Abstract
The transcription factor TCF4 was confirmed in several large genome-wide association studies as one of the most significant schizophrenia (SZ) susceptibility genes. Transgenic mice moderately overexpressing Tcf4 in forebrain (Tcf4tg) display deficits in fear memory and sensorimotor gating. As second hit, we exposed Tcf4tg animals to isolation rearing (IR), chronic social defeat (SD), enriched environment (EE), or handling control (HC) conditions and examined mice with heterozygous deletion of the exon 4 (Tcf4Ex4δ+/−) to unravel gene-dosage effects. We applied multivariate statistics for behavioral profiling and demonstrate that IR and SD cause strong cognitive deficits of Tcf4tg mice, whereas EE masked the genetic vulnerability. We observed enhanced long-term depression in Tcf4tg mice and enhanced long-term potentiation in Tcf4Ex4δ+/− mice indicating specific gene-dosage effects. Tcf4tg mice showed higher density of immature spines during development as assessed by STED nanoscopy and proteomic analyses of synaptosomes revealed concurrently increased levels of proteins involved in synaptic function and metabolic pathways. We conclude that environmental stress and Tcf4 misexpression precipitate cognitive deficits in 2-hit mouse models of relevance for schizophrenia.
Subject terms: Molecular neuroscience, Psychiatric disorders
Introduction
TCF4 encodes an ubiquitously expressed class I basic helix-loop-helix (bHLH) transcription factor which has been implicated in several neurodevelopmental disorders, mental retardation, intellectual disability, and schizophrenia (reviewed in ref. 1). The TCF4 gene has a complex structure harboring multiple 5′ initial exons that generate longer and shorter TCF4 protein variants2,3. TCF4 haploinsufficiency leads to Pitt–Hopkins syndrome (PTHS) characterized by mental and developmental retardation, episodic hyperventilation, and distinct facial features4–6. In most cases, PTHS causing chromosomal deletions or frame shift mutations map to the 3′ end of the gene and cause TCF4 protein variants that lack the bHLH domain and/or are incapable of DNA binding7. A mouse mutant haploinsufficient for Tcf4 3′ exons 16 and 17 (which encode the bHLH domain)8 has been considered a model for PTHS9. More 5′ located deletions in TCF4 have been identified in individuals with intellectual disability who lack other features of PTHS2,10.
Large genome-wide association studies (GWAS) consistently identified several non-coding single nucleotide polymorphisms (SNPs) in the 5′ located introns of the TCF4 gene contributing to an increased risk for SZ11–14 and more recently also to major depressive disorder (MDD)15,16. TCF4 transcript levels have been shown to be moderately increased in blood cells of SZ patients and in neurons derived from induced pluripotent stem cells (IPSCs)17,18 as well as in postmortem brain tissue of SZ patients19,20. Moreover, transgenic mice that slightly overexpress a long Tcf4 variant (corresponding to the TCF4B variant as described by Sepp et al.3 in the cortex and hippocampus (Tcf4tg mice) display deficits in sensory gating and impairments in fear-associated learning, attentional dysfunction, and delayed adaptation in a latent inhibition task21–23. TCF4 is considered a constitutive interaction partner of neuronal bHLH factors and displays pleiotrophic effects1 and has been shown to be regulated post-trancriptionally by neuronal activity24. Recent shRNA-mediated knock-down analyses have provided evidence that TCF4 modulates neuronal development and function by repressing neurexins and the ion channels SCN10A and KCNQ1, respectively25,26. In addition, high levels of overexpression of the long TCF4 variant TCF4B by in-utero electroporation has been shown to disturb cortical laminar development in an activity-dependent fashion27 and haploinsufficient mice display cortical malformations as well28. Thus, the view emerges that loss-of TCF4 is implicated in embryonic neurodevelopment as well as postnatal neuroplasticity. Studies in humans have shown that risk alleles associated with TCF4 may be implicated in cognitive performance and potentially also negative symptoms18,29,30 although sample sizes must be increased to allow definitive conclusions. According to the gene–environment interaction (GxE) model for psychiatric diseases31,32, it is likely that diverse environmental factors cooperate with TCF4 risk alleles and/or associated mechanisms. Thus far, only smoking has been identified as environmental risk factor modulating auditory sensory gating together with TCF4 risk alleles33. In patients and animal models, chronic social isolation and defeat are considered robust environmental risk factors, while intact social structures and support have been shown to ameliorate symptoms31,34–36. Moreover, an enriched environment positively influences rodent behavior and protects from psychopathologies36,37 whereas social isolation and social defeat (a model of chronic psychosocial stress) induce in mice a set of somatic, behavioral and molecular changes considered to be relevant endophenotypes of SZ35,38–40.
To unravel GxE interactions, we performed comprehensive behavioral phenotyping of Tcf4tg and wildtype mice subjected to environmental stress by post-weaning isolation rearing (IR) or social defeat (SD) in contrast to group housing in an enriched environment (EE) or handling control (HC). We also analyzed heterozygous Tcf4 loss-of-function mice (Tcf4Ex4δ+/−) in which heterozygous deletion of exon 4 reduces the expression of long Tcf4 transcript variants. These mice were subjected to IR to study gene-dosage effects. We complemented deep behavioral profiling by electrophysiological recordings of hippocampal plasticity, as well as structural and proteomic analyses characterizing gene-dosage dependent modulation of cognition and plasticity by TCF4.
Materials and methods
Behavioral profiling
The overall approach of the data calibration and dimensional reduction strategy and housing conditions were described previously35. Analyses were done in R software version 2.15.2 using the R-package nlme and R-functions gls and anova. Graphs were generated using R-package plotrix, exported as.eps files and edited in Adobe Illustrator CS5. Behavioral tests were in general following published procedures22,35. The investigator was blinded at the time of experimental procedures and the genotype decoding was performed at the end of the corresponding set of experiments. Further details and description of all mouse strains used are given in the Supplementary Information.
Electrophysiology
Microisland autaptic culture preparation, acute brain slice preparations, and electrophysiology were performed according to published procedures41. LTP and LTD measurements were performed with hippocampal slices obtained from 3 to 4 week old Tcf4tg and Tcf4Ex4δ+/− mice according to published procedures (see Supplementary Information for details).
STED nanoscopy and proteomics
The stimulated emission depletion (STED) nanoscopy experiment was performed essentially as described42. Synaptosomes were isolated according to refs. 43,44. All Proteome analysis of cytosolic fractions (S1) and synaptosomes (S4) and the western blots were performed according to established protocols42,45 (see Supplemental Information for details).
