Abstract
The use of imaging data has been reported to be useful for rapid diagnosis of COVID-19. Although computed tomography (CT) scans show a variety of signs caused by the viral infection, given a large amount of images, these visual features are difficult and can take a long time to be recognized by radiologists. Artificial intelligence methods for automated classification of COVID-19 on CT scans have been found to be very promising. However, current investigation of pretrained convolutional neural networks (CNNs) for COVID-19 diagnosis using CT data is limited. This study presents an investigation on 16 pretrained CNNs for classification of COVID-19 using a large public database of CT scans collected from COVID-19 patients and non-COVID-19 subjects. The results show that, using only 6 epochs for training, the CNNs achieved very high performance on the classification task. Among the 16 CNNs, DenseNet-201, which is the deepest net, is the best in terms of accuracy, balance between sensitivity and specificity, score, and area under curve. Furthermore, the implementation of transfer learning with the direct input of whole image slices and without the use of data augmentation provided better classification rates than the use of data augmentation. Such a finding alleviates the task of data augmentation and manual extraction of regions of interest on CT images, which are adopted by current implementation of deep-learning models for COVID-19 classification.
Subject terms: Viral infection, Computer science
Introduction
Image findings have been increasingly recognized as a useful tool for rapid diagnosis of COVID-191. The use of chest computed tomography (CT) to detect COVID-19 symptoms has been reported to have high sensitivity and can predate positive tests carried out in a laboratory2–7. Latest articles on image analysis of COVID-19 can be further found at the European Radiology webaite8. Because of the potential utilization of CT data, hospitals overloaded with admissions of patients are using CT imaging to diagnose COVID-19 infection and to decide the order of treatment of infected patients. In fact, chest CT has an important role in urgent clinical assessment and decision making for treatment of COVID-19 patients who suffer from severe and worsening respiratory symptoms. In other words, CT scans can be used to assess the severity of the infected lungs as well as progress of the disease, which tremendously help medical doctors in curbing the virus. It has been suggest that examinations and reports of CT findings can be used as a basis for improving the quality of care for COVID-19 patents9.
While CT imaging is useful for the diagnosis of COVID-19, manual reading of the scans is time-consuming and subject to human error. Therefore, the need for advanced artificial intelligence (AI)-based automated image analysis has the potential to analyze CT scans in the assessment of COVID-19. AI-based image analysis methods can provide accurate and rapid diagnosis of the disease to cope with the demand for a large number of patients10. For example, a manual assessment of a CT scan can take up to 15 minutes, while AI-based image analysis requires only a few seconds. Furthermore, AI can be useful in improving clinical workflow efficiency11.
A recent baseline study on AI for automated classification of COVID-19 using the largest publicly available CT dataset was reported in12, which will be described subsequently as the database used in this study. This work used the pretrained DenseNet for the classification task. These authors adopted transfer learning and data augmentation for training the pretrained deep-learning model with new image data. The rationale is that transfer learning can relieve the need for acquiring a large amount of training data by reusing a developed model as the starting point for training a new model with a different task. The data augmentation was performed by using a large dataset of chest X-ray images. The rationale for image data augmentation is to increase the size of the training dataset with plausible examples in order to improve the performance and ability of the deep-learning model to generalize the power of classification by getting familiar with samples of high variance.
Another recent work reported on the use of ten pretrained CNNs for classifying CT scans of COVID-19 and non-COVID-19 subjects13. These authors reported that ResNet-101 and Xception provided the best classification results on training and testing a CT dataset consisting 106 COVID-19 patients and 86 non-COVID-19 subjects. The CNNs were trained and tested with regions of interest extracted from the CT scans that were defined by a radiologist.
Other previous works on the classification of COVID-19 on CT scans were reported in14–16. A 3D deep-learning network was developed for the detection of COVID-19 from 4356 3D chest CT scans obtained from 3322 patients14. The network extracted both 2D local and 3D global features from the CT scans. This network, called COVNet, was built on the pretrained RestNet50. In15, the pretrained Inception was modified to detect COVID-19 using extracted regions of interest on CT scans obtained from 180 cases of COVID-19 and 79 cases of SARs-COV-2. In16, a total of 618 CT scans were used, consisting of 219 CT scans from 110 COVID-19 patients, 224 CT scans from 224 patients with Influenza-A viral pneumonia, and 175 CT scans from healthy people. Pulmonary regions of interest were extracted from the CT scans, and pretrained ResNet-18 was used for image feature extraction. Finally, the Noisy-or Bayesian function was used to classify the image regions into three types: COVID-19, Influenza-A-viral-pneumonia, and irrelevant-to-infection.
