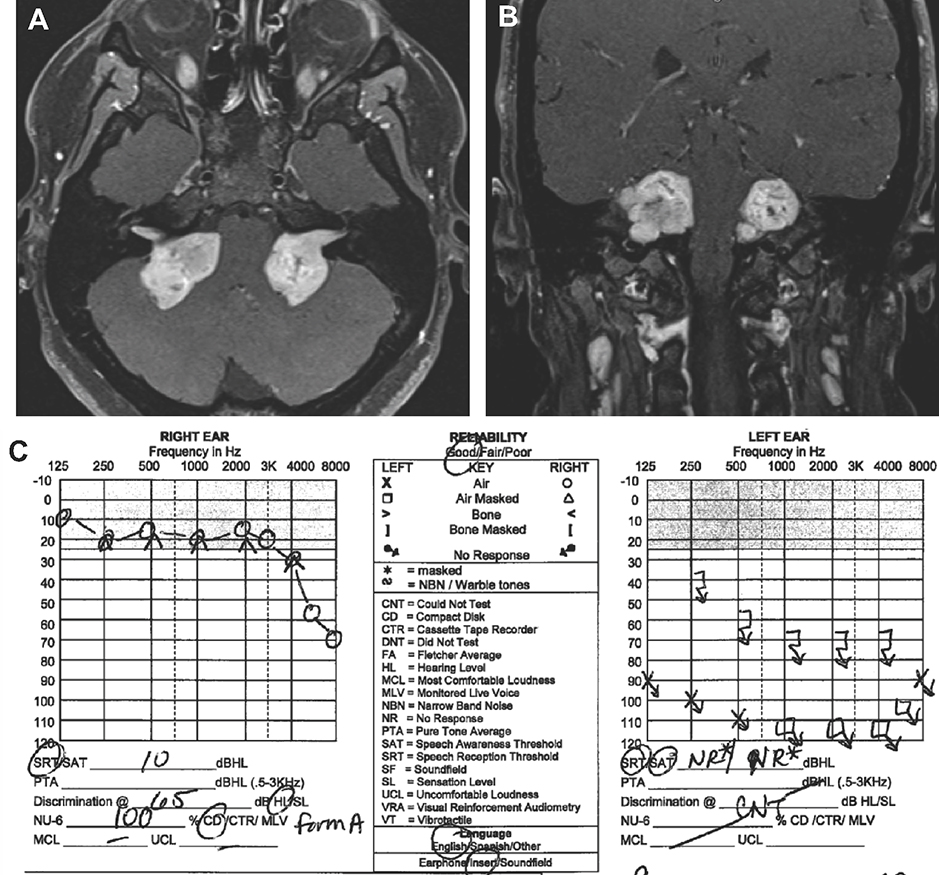
A broad spectrum of clinical findings can be seen in NF2 patients. Classically, individuals with NF2 develop multiple nervous system tumors, particularly bilateral vestibular schwannomas (VS) that arise from the Schwann cells of the cochleovestibular nerves.(4) VS are benign tumors that have a propensity to cause HL and disabling imbalance. As they grow, VS can compress the adjacent facial nerve and cause facial weakness. Life-threatening intracranial complications from brainstem and cerebellar compression can also occur. Figure 1A and 1B demonstrates bilateral VS on axial and coronal cuts of a T1-weighted and contrast-enhanced magnetic resonance imaging (MRI) study of a 25-year old female with NF2 (Figure 1A–B). The tumors are compressing the brainstem and cerebellum on both sides.
Figure 1. Magnetic Resonance Imaging (MRI) of Bilateral Vestibular Schwannoma (VS).
Axial [A] and coronal [B] cuts of a T1-weighted contrast-enhanced MRI demonstrate bilateral VS in a 25 year-old female with Neurofibromatosis Type 2. [C] The right VS is significantly larger than the left VS, however the hearing is overall preserved on the side with the larger tumor.
While NF2 is classically associated with the development of bilateral VS, other common clinical manifestations include: (1) schwannomas of other cranial, spinal, and peripheral nerves, (2) dermatological tumors, (3) presenile posterior subcapsular lenticular opacities (cataracts), (4) retinal hamartomas, (5) meningiomas, (6) ependymomas, and (7) peripheral neuropathy.(4–7)
NF2 affects approximately 1:60,000 people with an incidence of about 1:33,000.(8) However, the prevalence is likely higher as mosaic forms of NF2 have milder phenotypes, delayed onset of symptoms, and are more difficult to diagnosis.(9–11)
Genetic Origins of NF2
NF2 is caused by an inherited or de novo mutation in the NF2 gene (MIM #607379) on chromosome 22q12.2. The NF2 gene contains 17 coding exons spanning approximately 95 kilobases (kb) and encodes a protein called Merlin (moesin-ezrin-radixin-like protein), a tumor suppressor also referred to as Schwannomin. Merlin contains the FERM (four-point-one/ezrin/radixin/moesin) domain and regulates cell proliferation in response to adhesive signaling, activates anti-mitogenetic signaling at tight junctions, and inhibits oncogenic gene expression.(12–14) Thus, inactivation of merlin can lead to uncontrolled mitogenic signaling and tumorigenesis, particularly in the nervous system.
