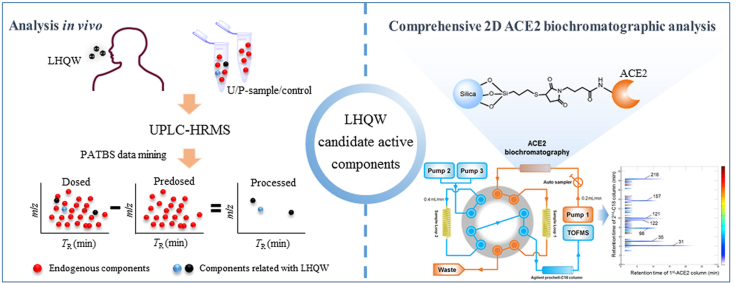
Abstract
Lianhuaqingwen (LHQW) capsule, a herb medicine product, has been clinically proved to be effective in coronavirus disease 2019 (COVID-19) pneumonia treatment. However, human exposure to LHQW components and their pharmacological effects remain largely unknown. Hence, this study aimed to determine human exposure to LHQW components and their anti-COVID-19 pharmacological activities. Analysis of LHQW component profiles in human plasma and urine after repeated therapeutic dosing was conducted using a combination of HRMS and an untargeted data-mining approach, leading to detection of 132 LHQW prototype and metabolite components, which were absorbed via the gastrointestinal tract and formed via biotransformation in human, respectively. Together with data from screening by comprehensive 2D angiotensin-converting enzyme 2 (ACE2) biochromatography, 8 components in LHQW that were exposed to human and had potential ACE2 targeting ability were identified for further pharmacodynamic evaluation. Results show that rhein, forsythoside A, forsythoside I, neochlorogenic acid and its isomers exhibited high inhibitory effect on ACE2. For the first time, this study provides chemical and biochemical evidence for exploring molecular mechanisms of therapeutic effects of LHQW capsule for the treatment of COVID-19 patients based on the components exposed to human. It also demonstrates the utility of the human exposure-based approach to identify pharmaceutically active components in Chinese herb medicines.
KEY WORDS: Lianhuaqingwen capsule, PATBS, COVID-19, ACE2, Biochromatography, Comprehensive 2D analysis, In vivo exposure, Surface plasma response, Molecular docking
Abbreviations: ACE2, angiotensin-converting enzyme 2; AT2, alveolar type II; COVID-19, corona virus disease 2019; ddMS2, data dependent tandem mass spectrometry 2; DMF, N,N-dimethylformamide; DMSO, dimethyl sulfoxide; ESI, electrospray ionization; GMBS, N-(4-maleimide butyryl oxide)succinimide; HPLC, high performance liquid chromatography; HRMS, high resolution mass spectrometry; LHQW, Lianhuaqingwen; MPTS, mercaptopropyltrimethoxysilane; NMPA, National Medical Products Administration; PATBS, precise-and-thorough background-subtraction; RAS, renin–angiotensin system; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SPR, surface plasmon resonance; TCM, traditional Chinese medicine; TIC, total ion chromatography; TOF/MS, time-of-flight mass spectrometry
Graphical abstract
This study provides chemical and biochemical evidence of LHQW capsule for the treatment of COVID-19 patients based on the components exposed to human.
1. Introduction
Recently, a new type of coronavirus disease 2019 (COVID-19), with rapid spread and high morbidity, broke out worldwide. Fortunately, a number of endeavors revealed that traditional Chinese medicine (TCM) treatment could effectively relieve symptoms and prevent the fatal deterioration of this disease1, 2, 3, 4. As a typical TCM formulation, Lianhuaqingwen (LHQW) capsule has been confirmed to show therapeutic effects by clinical research and observation5,6. More importantly, in April 2020, the “Pharmaceutical Supplement Application Document” issued by National Medical Products Administration (NMPA) showed that LHQW (granules formulation) were approved to treat fever, cough, or fatigue caused by the mild or common types of COVID-19. In details, the course of treatment is from 7 to 10 days, indicating the effectiveness of LHQW in treating COVID-19. However, the pharmaceutical components and pharmacological mechanism of LHQW in fighting COVID-19 are still largely unexplored.
LHQW is a patented Chinese medicine product developed for the treatment of cold and influenza7,8 and it was approved for marketing by the NMPA in 2004. LHQW has the functions of clearing away heat and detoxifying the lungs, and thus it is also clinically used to treat patients with the symptoms of fever and stagnation of the lung9. An anti-influenza A (H1N1) trial demonstrated that LHQW had better effect on the alleviation of fever, cough, sore throat, and fatigue, in comparison with oseltamivir. Furthermore, it showed comparative therapeutic effectiveness in reduction of illness duration and viral shedding duration8,10. Although molecular targets of LHQW remain unknown, pharmacological activities of LHQW have been explored in vitro. Ding et al.7 reported that the raw material of LHQW (dissolved in DMSO as stock solution) inhibited the proliferation of influenza viruses from various strains in vitro, with the 50% inhibitory concentration (IC50) ranging from 0.35 to 2 mg/mL. And it could also suppress virus-induced nuclear factor κB (NF-κB) activation and alleviate virus-induced gene expression of interleukin 6 (IL-6), interleukin 8 (IL-8), tumor necrosis factor-α (TNF-α), interferon-γ-inducible protein-10 (IP-10), and monocyte chemoattractant protein-1 (MCP-1). Using a similar in vitro approach, it was demonstrated that LHQW exerted anti-SARS-CoV-2 activity via inhibiting virus replication and reducing cytokine release from host cells5. Furthermore, networking analysis has been applied to identify main effective components in raw LHQW materials. For example, Wang et al.11 revealed 15 active components of LHQW and 61 related targets by using network pharmacology. Furthermore, You et al.12 found that the active components of LHQW might inhibit cytokine storm by regulating various inflammatory signal pathways by network pharmacology analysis. Just like a Western medicine product, it is well recognized that the efficacy and safety of TCM are associated with the chemical constituents of TCM including prototype components and their metabolites in the circulation, which are directly associated with the whole process of absorption, distribution, metabolism, and excretion (ADME)13,14. Identification of TCM components in animals or human circulation followed by activity testing of these components is another approach to study molecular mechanisms of pharmacological effects of TCM. For example, Wang et al.15 combined serum pharmacochemistry-based screening and high-resolution metabolomics analysis to find bioactive components. However, information on exposure, metabolism and disposition of LHQW in animals or human is not available in the literatures.
In addition to directly testing extracted raw materials from a TCM product or virtual network pharmacology analysis, phytochemical separation of individual components followed by pharmacological activity testing in vitro is a more common and useful practice in searching for active constitutes of an effective herbal medicine product16. However, this approach may not be able to focus on TCM components that are either absorbed in and exposed to human after oral administration or formed in human via biotransformation by metabolizing enzymes in human.