Results
Experimental design and Tcf4Ex4δ+/− mouse model
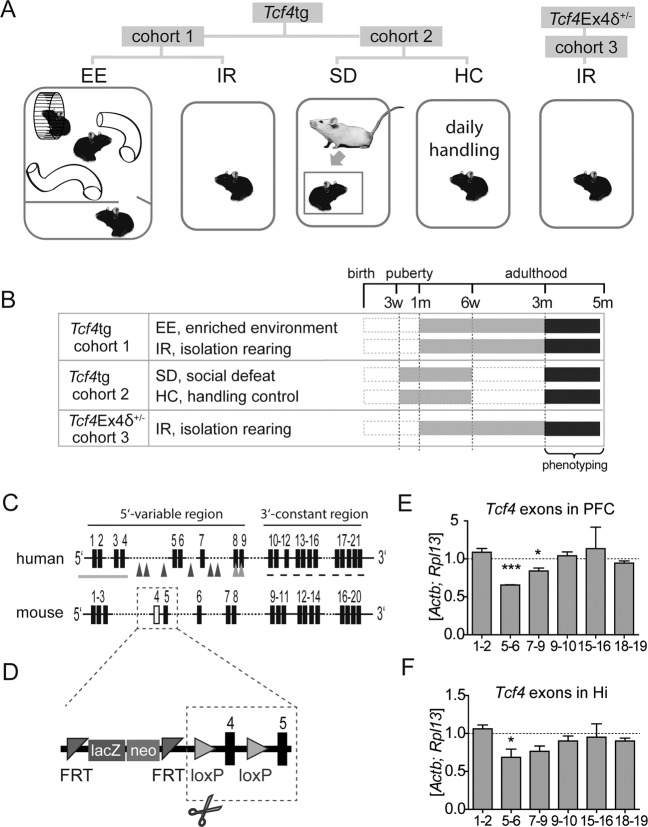
In this study, we combined analyses of TCF4 gain- and loss-of function mouse models: Tcf4tg mice, which mildly overexpress Tcf4 in postnatal forebrain; and Tcf4Ex4δ+/− mice, with heterozygous deletion of exon 4. To study the behavioral consequences of psychosocial stress in these mice, we analyzed three cohorts of wildtype (wt) and Tcf4 mouse models exposed to different environmental conditions starting from the peripubertal period (Fig. 1a–d). In cohort 1, wt and Tcf4tg mice were exposed to enriched environment (EE) and isolation rearing (IR); in cohort 2, wt and Tcf4tg mice were subjected to social defeat (SD) by a 3-weeks daily exposure to an aggressive resident mouse, and to daily handling as a control (HC). In cohort 3, wt and Tcf4Ex4δ+/ were subjected to IR. Subsequently, all animals were subjected to deep behavioral phenotyping in adulthood (Fig. 1b).
Fig. 1. Experimental design and Tcf4Ex4δ+/− mouse model.
a, b Housing conditions (a) and experimental timeline (b). Tcf4tg and wt mice were housed in enriched environment (EE) or isolation rearing (IR) from age 4 weeks onwards (cohort 1) or were kept individually and subjected to 3 weeks of social defeat (SD) or handling control conditions (HC) from age 3 weeks onwards (cohort 2). Tcf4Ex4δ+/− and wt animals (cohort 3) were housed in IR from age 4 weeks onwards. All mice were subjected to behavioral phenotyping from the age of 3 months. w, weeks; m, month(s). c Schematic representation of the human TCF4 and mouse Tcf4 gene. Black rectangles, exons; gray arrows, enrichment of SZ-associated risk single nucleotide polymorphisms (SNPs); gray line and gray arrow heads, 5′ region covering partial deletions associated with intellectual disability; dashed line, 3′ region with high occurrence of mutations causing mental retardation of the Pitt-Hopkins syndrome type; white rectangle, exon 4 excised in Tcf4Ex4δ+/− mice (see main text for citations). d The Tcf4Ex4 construct containing two marker genes (LacZ and neo) flanked by FRT sites and loxP sites flanking Tcf4 exon 4, excision of which leads to Tcf4Ex4δ+/− genotype. loxP, lox recognition sites, FRT, flippase recognition target. e, f Relative expression levels of indicated Tcf4 exons in prefrontal cortex (PFC; e) and hippocampus (Hi; f) of Tcf4Ex4δ+/− mice, normalized to wt levels (dashed line) and housekeeping genes Actb and Rpl13. Tcf4Ex4δ+/−, n = 5; wt, n = 6. Data represent mean ± SEM. ***, p < 0.001; *, p < 0.05.
By homology, exon 4 of the mouse Tcf4 gene corresponds to exon 5 in the human TCF4 gene which is located in the middle of the 5′ variable region (exons 1–9), which has been demonstrated to generate multiple mRNA isoforms by differential splicing and 5′ initial exons3 (Fig. 1c). The 3′ constant region encompasses exons 10–21 which are common to all known TCF4 splice variants3 (Fig. 1c). Deletions within the 5′ variable region have been shown to be associated with intellectual disability2,10, whereas most mutations associated with Pitt–Hopkins–Syndrome (PHS) are found in the 3′ constant region that encodes the bHLH dimerization and DNA binding domain46. Several SNPs thought to confer an increased risk of SZ are found in the 5′ variable region flanking human exon 5 and 6 (corresponding to murine exons 4 and 5)12 (Fig. 1c, d and Supplementary Fig. 1a). Therefore, we hypothesized that the Tcf4Ex4δ+/− mouse line might be of relevance in the context of cognitive disability and SZ. Molecular analysis showed that truncated exon 4 of Tcf4 was detectable only in Tcf4Ex4δ+/− and not in control mice (Supplementary Fig. 1b). To measure the mRNA level of different Tcf4 exons in Tcf4Ex4δ+/− mice, we performed qRT-PCR on transcripts from prefrontal cortex (PFC, Fig. 1e) and hippocampus (Hi, Fig. 1f). Expression levels of exons 1–2 in Tcf4Ex4δ+/− mice were comparable to wt in both brain regions whereas exons 5–6 containing transcripts, located directly downstream of the deleted exon 4, were clearly reduced in both PFC and Hi (p < 0.0001 and p = 0.0277, t-test, respectively). The expression levels of consecutive exons were higher with increasing distance from exon 4, i.e. exons 7–9 containing transcripts showed less pronounced decrease in FCx (p = 0.0174, t-test) and similar tendency in Hi (Fig. 1e, f). This shows that only the long Tcf4 transcript variants are reduced in Tcf4Ex4δ+/− mice. In wt mice, the exons most abundant in PFC are exons 7–9, and in hippocampus exons 5–6 (Supplementary Fig. 1c, d). Tcf4Ex4δ+/− hypomorphic mice are viable, breed well and show neither increased mortality nor major developmental impairments. We detect no abnormal facial or body features of Tcf4Ex4δ+/− mice that might reflect PHS-like phenotype (Supplementary Fig. 1e–i).
Tcf4 modulates cognition in a gene-dosage and environment-dependent manner
We profiled Tcf4 gain and loss-of-function mutants in the context of G×E interactions with a comprehensive test battery assessing various behaviors’ and condensed these to behavioral “domains” and “superdomains” adapted from selected research domain criteria (RDoc) for psychiatric disease models47. We focused on cognitive (spatial and flexibility learning, fear memory, working memory), affective (negative and positive valence) and activity-related domains according to a multivariate statistical strategy based on stepwise data calibration and reduction combined with multiple-testing corrections (see Table 1 and Supplementary Table 1 for details) as described previously35. The effects discussed below always refer to multiple-testing adjusted multivariate analyses unless stated otherwise.
Table 1.