While there is wide phenotypic variability, NF2 has nearly 100% penetrance by 60 years of age.(15) Approximately fifty percent of NF2 patients have another affected family member and possess a germline mutation that has an autosomal dominant inheritance pattern. The remaining 50% of NF2 patients have a spontaneous, or de novo, mutation and therefore have no family history of NF2.(1,16) These de novo mutations of the NF2 gene can either occur in the germline cells of the parents (prezygotic mutation), or in cells after fertilization (postzygotic mutation). Postzygotic mutations lead to two genetically distinct cellular populations to occur in an individual – cells with and without the mutation - an occurrence called mosaicism. Pedigree analysis and genetic tests in blood and tumor samples have shown that of the NF2 patients without a positive family history, about 25–30% are mosaic for an NF2 pathogenic variant, with the mutation often detected only in tumor specimens and not in blood.(1,10,15,16) Recent genetic tests employing next-generation sequencing (NGS) offer more sensitivity for detecting mosaicism in blood or saliva samples.
VS tumors in NF2 patients are thought to occur through the Knudson’s two-hit model.(17) In general, VS occur by either: (1) mutations in both NF2 alleles, or by (2) a mutation in one allele and loss of the other allele resulting in loss of heterozygosity (LOH) on chromosome 22.(18,19) Subsequently, merlin protein is not expressed or is dysfunctional, resulting in tumorigenesis in affected Schwann cells.(20)
Differential Diagnosis of NF2
It is important to mention the overlap between NF2 and a condition called “schwannomatosis.” Schwannomatosis is a genetic tumor disorder characterized by multiple schwannomas in the absence of VS. Patients with schwannomatosis often develop chronic localized or diffuse pain.(21) The incidence of schwannomatosis is unknown but it is estimated to be as common as NF2, affecting 1 in 40,000 individuals.(22) Approximately 10–15% of cases are familial and demonstrate an autosomal dominant pattern of inheritance, while the remainder are sporadic.(23–25) Schwannomatosis is caused by constitutional mutations in SMARCB1 and LZTR1 genes that are thought to encode tumor suppressor proteins.(23–26) However, some patients with schwannomatosis do not express mutations in these genes, which suggest that additional genes may also be implicated.(27) Furthermore, schwannoma formation requires the acquisition of a somatic mutation, which can involve the SMARCB1 and LZTR1 genes, but is commonly seen with biallelic somatic inactivation of the NF2 gene or a loss of chromosome 22.(24,27,28) This relationship is important because NF2, SMARCB1, and LZTR1 genes are all located on chromosome 22 and schwannomatosis requires a four-hit mechanism.(28) Therefore, if the presumed diagnosis is schwannomatosis, we should be vigilant about testing for NF2, LZTR1 and SMARCB1 genes. In patients with unilateral VS and ≥2 nondermal schwannomas, there was significant overlap in the NF2 and LZTR1-associated schwannomatosis diagnoses.(29) Therefore, in this subset of patients, adding LZTR1 genetic testing to NF2 mutational analyses would be beneficial to exclude LZTR1-associated schwannomatosis.
Clinical Subtypes of NF2
There is great variety in the clinical presentations of NF2 patients and historically NF2 was categorized as either congenital, Wishart, or Gardner clinical subtypes. Another category, called NF2 mosaicism, has emerged with the advancement of molecular genetic techniques for diagnosis. In the congenital form of NF2 – a very rare subtype that has only been described in a handful of case series and reports - infants present with bilateral VS detectable on MRI in the first days to early months of life, as well as early development of cutaneous NF2 plaques.(30,31) The disease is often clinically stable for the first one or two decades of life before sudden and rapid progression occurs, which in two cases was also accompanied by total regression of the NF2 plaques. In three cases – all of whom had no family history of NF2 - pathogenic NF2 mutations were identified (exon 3 Ins CCTT, exon 5 Ins CCTT, exon 13 Q470X).(30)
The Wishart subtype, also known as severe, prepubertal NF2, is characterized by multiple, rapidly progressive central nervous system tumors, often presenting before the development of VS and HL.(31,32) Primary presentation typically involves NF2 plaques, which are differentiated from skin lesions seen in Neurofibromatosis Type 1, by their raised, well-circumscribed, and pigmented features.(33) NF2 plaques are typically localized to the upper and lower limbs in the Wishart subtype, whereas plaques are more likely to appear on hands, feet, and face in the congenital form of NF2.(31,33,34) Ocular involvement is another early sign in 40–70% of NF2 patients and is often associated with the Wishart subtype. The most common ocular manifestation is the development of pediatric posterior capsular or cortical edge cataracts.(34–36)
Patients with the Gardner subtype, or the adult form of NF2, typically present with bilateral VS and associated symptoms of HL, tinnitus, and imbalance in young adulthood.(37,38) These symptoms are often the only clinical manifestation of the disease. If NF2 plaques are present, lesions are more likely to be localized to the trunk.(31)
NF2 mosaicism can present with symptoms of variable severity, although often with a milder phenotype than other NF2 subtypes. Baser et al. assessed the clinical differences between sporadic non-mosaic and sporadic mosaic NF2 patients and found that mosaic cases had a symptom onset 7.8 years later than non-mosaic NF2 patients.(39) In addition, patients with unilateral or no VS tumors were 7.1 times more likely to demonstrate mosaicism than not. NF2 mosaicism is also associated with ipsilateral meningiomas and many localized peripheral nervous system schwannomas.(31) The clinical manifestations of NF2 mosaicism overlap with schwannomatosis, a distinct and rare disease process which arises from germline or mosaic mutations in SMARCB1 or LZTR1 genes.(25,26) Unlike traditional NF2 patients, individuals with schwannomatosis develop multiple cutaneous schwannomas and central nervous system tumors without the development of VS.
These clinical subtypes, however, are unable to capture the broad spectrum of clinical manifestations seen in NF2, nor do they represent a particular group of NF2 mutations. Therefore, there has been a push to understand genotype-phenotype relationships and develop newer staging systems that are more representative of this genetic disorder.