However, it should be noted that the CT datasets used in the studies reported in13–16 are not publicly available. In this study, a comprehensive investigation on 16 pretrained CNNs for classification of COVID-19 using a publicly available CT database is presented. These pretrained CNNs reflect a variety of computational complexity and accuracy based on the training and testing of the ImageNet database17. Findings of this investigation would facilitate the timely deployment of AI-assisted tools to hospitals and clinics in terms of ease of both data preparation and software implementation for fighting against the pandemic.
Methods
COVID-19 CT database
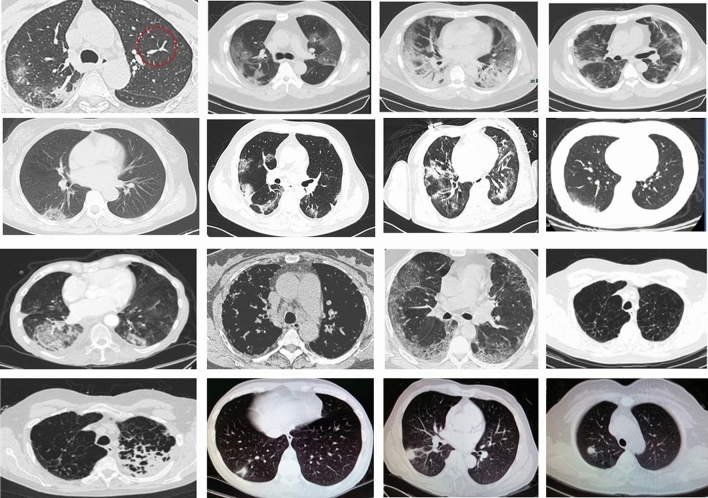
The COVID-19 CT database used in this study is publicly available18, and its details are described in12. The database consists of 349 CT images containing clinical findings of COVID-19 from 216 patients, and 397 CT images obtained from non-COVID-19 subjects. These CT images were collected from COVID19-related papers published in medRxiv, bioRxiv, NEJM, JAMA, Lancet, and others. Figure 1 shows CT images of COVID-19 and non-COVID-19. The usefulness of this dataset has been confirmed by a senior radiologist of Tongji Hospital, Wuhan, China. The radiologist has performed diagnosis and treatment of a large number of COVID-19 patients during the virus outbreak between January and April 202018.
Figure 1.
CT images: Rows 1 and 2: COVID-19, Rows 3 and 4: non-COVID-19 (aspect ratios of some images were rescaled to fit the figure frame).
Implementation of pretrained CNNs
Sixteen pretrained CNNs were investigated in this study for the classification of whole CT images to differentiate COVID-19 from non-COVID-19. These networks are: (1) SqueezeNet, (2) GoogLeNet, (3) Inception-v3, (4) DenseNet-201, (5) MobileNet-v2, (6) ResNet-18, (7) ResNet-50, (8) ResNet-101, (9) Xception, (10) Inception-ResNet-v2, (11) ShuffleNet, (12) NasNet-Mobile, (13) NasNet-Large, (14) AlexNet, (15) VGG-16, and (16) VGG-19. These pretrained networks were trained on more than a million images from the ImageNet database17. The pretrained networks can classify images into 1000 object categories, such as keyboard, mouse, pencil, and many animals. As a result, these networks have learned rich features representing a wide range of images. The properties of these networks are described in Table 1. To enable the reproduction of the results reported in this study, configurations for the transfer learning are described as follows.
Table 1.
