Abstract
Background
Non-hypermucoviscous carbapenem-resistant Klebsiella pneumoniae with enhanced virulence lacking hvKP-specific virulence factors is uncommon, and the virulence mechanisms of this organism are not understood.
Methods
Following a retrospective study of carbapenem-resistant K. pneumoniae based on core genome multilocus sequence typing (cgMLST), isolates that caused high mortality were investigated with a genome-wide association study (GWAS), proteome analysis and an animal model.
Results
The subclone of sequence type 11 (ST11) K. pneumoniae, which belongs to complex type 3176 (CT3176) and K-locus 47 (KL47), was highlighted due to the high mortality of infected patients. GWAS analysis showed that transcriptional regulatory gene ampR was associated with the CT3176 isolates. In a mouse model, the mortality, bacterial load and pathological changes of mice infected with ampR-carrying isolates were distinct from those infected with ampR-null isolates. The ampR gene that enhances the virulence of the non-hypermucoviscous KL47 strain was unable to enhance the virulence of hypermucoviscous KL1 strain. Proteome analysis showed that the expression of WcaJ in the ampR+ isolates was significantly higher than that in the ampR− isolates. Quantification of capsular polysaccharide confirmed that more capsule polysaccharide was produced by ampR+ and ampR-complementary strains compared to ampR− strains. It is suggested that the enhancement of the initial stage of capsule synthesis may be the cause of the enhanced virulence of these non-hypermucoviscous ST11 carbapenem-resistant K. pneumoniae isolates.
Conclusion
Non-hypermucoviscous ST11 carbapenem-resistant K. pneumoniae with enhanced virulence warrants continued surveillance and investigation.
Keywords: Klebsiella pneumonia, virulence, carbapenem-resistant, capsule synthesis
Introduction
Classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp) are two global K. pneumoniae pathotypes presently circulating.1,2 In North America and Europe, most K. pneumoniae infections are caused by cKp isolates, which are most commonly opportunistic pathogens causing infections primarily in the health care setting in immunocompromised hosts. The most problematic characteristic of this pathotype is the ability to acquire an increasing number of elements that confer antimicrobial resistance. Carbapenem-resistant K. pneumoniae (CRKP) associated with plasmid-encoded carbapenemases poses distinct clinical challenges and produces invasive infections with high mortality.3,4
Reports from Taiwan described a unique clinical syndrome of community-acquired, tissue-invasive K. pneumoniae infection in healthy individuals that often presented at multiple sites or subsequently spread, including pyogenic liver abscesses.5,6 hvKp was designated to distinguish this pathotype from cKp, and the incidence of infections due to hvKp has been steadily increasing over the last 3 decades in Asian Pacific countries.7–11 hvKp is usually susceptible to antimicrobials, but it has been found to be resistant, and even an extensively drug-resistant (XDR) cKp isolate that acquired part of an hvKp virulence plasmid caused a fatal nosocomial outbreak was reported.12–15
A characteristic that was initially believed to be sensitive and specific for hvKp isolates was a hypermucoviscous phenotype, which was defined by a positive string test.16 However, it created some confusion as not all hvKp isolates were determined as being hypermucoviscous, and some cKp isolates possessed this characteristic.17 Recently, multiple biomarkers including peg-344, iroB, iucA, plasmid-encoded rmpA and rmpA2 and quantitative siderophore production have been shown to accurately predict hvKp isolates; these biomarkers could be used to develop a diagnostic test for use by clinical laboratories for optimal patient care and for use in epidemiologic surveillance and research.18 However, in this study, we identified a novel group of non-hypermucoviscous CRKP isolates lacking most hvKp-specific genes. A pan genome-wide association study (Pan-GWAS), proteome analysis and virulence test in an animal model revealed that AmpR increases the virulence of these isolates by regulating the initiation of capsule synthesis. Moreover, we also found that ampR carried by K-locus 47 (KL47) strains in this study was unable to increase the virulence of KL1 hypermucoviscous K. pneumoniae.
Materials and Methods
Clinical Isolates and Data Collection
Non-repetitive clinical K. pneumoniae isolates were collected from routine detected and stored samples of 5 hospitals’ Department of Laboratory Medicine in Guangdong and Anhui provinces between 2018 and 2019. All isolates were stored at −80°C prior to use. Clinical information on patients was obtained. Species identification and antimicrobial susceptibility testing were performed with the VITEK-2 compact system (bioMérieux, Marcy-l’Étoile, France). The results were interpreted in accordance with guidelines published by the Clinical and Laboratory Standards Institute (CLSI; document M100-S26).19 The identified species of all isolates were confirmed with matrix-assisted laser desorption/ionization mass spectrometry (bioMérieux, Marcy-l’Étoile, France). CRKP was defined as resistant to imipenem or meropenem.