NF2 Genotype-Phenotype Relationships
Correlations between the NF2 genotype and phenotype have been identified in patients with NF2. In the United Kingdom, Halliday and colleagues developed the UK NF2 Genetic Severity Score that groups NF2 patients into severity categories based on clinical presentation and genotype, including mosaicism.(2)
When comparing NF2 patients assigned to the mild, moderate, and severe categories, Halliday et al. found statistically significant intergroup differences in age at diagnosis, quality of life, presence of VS and other intracranial tumors, ocular features, HL, and need for major interventions.(2) Mosaic cases had a milder phenotype as demonstrated by: (1) later age at diagnosis, (2) fewer cases of bilateral VS and CNS tumors, (3) fewer ocular defects, and (4) less severe HL. In the Heineman et al. analysis of missense mutations of the NF2 gene, severe NF2 was defined as an age at presentation less than 30 and the presence of a non-VS tumor.(40) Table 1 consolidates information about specific NF2 gene mutation types, locations, and phenotypic severity.
Table 1:
| Mutation type | Location | Mosaic* Mutation | Germline Mutation |
|---|---|---|---|
| Truncating mutation | Exon 1 | Mild NF2 | Mild NF2 |
| Exons 2–13 | Moderate NF2 | Severe NF2 | |
| Exons 14–15 | Mild NF2 | Moderate NF2 | |
| Splice site mutation | Exons 1–7 | Mild NF2 | Moderate NF2 |
| Exons 8–15 | Mild NF2 | Mild NF2 | |
| Large deletion | Excluding promoter or exon 1 | Mild NF2 | Moderate NF2 |
| Including promoter or exon 1 | Mild NF2 | Mild NF2 | |
| Small in–frame deletion or duplication | Mild NF2 | Mild NF2 | |
| Missense Mutation | Arg52Gln (exon 2) | ||
| Asn36Ser (exon 1) | Mild NF2 | ||
| Leu141Arg (exon 4) | |||
| Phe62Ser (exon 2) | |||
| Leu64Pro (exon 2) | Moderate- Severe NF2 | ||
| Met77Val (exon 2) | |||
| Lys149Asn (exon 4) | Severe NF2 | ||
Mosaic is defined as any NF2 mutation in less than 50% of cells as determined by next generation sequencing.
In particular, truncating mutations of the NF2 gene are associated with earlier age of symptom onset, earlier diagnosis of NF2, and lower average life expectancy than splice site mutations, large deletions, missense mutations, and somatic mosaicism.(2,41–47) Furthermore, individuals with truncating mutations of the 3’ region of exons 2 through 13 develop a more severe phenotype and have worse survival outcomes than patients with truncating mutations involving exons 1, 14 and 15.
Although splice site mutations are associated with milder disease, splice site mutations of the 5’ end of the NF2 gene are linked with worse clinical outcomes and higher mortality rates than those between exons 8 and 15.(9,44,48–51) In addition, frameshift mutations near the NF2 translation initiation codon are associated with better life expectancy.(2,44,48) Moreover, relevant to hearing, truncating mutations (non-sense or frameshift) of the NF2 gene are also associated with earlier age of HL than splice-site, missense, and large deletions.(52)
However, the molecular and cellular mechanisms that connect these genetic mutations to specific phenotypic presentations are currently poorly characterized. Stankovic and colleagues demonstrated that VS in patients with hearing loss express higher levels of inflammatory factors such as tumor necrosis factor alpha and components of the NRLP3 inflammasome, an example of a potential mechanistic bridge between genotype and phenotype.(53,54) In addition, Fernandez-Valle and colleagues showed that differences in aberrant DNA methylation across a variety of genes was correlated with varying levels of resistance to MEK1/2 inhibitors for NF2-associated VS, suggesting that epigenetics can contribute to the genotype-phenotype relationship as well.(55) More work is necessary to further identify and characterize these mechanisms, as they may represent targets for the development of novel treatment modalities.
Epigenetics
Epigenetics is a field of study that focuses on alterations in the expression of genes rather than changes to the gene sequence itself, and how that affects phenotype.(56) Examples of epigenetic modifications are DNA methylation, histone modifications, and non-coding RNAs. In mammals, almost all DNA methylation occurs at CpG dinucleotide sites.(57)
Welling et al. identified positive and negative regulatory elements in the 5’ flanking region of the NF2 gene promoter.(58) Methylation of these regulatory sites has been shown by Kino et al. to be a potential mechanism of gene silencing.(59) In this study, 14 of 23 VS tumors examined were found to have methylation in 3 CpG sites of the NF2 promoter region, which was associated with reduced mRNA expression of the NF2 gene. However, other studies have suggested that methylation of the NF2 gene and its promoter is actually rare and may not be a primary mechanism for formation of VS.(60–62)
A study by Kullar et al. showed that while hypermethylation of the NF2 gene is rare in sporadic VS, other genes, particularly THBS1, MGMT, TIMP3, and P73, were frequently found to be aberrantly hypermethylated in schwannomas.(63) Methylation of these genes has been shown to be tumor specific.(64)
In particular, epigenetics may also play a role in the development of HL in VS.(65) A focused exploration into the methylation of 16 tumor-related genes suggest abnormal methylation of THBS1, TP73, MGMT, NF2, TIMP3, FASSF1A, VHL, PTEN, TP16, CASP8, TIMP3, DAPK, HMLH1, and GTSP1 in VS.(64,66,67) In particular, aberrant methylation of tumor suppressor gene (TP73) that encodes tumor protein 73 (p73) was associated with tumor-mediated HL.(67) The corrected hearing thresholds for VS patients with and without methylated TP73 were 43 decibels (dB) and 17 dB (p=0.04), respectively. Moreover, the difference in speech discrimination scores was also striking between VS with methylated and unmethylated TP73 (47% versus 72%). Despite this correlation, the pathophysiology behind TP73 methylation and tumor-mediated HL is still unclear; however, it is known that TP73 plays a major role in apoptosis of neuronal tissue and VS.(68,69)