Properties of 16 pre-trained CNNs.
| CNN | Depth | Size (MB) | Parameters (millions) | Input image size |
|---|---|---|---|---|
| AlexNet | 8 | 227 | 61.0 | 227 227 |
| GoogLeNet | 22 | 27 | 7.0 | 224 224 |
| SqueezeNet | 18 | 4.6 | 1.24 | 227 227 |
| ShuffleNet | 50 | 6.3 | 1.4 | 224 224 |
| ResNet-18 | 18 | 44 | 11.7 | 224 224 |
| ResNet-50 | 50 | 96 | 25.6 | 224 224 |
| ResNet-101 | 101 | 167 | 44.6 | 224 224 |
| Xception | 71 | 85 | 22.9 | 299 299 |
| Inception-v3 | 48 | 89 | 23.9 | 299 299 |
| Inception-ResNet-v2 | 164 | 209 | 55.9 | 299 299 |
| VGG-16 | 16 | 515 | 138 | 224 224 |
| VGG-19 | 19 | 535 | 144 | 224 224 |
| DenseNet-201 | 201 | 77 | 20.0 | 224 224 |
| MobileNet-v2 | 53 | 13 | 3.5 | 224 224 |
| NasNet-Mobile | * | 20 | 5.3 | 224 224 |
| NasNet-Large | * | 360 | 88.9 | 331 331 |
*indicates NASNet-Mobile and NasNetLarge networks do not consist of a linear sequence of modules.
First, the layer graph from the pretrained network was extracted. If the network was a SeriesNetwork object, such as AlexNet, VGG-16, or VGG-19, then the list of layers was converted to a layer graph. In most pretrained networks, the last layer with learnable weights is a fully connected layer. This fully connected layer was replaced with a new fully connected layer with the number of outputs equal to the number of classes in the new data set, which is 2, in this study. In some pretrained networks, such as SqueezeNet, the last learnable layer is a 1-by-1 convolutional layer instead. In this case, the convolutional layer was replaced with a new convolutional layer with the number of filters equal to the number of classes.
For the option of data augmentation in this study, random reflection, translation, and scaling were carried out. Random reflection was done in the top-bottom direction, where each image was reflected vertically with probability = 0.5. The range of horizontal translation applied to the input image = [− 30, 30], where the translation distance was measured in pixels. The horizontal translation distance was selected randomly from a continuous uniform distribution within the specified range. Similarly, the interval of vertical translation applied to the input image in pixels = [− 30, 30]. The vertical translation distance was selected randomly from a continuous uniform distribution within the specified interval. The range of horizontal scaling was applied to the input image, where the horizontal scale factor was selected randomly from a continuous uniform distribution within the specified interval = [0.9, 1.1]. Similarly, the range of vertical scaling was applied to the input image, where the vertical scale factor was selected randomly from a continuous uniform distribution within the specified interval = [0.9, 1.1].
The original whole CT images were converted into RGB images and resized to fit into the input image size of each pretrained CNN. For the training options, the stochastic gradient descent with momentum optimizer was used, where the momentum value = 0.9000; gradient threshold method = norm; minimum batch size = 10; maximum number of epochs = 6; initial learning rate = 0.0003; the learning rate remained constant throughout training; the training data were shuffled before each training epoch, and the validation data were shuffled before each network validation; and factor for regularization (weight decay) = 0.0001.
Statistical measures of classification performance
Five statistical measures used for evaluating the two-class classification performance of the pretrained CNNs are accuracy, sensitivity, specificity, score, and the area under the receiver operating characteristic (ROC) curve (AUC).
Let the sensitivity (SEN) be the percentage of COVID-19 patients who are correctly identified as having the infection, and expressed as
| 1 |
where TP is called true positive, denoting the number of COVID-19 patients who are correctly identified as having the infection, FN false negative, denoting the number of COVID-19 patients who are misclassified as having no infection of COVID-19, and P the total number of COVID-19 patients.
Let the specificity (SPE) be the percentage of non-COVID-19 subjects who are correctly classified as having no infection of COVID-19:
| 2 |
where TN is called true negative and denotes the number of non-COVID-19 subjects who are correctly identified as having no infection of COVID-19, FP false positive, denoting the number of non-COVID-19 subjects who are misclassified as having the infection, and N the total number of non-COVID-19 subjects.