Hypermucoviscous Phenotypic Characterization
To identify the hypermucoviscous phenotype with the string test, isolates were inoculated onto agar plates containing 5% sheep blood and incubated at 37°C overnight. The string test was deemed positive when a viscous string longer than 5 mm could be generated by touching a single colony with a standard inoculation loop and pulling the colony upwards.2
Whole Genome Sequencing (WGS)
A 1 mL culture volume (optical density at 600 nm [OD600] of 0.6) was used for genomic DNA (gDNA) extraction (Sangon, Shanghai, China). gDNA was sequenced on an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, USA) using paired-end 150-bp reads. Genome assembly was performed using de novo SPAdes Genome Assembler (version 3.12.0).20 We performed capsule typing on the assembled sequences with Kaptive (version 0.5.1).21 Antimicrobial resistance genes and virulence factors were identified in the isolates by scanning the genome contigs against the ResFinder and VFDB databases using ABRicate (version 0.8.7). Multilocus sequence typing (MLST) and core genome MLST (cgMLST) genotyping analysis were performed with Ridom SeqSphere+ (version 5.1.0).22 Complex type (CT) was defined by following the K. pneumoniae cgMLST scheme (www.cgmlst.org/ncs/schema/2187931). Phylogenetic tree construction based on core genome single nucleotide variants (SNVs) was performed using the Harvest suite (version 1.2) with a 1000-bootstrap test.23 The online tool iTOL was used to display, manipulate and annotate the phylogenetic tree.24 We annotated the genome sequences with Prokka (version 1.13.3).25
Pan Genome-Wide Association Study (Pan-GWAS) Analysis
We used the pan genome pipeline Roary (version 3.12.0) to put annotated assemblies in GFF3 format (produced by Prokka) and calculated the pan genome.26 We used Scoary (version 1.6.16) to take the file from “https://sanger-pathogens.github.io/Roary/”, created a trait file and calculated the associations between all genes in the accessory genome and the traits. We reported a list of genes sorted by strength of association with P-value < 0.01 adjusted with Benjamini-Hochberg’s method for multiple comparisons correction.27
Proteome Analysis
A 1 mL culture volume (optical density at 600 nm [OD600] of 0.6) was used for protein extraction. The sample was sonicated three times on ice using a high-intensity ultrasonic processor (Scientz) in lysis buffer (8 M urea, 1% protease inhibitor cocktail). The remaining debris was removed by centrifugation at 12,000 g at 4°C for 10 min. Finally, the supernatant was collected, and the protein concentration was determined with a BCA kit according to the manufacturer’s instructions. For digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56°C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mM TEAB with urea at a concentration of less than 2M. Finally, trypsin was added at a 1:50 trypsin-to-protein mass ratio for the first digestion overnight and a 1:100 trypsin-to-protein mass ratio for a second 4 hours digestion. The tryptic peptides were dissolved in 0.1% formic acid (solvent A) and directly loaded onto a homemade reversed-phase analytical column (15-cm length, 75 μm i.d.). The gradient was performed as follows: an increase from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, an increase from 23% to 35% solvent B in 8 min, an increase to 80% solvent B in 3 min and holding at 80% solvent B for the last 3 min at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350 to 1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using the NCE setting of 28, and the fragments were detected in the Orbitrap at a resolution of 17,500. The data-dependent procedure alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control (AGC) was set at 5E4. The fixed first mass was set as 100 m/z. The resulting MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8). We used InterProScan (version 5.37–76.0) to annotate protein domains. A corrected p-value < 0.05 and a fold change > 1.2 were considered significant.
Construction of the ampR Complement Mutant
K. pneumonia ampR (Supplementary Materials) was synthesized and flanked with 5ʹXbaI and 3ʹHindIII restriction sites. This fragment was restriction digested and cloned into pUC57. Recombinant DNA was recovered in Escherichia coli DH5α with ampicillin selection. After restriction digestion, the ampR fragment was ligated to the pBAD33 expression vector (Invitrogen) according to the manufacturer’s instructions. pBAD33-ampR was used for transformation using electroporation as a standard for laboratory-isolated E. coli. The ampR complement strains were confirmed by polymerase chain reaction (PCR) (Table S1). The expression of ampR was induced by adding 100 μg/mL arabinose.
Mouse Pulmonary Infection Models
A pneumonia model of K. pneumoniae in mice was used to test the virulence of isolates. Six- to eight-week-old female BALB/c mice under specific pathogen-free grade were purchased from Hunan SJA Laboratory Animal Co., Ltd. The mice were intraperitoneally anaesthetized with pentobarbital sodium (75 mg/kg) and inoculated with 1.0 × 107 colony-forming units (CFU) of K. pneumoniae by non-invasive intratracheal instillation under direct vision. The survival of the mice was observed for 7 days post-infection. To assess bacterial burden in the lung, the organs from infected mice were homogenized and dissolved in 1 mL of phosphate buffer solution (PBS); 50 μL was inoculated onto liquid broth (LB) agar plates, and CFU enumeration was performed. For histopathology analysis, segments of the lungs were fixed with 10% neutral formalin, embedded in paraffin and stained with haematoxylin and eosin for visualization by light microscopy. Bacterial burden and histopathology analysis were performed after 24 hours of infection.
Extraction and Quantification of Capsular Polysaccharide
Capsular polysaccharides were extracted with Zwittergent 3–14 detergent. The amount of uronic acid was then measured according to the method described previously.28 In parallel, serial dilutions of the bacterial culture were plated to determine the number of CFU, and the concentration of capsular polysaccharide was expressed according to the amount of glucuronic acid (μg) for 109 CFU/mL of sample. Each experiment was performed in triplicate.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software version 8 (La Jolla, California).