Although not related to hearing, DNA methylation of CASP8 was found to be associated with patient age and tumor size. Furthermore, global hypomethylation of HOX genes have also been found in VS.(62) Aberrant expression of HOX genes has been linked to many different types of cancers.(70,71)
Aside from DNA methylation, there are other epigenetic mechanisms that have been linked to the development of VS: (1) post-transcriptional modifications of NF2 transcripts, (2) changes in lysine acetylation, and (3) dysregulation of miRNA expression.(72–75) While only a few studies to date have examined the epigenetic changes in VS, the much larger body of literature addressing epigenetic changes in meningiomas provides an example of what could be done for VS and NF2.(76,77) Meningioma classification using DNA methylation is more relevant clinically but it captures more clinically homogenous groups that are better at predicting tumor prognosis and recurrence than the traditional WHO classification.(78)
NF2 Diagnostic Criteria
The 1997 Manchester criteria for NF2 is the most commonly used criteria to make the clinical diagnosis of NF2.(4) NF2 diagnosis requires one of the three conditions: (1) bilateral VS, (2) ≥1 first degree relatives with NF2 and unilateral VS at <30 years of age, or (3) ≥2 of either meningioma, glioma, schwannoma, or juvenile posterior lenticular opacities. However, these clinical criteria resulted in the clinical diagnosis of NF2 later in life, in some cases delaying appropriate clinical management. As a result, there have been multiple revisions to the diagnostic criteria over the years to aid in NF2 diagnosis at younger ages. Most recently, updated diagnostic criteria were proposed at the Joint Global Neurofibromatosis Meeting in November 2018, with recommendations finalized in September 2019 at the NF Conference after further expert feedback. These are displayed in Table 2 and take into consideration the broad NF2 clinical spectrum and advent of molecular testing.(29,79,80) There is evidence that in patients 70+ years of age, bilateral VS may frequently occur spontaneously rather than as a manifestation of NF2.(29) However, in order to increase sensitivity in identifying all individuals with NF2, the decision was made to not specify an age range for bilateral VS as a diagnostic criteria.(79)
Table 2.
NF2 Diagnostic Criteria per 2019 NF Conference Revisions
| Finding | Additional Features Required for Diagnosis |
| Bilateral VS | None |
| Identical NF2 pathogenic variant in at least 2 anatomically distinct NF2-related tumors | None |
| One Major criteria (see below) | Additional Major criteria OR two additional Minor criteria (see below) |
| Major Criteria | Minor Criteria |
| Unilateral VS | Ependymoma, schwannoma (can count more than one of each type) |
| First degree relative other than sibling with NF2 | |
| Multiple meningiomas | Juvenile subcapsular/cortical cataract, retinal hamartoma, epiretinal membrane if age <40, meningioma (can count each type only once) |
| NF2 pathogenic variant in an unaffected tissue (e.g. blood) |
Due to the wide range of clinical presentation that can occur, NF2 diagnosis is often delayed in pediatric patients without a family history of NF2. For example, signs and symptoms that can present prior to the development of VS include visual impairment, unilateral optic atrophy, skin hyperpigmentation, plaques, seizures, and cranial nerve palsy.(81) Genetic testing plays an important role in NF2 diagnosis, particularly when obvious signs of NF2 are not yet present. However, multiple genetic tests may be necessary to identify a NF2 mutation and/or exclude other disorders with similar presentations, such as schwannomatosis.
Genetic tests that have been utilized to detect germline NF2 mutations should be able to detect sequence and copy number variants as well as mosaicism. NGS can simultaneously detect them, and Sanger sequencing and other techniques such as multiplex ligation-dependent probe amplification (MLPA), quantitative real-time PCR or comparative genomic hybridization can be used to confirm sequence and copy number variants, respectively.(10,19,82,83) More recently, mutational testing for LZTR1 and SMARCB1 genes has been incorporated into NF2 genetic testing to rule out similar conditions such as schwannomatosis.(84) NF2 mosaicism is more difficult to test with a genetic test. While NGS can detect mosaicism, low levels of mosaicism may be missed with a blood test. Furthermore, genetic testing of tumor tissue from multiple tumors may be necessary to confirm the diagnosis of NF2.(10)
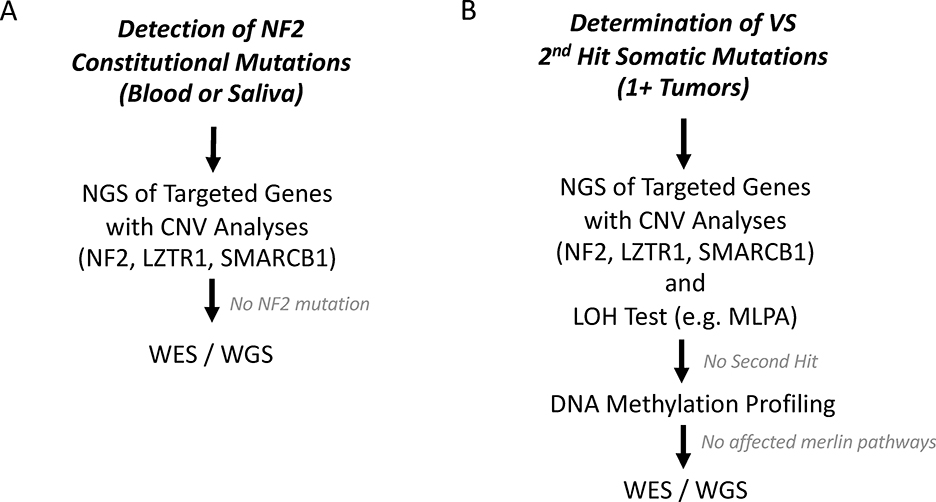
Based on this information, we have developed a genetic algorithm (Figure 2A) to assess for constitutional NF2 gene mutations and putative target genes located on other chromosomal regions. We support initial NF2 screening on blood or saliva samples using a NGS-based panel that includes analyses of NF2, LZTR1, and SMARCB1 genes to detect constitutional mutations. However, phenotypic variability within NF2 families and between monozygotic twins suggest that differences in second hit mutations, accumulation of other genetic mutations, and epigenetic factors such as DNA methylation may all have important impacts on phenotype and HL in VS.(65)
Figure 2. Simplified Genetic Workflow for Mutational Analysis for Clinical NF2 and Tumor Profiling.