The first primary objective of this study was to determine the component profiles of LHQW in human plasma and urine, after oral administration of multiple therapeutic doses to human subjects. The LHQW formula was composed by 13 herb components: lianqiao—Forsythiae Fructus; jinyinhua—Lonicerae Japonicae Flos; zhimahuang—Ephedrae Herba Praeparata Cum Melle; chaokuxingren—Armeniacae Semen Amarum Tostum; shigao—Gypsum Fibrosum; banlangen—Isatidis Radix; mianmaguanzhong—Dryopteridis Crassirhizomatis Rhizoma; yuxingcao—Houttuyniae Herba; guanghuoxiang—Pogostemonis Herba; dahuang—Rhei Radix et Rhizoma; hongjingtian—Rhodiolae Crenulatae Radix et Rhizoma; bohenao—l-menthol; and gancao—Glycyrrhizae Radix et Rhizoma. Analysis of major constitutes in LHQW was conducted by using liquid chromatography coupled with high-resolution mass spectrometry (LC–HRMS), leading to unambiguous confirmation or tentative identification of 61 compounds, including flavonoids, phenylpropanoids, anthraquinones, triterpenoids, iridoids, and other types of compounds. Among these compounds, salidroside, chlorogenic acid, forsythoside E, cryptochlorogenic acid, amygdalin, sweroside, hyperin, rutin, forsythoside A, phillyrin (forsythin), rhein, and glycyrrhizic acid (glycyrrhizin) are considered chemical markers17. Detection and structural characterization of TCM components in complex biological matrix represent great analytical challenges, which are heavily dependent on LC–HRMS instrumentation and data-mining technologies. Although targeted data mining tools, such as extracted ion chromatography, product ion filter and mass defect filters, are routinely employed in find TCM metabolites in vivo18, metabolomics approach has been increasingly used for untargeted detection of TCM components14. In this study, we applied a previously developed analytical strategy for studying ADME of TCM in vivo19,20, which used precise-and-thorough background subtraction (PATBS) for sensitive and comprehensive detection and structural characterization of TCM components in complex biological samples.
The second primary objective of this study was to identify potential anti-COVID-19 pharmacological components of LHQW, that were exposure to human after multiple oral doses. SARS-CoV-2 was reported to achieve human infections through the binding of its spike protein (S protein) to angiotensin converting enzyme 2 (ACE2) as receptor21, which is a negative regulator with the ability to maintain the steady state of renin-angiotensin system (RAS) and is critical to the physiology or pathology of all organs22. ACE2 has been found to be widely distributed in the heart, kidney, testis, adipose tissue, brain tissue, vascular smooth muscle cells, and gastrointestinal tract23. Zhao et al.24 applied high-throughput single-cell sequencing analysis technique to study 43,134 lung cells and found that ACE2 was expressed in 0.64% lung cells, and in 1.4% alveolar type II cells (AT2), which became the binding targets of SARS-CoV-2 as an invasion starting point. Furthermore, Xu et al.25 confirmed that ACE2 is a viral receptor by studying the binding capacity of the SARS-CoV-2 protein crystal structure to human ACE2 receptor. In general, the above studies indicated that screening drugs with ACE2 targeting ability are promising for the treatment of patients with SARS-CoV-2 infection. Hence, comprehensive 2D ACE2 biochromatography system was established for efficiently screening for active LHQW components exposed to human after repeated oral does. The ACE2 inhibitory activity of hits from the biochromatography screening was further confirmed by using the pharmacodynamics study of surface plasmon resonance (SPR) and analysis using ACE2 inhibitory activity assay kit as well as supported by results from docking simulation.
2. Methods and materials
2.1. Chemicals and materials
Methanol (batch No.: 19105007) and acetonitrile (batch No.: 10105007) were purchased from TEDIA (Fairfield, OH, USA). Formic acid (batch No.: 73C1707EN) was purchased from Anaqua Chemical Supply (Wilmington, DE, USA). Ultra-pure water was prepared by Milli-Q water purification system (Bedford, MA, USA). Oasis® HLB 3cc (60 mg) extraction cartridges (batch No.: 164A39127A) was from Waters (Milford, MA, USA). Silica gel (5 μmol/L, 200 Å) was purchased from Qingdao Meigao Chemical Co., Ltd. (Qingdao, China). N,N-Dimethylformamide (DMF), mercaptopropyltrimethoxysilane (MPTS), N-(4-maleimide butyryl oxide)succinimide (GMBS) and dimethyl sulfoxide (DMSO) were purchased from Sigma (St. Louis, MO, USA). ACE2 recombinant protein (human, His tag, Cat: 10108-H08H) was obtained from Sino Biological Co. (Beijing, China). ACE2 primary antibody (rabbit IgG) was purchased from Abmart Biological Co. (Shanghai, China). MLN-4760 (CAS#305335-31-3) was purchased from MedChemExpress (Monmouth Junction, NJ, USA). TCM reference substances, ephedrine and pseudoephedrine, were supplied by National Institutes for Food and Drug Control (Beijing, China); (R,S)-goitrin, salidroside, forsythoside E, amygdalin, secoxyloganin, liquiritin, forsythoside I, luteolin-7-O-glucoside, forsythoside B, forsythoside H, forsythoside A, isoquercitrin, kaempferol-3-O-neohesperidoside, quercitrin, forsythin, quercetin, phillygenin, aloe-emodin, glycyrrhizin, rhein, emodin, chrysophanol and physcion were supplied by Baoji Chenguang Biotechnology Co., Ltd. (Baoji, China); prunasin was supplied by BioBioPha (Yunnan, China); gallic acid, neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, rutin, kaempferol-3-O-rutinoside, emodin 8-O-β-d-glucoside, kaempferol were supplied by Chengdu Purechem-Standard Company (Chengdu, China).
2.2. Collection of clinical samples
A single-center, randomized, open, and multiple-dose trial was designed to enroll 14 healthy subjects. Each subject was confirmed (by physical examination, laboratory examination, or electrocardiogram) to have no heart, liver and kidney diseases; no history of allergies, and no LHQW or other drug or food administration, which might affect drug absorption, distribution, metabolism, and excretion within one month before the test. This study protocols were approved by the Ethics Committee of Hebei Yiling Hospital (Shijiazhuang, China), and all subjects signed a written informed consent before entering this study (the clinical trial registration number: 2019LS-002). This test was conducted by continuous administration (the dose was 4.2 g, 12 capsules), which was given on Days 1–8, except Day 2. Furthermore, the sampling (blood and urine) was taken on Days 1 and 2 after administration, while the blood samples were collected on Days 3–7 before each administration as well as on Day 8 (blood and urine). And the overall test period was 11 days. The subjects were requested to have their breakfast 1 h before drug administration, and to finish within 30 min before drug administration with 240 mL warm water. And the collected plasma and urine samples were stored in −80 °C refrigerator before testing.
2.3. Sample preparation
10 mg powder of LHQW capsule was dissolved in 2 mL 70% methanol, diluted one time and filtered by 0.22 μm filter membrane to obtain a final LHQW test solution of 2.5 mg/mL.
The plasma samples from 6 subjects (subject numbers 1, 3, 6, 8, 9, and 12) on Day 8 were prepared as follows. The plasma samples from subjects taken at 10, 30 min, 1, 3, 6, 9, 12, 16, 25, or 48 h after drug dosing (50 μL plasma at each time point) were mixed to obtain a 3-mL sample. After dilution for 3 times with ultrapure water, the samples were respectively enriched and purified by Oasis® HLB 3cc (60 mg) extraction cartridges. The methanol eluent was collected and dried with nitrogen. Reconstitutes were obtained by re-dissolving the residues using 100 μL 70% methanol. After 10 s of ultrasonication, vortex mixing, and 12,000 rpm centrifugation (D3024, DLAB Scientific Co., Beijing, China) for 5 min, the supernatants were collected for further analysis. Before administration, blank plasma samples were prepared in a similar manner.