The percent accuracy (ACC) of the classification is defined as
| 3 |
The score is defined as the balance between precision (TP divided by TP and FP) and sensitivity:
| 4 |
The ROC is a probability curve created by plotting the TP rate against the FP rate at various threshold settings, and the AUC represents the measure of performance of a classifier. The AUC value is within the range between 0.5 and 1, where the value = 0.5 represents the performance of a random classifier and the value = 1 indicates a perfect one. Thus, the higher the AUC is, the better the classifier performs. The AUC was calculated using the trapezoidal integration to estimate the area under the ROC curve.
Results
To compare the results with those obtained from previous reports, the dataset was randomly split into 80% for training and 20% for testing. The data splitting was repeated 5 times to obtain the average and standard deviation for each CNN. The whole CT images were used as the data input, which were resized to fit the input image size of each pretrained CNN, in both training and testing phases. The network training was performed for with and without data augmentation. Tables 2 and 3 show the classification results obtained from the 16 CNNs with and without data augmentation, respectively.
Table 2.
Classification results with data augmentation.
| CNN model | Accuracy (%) | Sensitivity (%) | Specificity (%) | score | AUC |
|---|---|---|---|---|---|
| AlexNet | 74.50 ± 4.40 | 70.46 ± 6.37 | 79.05 ± 8.61 | 0.75 ± 0.04 | 0.83 ± 0.04 |
| GoogLeNet | 78.97 ± 3.70 | 75.95 ± 13.69 | 82.38 ± 10.53 | 0.79 ± 0.06 | 0.91 ± 0.04 |
| SqueezeNet | 78.52 ± 7.56 | 91.56 ± 7.63 | 63.81 ± 23.79 | 0.82 ± 0.04 | 0.90 ± 0.01 |
| ShuffleNet | 86.13 ± 10.16 | 83.54 ± 19.89 | 89.05 ± 5.77 | 0.86 ± 0.12 | 0.93 ± 0.06 |
| ResNet-18 | 90.16 ± 2.36 | 89.45 ± 7.31 | 90.95 ± 9.29 | 0.91 ± 0.02 | 0.96 ± 0.05 |
| ResNet-50 | 92.62 ± 4.19 | 91.14 ± 3.35 | 94.29 ± 5.15 | 0.93 ± 0.04 | 0.98 ± 0.01 |
| ResNet-101 | 89.71 ± 10.05 | 82.28 ± 20.09 | 98.10 ± 2.18 | 0.89 ± 0.12 | 0.97 ± 0.03 |
| Xception | 85.68 ± 6.76 | 90.72 ± 4.79 | 80.00 ± 19.64 | 0.87 ± 0.05 | 0.94 ± 0.04 |
| Inception-v3 | 91.28 ± 8.25 | 90.30 ± 5.12 | 92.38 ± 11.98 | 0.92 ± 0.08 | 0.97 ± 0.02 |
| Inception-ResNet-v2 | 86.35 ± 5.71 | 88.19 ± 6.37 | 84.29 ± 14.50 | 0.87 ± 0.05 | 0.95 ± 0.05 |
| VGG-16 | 78.52 ± 10.02 | 74.68 ± 30.14 | 82.86 ± 15.91 | 0.76 ± 0.17 | 0.91 ± 0.04 |
| VGG-19 | 83.22 ± 5.85 | 90.72 ± 3.19 | 74.76 ± 12.96 | 0.85 ± 0.04 | 0.90 ± 0.05 |
| DenseNet-201 | 91.72 ± 6.52 | 88.61 ± 8.86 | 95.24 ± 4.36 | 0.92 ± 0.07 | 0.97 ± 0.03 |
| MobileNet-v2 | 87.25 ± 10.46 | 95.78 ± 2.64 | 77.62 ± 21.63 | 0.89 ± 0.08 | 0.95 ± 0.04 |
| NasNet-Mobile | 83.45 ± 7.36 | 84.81 ± 2.19 | 81.90 ± 17.46 | 0.85 ± 0.05 | 0.94 ± 0.04 |
| NasNet-Large | 85.23 ± 8.25 | 79.32 ± 16.28 | 91.90 ± 5.77 | 0.84 ± 0.10 | 0.93 ± 0.05 |
Table 3.