Genetic main effects in Tcf4tg and Tcf4Ex4δ+/− mice in different environments.
| Superdomain | Domain | Measure | Pglobal | EE environment tg/wt | IR environment tg/wt | SD environment tg/wt | IR environment Tcf4δEx4/wt† | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect tg | Statistic | P | Effect tg | Statistic | P | Effect tg | Statistic | P | Effect Tcf4δEx4 | Statistic | P | ||||
| COGNITIVE DOMAIN | m | * | −0.127 | t80 = −0.438 | 0.663 | −1.811 | t86 = −2.890 | 0.005 | −1.997 | t82 = −4.372 | <0.001 | −1.632 | t86 = −3.723 | <0.001 | |
| Spatial Learning | m | * | 0.015 | t76 = 0.040 | 0.969 | −4.103 | t82 = −2.397 | 0.019 | −3.899 | t85 = −6.578 | <0.001 | −4.112 | t82 = −6.670 | <0.001 | |
| Initial learning | m | * | −0.093 | t50 = −0.128 | 0.899 | −2.598 | t54 = −1.469 | 0.148 | −3.035 | t56 = −3.378 | 0.001 | −4.834 | t54 = −4.815 | <0.001 | |
| Flexibility learning | m | * | −0.621 | t50 = −0.849 | 0.400 | −7.877 | t54 = −2.919 | 0.005 | −4.698 | t56 = −6.020 | <0.001 | −6.862 | t54 = −4.632 | <0.001 | |
| Recall | m | * | 0.645 | t50 = 1.210 | 0.232 | 0.002 | t53 = 0.004 | 0.997 | −4.321 | t56 = −6.698 | <0.001 | −1.456 | t26 = −1.682 | 0.105 | |
| Fear memory | m | * | −0.413 | t54 = −0.939 | 0.352 | −0.807 | t58 = −2.056 | 0.044 | 0.068 | t48 = 0.179 | 0.858 | 0.806 | t53 = 1.433 | 0.158 | |
| Context memory | m | * | −0.417 | t52 = −0.830 | 0.410 | −0.066 | t58 = −0.144 | 0.886 | −0.318 | t23 = −1.129 | 0.271 | 0.763 | t46 = 1.512 | 0.137 | |
| Cue memory | m | * | −0.467 | t52 = −0.729 | 0.469 | −1.548 | t58 = −3.437 | 0.001 | 0.453 | t23 = 0.801 | 0.431 | 0.935 | t54 = 1.585 | 0.119 | |
| Social fear memory | m | ns | – | – | – | – | – | – | 1.153 | t56 = 2.099 | 0.040 | – | – | – | |
| Working memory | Working memory | u | * | 0.104 | t26 = 0.175 | 0.863 | 0.439 | t28 = 0.581 | 0.566 | −0.218 | t28 = −0.300 | 0.767 | −2.032 | t28 = −2.666 | 0.013 |
| AFFECTIVE DOMAIN | m | * | −0.922 | t110 = −3.359 | 0.001 | −0.389 | t121 = −0.682 | 0.497 | −0.159 | t88 = −0.537 | 0.593 | −0.018 | t118 = −0.028 | 0.978 | |
| Pain sensitivity | Pain sensitivity | u | * | −1.032 | t26 = −0.935 | 0.358 | −0.773 | t28 = −0.348 | 0.730 | – | – | – | 0.120 | t28 = 0.098 | 0.923 |
| Anxiety | Anxiety | m | * | −0.492 | t54 = −1.251 | 0.216 | −0.115 | t58 = −0.490 | 0.626 | 0.371 | t87 = 0.641 | 0.523 | −0.829 | t83 = −1.271 | 0.207 |
| Curiosity | Curiosity | m | * | −1.994 | t54 = −2.742 | 0.008 | −1.560 | t58 = −1.238 | 0.221 | −0.486 | t57 = −0.828 | 0.411 | – | F1.0,27.2 = 0.251 | 0.621 |
| Motivation | Motivation | m | * | −0.246 | t54 = −0.403 | 0.689 | 0.115 | t59 = 0.086 | 0.932 | −0.439 | t58 = −2.045 | 0.045 | −0.142 | t53 = −0.287 | 0.775 |
| ACTIVITY DOMAIN | m | * | −0.418 | t54 = −0.824 | 0.413 | −0.224 | t60 = −0.266 | 0.791 | 0.163 | t58 = 0.560 | 0.577 | 0.490 | t58 = 1.070 | 0.289 | |
| Hyperactivity | m | * | −0.418 | t54 = −0.824 | 0.413 | −0.224 | t60 = −0.266 | 0.791 | 0.163 | t58 = 0.560 | 0.577 | 0.490 | t58 = 1.070 | 0.289 | |
| Ambulation | m | * | 0.409 | t110 = 0.890 | 0.376 | −0.768 | t119 = −0.919 | 0.360 | 0.776 | t117 = 1.205 | 0.231 | 0.447 | t113 = 0.832 | 0.407 | |
| Speed | m | * | −1.238 | t52 = −1.553 | 0.127 | −0.087 | t55 = −0.071 | 0.944 | −0.413 | t56 = −1.180 | 0.243 | 0.380 | t56 = 0.944 | 0.349 | |
Significant differences are marked in bold and with stars in the case of Pglobal. Numbers of degrees of freedom are marked for each statistic in subscript. Statistics depicted are from the multivariate analyses (see Supplementary Table 1 for all details).
A global test revealed highly significant effects of Tcf4 expression and environmental stress factors IR and SD, with p < 0.001 for all symptom classes (Supplementary Table 1). Most behavioral subdomains were significantly affected by environment, although marked differences were observed for cohort 1 (IR vs EE) and cohort 2 (HC vs SD) (see details under “Environmental Effects” column in Supplementary Table 1 and for selected tests Supplementary Fig. 2). We examined genetic main effects of Tcf4tg in EE, IR, SD, HC, and genetic main effects of Tcf4Ex4δ+/− in IR, in comparison to respective wt controls. To comprehensively analyze these effects towards clinical symptom groups, we collapsed all individual behavioral tests (Supplementary Table 1) into behavioral domains that are depicted in Table 1 (for brevity, genetic main effects in HC (see Supplementary Table 1) are not displayed in Table 1 as they did not reach significance at the symptom class level). In EE, we obtained a significant difference between Tcf4tg and wt mice in the negative symptom class (p = 0.001), which was exclusively driven by the curiosity tests (rearing in the open field and hole board test, indicating reduced curiosity of Tcf4tg mice) (Supplementary Table 1). Note, that the effects for the individual tests failed to pass the significance threshold after Bonferroni correction whereas the multivariate and sum score analyses reached significance (Table 1 and Supplementary Table 1).
In IR (Tcf4tg and Tcf4Ex4δ+/−) and SD (Tcf4tg), we did not obtain significant genetic main effects of Tcf4 alterations at the level of affective and activity domains (Table 1). Nonetheless, given the recent association of TCF4 with depression16, we further inspected the subdomain level and identified that Tcf4tg mice exhibited under HC and SD significantly reduced fighting times in the tail suspension test monitoring depressive-like behavior (p < 0.001, Supplementary Table 1, Supplementary Fig. 3). The effect size of these observations, however, was not sufficient to obtain a significant result in the subdomain “Motivation” (Table 1, Supplementary Table 1).
In contrast to the affective and activity domains, the genotype significantly influenced cognitive symptoms. Tcf4tg mice displayed more severe cognitive impairments under IR (Tcf4tg: p = 0.005; Tcf4Ex4δ+/−: p < 0.001) and SD (Tcf4tg: p < 0.001) than respective wt controls (Table 1, Supplementary Table 1). Housing mice in EE prevented cognitive deficits in Tcf4tg mice (p = 0.663) while daily handling (HC) partially ameliorated them (p = 0.068) (Table 1, Supplementary Table 1). In contrast to EE, HC was insufficient to prevent deficits in spatial learning (which became most pronounced under SD, Dataset Supplementary Fig. 1), possibly because of the missing social and sensory stimulation. We further inspected the genotype effects on the cognitive subdomains by visualizing the complete spectrum of cognition-related tests in radar charts (Supplementary Fig. 4).