Results
Isolate Characteristics
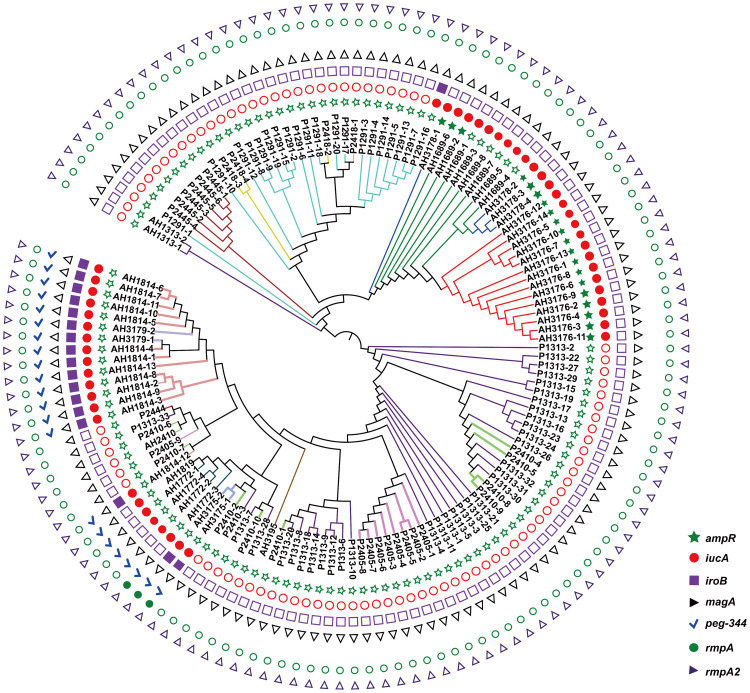
Non-repetitive CRKP (n = 135) isolates were used in this study. All isolates were sequence type 11 (ST11) and expressed K. pneumoniae carbapenemase (KPC-2), and 1 isolate carried both OXA-23 and KPC-2 (Figure S1). Isolates were assigned to 16 CTs using the cgMLST scheme. The most common CTs were CT1313 (n = 35), CT1291 (n = 20), CT3176 (n = 14), CT1814 (n = 13), and CT2410 (n = 11), followed by 11 other CTs identified in less than 10 isolates each (Figure 1). All isolates were assigned to two KL types, KL64 (n = 86) and KL47 (n = 49) (Figure S2). No hypermucoviscous isolates were found. The virulence gene analysis showed that no isolates carrying all biomarkers for differentiation of the hvKp and cKp isolates had been reported previously (Figure 1, Figure S2).18
Figure 1.
Phylogenetic analysis of 135 isolates in this study. The branches of different CT type isolates are shown in colours. The corresponding isolates of each CT type are CT1291 (P1291-1 to P1291-20), CT1313 (P1313-1 to P1313-33, AH1313-1 to AH1313-2), CT1689 (AH1689-1 to AH1689-8), CT1772 (AH1772-1 to AH1772-4), CT1814 (AH1814-1 to AH1814-13), CT1819 (AH1819), CT2405 (P2405-1 to P2405-9), CT2410 (P2410-1 to P2410-11), CT2418 (P2418-1 to P2418-4), CT2444 (P2444), CT2445 (P2445-1 to P2445-6), CT3175 (AH3175-1 to AH3175-2), CT3176 (AH3176-1 to AH3176-14), CT3178 (AH3178-1 to AH3178-4), CT3179 (AH3179-1 to AH3179-2), and CT3195 (AH3195). rmpA, rmpA2, magA iroB, peg-344 and iucA were identified for the differentiation of hypervirulent K. pneumoniae from classical K. pneumoniae in a previous study.18
Abbreviations:rmpA/rmpA2, cps transcriptional activator; magA, outer membrane protein; iroB, iron acquisition system; peg-344, metabolite transporter; iucA, aerobactin-related genes.
Clinical Data Analysis
There were no significant differences between the CTs in terms of demographics, infection types, main comorbidities, invasive operations, Charlson comorbidity index and Pitt bacteraemia score, except for central venous catheter, and all cause in-hospital mortality (P = 0.035 and P = 0.001, Fisher’s exact test). Patients infected with CT3176 had significantly higher mortality rates (78.6% vs 37.1%, P = 0.012; 78.6% vs 20.0%, P = 0.001; 78.6% vs 7.7%, P = 0.0003; 78.6% vs 31.0%, P = 0.004; Fisher’s exact test) compared with the CT1313, CT1291, CT1814 and the other CT groups, respectively. The utilization of central venous catheters in CT1814 was significantly higher than that in CT1313 and CT1291 (84.6% vs 51.4%, P = 0.049; 84.6% vs 35.0%, P = 0.011, Fisher’s exact test) (Table 1).
Table 1.