The urine samples from 6 subjects (subject numbers 1, 3, 6, 8, 9, and 12) on Day 8 were prepared as follows. The urine samples from subjects collected in the periods of 0–4, 4–10, 10–24, 24–32, or 32–48 h after drug dosing (100 μL urine at each time point) were mixed to obtain a 3-mL sample. Furthermore, the samples were respectively enriched and purified by Oasis® HLB 3cc (60 mg) extraction cartridges. The methanol eluent was collected and dried with nitrogen at 37 °C. Reconstitutes were obtained by re-dissolving the residues using 100 μL 70% methanol. After 10 s of ultrasonication, vortex mixing, and 12,000 rpm centrifugation (D3024, DLAB Scientific Co., Beijing, China) for 5 min, the supernatants were collected for further analysis. Before administration, blank urine samples were prepared in a similar manner.
2.4. UPLC–HRMS analysis
To determine component profiles of LHQW in human body, human plasma and urine samples were detected with Thermo Fisher Q Exactive Orbitrap LC–MS/MS equipped with electrospray ionization (ESI, Thermo Fisher Scientific, Waltham, MA, USA) in positive and negative ion modes which was controlled by Thermo Xcalibur 3.0.63 (Thermo Fisher Scientific). An ACQUITY UPLC CSH C18 column (50 mm × 2.1 mm, 1.7 μm) was used to separate the sample with the temperature set at 35 °C. Mobile phase A was H2O with 0.1% formic acid, and mobile phase B was 100% acetonitrile with a flow rate of 0.3 mL/min. The gradient was set as follows: 0–3 min (2% B); 3–4 min (2%–5% B); 4–6 min (5% B); 6–9 min (5%–10% B); 9–14 min (10%–15% B); 14–18 min (15%–20% B); 18–25 min (20%–30% B); 25–28 min (30%–95% B); 28–32 min (95% B); 32.01–37 min (5% B). The injection volume was set as 3 μL, and the eluent was monitored at λ 205, 254 and 310 nm.
In order to optimize the mass spectrometry conditions, 8 reference materials with different structures were selected (ephedrine and (R,S)-goitrin were chosen in positive ion mode; while forsythoside A, forsythin, rhein, glycyrrhizin, amygdalin, and luteolin-7-O-glucoside were chosen in negative ion mode). These references at the concentration of 10 μg/mL and velocity of 3 μL/min were utilized to optimize the MS conditions. The optimizations included the proper ionization mode (positive ion mode/negative ion mode), spray voltage, capillary temperature, capillary voltage, RF lens, auxiliary gas, and sweep gas flow rate. Full MS and data dependent tandem mass spectrometry 2 (ddMS2) in both positive and negative ion mode were used to acquire full scan MS and MS data of LHQW raw materials, plasma and urine samples. The scan mass range 100–1200 Da was selected. High resolution full-scan MS and MS2 data were collected at resolving power of 35,000 and 17,500, respectively. Stepped HCD collision energy of 35% was used. The temperature of capillary was set at 320 °C, and the spray voltage was 3.5 kV for negative ion mode, and 3.8 kV for positive ion mode. The flow rate of sheath gas (N2) and auxiliary gas (N2) were 40 and 10 arb, respectively. The data obtained were analyzed by Thermo Xcalibur 3.0.63 Qual Browser (Thermo Fisher Scientific).
After collection of MS and MS/MS data of the LHQW test solution, structural characterization and confirmation of LHQW constituents were carried out by using Compound Discoverer 3.1 software (Thermo Fisher Scientific). The main parameters of this software were set as follows: align retention times, maximum shift 2 min; mass tolerance, 5 ppm; detect compound, mass tolerance 5 ppm; intensity tolerance, 30%; minimal peak intensity, 500,000; data sources, mzCloud, mzVault, MassList, and ChemSpider Search; and S/N threshold, 3, after which the compounds searching and matching were conducted. And then, the software searching report was combined with the fragmentation rule of reference MS as well as the corresponding characteristic fragment ion information. In addition, 34 reference standards were used for structural confirmation.
Detection and structural characterization of LHQW components in human plasma and urine were carried out by a LC–HRMS workflow, which was previously developed for ADME study of a TCM product in vivo19. Briefly, the workflow used PATBS, an untargeted data-mining technology, to detect LHQW components in a plasma or urine sample. Once the LHQW components were found in full scan LC–HRMS dataset, their MS/MS spectral data were either retrieved from the MS/MS datasets acquired by ddMS2 methods or acquired via targeted production ion scanning analysis. And then, structural conformation of prototype components of LHQW in the plasma and urine samples was accomplished by comparing their LC–HRMS data with LHQW test solution. Meanwhile, metabolites of LHQW components were structurally characterized using some functions of Compound Discoverer 3.1 software, including biotransformation matching, spectral similarity comparison and fragmentation interpretation.
2.5. Synthesis of ACE2 biochromatographic stationary phase
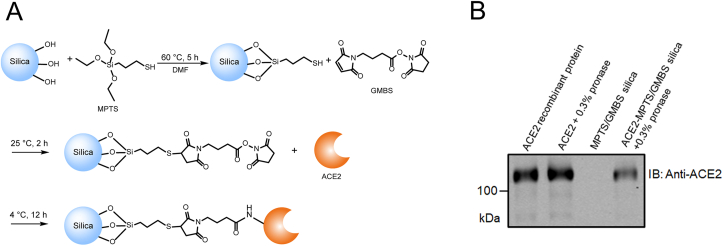
The reaction scheme was shown in Fig. 1A, according to the method developed in our previous research with slight modification26. Briefly, 1 mL MPTS was added to 1 g silica gel and stirred for 5 h in 100 mL DMF under the condition of 60 °C and nitrogen protection. The suspension was cleaned with DMF at 5000×g for three times. Then the precipitate was obtained and reacted with 5% GMBS in 500 mL DMSO solution for 2 h. The MPTS/GMBS modified silica gel was cleaned with DMSO at 5000×g for three times and dried in the vacuum drying pump for 48 h before use. ACE2 recombinant protein (20 μg) was mixed with MPTS/GMBS-modified silica gel (40 mg) in PBS for continuous stirring for 12 h at 4 °C, realizing the covalent immobilization of ACE2 protein on silica stationary phase.
Figure 1.
(A) Synthetic route of MPTS/GMBS-modified silica gel and ACE2 biochromatographic stationary phase; (B) ACE2 biochromatographic stationary phase was digested by 0.3% pronase and lysed by RIPA buffer, then immunoblotted with the anti-ACE2 antibody.
2.6. Comprehensive two-dimensional (2D) ACE2 column/C18 column/time-of-flight mass spectrometry (TOFMS) system
The comprehensive 2D ACE2 column/C18 column/TOFMS system was performed on an Agilent 1200 series HPLC system coupled with a 6220 TOF mass spectrometry consisting of an auto-sampler, which was controlled by Agilent MassHunter Workstation (Agilent Technologies, Palo Alto, CA, USA). As shown in Supporting Information Fig. S1, the ACE2 column (10 mm × 2.0 mm i.d., 5 μm) was equipped as the first dimensional column with 10 mmol/L ammonia acetate as the mobile phase, and the flow rate set at 0.2 mL/min. For the second dimension, an Agilent Poroshell 120 EC-C18 column (150 mm × 3.0 mm i.d., 2.7 μm) was applied with the mobile phase composed of solvent A (0.1% formic acid) and solvent B (acetonitrile) at 0.4 mL/min. The linear gradient elution program is: 0–12 min, 5%–20% B; 12.01–17 min, 20%–70% B; 17.01–20 min, 5% B. Other detailed parameters, modules and operations of the comprehensive 2D system were described in our previous studies27,28.