[A] To detect constitutional NF2 mutations, next generation sequencing (NGS) for target genes (NF2, LZTR1, SMARC1) should be performed along with copy number variation analyses (CNV). If a NF2 mutation cannot be determined, then whole exome sequencing (WES) or whole genome sequencing (WGS) should be considered. [B] To determine 2nd hit events in the NF2 gene, we advocate processing tumor chunks with NGS for target genes with CNV analyses followed by loss of heterozygosity (LOH) tests (e.g. multiplex ligation dependent probe amplification (MLPA). DNA methylation profiling should be considered to assess epigenetic causes if two hits of the NF2 gene cannot be identified. If a 2nd hit NF2 mutation cannot be found, WES and WGS should also be considered. Multiple tumors from the same patient need to be tested to assess for NF2 mosaicism.
Because tumors express two hits in the NF2 gene versus the single hit detectable in blood, tumor samples may also be analyzed to link mutational profile with HL and other phenotypic manifestations in NF2 patients (Figure 2B). For example, differences in second hit mutation may potentially account for differences in HL and tumor size between multiple VS within the same patient (Figure 1C). In order to detect second hit mutations in tumor samples and assess for NF2 mosaicism, NGS for NF2, LZTR1, and SMARCB1 genes should also be performed and include LOH testing and copy number variant analysis. To look for accumulation of multiple non-NF2 mutations or for those cases who do not have an identifiable mutation, a more comprehensive test such as whole exome sequencing (WES) or whole genome sequencing (WGS) can be considered for discovering novel genes leading to VS and phenotypic variability.(82,85,86) Methylation analysis in the tumor tissue using bisulfite sequencing of the NF2 gene or broader assessments may also identify methylation patterns that lead to NF2 phenotypes.(62,85)
Future Genetic and Epigenetic Directions for NF2 and VS
There are limitations to older algorithms using direct sequencing of the NF2 gene to diagnose and stratify clinical NF2. As eluded by the most recent guidelines for NF2 diagnosis, NGS should be implemented as a standard of care to improve diagnostic accuracy to more comprehensively analyze the NF2 genes and exclude the possibility of schwannomatosis. Further work is also necessary to improve diagnostic sensitivity in the subset of patients with a low degree of mosaicism. In patients that meet the clinical criteria for NF2 that do not have an identifiable NF2 mutation, further testing with WES and WGS can identify new genes that can become a target for further research. Lastly, genetic testing must be accompanied with pre and post-test genetic counseling in order to give patients context on their syndrome and allow them to make informed decisions.
However, constitutional NF2 mutations can only provide basic information about NF2 severity. It does not provide comprehensive prognostic data that can better guide clinical management. Comprehensive genetic and epigenetic testing at the level of the tumor may provide important prognostic information that can potentially guide clinical management in the future. Therefore, to fully understand how NF2-associated VS cause HL, more research is needed to identify the genetic and epigenetic variations that can lead to molecular and phenotypic differences both within and between patients and families. Elucidating these relationships can also help us stratify NF2 patients toward early management that can potentially improve clinical outcomes and determine targets for new therapeutic interventions.