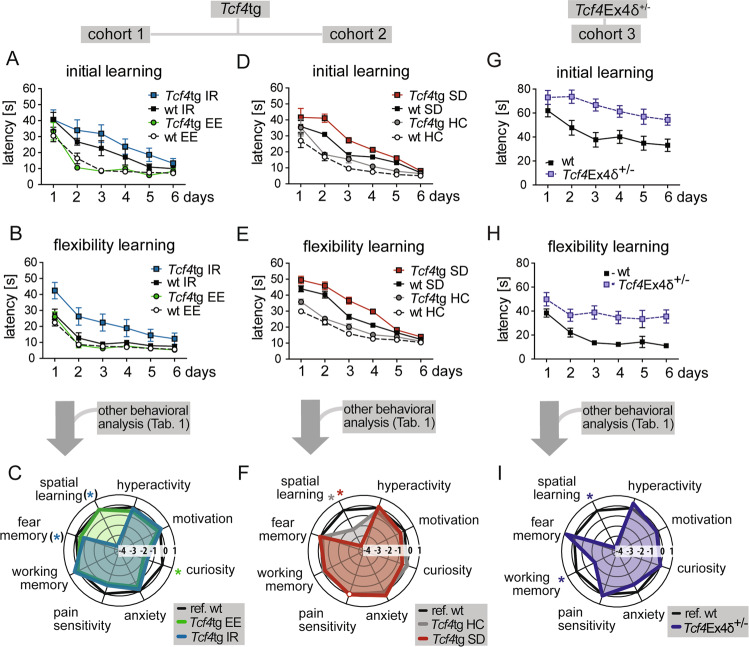
The most pronounced environment-dependent cognitive deficits of the Tcf4 mouse models manifested in spatial learning, as assessed in Morris Water Maze, in the initial learning (Tcf4tg in SD, p = 0.001, and Tcf4Ex4δ+/− in IR, p < 0.001) and flexibility learning (Tcf4tg in IR, p = 0.005; Tcf4tg in SD, p < 0.001; and Tcf4Ex4δ+/− in IR, p < 0.001)(Fig. 2, Supplementary Fig. 4, Table 1). The strong impact on these cognitive “superdomains” (as defined in Table 1) was visualized in radar charts for all conditions (Fig. 2c, f, i).
Fig. 2. Tcf4 gene-dosage modulates cognition in environment-dependent manner.
Tcf4tg mice (cohort 1, a, b; cohort 2, d, e) and Tcf4Ex4δ+/− mice (cohort 3, g, h) were submitted to spatial learning in the water maze task. Cumulated results from water maze and other behavioral profiling (Table 1) are depicted as radar charts for Tcf4tg (c, f) and Tcf4Ex4δ+/− (i) mice. a In the initial learning task in Morris Water Maze, Tcf4tg and wt mice in isolation rearing (IR) learned slower than mice in enriched environment (EE) (p < 0.001, environmental effect), as measured by the latency to reach a hidden platform. The effect was independent of genotype (p = 0.210, G×E interaction test). b During reversal learning, Tcf4tg mice subjected to IR needed more time to reach the platform than IR subjected wt littermates (p = 0.005), indicating an impairment of cognitive flexibility. c Behavioral profiles show impaired spatial learning upon IR (blue) and no cognitive deficits upon EE (green) in Tcf4tg mice compared to wt mice from the corresponding environment (black). Green and blue stars indicate significant differences (see Table 1 and Dataset Supplementary Fig. 1 for details) between Tcf4tg EE (green) vs. reference (wt EE, black) and between Tcf4tg IR (blue) and reference (wt IR, black), respectively. IR significantly impaired cognition in Tcf4tg mice (cognitive symptom class: p = 0.005, cognitive domain flexibility learning: p = 0.005, cognitive trait cue memory: p = 0.001; all passing multiple-tesing correction). However, at the superdomain level displayed here, only nominal significance was reached (spatial learning p = 0.019, fear memory p = 0.044, blue stars in brackets). d During initial learning, socially defeated (SD) Tcf4tg mice displayed longer platform latencies than mutants from the handling control (HC) group or wt animals (p ≤ 0.001). Tcf4tg HC mice showed slightly delayed platform latencies than wt HC animals (p = 0.007, not reaching significance after Bonferroni correction). e In reversal learning (i.e. flexibility learning), Tcf4tg mice needed significantly more time to reach the platform than wt animals in both SD (p < 0.001) and HC groups (p = 0.002). f Behavioral profiles of Tcf4tg mice from SD and HC. Spatial learning in Tcf4tg mice is significantly impaired upon SD (red) and mildly in HC (gray) compared to wt mice in the corresponding conditions (black) as indicated by stars of corresponding colors (multiple-testing adjusted significance, see Table 1 and Dataset Supplementary Fig. 1 for details). Pain sensitivity was not assessed in this cohort (as indicated by white circle). g, h Tcf4Ex4δ+/− mice housed in IR displayed higher platform latencies than wt controls in initial learning (p < 0.001)(g) and flexibility learning (p < 0.001)(h). i Behavioral profiles of Tcf4Ex4δ+/− mice (blue) show that spatial learning and working memory are impaired compared to wt mice (black), as indicated by blue stars (multiple-testing adjusted significance, see Table 1 and Supplementary Table 1 for details). a, b, d, e, g, h Data represent mean ± SEM. n = 12–16 mice per genotype and housing conditions. See Table 1 and Dataset Supplementary Fig. 1 for detailed statistics.
In summary, the behavioral profiling shows that (i) the cognitive performance is cooperatively compromised in gain and loss-of-function Tcf4 models subjected to chronic stress and that (ii) cognitive deficits of Tcf4tg mice upon SD stress are not observed upon group housing in an EE.
The impact of Tcf4 gene dosage on neuronal plasticity
Based on the observed cognitive deficits in Tcf4tg and Tcf4Ex4δ+/− mice, we hypothesized that basal synaptic transmission and/or neuronal plasticity may be compromised upon gain and loss of TCF4 function. To study glutamatergic neurotransmission at the single-cell level, we used the well-established autaptic culture paradigm of primary hippocampal neurons41 isolated from wt, Tcf4tg and Tcf4Ex4δ+/− mice. With this system, we studied several pre- and postsynaptic properties including short-term plasticity but none of these measurements revealed a genotype-dependent alteration (Supplementary Fig. 5).
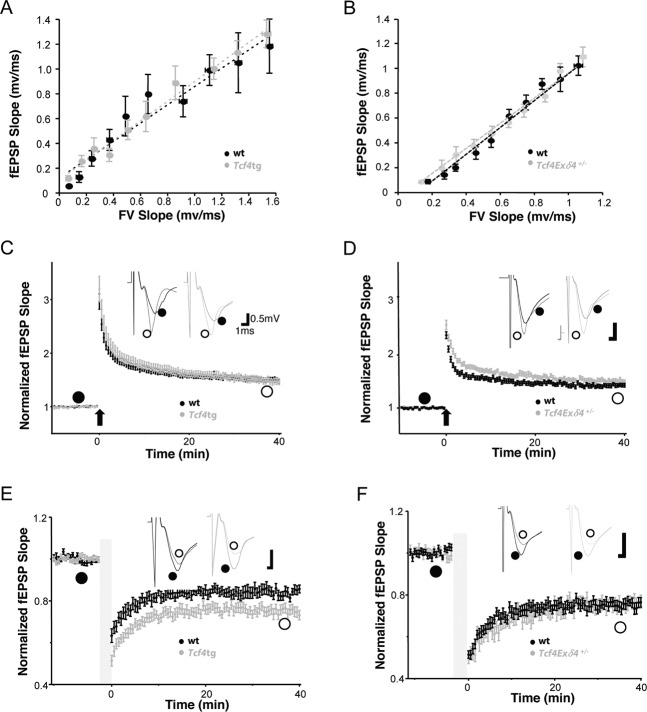
To address the functional consequences of altered Tcf4 expression levels for synaptic transmission at the network level, we next studied synaptic transmission and plasticity at Schaffer collateral-CA1 pyramidal (SC-CA1) synapses in acute hippocampal slices from 4–5-week-old Tcf4tg and Tcf4Ex4δ+/− mice and their wildtype littermates (Fig. 3). First, we measured input–output curves at the Schaffer collateral synapses by extracellular field recordings to reveal the impact of TCF4 on the basal synaptic transmission. There was no difference detected between slices obtained from Tcf4tg and Tcf4Ex4δ+/− mice and corresponding wt controls (Fig. 3a, b), showing that different expression levels of TCF4 have no effect on basal synaptic transmission. Next, to understand the function of TCF4 on long-term synaptic plasticity, we measured hippocampal long-term potentiation (LTP) induced by high-frequency stimulation (100 Hz, 1 s) and long-term depression (LTD) induced by low-frequency stimulation (1 Hz, 15 min) at SC-CA1 synapses (Fig. 3c–f). LTP was unaltered in slices from Tcf4tg mice (mean at 35–40 min in Tcf4tg 150 ± 5% and in wt 151 ± 3%; p = 0.186) (Fig. 3c) but was significantly elevated in slices obtained from Tcf4Ex4δ+/− mice (mean at 35–40 min in Tcf4Ex4δ+/− 151 ± 3% versus wt 141 ± 3%; p < 0.001)(Fig. 3d). Moreover, Tcf4tg slices displayed a profound increase in LTD level (mean at 35 to 40 min in Tcf4tg 73 ± 2% versus wt 83 ± 2%; p < 0.001)(Fig. 3e), whereas in the Tcf4Ex4δ+/− slices LTD was unchanged (mean at 35–40 min in Tcf4Ex4δ+/− 79 ± 1% versus wt 79 ± 2%; p = 0.143) (Fig. 3f).