2.7. SPR analysis
SPR assays were performed on Biacore T200 system (GE Healthcare Life Sciences, Marlborough, MA, USA). ACE2 recombinant protein with HIS label was diluted with acetate (pH = 4.0) and conjugated to a Series S Sensor Chip CM5 (BR100530, GE Healthcare Life Sciences) by EDC/NHS cross-linking reaction according to manufacturer's protocols. The target immobilization level of ACE2 protein was 8000 RU. Small molecules were diluted with a running buffer containing 5% DMSO from 0.024 to 50 μmol/L, and was injected into the reference channel and ACE2 protein channel, respectively, at a flow rate of 30 μL/min. The coupling and dissociation time were both 120 s. Biacore T200 evaluation software was used to fit the affinity curves by the steady-state affinity model (1:1), and the equilibrium dissociation constant (KD) was calculated.
2.8. Test of ACE2 inhibitory activity
The ACE2 inhibitory activities of the candidate active components were tested by ACE2 Inhibitor Screening Kit (Catalog #K310; BioVision Inc., Mountain View, CA, USA) according to the manufacturer's written instructions. All data points were tested with one duplication and were expressed by mean values. The IC50 was calculated by the nonlinear fit tool of GraphPad Prism 7.00.2.9 (La Jolla, CA, USA).
2.9. Molecular docking assay
X-ray crystal structure of protein ACE2 was downloaded from the Protein Data Bank (PDB code: 6M0J) database. Glide in Schrödinger v2019.03 (Schrödinger Inc., Plainview, NY, USA) was used for molecular docking simulations of ligand–protein interactions and predicting the binding affinity and sites with ACE2. The 2D structures of the molecules were chemically standardized (including adding hydrogen atoms, ionizing at the pH range from 5.1 to 9.1, and generating stereoisomers and valid single 3D conformers) by means of the LigPrep module in Maestro. The structure of ACE2 was manipulated with the “Protein Preparation Wizard” workflow in Maestro. The main manipulations are removing all water molecules, protonation, and optimization based on OPLS_2005 force field. The Glide module is used to generate a protein receptor's grid file to determine the active site of the protein in order to mimic the binding of ligands and proteins. The grid encloses a box centered on the binding sites of SARS-CoV-2 S protein or active sites of ACE2 peptidase with a dimension of 10×10×10 (x×y×z, Å). The scaling factor of 0.8 was set for van der Waals radii of receptor atoms with a partial atomic charge less than 0.15. Extra precision of Glide docking procedure (Glide-XP) was employed for docking the compounds to the binding site of protein and the best docking pose of the compound was kept based upon Glide scoring function (G-score).
3. Results
3.1. UPLC–HRMS analysis of chemical constituents of LHQW
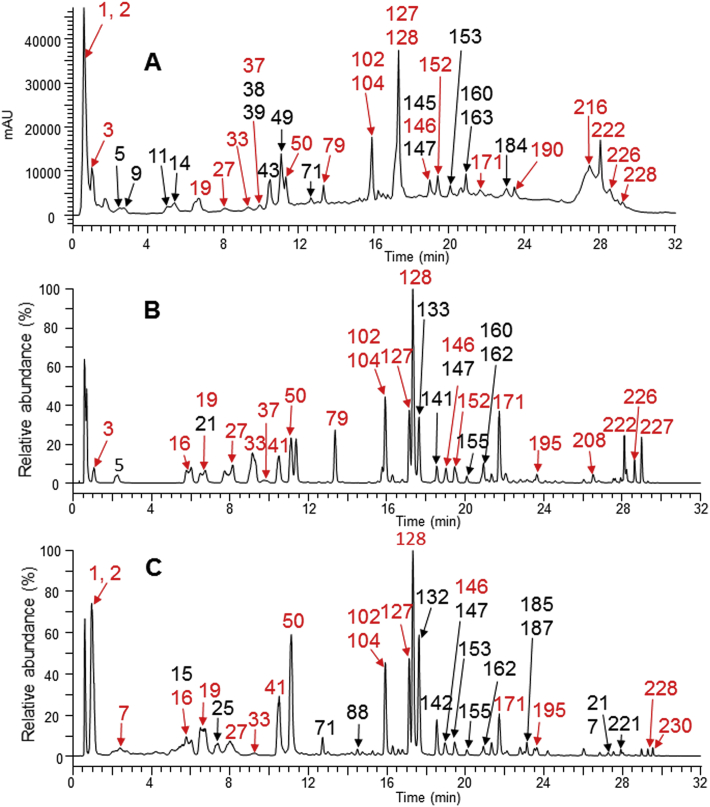
To facilitate the identification of LHQW components in human plasma and urine, LHQW test solution was first analyzed by the UHPLC–HRMS. As shown in Fig. 2, a total of 185 significant constituents were found in the LHQW test solution. Their accurate MS and MS/MS data were summarized in Supporting Information Table S1. Based on spectral interpretation and database search, 126 constituents were structurally characterized, among which 34 constituents have been confirmed in comparison with the references (Supporting Information Fig. S2).
Figure 2.
Analysis of LHQW components in the test solution by UPLC-UV–HRMS. (A) UPLC–UV chromatogram (254 nm); (B) Negative ion mode high resolution total ion current chromatogram; (C) Positive ion mode high resolution total ion current chromatogram. The peaks with red labels show that the constituents have been confirmed in comparison with the references.