References
- 1.Evans DG. Neurofibromatosis 2 In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, eds. GeneReviews((R)). Seattle (WA): University of Washington, Seattle, 1993. [Google Scholar]
- 2.Halliday D, Emmanouil B, Pretorius P et al. Genetic Severity Score predicts clinical phenotype in NF2. Journal of medical genetics 2017;54:657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JM, Chang JW, Choi JY et al. Hearing Restoration in Neurofibromatosis Type II Patients. Yonsei Med J 2016;57:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans DG, Huson SM, Donnai D et al. A clinical study of type 2 neurofibromatosis. The Quarterly journal of medicine 1992;84:603–18. [PubMed] [Google Scholar]
- 5.Parry DM, Eldridge R, Kaiser-Kupfer MI et al. Neurofibromatosis 2 (NF2): clinical characteristics of 63 affected individuals and clinical evidence for heterogeneity. American journal of medical genetics 1994;52:450–61. [DOI] [PubMed] [Google Scholar]
- 6.MacCollin M, Mautner VF. The diagnosis and management of neurofibromatosis 2 in childhood. Seminars in pediatric neurology 1998;5:243–52. [DOI] [PubMed] [Google Scholar]
- 7.Mautner VF, Lindenau M, Baser ME et al. Skin abnormalities in neurofibromatosis 2. Archives of dermatology 1997;133:1539–43. [PubMed] [Google Scholar]
- 8.Evans DG, Howard E, Giblin C et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. American journal of medical genetics. Part A 2010;152a:327–32. [DOI] [PubMed] [Google Scholar]
- 9.Baser ME, Kuramoto L, Joe H et al. Genotype-phenotype correlations for nervous system tumors in neurofibromatosis 2: a population-based study. American journal of human genetics 2004;75:231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans DG, Ramsden RT, Shenton A et al. Mosaicism in neurofibromatosis type 2: an update of risk based on uni/bilaterality of vestibular schwannoma at presentation and sensitive mutation analysis including multiple ligation-dependent probe amplification. Journal of medical genetics 2007;44:424–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyhuddin A, Baser ME, Watson C et al. Somatic mosaicism in neurofibromatosis 2: prevalence and risk of disease transmission to offspring. Journal of medical genetics 2003;40:459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper J, Giancotti FG. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett 2014;588:2743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouleau GA, Merel P, Lutchman M et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 1993;363:515–21. [DOI] [PubMed] [Google Scholar]
- 14.Trofatter JA, MacCollin MM, Rutter JL et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 1993;75:826. [DOI] [PubMed] [Google Scholar]
- 15.Evans DG, Huson SM, Donnai D et al. A genetic study of type 2 neurofibromatosis in the United Kingdom. I. Prevalence, mutation rate, fitness, and confirmation of maternal transmission effect on severity. Journal of medical genetics 1992;29:841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kluwe L, Mautner V, Heinrich B et al. Molecular study of frequency of mosaicism in neurofibromatosis 2 patients with bilateral vestibular schwannomas. Journal of medical genetics 2003;40:109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knudson AG. Two genetic hits (more or less) to cancer. Nature reviews. Cancer 2001;1:157–62. [DOI] [PubMed] [Google Scholar]
- 18.Giovannini M, Robanus-Maandag E, van der Valk M et al. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes & development 2000;14:1617–30. [PMC free article] [PubMed] [Google Scholar]
- 19.Hadfield KD, Smith MJ, Urquhart JE et al. Rates of loss of heterozygosity and mitotic recombination in NF2 schwannomas, sporadic vestibular schwannomas and schwannomatosis schwannomas. Oncogene 2010;29:6216–21. [DOI] [PubMed] [Google Scholar]
- 20.Petrilli AM, Fernandez-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 2016;35:537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhamija R, Plotkin S, Asthagiri A et al. Schwannomatosis In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, eds. GeneReviews((R)). Seattle (WA): University of Washington, Seattle, 1993. [Google Scholar]
- 22.Kresak JL, Walsh M. Neurofibromatosis: A Review of NF1, NF2, and Schwannomatosis. Journal of pediatric genetics 2016;5:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd C, Smith MJ, Kluwe L et al. Alterations in the SMARCB1 (INI1) tumor suppressor gene in familial schwannomatosis. Clinical genetics 2008;74:358–66. [DOI] [PubMed] [Google Scholar]
- 24.Hadfield KD, Newman WG, Bowers NL et al. Molecular characterisation of SMARCB1 and NF2 in familial and sporadic schwannomatosis. Journal of medical genetics 2008;45:332–9. [DOI] [PubMed] [Google Scholar]
- 25.Hulsebos TJ, Plomp AS, Wolterman RA et al. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. American journal of human genetics 2007;80:805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piotrowski A, Xie J, Liu YF et al. Germline loss-of-function mutations in LZTR1 predispose to an inherited disorder of multiple schwannomas. Nature genetics 2014;46:182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plotkin SR, Blakeley JO, Evans DG et al. Update from the 2011 International Schwannomatosis Workshop: From genetics to diagnostic criteria. American journal of medical genetics. Part A 2013;161a:405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sestini R, Bacci C, Provenzano A et al. Evidence of a four-hit mechanism involving SMARCB1 and NF2 in schwannomatosis-associated schwannomas. Human mutation 2008;29:227–31. [DOI] [PubMed] [Google Scholar]
- 29.Evans DG, King AT, Bowers NL et al. Identifying the deficiencies of current diagnostic criteria for neurofibromatosis 2 using databases of 2777 individuals with molecular testing. Genetics in medicine : official journal of the American College of Medical Genetics 2019;21:1525–33. [DOI] [PubMed] [Google Scholar]