Classification results without data augmentation.
| CNN model | Accuracy (%) | Sensitivity (%) | Specificity (%) | score | AUC |
|---|---|---|---|---|---|
| AlexNet | 86.85 ± 13.66 | 80.25 ± 22.49 | 94.29 ± 4.84 | 0.85 ± 0.16 | 0.94 ± 0.04 |
| GoogLeNet | 93.83 ± 6.97 | 96.71 ± 4.06 | 90.57 ± 10.53 | 0.94 ± 0.06 | 0.96 ± 0.04 |
| SqueezeNet | 87.52 ± 6.45 | 86.84 ± 10.11 | 88.29 ± 12.01 | 0.88 ± 0.06 | 0.94 ± 0.06 |
| ShuffleNet | 95.97 ± 5.09 | 95.44 ± 7.47 | 96.57 ± 2.96 | 0.96 ± 0.05 | 0.97 ± 0.03 |
| ResNet-18 | 95.44 ± 8.02 | 98.99 ± 1.65 | 91.43 ± 15.25 | 0.96 ± 0.07 | 0.98 ± 0.03 |
| ResNet-50 | 93.62 ± 6.17 | 95.57 ± 6.27 | 91.43 ± 6.06 | 0.94 ± 0.06 | 0.98 ± 0.02 |
| ResNet-101 | 93.29 ± 5.69 | 96.20 ± 1.79 | 90.00 ± 10.10 | 0.94 ± 0.05 | 0.98 ± 0.02 |
| Xception | 91.11 ± 10.14 | 89.56 ± 12.55 | 92.86 ± 7.80 | 0.91 ± 0.10 | 0.96 ± 0.03 |
| Inception-v3 | 93.62 ± 5.22 | 96.20 ± 0.00 | 90.71 ± 11.11 | 0.94 ± 0.07 | 0.97 ± 0.04 |
| Inception-ResNet-v2 | 88.59 ± 7.59 | 89.24 ± 2.69 | 87.86 ± 13.13 | 0.89 ± 0.07 | 0.96 ± 0.05 |
| VGG-16 | 89.26 ± 8.80 | 92.83 ± 6.24 | 85.24 ± 14.45 | 0.90 ± 0.08 | 0.96 ± 0.03 |
| VGG-19 | 90.16 ± 7.72 | 87.34 ± 10.36 | 93.33 ± 5.77 | 0.90 ± 0.08 | 0.97 ± 0.03 |
| DenseNet-201 | 96.20 ± 4.95 | 95.78 ± 5.27 | 96.67 ± 4.59 | 0.96 ± 0.05 | 0.98 ± 0.03 |
| MobileNet-v2 | 95.97 ± 7.18 | 96.71 ± 6.04 | 95.14 ± 8.55 | 0.96 ± 0.07 | 0.97 ± 0.05 |
| NasNet-Mobile | 89.26 ± 8.14 | 91.56 ± 5.12 | 86.67 ± 13.27 | 0.90 ± 0.07 | 0.95 ± 0.06 |
| NasNet-Large | 88.59 ± 7.59 | 90.51 ± 0.90 | 86.43 ± 17.17 | 0.90 ± 0.06 | 0.96 ± 0.03 |
For the case of training the networks without data augmentation, DenseNet-201, MobileNet-v2, ShuffleNet, and ResNet-18 provide the average accuracy > 95%, while DenseNet-201 achieves the highest average accuracy (96.20%). GoogLeNet, ShuffleNet, ResNet-18, ResNet-50, ResNet-101, Inception-v3, DenseNet-201, and MobileNet-v2 result in the average sensitivity > 95%, while ResNet-18 has the highest average sensitivity (98.99%). ShuffleNet, DenseNet-201, and MobileNet-v2 provide the average specificity > 95%, while DenseNet-201 gives the highest average specificity (96.67%). ShuffleNet, ResNet-18, DenseNet-201, and MobileNet-v2 result in the highest average score = 0.96. The top three CNNs that achieve sensitivity > 95%, specificity > 95%, and score > 0.95 are ShuffleNet, DenseNet-201, and MobileNet-v2. DenseNet-201 is the best model for the classification of COVID-19 CT data. The networks that have the highest average AUC (0.98) are the ResNet family and DenseNet-201.