Demographics, Comorbidities, Invasive Procedures and Mortality of Patients with Carbapenem-Resistant Klebsiella pneumoniae Infections
| n = 135 | CT1313 (n=35) |
CT1291 (n=20) |
CT3176 (n=14) |
CT1814 (n=13) |
CT2410 (n=11) |
The other CTs (n=42) |
p value* |
|---|---|---|---|---|---|---|---|
| Age in years, mean ± SD | 68.63±20.65 | 65.85±19.14 | 72.64±15.29 | 64.92±15.14 | 67.55±15.79 | 64.81±15.73 | 0.994 |
| Gender (Male) | 28(80.0%) | 14(70.0%) | 12(85.7%) | 11(84.6%) | 6(54.5%) | 30(71.4%) | 0.423 |
| Bacteremia | 9(25.7%) | 8(40.0%) | 2(14.3%) | 1(7.7%) | 1(9.0%) | 5(11.9%) | 0.075 |
| Pulmonary infection | 30(85.7%) | 18(90.0%) | 10(71.4%) | 12(92.3%) | 9(81.8%) | 37(88.1%) | 0.629 |
| Urinary infection | 6(17.1%) | 6(30.0%) | 2(14.3%) | 1(7.7%) | 3(27.3%) | 3(7.1%) | 0.200 |
| Abdominal infection | 5(14.3%) | 3(15%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 5(11.9%) | 0.323 |
| Cerebral vascular disease | 22(62.9%) | 12(60%) | 4(28.6%) | 7(53.8%) | 5(45.5%) | 22(52.4%) | 0.372 |
| Hypertension | 21(60.0%) | 13(65.0%) | 9(64.3%) | 8(61.5%) | 5(45.5%) | 32(76.2%) | 0.463 |
| Diabetes | 11(31.4%) | 6(30.0%) | 8(57.1%) | 7(53.8%) | 4(36.4%) | 17(40.5%) | 0.451 |
| Mechanical ventilation | 27(77.1%) | 14(70.0%) | 9(64.3%) | 10(76.9%) | 6(54.5%) | 27(64.3%) | 0.681 |
| Central venous catheter | 18(51.4%)a | 7(35.0%)a | 11(78.6%) | 11(84.6%)a | 6(54.5%) | 27(64.3%) | 0.035+ |
| Foley catheter | 23(65.7%) | 16(80.0%) | 13(92.9%) | 12(92.3%) | 8(72.7%) | 32(76.2%) | 0.264 |
| Nasogastric tube | 24(68.6%) | 11(55.0%) | 12(85.7%) | 7(53.8%) | 7(63.6%) | 27(64.3%) | 0.486 |
| Continuous renal replacement therapy | 6(17.1%) | 4(20.0%) | 0(0.0%) | 2(15.4%) | 2(18.2%) | 3(7.1%) | 0.391 |
| All cause in-hospital mortality | 13(37.1%)a | 4(20.0%)a | 11(78.6%)a | 1(7.7%)a | 6(54.5%) | 13(31.0%)a | 0.001+ |
| Charlson comorbidity indexb | 4.72±0.81 | 3.55±0.61 | 3.91±0.63 | 4.41±0.69 | 3.88±0.46 | 2.74±0.77 | 0.232 |
| Pitt Bacteremia Scorec | 3.50±0.51 | 2.80±0.64 | 2.64±0.59 | 3.92±0.54 | 3.55±0.79 | 2.81±0.73 | 0.211 |
Notes: aThese data were significant using pairwise analyses: CT3176 vs CT1313 (All cause in-hospital mortality; P = 0.012); CT3176 vs CT1291 (All cause in-hospital mortality; P = 0.001); CT3176 vs CT1814 (All cause in-hospital mortality; P = 0.0003); CT3176 vs The other CTs (All cause in-hospital mortality; P = 0.004); CT1814 vs CT1313 (Central venous catheter; P = 0.049); CT1814 vs CT1291 (Central venous catheter; P = 0.011); bCharlson comorbidity index. The charlson comorbidity index can range from 0 to 37. (A patient <40 who has no other medical or surgical conditions would have a score of 0; an 81-year-old with AIDS and cancer would have a score of 37). cA Pitt bacteremia score >4 indicates severe illness. The Pitt bacteremia score is based on temperature, hypotension, mental status, and presence or absence of mechanical ventilation. *p values are for comparisons between CTs. Dichotomous variables are analyzed using Fisher’s exact test and continuous variables are analyzed using one-way ANOVA. +The bolded numbers correspond to highlighted parameters in the article.
Abbreviation: CT, complex type.