- 30.Ruggieri M, Gabriele AL, Polizzi A et al. Natural history of neurofibromatosis type 2 with onset before the age of 1 year. Neurogenetics 2013;14:89–98. [DOI] [PubMed] [Google Scholar]
- 31.Ruggieri M, Pratico AD, Evans DG. Diagnosis, Management, and New Therapeutic Options in Childhood Neurofibromatosis Type 2 and Related Forms. Seminars in pediatric neurology 2015;22:240–58. [DOI] [PubMed] [Google Scholar]
- 32.Wishart JH. Case of Tumours in the Skull, Dura Mater, and Brain. Edinburgh medical and surgical journal 1822;18:393–7. [PMC free article] [PubMed] [Google Scholar]
- 33.Ruggieri M, Iannetti P, Polizzi A et al. Earliest clinical manifestations and natural history of neurofibromatosis type 2 (NF2) in childhood: a study of 24 patients. Neuropediatrics 2005;36:21–34. [DOI] [PubMed] [Google Scholar]
- 34.Evans DG, Birch JM, Ramsden RT. Paediatric presentation of type 2 neurofibromatosis. Archives of disease in childhood 1999;81:496–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosch MM, Boltshauser E, Harpes P et al. Ophthalmologic findings and long-term course in patients with neurofibromatosis type 2. American journal of ophthalmology 2006;141:1068–77. [DOI] [PubMed] [Google Scholar]
- 36.Matsuo M, Ohno K, Ohtsuka F. Characterization of early onset neurofibromatosis type 2. Brain & development 2014;36:148–52. [DOI] [PubMed] [Google Scholar]
- 37.Gardner WJMD, Frazier CHMDSD. HEREDITARY BILATERAL ACOUSTIC TUMORS*: A Survey of a Family of Five Generations with Bilateral Deafness in Thirty-eight Members. Journal of Heredity 1931;22:7–8. [Google Scholar]
- 38.Ruggieri M The different forms of neurofibromatosis. Child’s Nerv Syst 1999;15:295–308. [DOI] [PubMed] [Google Scholar]
- 39.Baser ME, Wallace AJ, Strachan T et al. Clinical and molecular correlates of somatic mosaicism in neurofibromatosis 2. Journal of medical genetics 2000;37:542–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heineman TE, Evans DG, Campagne F et al. In Silico Analysis of NF2 Gene Missense Mutations in Neurofibromatosis Type 2: From Genotype to Phenotype. Otol Neurotol 2015;36:908–14. [DOI] [PubMed] [Google Scholar]
- 41.Abo-Dalo B, Kutsche K, Mautner V et al. Large intragenic deletions of the NF2 gene: breakpoints and associated phenotypes. Genes, chromosomes & cancer 2010;49:171–5. [DOI] [PubMed] [Google Scholar]
- 42.Baser ME, Friedman JM, Aeschliman D et al. Predictors of the risk of mortality in neurofibromatosis 2. American journal of human genetics 2002;71:715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baser ME, Makariou EV, Parry DM. Predictors of vestibular schwannoma growth in patients with neurofibromatosis Type 2. Journal of neurosurgery 2002;96:217–22. [DOI] [PubMed] [Google Scholar]
- 44.Hexter A, Jones A, Joe H et al. Clinical and molecular predictors of mortality in neurofibromatosis 2: a UK national analysis of 1192 patients. Journal of medical genetics 2015;52:699–705. [DOI] [PubMed] [Google Scholar]
- 45.Mautner VF, Baser ME, Thakkar SD et al. Vestibular schwannoma growth in patients with neurofibromatosis Type 2: a longitudinal study. Journal of neurosurgery 2002;96:223–8. [DOI] [PubMed] [Google Scholar]
- 46.Otsuka G, Saito K, Nagatani T et al. Age at symptom onset and long-term survival in patients with neurofibromatosis Type 2. Journal of neurosurgery 2003;99:480–3. [DOI] [PubMed] [Google Scholar]
- 47.Selvanathan SK, Shenton A, Ferner R et al. Further genotype--phenotype correlations in neurofibromatosis 2. Clinical genetics 2010;77:163–70. [DOI] [PubMed] [Google Scholar]
- 48.Baser ME, Kuramoto L, Woods R et al. The location of constitutional neurofibromatosis 2 (NF2) splice site mutations is associated with the severity of NF2. Journal of medical genetics 2005;42:540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans DG, Bowers N, Huson SM et al. Mutation type and position varies between mosaic and inherited NF2 and correlates with disease severity. Clinical genetics 2013;83:594–5. [DOI] [PubMed] [Google Scholar]
- 50.Kluwe L, Mautner VF. Mosaicism in sporadic neurofibromatosis 2 patients. Hum Mol Genet 1998;7:2051–5. [DOI] [PubMed] [Google Scholar]
- 51.Smith MJ, Higgs JE, Bowers NL et al. Cranial meningiomas in 411 neurofibromatosis type 2 (NF2) patients with proven gene mutations: clear positional effect of mutations, but absence of female severity effect on age at onset. Journal of medical genetics 2011;48:261–5. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Y, Kumar RA, Baser ME et al. Intrafamilial correlation of clinical manifestations in neurofibromatosis 2 (NF2). Genetic epidemiology 2002;23:245–59. [DOI] [PubMed] [Google Scholar]
- 53.Sagers JE, Sahin MI, Moon I et al. NLRP3 inflammasome activation in human vestibular schwannoma: Implications for tumor-induced hearing loss. Hearing Research 2019;381:107770. [DOI] [PubMed] [Google Scholar]
- 54.Ren Y, Stankovic KM. The Role of Tumor Necrosis Factor Alpha (TNFα)in Hearing Loss and Vestibular Schwannomas. Curr Otorhinolaryngol Rep 2018;6:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuse MA, Dinh CT, Vitte J et al. Preclinical assessment of MEK1/2 inhibitors for neurofibromatosis type 2–associated schwannomas reveals differences in efficacy and drug resistance development. Neuro-oncology 2019;21:486–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ajonijebu DC, Abboussi O, Russell VA et al. Epigenetics: a link between addiction and social environment. Cellular and molecular life sciences : CMLS 2017;74:2735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lister R, Pelizzola M, Dowen RH et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welling DB, Akhmametyeva EM, Daniels RL et al. Analysis of the human neurofibromatosis type 2 gene promoter and its expression. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery 2000;123:413–8. [DOI] [PubMed] [Google Scholar]