3.2. Detection and characterization of LHQW components in human plasma and urine
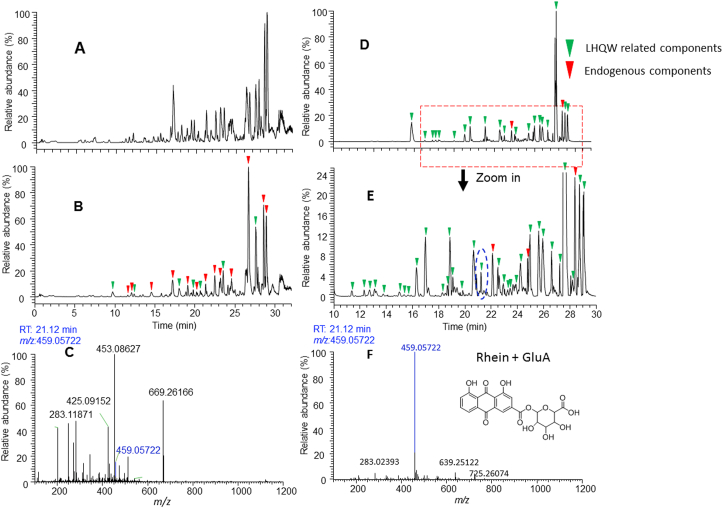
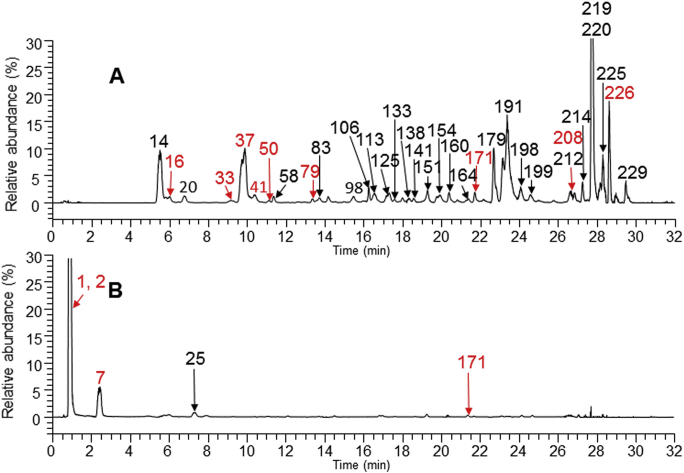
An unprocessed chromatographic profile of a human urine sample after LHQW administration (Fig. 3B) shows that a majority of peaks were endogenous components as those in a control urine sample (Fig. 3A) although several LHQW components were detected. In contrast, a PATBS-processed chromatographic profile of the same urine sample was able to reveal about 106 LHQW-related components (Fig. 3D and E). In addition, PATBS processing significantly improved quality of full-scan MS dataset. For an example, full-scan MS spectrum of a LHQW component (Rhein-GluA) shows a single molecular ion of Rhein-GluA without any significant interference ion species (Fig. 3C and F). Consequently, accurate MS/MS spectral data of the detected LHQW components in the urine sample were retrieved from recorded MS/MS datasets or acquired by additional injections. Structural characterization of the LHQW-related components were further accomplished by comparing their LC–HRMS data with those of LHQW test solution (Fig. 2), reference standards, as well as spectral interpterion and biotransformation marching. As a result, 70 LHQW components in the human urine were tentatively identified or confirmed (Table S1).
Figure 3.
Analysis of LHQW components in a human urine sample using UPLC–HRMS (negative ion mode) and PATBS data-processing. (A) Chromatography profile of full scan MS analysis a blank urine sample (predosed sample). (B) Chromatography profile of full scan MS analysis the human urine sample. (C) Full scan MS spectrum of a LHQW component (Rhein+GluA (164)) at 21.12 min from unprocessed LC–MS dataset of the urine sample. (D) PATBS-processed chromatography profile from full scan MS analysis of the urine sample. (E) The partially enlarged view of (D). (F) Full scan MS spectrum of the LHQW component [Rhein+GluA (164)] from PATBS-processed LC–MS dataset of the urine sample. Green triangle indicates LHQW prototype or metabolite components in urine, while the red triangle indicates the endogenous constituents.
Fig. 4A and B are representatively extracted ion chromatograms of LHQW components in negative ion and positive ion modes, respectively, in a human plasma sample after oral administration of repeated doses. A total of 107 LHQW compounds were found in human plasma, including 66 prototype components and 41 metabolite components, among which 72 components were structurally characterized (Table S1). In addition to detection and structural characterization, the UPLC–HRMS profiles provides semi-quantitative information on the LHQW components, while LC–UV profile of the plasma does not exhibit a majority of these LHQW components due to low concentration in human plasma. Based on the results of analyzing urine (Supporting Information Fig. S3) and plasma samples, 87 significant LHQW prototype components were absorbed after repeated oral administration, and 45 LHQW metabolite components were formed in humans (Table S1). Thus, we considered that a total of 132 LHQW related components were exposed to human after repeated oral administration of LHQW.
Figure 4.
High-resolution extracted ion chromatograms of LHQW components in human plasma sample. (A) Data from UPLC–HRMS analysis in negative ion mode. (B) Data from UPLC–HRMS analysis in positive ion mode. The peaks with red labels show that the constituents have been confirmed in comparison with the references.
3.3. Synthesis and characterization of ACE2 biochromatographic stationary phase
MPTS and GMBS were applied for biochromatographic stationary phase modification, in order to achieve covalent immobilization of ACE2 recombinant protein for screening potential anti-COVID-19 components from LHQW. Using this strategy, the interaction yield of mercapto with the maleamide group was high under mild reaction conditions and the self-polymerization could be avoided26. The exposed N-hydroxysuccinimide groups were practical linkages for amine coupling of target proteins with high recovery and stable chemical environment29. As shown in Fig. 1B, ACE2 antibody-based immunoblotting shows that the immobilized ACE2 could be digested by 0.3% pronase from the ACE2 biochromatographic stationary phase, indicating the successful covalent immobilization of ACE2. The content of immobilized membrane protein on 40 mg MPTS/GMBS modified silica gel was measured as 18.24 ± 1.05 μg, while a good protein recovery of >90% was achieved.
3.4. Suitability of the comprehensive 2D ACE2 column/C18 column/TOFMS system
As shown in Fig. S1A, the first fraction from the ACE2 biochromatography column was enriched into a 500 μL sample loop 1 at the 10-port valve switched at position 1. After switching to position 2 (Fig. S1B), the enriched components in sample loop 1 were pumped into an Agilent Poroshell EC-C18 column and TOFMS detector, for further separation and qualitative analysis of chemical components. Meanwhile, the second fraction of ACE2 column was pumped into sample loop 2 and waited for analysis, with the alternate switching of 10-port valve. In order to get comprehensive 2D biochromatographic analysis results, it was necessary to collect at least two fractions across a retention peak. Hence, the collection period was set at 2.5 min for each round of analysis. This novel comprehensive 2D ACE2 column/C18 column/TOFMS system realizes the automated and high-throughput analytical process, rapid separation, and accurate identification, which are helpful in discovering potential ACE2 target ligands from complex chemical samples.
3.5. Screening of ACE2 binding components from enriched urine sample and extract of LHQW
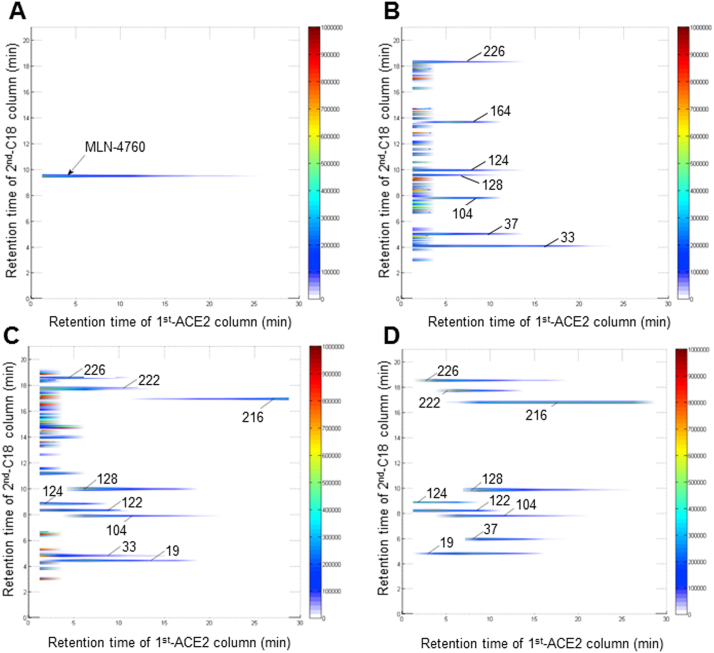
The comprehensive 2D ACE2 column/C18 column/TOFMS system was applied into screening potential ACE2 target components from enriched urine sample and extract of LHQW. As shown in Fig. 5A, MLN-4760, a selective ACE2 inhibitor, had significant retention on ACE2 biochromatography, indicating the specificity of this screening model. Furthermore, urine samples of LHQW were combined and enriched for biochromatographic analysis. Interestingly, 7 significant retention components were screened out after deducting the blank urine background (Fig. 5B). More importantly, the complex components from the extract could also be characterized by the 2D spectrum, obtained from the comprehensive 2D ACE2 biochromatography system (Fig. 5C). In details, these prototype components and their metabolites were tentatively identified by TOFMS according to the “generate formula from mass peak” function of Agilent MassHunter Qualitative Analysis software and the data of fragment ions. As shown in Supporting Information Table S2, 10 candidate active components (8 prototype components and 2 metabolites) with retention behaviors from 1st dimensional ACE2 biochromatography column were listed. Furthermore, as shown in Fig. 5D, 8 prototype components and prunasin (metabolite of amygdalin) were validated by authentic standard compounds in this model, which had similar retention behaviors with that in urine sample and LHQW extract, indicating these components could have binding affinity to ACE2 protein. Experimental results reveal that the comprehensive 2D ACE2 bichromatography system is a practical approach for rapid recognizing ACE2 target components from complex TCM extract. It could also be applied to other TCM formulas or chemical libraries for discovering anti-SARS-CoV-2 virus candidate drugs. However, this method is a preliminary screening tool for complex chemical sample, which could not obtain the definite interaction parameters between ligand and protein. Thus, the detailed parameters of the interactions between candidate components with ACE2 required further validation.