Pan-GWAS Analysis
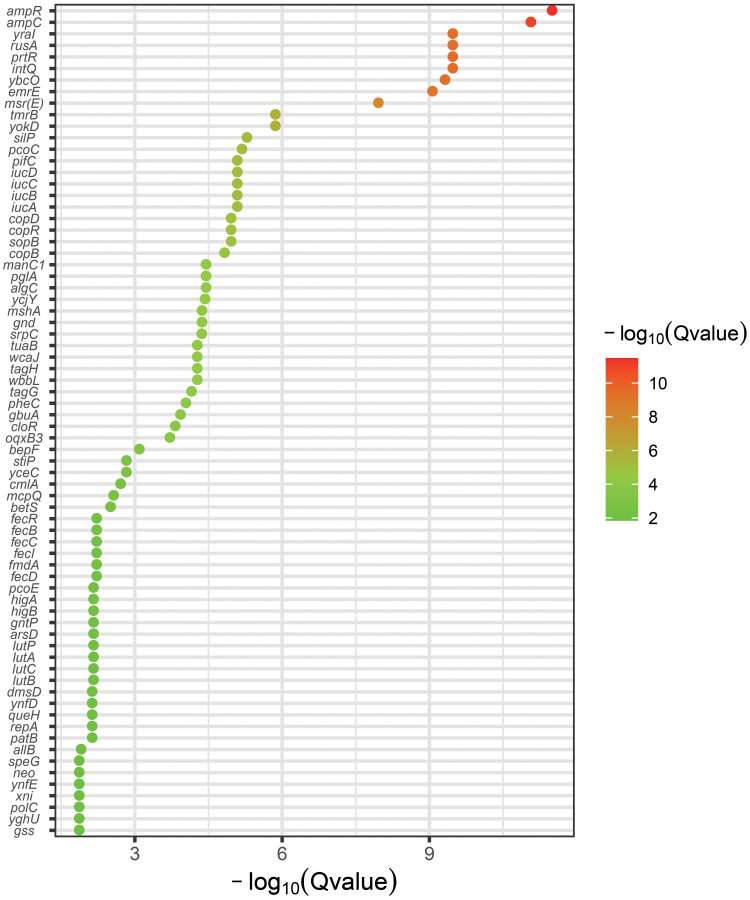
To identify the genetic basis of CT3176, we performed a Pan-GWAS analysis between the CT3176 and non-CT3176 groups. We identified 39 genes with known functions associated with CT3176 (P < 0.01 adjusted with Benjamini-Hochberg’s method) (Figure 2), including virulence factors, capsule synthesis genes, antimicrobial resistance genes and multidrug efflux transporters. Among them, ampR had the greatest association with CT3176. However, we noted that other CTs, such as CT3178 and some CT1689 isolates (AH1689-2 and AH1689-6), also carried the ampR gene (Figure 1). All isolates carrying the ampR gene belong to KL47.
Figure 2.
Pan-GWAS analysis between the CT3176 and non-CT3176 groups. The pangenome and associations were calculated with Roary and Scoary. A plot map was generated with ggplot2.
Proteome Analysis
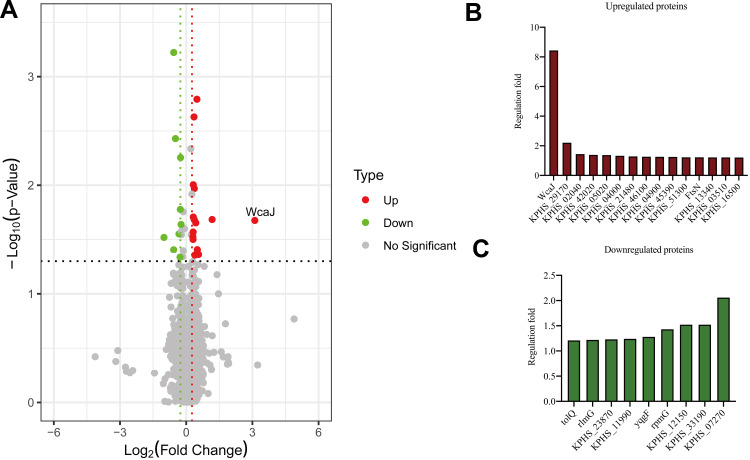
Three ampR+ (AH3176-1, AH3178-1 and AH1689-2) and three ampR− (AH1689-3, AH1689-4 and AH1689-5) isolates were selected for proteome analysis. A total of 3093 proteins were identified in both groups based on 36,727 unique peptides. Finally, we identified 24 differentially expressed proteins (DEPs). Among them, 15 were upregulated, and 9 were downregulated in the ampR+ isolates compared to the ampR− isolates (Figure 3A–C, Table S2). WcaJ had the highest fold change and was therefore highlighted.
Figure 3.
Proteome analysis and capsule staining. (A) Volcano plot showing the comparison of quantitative protein expression between ampR+ (AH1689-2, AH3176-1 and AH3178-1) and ampR− (AH1689-3, AH1689-4, and AH1689-5) Klebsiella pneumoniae isolates. (B, C) Differentially expressed proteins with a fold change over 1.2 are marked in colour.
Virulence in Animal Models and Quantification of Capsule
With aim to identify the role of ampR in the virulence of non-hypermucoviscous CRKP isolates, we selected three isolates (AH3176-1, AH1689-2 and AH1689-3), and tested the virulence in a mouse model. AH3176-1 and AH1689-2 were two ampR+ isolates, while AH1689-3 was the ampR− isolate (Figure 1). In addition, we also selected two hypermucoviscous strains GM2 and GM6 from our collection to test the virulence. Both GM2 and GM6 belong to KL1, among which GM6 carries ampR gene while GM2 does not. However, the ampR sequence of KL47 non-hypermucoviscous strain was different from that of KL1 hypermucoviscous strain, so they were named ampRKL1 and ampRKL47 respectively in this study.