The results obtained from CNNs without data augmentation are all better than those with data augmentation in terms of accuracy (compare results between Tables 2 and 3). For the case of training the networks with data augmentation, models that have average accuracy > 90% are ResNet-18 (90.16%), ResNet-50 (92.62%), Inception-v3 b(91.28%), and DenseNet-201 (91.72%); models that have average sensitivity > 90% are SqueezeNet (91.56%), ResNet-50 (91.14%), Xception (90.72%), Inception-v3 (90.30%), VGG-19 (90.72%), and MobileNet-v2 (95.78%); models that have average specificity > 90% are ResNet-18 (90.95%), ResNet-50 (94.29%), ResNet-101 (98.10%), Inception-v3 (92.38%), DenseNet-201 (95.24%), and NasNet-Large (91.90%); and models that have average score > 0.9 are ResNet-18 (0.91), ResNet-50 (0.93), Inception-v3 (0.92), and DenseNet-201 (0.92). Two CNNs using data augmentation that have accuracy > 90%, sensitivity > 90%, specificity > 90%, and score > 0.9 are ResNet-50 and Inception-v3. The networks without data augmenttaion have higher or equal values for the average AUC than or to those with data augmentation.
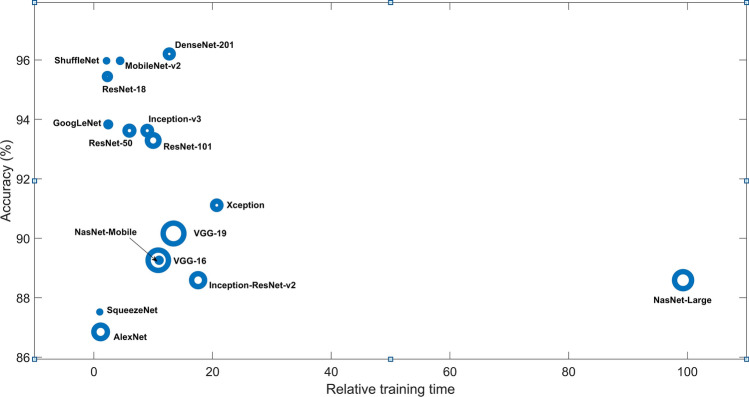
In summary, without data augmentation, the best classifier is DenseNet-201, which has the best accuracy, best balance between sensitivity and specificity, top score, and top AUC. Figure 2 shows the plot of accuracy versus relative training time obtained from the 16 pretrained CNNs without data augmentation.
Figure 2.
Plots of accuracy vs. relative training time (ratio of training time of a network to the training time of the SqueezeNet) of 16 pretrained CNNs using COVID-19 CT database, where the circle size indicates the magnitude of memory in MB.
Discussion
The benchmark results using the same database reported in12, with a fixed split data of about 80% for training and 20% for testing, have accuracy = 84.7%, sensitivity = 76.2%, and score = 0.85, using a fine-tuned pretrained DenseNet with data augmentation. The results obtained from the 16 CNNs without data augmentation are better than these benchmark results.
The study published in13 applied 10 pretrained CNN using a different COVID-19 database with the same ratio of training and testing data, which is not publicly available, reported among all the 10 networks, ResNet-101 was the best classification model. ResNet-101 achieved accuracy = 99.51%, sensitivity = 100%, and specificity = 99.02%. Although using a different database, the results obtained in this study are comparable. However, the input data processing and training reported in13 requires much effort by requiring the extraction of regions of interest by a radiologist, which is subjective, time-consuming, and likely hinders the real-time application of the pretrained networks.
The work reported in14 requires the pre-processing of 3D CT scans by extracting the regions of interest using a U-Net for image segmentation. The pre-processed images were then passed to the COVNet for the prediction. The sensitivity and specificity obtained from COVNet were 87% and 92%, respectively, using a dataset that is not publicly available. Another work on the classification of COVID-19 CT images collected from 259 patients reported in15 modified the pretrained Inception that achieved accuracy = 79.3%, sensitivity = 67%, and specificity = 83%, and another test achieving accuracy = 85.2%. Similarly, the input images are extracted regions of interest such as small patchy shadows and interstitial changes, multiple ground glass and infiltrates in both lungs. The study reported in16 used the concatenation of two pretrained ResNet-based networks and the Bayesian function for screening COVID-19 patients using CT imaging. The data pre-processing of classification procedure requires 3D segmentation, extraction of regions of interest (such as ground-glass appearance, striking peripheral distribution along with the pleura, and independent focus of infections), and data augmentation. The overall accuracy obtained was 86.7%. The classification results obtained in this study are preferable to those reported in14–16 in terms of accuracy and implementation of input data.