Fig. 3. The expression level of Tcf4 regulates long-term synaptic plasticity without change in basal synaptic transmission.
Electrophysiological characterization of Tcf4tg (a, c, e) and Tcf4Ex4δ+/− mice (b,d,f). Values from Tcf4tg and Tcf4Ex4δ+/− are plotted in gray and those from littermate wt controls in black, respectively. a, b) Input/output relationship of excitatory synapses examined using extracellular field potential recording in the stratum radiatum layer CA1 regions of acute hippocampal slices from Tcf4tg and wt (a) and Tcf4Ex4δ+/− and wt mice (b). A summary plot of the slope of the field excitatory postsynaptic potential (fEPSP) in mV/ms versus the fiber volley (FV) indicates no differences between the genotypes. Tcf4tg n = 28, wt n = 32; (a) and Tcf4Ex4δ+/− n = 24, wt n = 15; (b). c Superimposed pooled LTP data showing the normalized changes in fEPSP slope. LTP induced by strong stimulation (100 Hz x 1) for 1 s in CA1. The fEPSP slope was measured and expressed as a mean percentage against time. The levels of LTP are unchanged in Tcf4tg mice (effect of genotype, p = 0.186). Summary graphs of LTP obtained by extracellular field recordings in CA1 stratum radiatum of acute hippocampal slices from Tcf4tg and wt mice (n = 28 and 32 slices, respectively). d Superimposed pooled LTP data showing the normalized changes in fEPSP slope. LTP induced by strong stimulation (100 Hz x 1) for 1 s in CA1. Summary graphs of LTP obtained by extracellular field recordings in CA1 stratum radiatum of acute hippocampal slices from Tcf4Ex4δ+/− mice and wt controls (n = 22 and 34 slices, respectively) showing significantly increased LTP in Tcf4Ex4δ+/− mice (effect of genotype, p < 0.001). e Superimposed pooled LTD data showing the normalized changes in fEPSP slope. LTD triggered by low stimulation (1 Hz) for 15 min. Summary graphs of LTD in the acute hippocampal slices from Tcf4tg and wt mice (n = 24 and 15 slices, respectively). LTD is significantly increased in Tcf4tg mice (effect of genotype, p < 0.001). f Superimposed pooled LTD data showing the normalized changes in fEPSP slope. LTD triggered by low stimulation (1 Hz) for 15 min. Summary graphs of LTD in the acute hippocampal slices from Tcf4Ex4δ+/− and wt mice (n = 29 and 20 slices, respectively). LTD is unchanged in Tcf4Ex4δ+/− mice (effect of genotype, p = 0.143). Data represent mean ± SEM. LTP, long-term potentiation; LTD, long-term depression; fEPSP, field excitatory postsynaptic potentials. wt, filled black circles; open gray circles represent Tcf4tg, (c, e) or Tcf4Ex4δ+/− (d, f) vs littermate wt animals 4–5 weeks of age, respectively.
Thus, slight dysregulation of Tcf4 expression levels does not alter basic neurotransmission but shapes long-term plasticity in hippocampal neuronal networks. Moreover, changes in Tcf4 gene-dosage can mediate a differential impact on LTD and LTP.
Synapse-related alterations in Tcf4tg mice
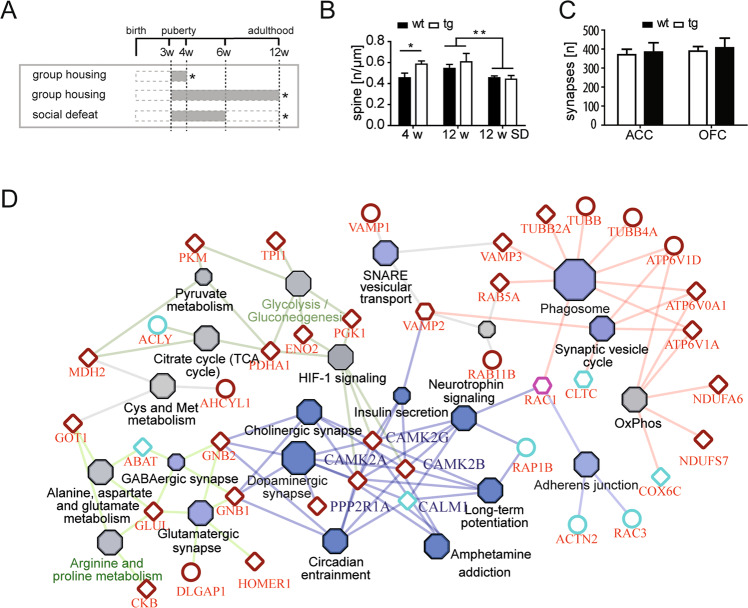
Based on the altered synaptic plasticity and the evident cognitive deficits in adult Tcf4tg mice which were particularly prominent in the reversal “flexibility” learning paradigm (Fig. 2b, c), we aimed at identifying cellular and molecular correlates in synaptic structures. For these analyses, we focused on frontal cortical structures (anterior cingulate cortex, ACC; and medial-prefrontral/orbitofrontal cortex, OFC) implicated in associative and flexibility learning48 and known for its importance in schizophrenia49. First, we applied super-resolution STED microscopy to quantitatively assess spine morphologies and densities on dendrites from Tcf4tg and wt control mice where sparse numbers of neurons were genetically labeled with EYFP42,50. Spine morphology was analyzed in 4- and 12-week-old Tcf4tg and wt control mice in EE and 12-week-old Tcf4tg and wt mice subjected to SD (Fig. 4a). Tcf4tg mice showed increased total spine density at 4 weeks of age (p = 0.031), but no such difference was observed in 12-week-old animals (Fig. 4b). SD stress during puberty caused a significant decrease in spine density in 12-week-old mice (p = 0.003) independent of the genotype (Fig. 4b). We further analyzed the relative distributions and densities of five subtypes of spines with increasing morphological complexity: filopodium/stripe, stubby/stump, mushroom/racket, cup/sickle, and branched spines, as previously reported42 (Supplementary Fig. 6a). The relative distribution of different spine types was similar across the genotypes and experimental groups (Supplementary Fig. 6b, d, f). In accordance with the total spine analysis (Fig. 4b), spine densities of all subtypes showed a subtle yet overall significant increase in 4 weeks old Tcf4tg mice (p = 0.0055, two-way ANOVA) with the most prominent difference detected at immature “stubby/stump” like spines (p < 0.01, post hoc)(Supplementary Fig. 6c). Again, no significant differences between the genotypes were observed at 12-week-old animals independent of stress conditions (Supplementary Fig. 6e, g).