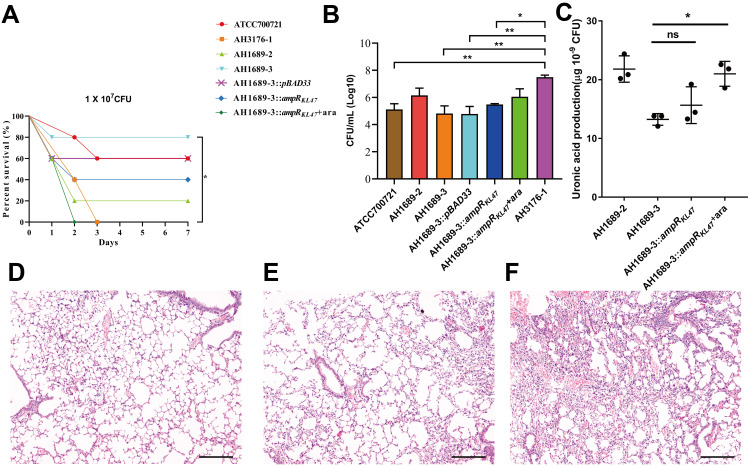
As shown in Figure 4A, AH3176-1 and AH1689-3:ampRKL47 plus arabinose had a survival of 0% with an inoculum of 1 × 107 colony-forming units (CFU) at 4 d post-infection, while 20% survival with AH1689-2, 40% survival with AH1689-3:ampRKL47 without arabinose, 60% survival with AH1689-3:pBAD33 and ATCC70721, and 80% survival with AH1698-3 was observed at 7 d post-infection. Survival rate was significantly decreased after complementation with the ampRKL47 gene in AH1689-3 (n = 10; P = 0.033, log-rank Mantel-Cox test), suggesting the important role of ampR in virulence. Infection of mice with AH3176-1 resulted in approximately 10–100-fold higher CFU in the lungs compared to other isolates (Figure 4B). The pathology of the lungs showed varying degrees of alveolar wall thickening, lymphocyte infiltration, and intravascular bleeding in the infection group. Among them, the pulmonary pathological changes caused by AH3176-1 were the most serious, presenting as extensive pulmonary consolidation and intra-alveolar haemorrhage (Figure 4D–F).
Figure 4.
Virulence potential of non-hypermucoviscous Klebsiella pneumoniae isolates in a mouse pulmonary infection model. (A) The effect of 1 × 107 colony-forming units (CFU) of AH3176-1, AH1689-2, AH1689-3 and AH1689-3 ampRKL47 complement mutant on survival was assessed. (B) At 24 hours post-infection (hpi), the lungs of infected mice were harvested, and the bacterial burden was determined by CFU enumeration (n = 10; *P < 0.05, **P < 0.01, one-way ANOVA). Data are representative of 3 independent experiments. (C) Uronic acid production of different strains. An unpaired two-sided Student’s t-test was performed. Each data point was repeated three times (n = 3). Data are presented as the mean ± s.e.m. *P < 0.05, ns = no significance. The lungs of mice without infection (D) or infected with ATCC700721 (E) and AH3176-1 (F) were collected and stained with haematoxylin and eosin. All images are at ×40 magnification. Scale bars: 250 μM.
Abbreviation: ara, arabinose.
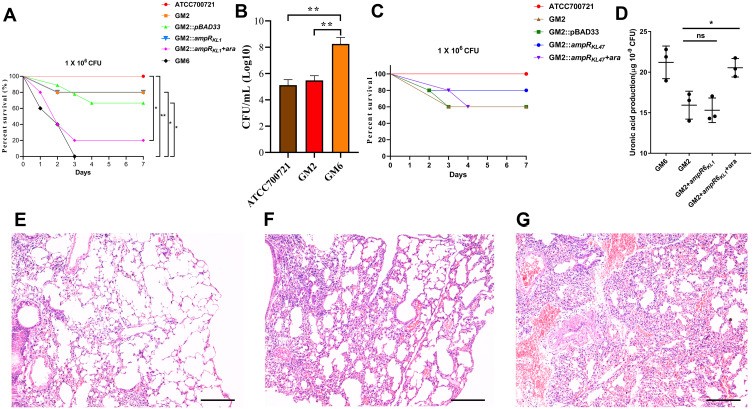
As shown in Figure 5A, the survival rate of mice infected with GM6 was significantly lower than that of GM2 and ATCC700721. A decrease of survival rate was observed after complementation of ampRKL1 in GM2. In addition, the bacterial load in the lungs of mice infected with GM6 was significantly higher than that of GM2 and ATCC700721 (Figure 5B); HE staining showed that GM6 infection caused extensive consolidation of the lungs and intra alveolar haemorrhage (Figure 5E-G). However, the survival rate was not affected if ampRKL47 was introduced into GM2 (Figure 5C).
Figure 5.