Figure 5.
2D contour plots of (A) Positive drug, MLN-4760, (B) enriched urine samples, (C) LHQW extracts and (D) standard compounds of potential active components obtained by the comprehensive 2D ACE2 column/C18 column/TOFMS system.
Based on the results of in vivo exposure and preliminary screening of ACE2 bichromatography, 8 candidate active components with potential ACE2 binding activity were obtained, including neochlorogenic acid and its isomers (19), amygdalin (33), prunasin (37), forsythoside I (104), rutin (124), forsythoside A (128), glycyrrhizin (222) and rhein (226).
3.6. SPR analysis of candidate constituents
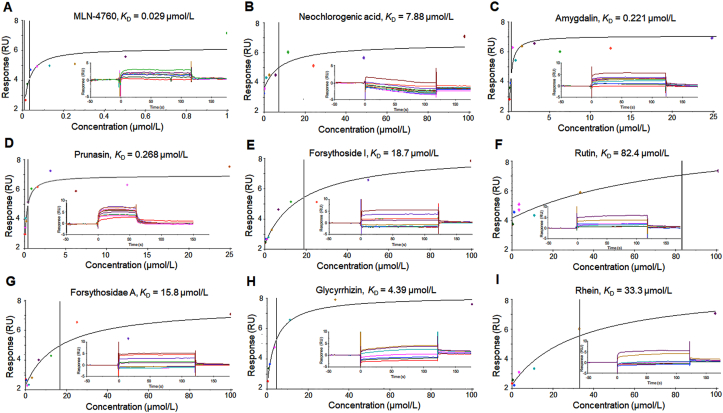
SPR affinity assays were performed to validate the affinities of 8 candidate active components to ACE2 recombinant protein. As shown in Fig. 6A, MLN-4760 showed a strong affinity to ACE2 recombinant protein on SPR chips with 0.029 μmol/L, indicating the SPR analytical system has good selectivity for screening potential ACE2 inhibitors. The sensorgrams and KD fitting curves of neochlorogenic acid (19), amygdalin (33), prunasin (37), forsythoside I (104), rutin (124), forsythoside A (128), glycyrrhizin (222) and rhein (226) are shown in Fig. 6B–I, respectively. The detailed SPR assay results of candidate active components were listed in Table 1. We found that amygdalin and prunasin had strong binding affinities of KD = 0.221 and 0.268 μmol/L. Prunasin is the major metabolite of amygdalin in vivo, and has very high exposure in human serum. Interestingly, glycyrrhizin has been considered as an efficacious therapeutic agent for COVID-19 with potential ACE2 binding activity30. In this study, we confirmed that the KD value of glycyrrhizin was 4.39 μmol/L. Forsythoside A and forsythoside I were the major components in Forsythiae Fructus, their affinities to ACE2 were assayed as 15.8 and 18.7 μmol/L, respectively. Rhein was another component with high exposure in human serum, which was also found having a moderate affinity to ACE2 with KD value calculated as 33.3 μmol/L. Due to the high exposure concentration of rhein in human body, physcion, aloe-emodin and emodin 8-O-β-d-glucoside, with structural similarity with rhein, were also analyzed by SPR. The experimental results show that they could also exhibit ACE2 binding activity, indicating ACE2 binding potentials of rhubarb anthraquinone compounds. Furthermore, rutin and its deglycosylated compounds (quercetin) had also been proved to show certain binding ability with ACE2. The sensorgrams of another five components with affinities to ACE2 protein were shown in Supporting Information Fig. S4A–S4E. According to the general criteria31, the situation of Rmax value exceeding 2 times of the response of compounds should be considered as nonspecific binding ligands. As shown in Table 1, the Rmax values of all positive components were less than 25 RU, conforming to the characteristic of specific binding. The χ2 values were all less than 10, indicating the accuracy of the fitting curves of each compound. As SPR is the gold standard for detecting drug–target interactions, these results indicate that all the candidate active components screened by the comprehensive 2D ACE2 biochromatography system could directly bind to ACE2 recombinant protein with ideal affinities, which reveals the efficiency of screening target constituents from complex chemical samples by this system. It is worth mentioning that, although 13 main constituents of LHQW was screened [ephedrine (1), pseudoephedrine (2), gallic acid (3), (R,S)-goitrin (7), salidroside (16), forsythoside E (27), cryptochlorogenic acid (41), secoxyloganin (79), liquiritin (102), forsythoside B (122), quercitrin (152), forsythin (171) and emodin (227)] by SPR, none of these compounds exhibited ACE2 binding ability, except forsythoside B, a structural analogue of forsythoside A, shows low binding activity (Fig. S4C). This result might indicate the high efficiency of using biochromatography as analysis strategy from the perspective of human exposure in the discovery of active compounds of TCM. Among these 13 constituents, ephedrine (1), pseudoephedrine (2), (R,S)-goitrin (7), salidroside (16), quercitrin (152) and forsythin (171) have higher human exposure. Although they did not show ACE2 binding activity, their higher in vivo exposure reveals that the pharmacological effects of LHQW might also be synergistically exerted from other targets, indicating the characteristics of the TCM's multi-target effects.
Figure 6.
SPR sensorgrams and the fitting curves of equilibrium dissociation constant (KD) of (A) MLN-4760, (B) neochlorogenic acid, (C) amygdalin, (D) prunasin, (E) forsythoside I, (F) rutin, (G) forsythoside A, (H) glycyrrhizin, and (I) rhein.
Table 1.