Fig. 4. Tcf4 overexpression leads to an increase of premature spine numbers and proteome alterations in peripubertal mice.
a Experimental design. Group-housed Tcf4tg and wt mice were analyzed (*) at the age of 4 weeks (upper bar) (n = 4, each). Independent cohorts of Tcf4tg and wt mice were group-housed (middle bar) or exposed to chronic social defeat (SD, bottom bar) and analyzed at the age of 12 weeks (*). b Total spine density in the frontal cortex of Tcf4tg and wt mice was analyzed with STED nanoscopy. Spine density was significantly increased in 4-week-old Tcf4tg mice (p = 0.031). There was no difference in total spine density between the genotypes upon group housing (p = 0.686) or SD (p = 0.786) in 12-week-old mice. Subjecting mice to SD during puberty reduced total spine densities significantly (p = 0.003) without genotype differences. Data represent mean ± SEM. See Supplementary Fig. 6 for detailed analysis of spine types. c The analysis of electron-microscopic pictures for asymmetric/glutamatergic synapses reveals no differences between Tcf4tg and wt mice (n = 5, each). in the anterior cingulate cortex (ACC) nor the orbitofrontal cortex (OFC) at 4 weeks of age (p > 0.400). d Visualization of proteomic data from cytoplasmic and synaptosomal fractions isolated from frontal cortex of 4 weeks old Tcf4tg mice and wt controls (n = 4, each). Network graph of differentially regulated proteins in cytoplasmic (circular nodes), synaptosomal (diamond-shape nodes) and in both (hexagonal nodes) fractions. KEGG networks and overrepresented pathways visualized with ClueGo and CluePedia in Cytoscape are depicted as filled hexagons (blue = synapse associated pathways; grey = metabolic pathways). Primary data are from Supplementary Table 2.
We performed electron microscopy on tissue sections from 4-week-old Tcf4tg and wt controls to assess abundance and morphologies of synapses in the ACC and OFC at the ultrastructural level (Supplementary Fig. 7). The total number of asymmetric (i.e. mature glutamatergic) synapses was not altered between the genotypes and regions (Fig. 4c). Moreover, a close inspection of several ultrastructural characteristics of synapses (shape of the postsynaptic density and active zone, number and distribution of synaptic vesicles) did not reveal any quantitative differences (Supplementary Fig. 7d–l), as was the number of mitochondria and myelinated axons unchanged (Supplementary Fig. 7m–q).
Finally, we applied a quantitative isotope-coded mass spectrometric proteomic analysis of cytoplasmic fractions and synaptosomes (enriched for pre- and postsynaptic components associated with the postsynaptic density) isolated from frontal cortex tissue of 4 weeks old Tcf4tg and wt littermates. After stringent filtering (fold-change >1.5/<-1.5, corr. p-val<0.05, ≥2 peptides), we identified 38 up- and 10 down-regulated proteins in the synaptosomal fraction of Tcf4tg mice (Supplementary Table 2). In the cytoplasmic fraction, we detected 13 up- and 24 down-regulated proteins in Tcf4tg (Supplementary Table 2). As expected, the overlap was low with only 4 proteins detected in both subcellular fractions (Supplementary Table 2). We performed a gene ontology pathway enrichment analysis and visualized the results as a highly connected network of pathway-node associations (Fig. 4d). This analysis identified synapse-function and interconnected metabolism associated pathways as the two major subclusters (Fig. 4d).
Taken together, our results imply that moderately dysregulated levels of TCF4 expression—50% increase in Tcf4tg mice22 and 50% decrease of the “long isoforms” in Tcf4Ex4δ+/− mice—have a profound impact on higher order cognition likely caused by disturbed synaptic plasticity rather than severe structural alterations in the corresponding neuronal networks.
Discussion
In this study, we investigated gene–environment interactions in two mouse models with slightly altered Tcf4 transcript levels. This may mimic the changes in human TCF4 expression levels linked to risk variants in intronic TCF4 regions associated with SZ, MDD, and intellectual disability. By applying a “domain-oriented” deep behavioral profiling, we detected behavioral deficits in these mice, which were induced by the combination of the genetic and psychosocial risk factors. We saw that isolation rearing (IR) was more detrimental for Tcf4-overexpressing (Tcf4tg) than for wt mice, while enriched environment (EE) masked this genetic vulnerability. Similar effects were obtained when we compared Tcf4tg and wt mice subjected to social defeat (SD), a chronic psychosocial stress paradigm, with mice handled once daily but otherwise kept in isolation (HC). In both comparisons, most behavioral domains were affected except activity, which was assessed by “ambulation” (i.e. speed and distance running) in several behavioral tests. These were used as the only surrogate parameter of a hyperactive state often observed during a psychosis-associated phase in patients, possibly reflecting an aspect of positive symptoms51. This clearly indicates a limitation of rodent models because prototypic positive symptoms cannot be investigated. Nonetheless, we believe that our findings of strong GxE interaction provide evidence that the “2-hit” Tcf4/SD and Tcf4/IR approaches represent valid mouse models for the cognitive subdomain of relevance for SZ and possibly also MDD. Tcf4tg mice used for behavior were on F1 hybrid background while Tcf4Ex4δ+/− were kept on a pure C57bl6 background. Thus, we cannot formally rule that this may have an impact on the direct comparison of the different lines, we consider this as rather unlikely since we have previously shown that Tcf4tg mice backcrossed for several generations on the C57bl6 background performed highly similar in behavioral testing compared to F1 hybrids22. The “pairing” of risk gene alterations with environmental stress conditions has been suggested as promising strategy to improve validity of psychiatric mouse models52. Given the strong associations of TCF4 and psychosocial stress with schizophrenia and MDD, it seems likely that relevant mechanisms are affected in our 2-hit Tcf4 gain- and loss-of-function mouse models.
TCF4 has recently been identified by the PsychEncode consortium as a key “hub” gene in human brain development28, and was found to be expressed at increased levels in the Hi and PFC of SZ patients from independent postmortem sample collections53. On the other hand, both increased and reduced TCF4 expression in blood has been detected in independent studies and subject groups suffering from psychosis18,54, and reduced levels were found in recurrent MDD55. However, this variability of disease-associated changes of TCF4 expression in postmortem brain and blood samples does not allow conclusions on causal mechanisms and may be secondary or independent of the proposed neurodevelopmental alterations caused by TCF4 misexpression. Nonetheless, it may be possible that dichotomous patterns of deregulated expression of risk genes in SZ and MDD, including TCF4, may occur during later neurodevelopmental stages, converging at the level of synaptic dysfunctions, which are major molecular pathways detected in the recent comprehensive GWAS analyses of SZ and MDD11,15. Our analysis supports this assumption, because it demonstrates altered structural and functional synaptic plasticity potentially at the level of developmental spine dynamics, enhanced hippocampal LTD (Tcf4tg) and LTP (Tcf4Ex4δ+/−) and changes in the molecular composition of synaptosomes in Tcf4 gain-of-function (Tcf4tg) mouse model. The enhanced LTP in Tcf4Ex4δ+/− mice is in agreement with the same phenotype observed in other Tcf4 loss-of-function mutants9,56. Moreover, reduction of spine densities has recently been described in the cortex and Hi in loss-of-function mice57. In these animals, more immature synapses of the stubby subtype were affected in the cortex, which may indicate an inverse gene-dosage dependent phenotype when compared to the increased number of stubby-like synapses that we observed in 4 weeks old Tcf4tg mice (Supplementary Fig. 6c). However, we detected no changes at the ultrastructural level of pre- and post-synapses and no alterations of electrophysiological features in individual neurons when cultured in isolation. This indicates that a slight deregulation of Tcf4 expression causes rather subtle changes in neuronal plasticity and during subsequent network refinement upon interaction with psychosocial stressors. Since heterozygous null mutant mice also display structural as well as neuroanatomical aberrations likely changing micro- and macroconnectivity, it may as well be possible that structural changes also contribute to the cognitive alterations observed in our study28,58. Our study thus provides the basis to further dissect the molecular and cellular mechanisms misrouted upon deregulated Tcf4 expression, e.g. by identifying gene regulatory networks with transcriptome profiling. The switch from enhanced LTP to increased LTD in Tcf4 gain- versus loss-of-function mouse models is to our knowledge the first example where such a gene-dosage dependent relationship has been described.