Although the use of regions of interest or cropped images is widely adopted for deep learning, including other classification problems19–22, this study finds that the direct input of CT images, which are then resized to fit the input size of the pretrained CNN, and transfer learning without data augmentation can achieve very high and better classification performance than those using data augmentation. Such findings are useful for the rapid deployment of AI tools to meet the urgent demand for curbing the pandemic, because it can relieve the task of manual detection of regions of interest carried out by experienced radiologists, employment of image segmentation methods, and more data collection.
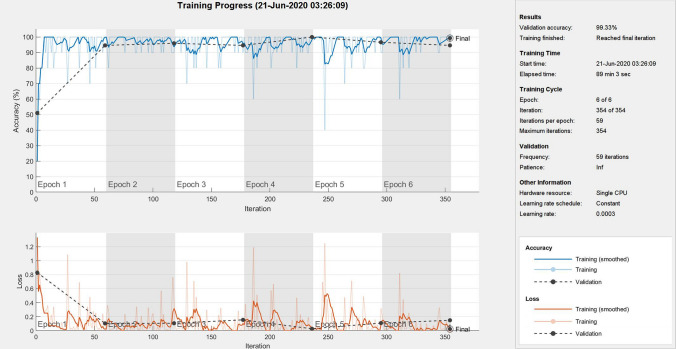
Using the described network-training configuration with only 6 epochs, the CNNs could provide a very high performance of classification. Figures 3 and 4 show one of the training processes of DenseNet-201 (best network) and some features obtained from the deep learning of the best network, respectively.
Figure 3.
A training process of DenseNet-201.
Figure 4.

Features learned by DenseNet-201: 36 features in layer ’conv1|conv’(convolution) (top left), layer ‘conv4_block7_1_conv’ (convolution) (top right), layer ‘conv5_block9_1_conv’ (convolution) (bottom left), and 2 features in layer ‘new_fc’ (fully connected) (bottom right).
As the numbers of COVID-19 and non-COVID-19 CT images used in this study are 349 and 397, respectively, the binary classification in this study was not much disadvantaged from the class imbalance problem, where the class distributions are highly imbalanced. Due to imbalanced data, classifiers tend to result in low predictive accuracy for the minority class. Medical datasets are often not balanced in the class labels because of limited samples collected from patients and cost for acquiring annotated data. There are many techniques proposed for addressing class imbalance , which can be applied to medical imaging, such as the “deep domain adaptation”23 for handling the shortage of large amounts of labeled data, weighted loss method by updating the loss function to result in the same loss for all classes, downsampling by removing images from the majority class, and oversampling by adding more images to minority classes using artificial data augmentation24,25. Open challenges in imbalance data and exploration for solutions can be found in26.
Conclusions
AI-based medical diagnosis systems based on deep learning of medical imaging are increasingly recognized to be clinically useful. However, development of suitable deep-learning networks and effective training strategy for clinical applications is a topic of research that needs to be explored27. Through a comprehensive investigation of 16 pretrained CNNs using certain parameter specification and training strategy for the networks, this study discovers the very high performance of several of these networks for COVID-19 diagnosis using CT images. The network configuration of the pretrained models can be implemented for classification of other image modality, such as X-ray, for the detection of COVID-19.
Most AI studies on chest CT used for differentiating COVID-19 pneumonia from other causes of pneumonia consider both three-class classification problems (COVID-19 pneumonia, non-COVID-19 pneumonia, and healthy) and two-class classification (COVID-19 pneumonia and healthy)2. Due to the limit of publicly available data, this study concerns with the two-class classification. However, extension of the use of pretrained CNNs to the three-class classification of COVID-19 imaging data is straightforward.
The findings reported from this study bring benefits to the development of fast and efficient diagnostic tools using imaging data and contribute to further leading into the development of more accurate point-of-care diagnostic and detection tools for containing the coronavirus pandemic.