Affinities of candidate active components targeting ACE2 protein obtained by SPR analysis.
| Compd. | KD (μmol/L) | Rmax (RU) | χ2 |
|---|---|---|---|
| Neochlorogenic acid (19) | 7.880 | 6.894 | 1.260 |
| Amygdalin (33) | 0.221 | 6.856 | 0.462 |
| Prunasin (37) | 0.268 | 3.814 | 0.244 |
| Forsythoside I (104) | 18.700 | 19.020 | 1.910 |
| Rutin (124) | 82.400 | 12.770 | 1.290 |
| Forsythoside B (125) | 61.000 | 10.620 | 0.328 |
| Forsythoside A (128) | 15.800 | 21.490 | 5.280 |
| Quercetin (190) | 21.100 | 12.780 | 1.580 |
| Emodin 8-O-β-d-glucoside (195) | 21.270 | 16.930 | 3.190 |
| Aloe-emodin (216) | 6.020 | 16.120 | 7.720 |
| Glycyrrhizin (222) | 4.390 | 9.614 | 0.321 |
| Rhein (226) | 33.300 | 13.200 | 0.669 |
| Physcion (230) | 0.379 | 18.390 | 1.630 |
3.7. Inhibition of ACE2 activity by ACE2 binding components
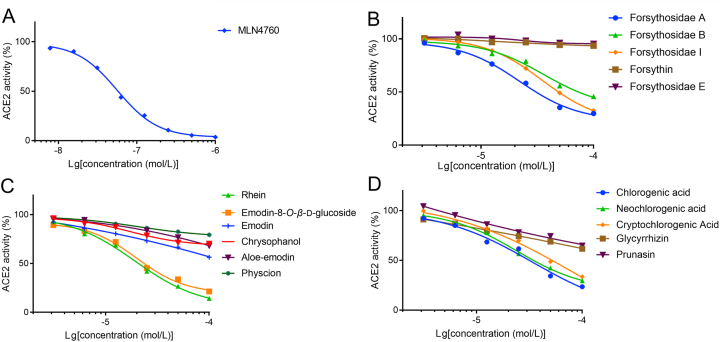
The effects of positive drug and all candidate components on the activity of ACE2 were tested using the ACE2 Inhibitor Screening Kit. Based on the SPR results, rhubarb anthraquinone compounds, forsythiaside compounds, neochlorogenic acid and its isomers, as well as prunasin and glycyrrhizin with high in vivo exposure levels were selected. As shown in Fig. 7A, the selective ACE2 inhibitor MLN-4760 has strong inhibitory effect with IC50 at 0.063 μmol/L. The three forsythosides A, B and I screened out by biochromatography and SPR analysis all have ACE2 inhibitory effects and the molecular docking confirmed that they could bind to the typical peptidase active sites. While two compounds, forsythin and forsythoside E, with similar structures but no affinities, have no ACE2 inhibitory activities (Fig. 7B). Rhein and emodin 8-O-β-d-glucoside showed good ACE2 inhibitory effects with IC50 at 18.33 and 22.50 μmol/L, respectively, while the other major components in Rhei Radix et Rhizoma did not show significant inhibitory effects (Fig. 7C). Moreover, as shown in Fig. 7D, three chlorogenic acid isomers all reveal moderate ACE2 inhibitory activities with IC50 at about 40 μmol/L, while the two components prunasin and glycyrrhizin, with strong ACE2 binding affinities, did not show significant peptidase inhibitory effects. They failed to bind with any of the peptidase active site, which was confirmed by molecular docking assays. It is worth mentioning that the in vivo exposure concentration of rhein is relatively high, with good activity, indicating that it might be the main active ingredient of LHQW to exert ACE2 inhibition ability. Other active compounds, such as prunasin and glycyrrhizin, had high in vivo exposure concentrations but low inhibitory activity; while forsythosides A and I, or neochlorogenic acid and its isomers had good in vitro activity, but low in vivo exposure, which might show synergistic effects with rhein. This might also reflect the combined synergistic effect of TCM.
Figure 7.
The inhibitory effects of candidate components on ACE2 enzymatic activity. (A) positive drug MLN-4760, (B) candidate components from Forsythiae Fructus, (C) Rhei Radix et Rhizoma, and (D) Lonicerae Japonicae Flos/Armeniacae Semen Amarum Tostum/Glycyrrhizae Radix et Rhizoma.
3.8. Molecular docking assays
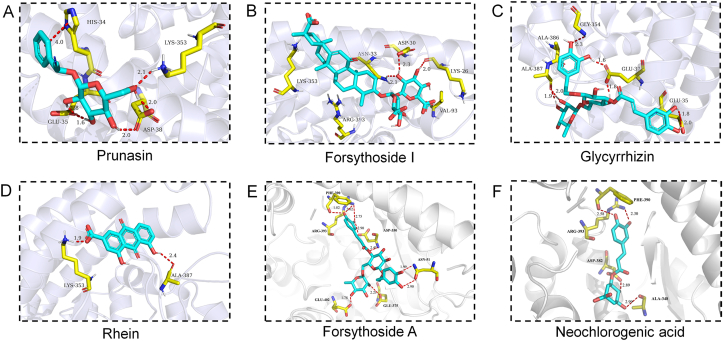
Docking simulation studies were carried out to investigate the binding sites of six representative components. Previous studies showed that the main interaction interface of ACE2 that binding with the RBD region of S protein are Gln24 to Tyr83 and Gln325 to Arg39332,33. It is worth mentioning that the peptidyl dipeptidase activity sites were widely distributed in various regions at the N-terminal of ACE2 protein, including Trp271 to Arg273, His345 to Ala387, and His505 to Arg51434,35. The potential intersection region of peptidase activity and S protein binding is around His345 to Arg393. Thus, the compounds that block S protein binding to ACE2 may not have strong catalytic activity to inhibit ACE2 peptidase36. Recent reports showed that some potential ACE2 inhibitors screened by computer simulations do not directly bind to the interface between ACE2 and spike protein complex37, indicating that these compounds were not suitable for inhibiting SARS-CoV-2 infection. In this case, it may be worthy to conduct further exploration to identify whether their specific binding site is directly bound to the contact surface between ACE2 and spike complex or ACE2 peptidase active sites, as this interaction might determine the compound's molecular mechanism in inhibiting SARS-CoV-2 infection21. As shown in Fig. 8A–F, six components with strong affinities, prunasin, forsythoside I, glycyrrhizin, rhein, forsythoside A and neochlorogenic acid, could all form suitable steric complementarities with the binding interface of ACE2 with SARS-CoV-2 spike protein. Prunasin, a major metabolite of amygdalin in serum, its glucoside and alkinyl groups contribute to the affinity of the key binding interface of ACE2 and S protein (His34 to Asp38). Forsythosides A and I, a pair of isomers, all have good affinities and inhibitory effects to ACE2 protein. The docking results show that they could bind to the intersection region of peptidase activity and S protein binding (Asp350 to Arg393), while forsythoside B with an additional pentose showed lower affinity and docking score probably due to the steric hindrance. Glycyrrhizin also showed good affinity to ACE2 protein but poor inhibitory effect. The docking result indicates glycyrrhizin could only bind to the region (Lys26 to Asp30) and not bind to any of the peptidase region. Rhein showed the best ACE2 inhibitory effect, and was confirmed by the molecular docking that the carboxyl could form hydrogen bond to Ala387, a key peptidase active site. The similar situation was observed on emodin 8-O-β-d-glucoside. However, aloe-emodin showed good affinity to ACE2 protein but no inhibitory effect. The docking result reveals it could bind to the sites of Glu35 and Gln76 with phenolic hydroxyl group, not peptidase sites (Supporting Information Fig. S5). Neochlogrogenic acid and its isomers could form 4 hydrogen bonds to the intersection region of peptidase activity and S protein binding (Ala348 to Arg393), indicating the potential double inhibition of S protein binding and ACE2 enzyme activity.