- 59.Kino T, Takeshima H, Nakao M et al. Identification of the cis-acting region in the NF2 gene promoter as a potential target for mutation and methylation-dependent silencing in schwannoma. Genes Cells 2001;6:441–54. [DOI] [PubMed] [Google Scholar]
- 60.Koutsimpelas D, Ruerup G, Mann WJ et al. Lack of neurofibromatosis type 2 gene promoter methylation in sporadic vestibular schwannomas. ORL; journal for oto-rhino-laryngology and its related specialties 2012;74:33–7. [DOI] [PubMed] [Google Scholar]
- 61.Lee JD, Kwon TJ, Kim UK et al. Genetic and epigenetic alterations of the NF2 gene in sporadic vestibular schwannomas. PLoS One 2012;7:e30418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torres-Martin M, Lassaletta L, de Campos JM et al. Genome-wide methylation analysis in vestibular schwannomas shows putative mechanisms of gene expression modulation and global hypomethylation at the HOX gene cluster. Genes, chromosomes & cancer 2015;54:197–209. [DOI] [PubMed] [Google Scholar]
- 63.Kullar PJ, Pearson DM, Malley DS et al. CpG island hypermethylation of the neurofibromatosis type 2 (NF2) gene is rare in sporadic vestibular schwannomas. Neuropathology and applied neurobiology 2010;36:505–14. [DOI] [PubMed] [Google Scholar]
- 64.Gonzalez-Gomez P, Bello MJ, Alonso ME et al. CpG island methylation in sporadic and neurofibromatis type 2-associated schwannomas. Clinical cancer research : an official journal of the American Association for Cancer Research 2003;9:5601–6. [PubMed] [Google Scholar]
- 65.Celis-Aguilar E, Lassaletta L, Torres-Martin M et al. The molecular biology of vestibular schwannomas and its association with hearing loss: a review. Genetics research international 2012;2012:856157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bello MJ, Martinez-Glez V, Franco-Hernandez C et al. DNA methylation pattern in 16 tumor-related genes in schwannomas. Cancer Genet Cytogenet 2007;172:84–6. [DOI] [PubMed] [Google Scholar]
- 67.Lassaletta L, Bello MJ, Del Rio L et al. DNA methylation of multiple genes in vestibular schwannoma: Relationship with clinical and radiological findings. Otol Neurotol 2006;27:1180–5. [DOI] [PubMed] [Google Scholar]
- 68.Ahmad ZK, Altuna X, Lopez JP et al. p73 expression and function in vestibular schwannoma. Arch Otolaryngol Head Neck Surg 2009;135:662–9. [DOI] [PubMed] [Google Scholar]
- 69.Pozniak CD, Radinovic S, Yang A et al. An Anti-Apoptotic Role for the p53 Family Member, p73, During Developmental Neuron Death. Science 2000;289:304. [DOI] [PubMed] [Google Scholar]
- 70.Jelinek J, Gharibyan V, He R et al. DNA methylation of HOX genes in leukemia and myeloproliferative disorders. Cancer Research 2007;67:235. [Google Scholar]
- 71.Tommasi S, Karm DL, Wu X et al. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast cancer research : BCR 2009;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang LS, Akhmametyeva EM, Wu Y et al. Multiple transcription initiation sites, alternative splicing, and differential polyadenylation contribute to the complexity of human neurofibromatosis 2 transcripts. Genomics 2002;79:63–76. [DOI] [PubMed] [Google Scholar]
- 73.Cioffi JA, Yue WY, Mendolia-Loffredo S et al. MicroRNA-21 overexpression contributes to vestibular schwannoma cell proliferation and survival. Otol Neurotol 2010;31:1455–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petrilli A, Bott M, Fernández-Valle C. Inhibition of SIRT2 in merlin/NF2-mutant Schwann cells triggers necrosis. Oncotarget 2013;4:2354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torres-Martin M, Lassaletta L, de Campos JM et al. Global profiling in vestibular schwannomas shows critical deregulation of microRNAs and upregulation in those included in chromosomal region 14q32 . PLoS One 2013;8:e65868-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katz LM, Hielscher T, Liechty B et al. Loss of histone H3K27me3 identifies a subset of meningiomas with increased risk of recurrence. Acta neuropathologica 2018;135:955–63. [DOI] [PubMed] [Google Scholar]
- 77.San-Miguel T, Navarro L, Megias J et al. Epigenetic changes underlie the aggressiveness of histologically benign meningiomas that recur. Human pathology 2019;84:105–14. [DOI] [PubMed] [Google Scholar]
- 78.Sahm F, Schrimpf D, Stichel D et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. The Lancet. Oncology 2017;18:682–94. [DOI] [PubMed] [Google Scholar]
- 79.Evans DG, Huson SM, Legius E et al. Revision of Neurofibromatosis DIagnostic Criteria Joint Global Neurofibromatosis Meeting Paris, France 2018. [Google Scholar]
- 80.Evans DG, Huson SM, Legius E et al. Revision of Neurofibromatosis DIagnostic Criteria NF Conference San Francisco, California 2019. [Google Scholar]
- 81.Anand G, Vasallo G, Spanou M et al. Diagnosis of sporadic neurofibromatosis type 2 in the paediatric population. Archives of disease in childhood 2018;103:463–9. [DOI] [PubMed] [Google Scholar]
- 82.Havik AL, Bruland O, Myrseth E et al. Genetic landscape of sporadic vestibular schwannoma. Journal of neurosurgery 2018;128:911–22. [DOI] [PubMed] [Google Scholar]
- 83.Pasmant E, Louvrier C, Luscan A et al. Neurofibromatosis type 2 French cohort analysis using a comprehensive NF2 molecular diagnostic strategy. Neuro-Chirurgie 2018;64:335–41. [DOI] [PubMed] [Google Scholar]
- 84.Smith MJ, Bowers NL, Bulman M et al. Revisiting neurofibromatosis type 2 diagnostic criteria to exclude LZTR1-related schwannomatosis. Neurology 2017;88:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Agnihotri S, Jalali S, Wilson MR et al. The genomic landscape of schwannoma. Nature genetics 2016;48:1339–48. [DOI] [PubMed] [Google Scholar]
- 86.Louvrier C, Pasmant E, Briand-Suleau A et al. Targeted next-generation sequencing for differential diagnosis of neurofibromatosis type 2, schwannomatosis, and meningiomatosis. Neuro-oncology 2018;20:917–29. [DOI] [PMC free article] [PubMed] [Google Scholar]