Calcium signals from ligand and voltage-gated channels play a central role in coupling synaptic activity to the downstream signaling cascades, and several members of both groups (NMDA- and AMPA-receptors and L-type Ca-channels, in particular) have been strongly associated with psychotic disorders11,15. The relationship between the strength of synaptic activity and the equivalent calcium signal on one hand and the sign (potentiation versus depression) and degree on the other is described by the Bienenstock-Cooper-Munro (BCM) model59,60. This model is in agreement with the findings that sliding calcium levels activate different calcium/calmodulin-dependent protein kinases and/or other calcium-modulated synaptic proteins, which partially determine the differential induction of LTP or LTD61. Several such candidates have been identified in our proteome analysis. Based on our findings, follow up experiments are needed to substantiate the hypothesis that Tcf4 gene dosage determines postsynaptic Ca2+ levels and to identify the responsible target genes of TCF4. We conclude that the presented 2-hit mouse models represent valid tools for pre-clinical treatment trials, e.g. with cognitive enhancers that target cognitive processing units of relevance for affective and non-affective psychoses.
Supplementary information
Acknowledgements
The authors would like to acknowledge Harry Scherer for expert technical assistance with behavioral setups and Torben Ruhwedel for the preparation of the tissue samples for electron microscopy. This work was supported by the Max-Planck-Society, and the German Research Foundation (Deutsche Forschungsgesellschaft) (PsyCourse, RO 576/5-1 to M.J.R., and Klinische Forschergruppe (KFO) 241, BI 576/5-1). D.B. has been a doctoral student of the Ph.D. program “Neurosciences”—International Max Planck Research School and the Göttingen Graduate School for Neurosciences and Molecular Biosciences (GGNB) (DFG grant GSC 226/1) at the Georg August University Göttingen. Open access funding provided by Projekt DEAL.
Author contributions
D.M.B., M.M.B., and A.C. performed behavioral analyses. D.M.B. performed electron-microscopic analyses. N.K. performed bioinformatic analyses of omics data. P.D., H.S., and K.W. performed S.T.E.D. imaging and analysis. M.M.B., D.M., and C.W.T. performed synaptosome isolations and proteomic profiling. C.T. and J.S.R. performed electrophysiological measurements. D.M. performed multiparametric statistical analysis. P.F., A.S., and S.P. contributed conceptually. D.M.B., V.S., D.M., and M.J.R. wrote the paper. M.J.R. conceived the study.
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: D. M. Badowska, M. M. Brzózka
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-020-01026-7).
References
- 1.Quednow BB, Brzózka MM, Rossner MJ. Transcription factor 4 (TCF4) and schizophrenia: integrating the animal and the human perspective. Cell Mol. Life Sci. CMLS. 2014;71:2815–2835. doi: 10.1007/s00018-013-1553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kharbanda M, et al. Partial deletion of TCF4 in three generation family with non-syndromic intellectual disability, without features of Pitt-Hopkins syndrome. Eur. J. Med. Genet. 2016;59:310–314. doi: 10.1016/j.ejmg.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Sepp M, Kannike K, Eesmaa A, Urb M, Timmusk T. functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5’ exon usage and splicing. PloS ONE. 2011;6:e22138. doi: 10.1371/journal.pone.0022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amiel J, et al. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am. J. Hum. Genet. 2007;80:988–993. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweatt JD. Pitt-Hopkins Syndrome: intellectual disability due to loss of TCF4-regulated gene transcription. Exp. Mol. Med. 2013;45:e21. doi: 10.1038/emm.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zweier C, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am. J. Hum. Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whalen S, et al. Novel comprehensive diagnostic strategy in Pitt-Hopkins syndrome: clinical score and further delineation of the TCF4 mutational spectrum. Hum. Mutat. 2012;33:64–72. doi: 10.1002/humu.21639. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol. Cell Biol. 1996;16:2898–2905. doi: 10.1128/MCB.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy AJ, et al. Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Rep. 2016;16:2666–2685. doi: 10.1016/j.celrep.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalscheuer VM, et al. Disruption of the TCF4 gene in a girl with mental retardation but without the classical Pitt-Hopkins syndrome. Am. J. Med. Genet. A. 2008;146A:2053–2059. doi: 10.1002/ajmg.a.32419. [DOI] [PubMed] [Google Scholar]
- 11.Pardiñas AF, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefansson H, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci.10.1038/s41593-018-0326-7 (2019). [DOI] [PMC free article] [PubMed]
- 16.Wray NR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol. Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirgenes KV, et al. TCF4 sequence variants and mRNA levels are associated with neurodevelopmental characteristics in psychotic disorders. Transl. Psychiatry. 2012;2:e112. doi: 10.1038/tp.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guella I, et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J. Psychiatr. Res. 2013;47:1215–1221. doi: 10.1016/j.jpsychires.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudge J, et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PloS ONE. 2008;3:e3625. doi: 10.1371/journal.pone.0003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brzózka MM, Rossner MJ. Deficits in trace fear memory in a mouse model of the schizophrenia risk gene TCF4. Behav. Brain Res. 2013;237:348–356. doi: 10.1016/j.bbr.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Brzózka MM, Radyushkin K, Wichert SP, Ehrenreich H, Rossner MJ. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol. Psychiatry. 2010;68:33–40. doi: 10.1016/j.biopsych.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Brzózka, M. M., Rossner, M. J., de Hoz, L. Tcf4 transgenic female mice display delayed adaptation in an auditory latent inhibition paradigm. Eur. Arch. Psychiatry Clin. Neurosci.10.1007/s00406-015-0643-8 (2015). [DOI] [PubMed]
- 24.Sepp M, et al. The intellectual disability and schizophrenia associated transcription factor TCF4 Is regulated by neuronal activity and protein kinase A. J. Neurosci. J. Soc. Neurosci. 2017;37:10516–10527. doi: 10.1523/JNEUROSCI.1151-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Rozario M, et al. Type I bHLH proteins daughterless and Tcf4 restrict neurite branching and synapse formation by repressing neurexin in postmitotic neurons. Cell Rep. 2016;15:386–397. doi: 10.1016/j.celrep.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rannals MD, et al. Psychiatric risk gene transcription factor 4 regulates intrinsic excitability of prefrontal neurons via repression of SCN10a KCNQ1. Neuron. 2016;90:43–55. doi: 10.1016/j.neuron.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page, S. C. et al. The schizophrenia- and autism-associated gene, transcription factor 4 regulates the columnar distribution of layer 2/3 prefrontal pyramidal neurons in an activity-dependent manner. Mol. Psychiatry,10.1038/mp.2017.37 (2017). [DOI] [PMC free article] [PubMed]