Author contributions
T.D.P. conceived and designed the research. T.D.P. performed the study of AI, carried out the computer experiments, and analyzed the results. T.D.P. wrote the manuscript.
Data availability
The MATLAB code used in this study is available at the author’s personal homepage: https://sites.google.com/view/tuan-d-pham/codes.
Competing interests
The author declares no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang W, et al. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19) Eur. Radiol. 2020 doi: 10.1007/s00330-020-06827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kundu S, Elhalawani H, Gichoya JW, Kahn CE. How might AI and chest imaging help unravel COVID-19’s mysteries? Radiol. Artif. Intell. 2020;2:3. doi: 10.1148/ryai.2020200053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li K, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur. Radiol. 2020 doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, et al. CT imaging changes of corona virus disease 2019 (COVID-19): A multi-center study in Southwest China. J. Transl. Med. 2020;18:154. doi: 10.1186/s12967-020-02324-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CT Image Analytics for COVID-19. Alibaba Cloud. https://www.alibabacloud.com/solutions/ct-image-analytics.
- 6.Automated diagnosis and quantitative analysis of COVID-19 on imaging. European Society of Medical Imaging Informatics. https://imagingcovid19ai.eu/.
- 7.Ojha V, et al. CT in coronavirus disease 2019 (COVID-19): A systematic review of chest CT findings in 4410 adult patients. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latest articles on COVID-19. European Radiology. https://www.european-radiology.org/highlights/covid-19/.
- 9.Salehi S, et al. Coronavirus disease 2019 (COVID-19) imaging reporting and data system (COVID-RADS) and common lexicon: A proposal based on the imaging data of 37 studies. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mei X, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCall B. COVID-19 and artificial intelligence: Protecting health-care workers and curbing the spread. The Lancet. 2020;2:e166–e167. doi: 10.1016/S2589-7500(20)30054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao, J., Zhang, Y., He, X., & Xie, P. COVID-CT-Dataset: A CT scan dataset about COVID-19. arXiv:2003.13865 (2020).
- 13.Ardakani AA, et al. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19) medRxiv. 2020 doi: 10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu, X. et al. Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv:2002.09334.
- 17.ImageNet. http://www.image-net.org.
- 18.COVID-CT. https://github.com/UCSD-AI4H/COVID-CT. Accessed 05 May 2020.
- 19.Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J. Big Data. 2019;6:60. doi: 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahbod A, et al. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Methods Progr. Biomed. 2020;193:105475. doi: 10.1016/j.cmpb.2020.105475. [DOI] [PubMed] [Google Scholar]
- 21.Cirillo MD, Mirdell R, Sjoberg F, Pham TD. Time-independent prediction of burn depth by deep convolutional neural networks. J. Burn Care Res. 2019;40:857–863. doi: 10.1093/jbcr/irz103. [DOI] [PubMed] [Google Scholar]
- 22.Pham TD. Geostatistical simulation of medical images for data augmentation in deep learning. IEEE Access. 2019;7:68752–68763. doi: 10.1109/ACCESS.2019.2919678. [DOI] [Google Scholar]
- 23.Wang M, Deng W. Deep visual domain adaptation: A survey. Neurocomputing. 2018;312:135–153. doi: 10.1016/j.neucom.2018.05.083. [DOI] [Google Scholar]
- 24.He H, Garcia EA. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009;21:1263–1284. doi: 10.1109/TKDE.2008.239. [DOI] [Google Scholar]
- 25.Rahman MM, Davis DN. Addressing the class imbalance problem in medical datasets. Int. J. Mach. Learn. Comput. 2013;3:224–228. doi: 10.7763/IJMLC.2013.V3.307. [DOI] [Google Scholar]
- 26.Krawczyk B. Learning from imbalanced data: Open challenges and future directions. Prog. Artif. Intell. 2016;5:221–232. doi: 10.1007/s13748-016-0094-0. [DOI] [Google Scholar]
- 27.Liang G, Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Methods Progr. Biomed. 2020;187:104964. doi: 10.1016/j.cmpb.2019.06.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MATLAB code used in this study is available at the author’s personal homepage: https://sites.google.com/view/tuan-d-pham/codes.