Virulence potential of hypermucoviscous Klebsiella pneumoniae isolates in a mouse pulmonary infection model. (A) The effect of 1 × 106 colony-forming units (CFU) of GM2, GM6 and GM2 ampRKL1 complement mutant on survival was assessed (n = 10; *P < 0.05, **P < 0.01, log-rank Mantel-Cox test),). (B) At 24 hours post-infection (hpi), the lungs of infected mice were harvested, and the bacterial burden was determined by CFU enumeration (n = 10; **P < 0.01, one-way ANOVA). Data are representative of 3 independent experiments. (C) The effect of 1 × 106 CFU of GM2 and GM2 ampRKL47 complement mutant on survival was assessed. (D) Uronic acid production of different strains. An unpaired two-sided Student’s t-test was performed. Each data point was repeated three times (n = 3). Data are presented as the mean ± s.e.m. *P < 0.05, ns = no significance. The lungs of mice without infection (E) or infected with GM2 (F) and GM6 (G) were collected and stained with haematoxylin and eosin. All images are at ×40 magnification. Scale bars: 250 μM.
Abbreviation: ara, arabinose.
Since the capsule is the main virulence factor of K. pneumonia and WcaJ which was highlighted according to proteome analysis result regulates the initial step of capsule synthesis, we investigated the amounts of capsular polysaccharide production. The result showed that the ampR-complementary strains AH1689-3:ampRKL47 and GM2:ampRKL1 plus arabinose produced significantly more capsule polysaccharide (uronic acid) than AH1689-3 and GM2, respectively (Figure 4C, Figure 5D).
Discussion
MLST has been the most commonly used technique for defining K. pneumoniae populations. With fast and affordable WGS, it is possible to compare whole genomes for isolate typing rather than just a few loci, as in traditional MLST. Genotyping with cgMLST uses thousands of alleles across the genome, resulting in a higher level of isolate discrimination. In this study, we used cgMLST to classify 135 clinical CRKP isolates and found differences between CT types based on clinical information. Due to the limited number of cases, we were unable to obtain an association between CT types and clinical outcome. However, the differences in mortality rates allowed us to further investigate the isolates belonging to CT3176. GWAS analysis showed that the ampR gene was significantly associated with the CT3176 isolates.
Previous studies have shown that the AmpC β-lactamase regulator AmpR, a member of the LysR family of transcription factors, also controls multiple virulence mechanisms in Pseudomonas aeruginosa.29,30 To date, only one article has reported the role of AmpR in regulating the virulence of K. pneumoniae. A clonal isolate of K. pneumoniae showed that few known virulence genes were responsible for severe infections. AmpR in these isolates was involved in the upregulation of capsule synthesis, modulated biofilm formation and type 3 fimbrial gene expression, as well as colonization of the murine gastrointestinal tract.31 However, the virulence level of these isolates remains unclear due to the lack of data on lethal infection models. In this study, we used a pneumonia model to confirm the enhanced virulence of these ampR-carrying isolates. This would explain why patients infected with these isolates had a high mortality rate.
Previously, it was common for ST11-type K. pneumoniae to be resistant to carbapenems but not hypervirulent. However, in 2017, an outbreak of hospital infections caused by ST11 carbapenem-resistant hypervirulent K. pneumoniae isolates was reported. The hypervirulence phenotype of these isolates was due to the acquisition of an approximately 170 kbp pLVPK-like virulence plasmid by classic CRKP isolates belonging to ST11 and serotype K47.15 In contrast to the above report, no virulence plasmids were found in ST11 CRKP isolates with enhanced virulence in this study, suggesting that the virulence enhancement was not due to the classical mechanism.32 Unlike the virulence plasmid, which contains multiple virulence factors, AmpR can increases virulence independently. This was confirmed by virulence testing of the ampR complement strain.
WcaJ is the enzyme that initiates colanic acid synthesis and loads the first sugar (glucose-1-P) on the lipid carrier undecaprenyl phosphate. The potential role of WcaJ in the virulence of K. pneumoniae was defined recently.33 The results of proteome analysis suggest that AmpR enhances the virulence of K. pneumoniae by regulating the initial step of capsule synthesis. As a regulator, AmpR needs to bind to the gene promoter to exert its regulatory role. To date, many capsular types have been identified in K. pneumoniae,34 and the absence of AmpR binding sites in some capsule types may explain why the virulence of KL1 isolates cannot be enhanced by the AmpR found in KL47 isolates.
New evidence has suggested that hypermucoviscosity and hypervirulence are different phenotypes that should not be used synonymously. Moreover, it is important to establish that a negative string test is insufficient in determining whether an isolate is hypervirulent.35 A key finding of our work was that we identified non-hypermucoviscous ST11 CRKP subclone with enhanced virulence.
The main limitation of this study is that there are only a small number of cases; therefore, we are unable to study the relationship between the clinical outcomes and non-hypermucoviscous hypervirulent CRKP infection. We were also unable to construct ampR knockout strain, and this may be explained by genomic analysis showed that ampR was putatively located on plasmid (Figure S3). Since strains with the non-hypermucoviscous phenotype are hardly distinguished in the clinic, continued surveillance and investigation of these novel strains are urgently needed.