- 28.Li H, et al. Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol. Psychiatry. 2019;24:1235–1246. doi: 10.1038/s41380-019-0353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albanna A, et al. TCF4 gene polymorphism and cognitive performance in patients with first episode psychosis. Schizophr. Res. 2014;152:124–129. doi: 10.1016/j.schres.2013.10.038. [DOI] [PubMed] [Google Scholar]
- 30.Lennertz L, et al. Novel schizophrenia risk gene TCF4 influences verbal learning and memory functioning in schizophrenia patients. Neuropsychobiology. 2011;63:131–136. doi: 10.1159/000317844. [DOI] [PubMed] [Google Scholar]
- 31.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 32.Tost H, Meyer-Lindenberg A. Puzzling over schizophrenia: schizophrenia, social environment and the brain. Nat. Med. 2012;18:211–213. doi: 10.1038/nm.2671. [DOI] [PubMed] [Google Scholar]
- 33.Quednow BB, et al. Schizophrenia risk polymorphisms in the TCF4 gene interact with smoking in the modulation of auditory sensory gating. Proc. Natl Acad. Sci. USA. 2012;109:6271–6276. doi: 10.1073/pnas.1118051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adamcio B, Havemann-Reinecke U, Ehrenreich H. Chronic psychosocial stress in the absence of social support induces pathological pre-pulse inhibition in mice. Behav. Brain Res. 2009;204:246–249. doi: 10.1016/j.bbr.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 35.Badowska, D. M., Brzózka, M. M., Chowdhury, A., Malzahn, D. & Rossner M. J. Data calibration and reduction allows to visualize behavioural profiles of psychosocial influences in mice towards clinical domains. Eur. Arch. Psychiatry Clin. Neurosci. 1–14 (2014). [DOI] [PubMed]
- 36.Fox C, Merali Z, Harrison C. Therapeutic and protective effect of environmental enrichment against psychogenic and neurogenic stress. Behav. Brain Res. 2006;175:1–8. doi: 10.1016/j.bbr.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Nithianantharajah J, Hannan AJ. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat. Rev. Neurosci. 2006;7:697–709. doi: 10.1038/nrn1970. [DOI] [PubMed] [Google Scholar]
- 38.Valzelli L. The ‘isolation syndrome’ in mice. Psychopharmacologia. 1973;31:305–320. doi: 10.1007/BF00421275. [DOI] [PubMed] [Google Scholar]
- 39.Selten J-P, Cantor-Graae E. Social defeat: risk factor for schizophrenia? Br. J. Psychiatry J. Ment. Sci. 2005;187:101–102. doi: 10.1192/bjp.187.2.101. [DOI] [PubMed] [Google Scholar]
- 40.Holtzman CW, et al. Stress and neurodevelopmental processes in the emergence of psychosis. Neuroscience. 2013;249:172–191. doi: 10.1016/j.neuroscience.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgalossi A, et al. Analysis of neurotransmitter release mechanisms by photolysis of caged Ca2+ in an autaptic neuron culture system. Nat. Protoc. 2012;7:1351–1365. doi: 10.1038/nprot.2012.074. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal, A. et al. Dysregulated expression of neuregulin-1 by cortical pyramidal neurons disrupts synaptic plasticity. Cell Rep.10.1016/j.celrep.2014.07.026 (2014). [DOI] [PubMed]
- 43.Gray EG, Whittaker VP. The isolation of nerve endings from brain: an electron-microscopic study of cell fragments derived by homogenization and centrifugation. J. Anat. 1962;96:79–88. [PMC free article] [PubMed] [Google Scholar]
- 44.Biesemann C, et al. Proteomic screening of glutamatergic mouse brain synaptosomes isolated by fluorescence activated sorting. EMBO J. 2014;33:157–170. doi: 10.1002/embj.201386120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maccarrone G, Turck CW, Martins-de-Souza D. Shotgun mass spectrometry workflow combining IEF and LC-MALDI-TOF/TOF. Protein J. 2010;29:99–102. doi: 10.1007/s10930-009-9227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sepp, M., Pruunsild, P. & Timmusk, T. Pitt–Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum. Mol. Genet. 10.1093/hmg/dds112 (2012). [DOI] [PubMed]
- 47.Anderzhanova E, Kirmeier T, Wotjak CT. Animal models in psychiatric research: The RDoC system as a new framework for endophenotype-oriented translational neuroscience. Neurobiol. Stress. 2017;7:47–56. doi: 10.1016/j.ynstr.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAlonan K, Brown VJ. Orbital prefrontal cortex mediates reversal learning and not attentional set shifting in the rat. Behav. Brain Res. 2003;146:97–103. doi: 10.1016/j.bbr.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 49.Murray GK, et al. Reinforcement and reversal learning in first-episode psychosis. Schizophr. Bull. 2008;34:848–855. doi: 10.1093/schbul/sbn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willig KI, et al. Nanoscopy of filamentous actin in cortical dendrites of a living mouse. Biophys. J. 2014;106:L01–L03. doi: 10.1016/j.bpj.2013.11.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Tuathaigh CMP, Moran PM, Waddington JL. Genetic models of schizophrenia and related psychotic disorders: progress and pitfalls across the methodological ‘minefield’. Cell Tissue Res. 2013;354:247–257. doi: 10.1007/s00441-013-1652-4. [DOI] [PubMed] [Google Scholar]
- 52.Burrows EL, Hannan AJ. Cognitive endophenotypes, gene-environment interactions and experience-dependent plasticity in animal models of schizophrenia. Biol. Psychol. 2016;116:82–89. doi: 10.1016/j.biopsycho.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Ma C, Gu C, Huo Y, Li X, Luo X-J. The integrated landscape of causal genes and pathways in schizophrenia. Transl. Psychiatry. 2018;8:67. doi: 10.1038/s41398-018-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurian SM, et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol. Psychiatry. 2011;16:37–58. doi: 10.1038/mp.2009.117. [DOI] [PubMed] [Google Scholar]
- 55.Mossakowska-Wójcik J, Orzechowska A, Talarowska M, Szemraj J, Gałecki P. The importance of TCF4 gene in the etiology of recurrent depressive disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;80:304–308. doi: 10.1016/j.pnpbp.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 56.Thaxton C, et al. Common pathophysiology in multiple mouse models of Pitt-Hopkins syndrome. J. Neurosci. J. Soc. Neurosci. 2018;38:918–936. doi: 10.1523/JNEUROSCI.1305-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crux S, Herms J, Dorostkar MM. Tcf4 regulates dendritic spine density and morphology in the adult brain. PloS ONE. 2018;13:e0199359. doi: 10.1371/journal.pone.0199359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung M, et al. Analysis of the expression pattern of the schizophrenia-risk and intellectual disability gene TCF4 in the developing and adult brain suggests a role in development and plasticity of cortical and hippocampal neurons. Mol. Autism. 2018;9:20. doi: 10.1186/s13229-018-0200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooper LN, Bear MF. The BCM theory of synapse modification at 30: interaction of theory with experiment. Nat. Rev. Neurosci. 2012;13:798–810. doi: 10.1038/nrn3353. [DOI] [PubMed] [Google Scholar]
- 60.Shouval HZ, Bear MF, Cooper LN. A unified model of NMDA receptor-dependent bidirectional synaptic plasticity. Proc. Natl Acad. Sci. USA. 2002;99:10831–10836. doi: 10.1073/pnas.152343099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hulme SR, Jones OD, Abraham WC. Emerging roles of metaplasticity in behaviour and disease. Trends Neurosci. 2013;36:353–362. doi: 10.1016/j.tins.2013.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.