Acknowledgment
We thank Shuye Xu, Chunxia Hu, Qiuyang Deng, Zhou Liu, Xiaoqian Hu, Wenhui Zhang, Nan Wang, Shiyi Liu, Ruiqin Cui, Zhen Hui, Yuxin Zhong, Yutian Luo, Huaisheng Chen, Weiyuan Wu, Jinsong Wu and Yuemei Lu for their help in this study.
Funding Statement
This work was supported by the International Collaborative Research Fund (GJHZ20180413181716797) and Free Inquiry Fund (JCYJ20180305163929948) of Shenzhen Science and Technology Innovation Commission.
Data Sharing Statement
We deposited the genome sequences in GenBank under BioProject PRJNA517992. The genome sequence of GM2 and GM6 were deposited in GenBank under BioProject PRJNA556307.
Ethics Statement
All animal care and use protocols in this study were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China. All animal experiments in this study were approved by the Animal Ethical and Experimental Committee of the Army Military Medical University (Chongqing, Permit No. 2011-04) in accordance with their rules and regulations.
Disclosure
The authors state that they have no conflicts of interest for this work.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeon JH, Lee JH, Lee JJ, et al. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int J Mol Sci. 2015;16:9654–9692. doi: 10.3390/ijms16059654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- 5.Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146:1913–1916. doi: 10.1001/archinte.1986.00360220057011 [DOI] [PubMed] [Google Scholar]
- 6.Fang CT, Lai SY, Yi WC, et al. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45:284–293. doi: 10.1086/519262 [DOI] [PubMed] [Google Scholar]
- 7.Wang JL, Chen KY, Fang CT, et al. Changing bacteriology of adult community-acquired lung abscess in Taiwan: klebsiella pneumoniae versus anaerobes. Clin Infect Dis. 2005;40:915–922. doi: 10.1086/428574 [DOI] [PubMed] [Google Scholar]
- 8.Chung DR, Lee SS, Lee HR, et al. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J Infect. 2007;54:578–583. doi: 10.1016/j.jinf.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 9.Tsai FC, Huang YT, Chang LY, et al. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis. 2008;14:1592–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YT, Jeng YY, Chen TL, et al. Bacteremic community acquired pneumonia due to Klebsiella pneumoniae: clinical and microbiological characteristics in Taiwan, 2001–2008. BMC Infect Dis. 2010;10:307. doi: 10.1186/1471-2334-10-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang WN, Huang CR, Lu CH, et al. Adult Klebsiella pneumoniae meningitis in Taiwan: an overview. Acta Neurol Taiwan. 2012;21:87–96. [PubMed] [Google Scholar]
- 12.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siu LK, Huang DB, Chiang T. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae.. BMC Infect Dis. 2014;14:176. doi: 10.1186/1471-2334-14-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2013;58:225–232. doi: 10.1093/cid/cit675 [DOI] [PubMed] [Google Scholar]
- 15.Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- 16.Fang CT, Chuang YP, Shun CT, et al. A novel virulence gene in Klebsiella pneumoniae isolates causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697–705. doi: 10.1084/jem.20030857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. Phenotypes Virulence. 2017;8:1111–1123. doi: 10.1080/21505594.2017.1317412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56:e00776–18. doi: 10.1128/JCM.00776-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Sixth Informational Supplement. CLSI Document M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 20.Antipov D, Korobeynikov A, McLean JS, et al. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 2016;32:1009−15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyres KL, Wick RR, Gorrie C, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jünemann S, Sedlazeck FJ, Prior K, et al. Updating benchtop sequencing performance comparison. Nat Biotechnol. 2013;31:294–296. doi: 10.1038/nbt.2522 [DOI] [PubMed] [Google Scholar]
- 23.Treangen TJ, Ondov BD, Koren S, et al. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:1–15. doi: 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:256–259. doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 26.Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brynildsrud O, Bohlin J, Scheffer L, et al. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:238. doi: 10.1186/s13059-016-1108-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domenico P, Schwartz S, Cunha BA. Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect Immun. 1989;57(12):3778–3782. doi: 10.1128/IAI.57.12.3778-3782.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong KF, Jayawardena SR, Indulkar SD, et al. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother. 2005;49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balasubramanian D, Kumari H, Jaric M, et al. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat shock and oxidative stress response. Nucleic Acids Res. 2014;42:979–998. doi: 10.1093/nar/gkt942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hennequin C, Robin F, Cabrolier N, et al. Characterization of a DHA-1-producing Klebsiella pneumoniae isolate involved in an outbreak and role of the AmpR regulator in virulence. Antimicrob Agents Chemother. 2012;56:288–294. doi: 10.1128/AAC.00164-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng HY, Chen YS, Wu CY, et al. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192:3144–3158. doi: 10.1128/JB.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai R, Wang G, Le S, et al. Three Capsular Polysaccharide Synthesis-Related Glucosyltransferases, GT-1, GT-2 and WcaJ, Are Associated With Virulence and Phage Sensitivity of Klebsiella pneumoniae. Front Microbiol. 2019;10:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan YJ, Lin TL, Chen CT, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep. 2015;5:15573. doi: 10.1038/srep15573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32:e00001–19. [DOI] [PMC free article] [PubMed] [Google Scholar]