Figure 8.
Molecular docking results of (A) prunasin, (B) forsythoside I, (C) glycyrrhizin, (D) rhein, (E) forsythoside A, and (F) neochlorogenic acid.
4. Discussion
A TCM product usually contains from several dozens to a few hundreds of prototype components, while each constituent might change quantitatively with environmental factors or processing conditions14,38. TCM component profiles in human and animals become much complicated due to biotransformation by metabolizing enzymes, which makes identification of in vivo TCM metabolite components and their formation pathways very challenging.
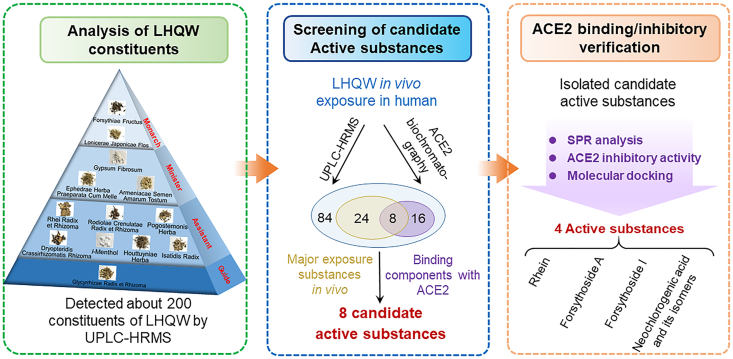
A novel strategy for the discovery of ACE2 inhibitory active components in LHQW that may play important roles in COVID-19 pneumonia treatment was established in this study (Fig. 9). In the first step, UPLC–HRMS was utilized to comprehensively analyze and identify the chemical compositions of LHQW. Resultant information was utilized for subsequent identification of LHQW components exposed to human. In the second step, major LHQW components in human urine were detected and structurally characterized followed by their inhibitory effects on ACE2 using 2D biochromatography. In the last step, SPR and ACE2 inhibitory activity assay were applied to confirm the pharmacodynamics of active components from the screening, while computer docking was used to elaborate on the possible sites of pharmacology action. The most important element of this strategy was to combine in human exposure study with 2D-biochromatography analysis, which was proved to be efficient in revealing the active components from LHQW that may exhibit inhibitory effects on ACE2 in human.
Figure 9.
The main process and results of the study on the potential active components of LHQW against COVID-19.
In this study, a LHQW test solution sample was initially used for setting up for UPLC–HRMS method as well as characterizing structures of LHQW prototype components (Fig. 2) to support identification of LHQW component in vivo. As shown in Table S1, 126 prototypes in the LHQW test solution were tentatively identified and 34 prototypes of them were fully confirmed by using reference standards, which were significantly more than the total prototype components found in LHQW in a previous study although major components identified from the two studies are similar17. Furthermore, the HRMS-based PATBS technique was applied to detection of LHQW components in human urine and plasma samples using a workflow previously developed for studying ADME of TCM in vivo19. PATBS is an untargeted data-mining tool capable of finding TCM components or other types of xenobiotics in a test in vivo sample regardless of their molecular weights, mass defects, isotope patterns and fragmentations since these xenobiotics were not present in a corresponding control sample such as a pre-dosing plasma and urine sample. As shown in Fig. 3, PATBS not only removed large portions of endogenous components and background noise to reveal minor LHQW components (Fig. 3D and E), but also significantly cleaned full-scan MS spectral data to make molecular ions of interest as dominant ion species (Fig. 3F). Both metabolomics approach and PATBS are untargeted data mining tools and their effectiveness of finding xenobiotics is comparable. However, PATBS has two significant advantages39. First, PATBS can be used in a combination with targeted or semi-targeted data mining tools, such as extracted ion chromatographic processing or mass defect filters, to increase detection sensitivity and selectivity20. Second, PATBS is capable of processing a single test sample by using a single control sample, while the metabolomics approach requires analysis two groups of test and control samples and each of the group has three or more samples. Results from this study demonstrate that HRMS combined with PATBS is a superior data-mining tool for studying ADME of a TCM product in human.
A good understanding of human exposure to TCM components after oral administration of a TCM product is considered as a very important step to investigate molecular mechanisms of its pharmaceutical effects in human. In this study, we detected and characterize a total of 107 LHQW components (66 prototypes and 41 metabolites) in human plasma after oral administration of therapeutic doses (Table S1). In addition, UPLC–HRMS profiles of plasma samples provided semi-quantitative estimation on the LHQW components in the human circulation (Fig. 4A and B). Based on the information, more accurate quantification of circulating components in human can be performed using LC–MS and their reference standards. Furthermore, we found additional 21 LHQW prototype components that were absorbed and then underwent rapid metabolic clearance or renal excretion. These LHQW components were also exposed to human even without the significant presence in the circulation. The approach significantly reduced cost and time in studying pharmacological mechanism of LHQW by avoiding testing over a large number of individual prototype components that were found in LHQW test solution, but unlikely exposed to human.
The experimental results from this study show that many of the main exposed constituents of LHQW in the human body have good affinity with ACE2. However, our results show that not all compounds with high affinities to ACE2 recombinant protein presented significant enzymatic inhibitory effect, which was owing to the difference of peptidase active sites and S protein binding sites of ACE2 protein. Several components were screened out with strong affinities to S protein binding sites, or binding to the intersection region of peptidase activity and S protein binding sites. This result might indicate an advantage of TCM with multiple drugs, as these multiple drug combination might show superimposed effects and exert their effects together. In this case, the clinical toxic side effects caused by single potent ACE2 inhibitor might be avoided. Previous studies have shown that LHQW could significantly inhibit the replication of SARS-CoV-2 in cells, and the number of virus particles was significantly reduced after LHQW treatment5. On the other side, cytokine storm (or inflammatory storm) is an excessive immune response of the body to external stimuli such as viruses and bacteria, which represents an important node in the development of COVID-19 from mild to severe or critical status. LHQW could significantly inhibit the gene overexpression of the inflammatory factors (for example, TNF-a, IL-6, MCP-1, or IP-10), caused by SARS-CoV-2 infection40, 41, 42, 43. It is worth mentioned that the current studies on the combination of the main exposed components of LHQW in the body and its possible interaction with ACE2 have been rarely reported. This interesting result also illustrates that it is necessary to conduct research on the efficacy of TCM from an in vivo exposure perspective.
5. Conclusions
In this study, human exposure to LHQW components after 8 repeated therapeutic doses were evaluated by determining LHQW component profiles in human plasma and urine, which were accomplished mainly by acquiring accurate MS and MS datasets using UPLC–HRMS and processing the MS dataset using PATBS for untargeted detection of LHQW components. As a result, a total of 86 LHQW components in human were tentatively identified based on spectral data interpretation or marching, among which 22 constituents were confirmed using reference standards. Furthermore, a novel ACE2 biochromatographic stationary phase was synthesized to screen for potential ACE2-target components in human urine sample and LHQW extract. Based on the data obtained from human exposure and comprehensive 2D ACE2 biochromatography system, selected LHQW components were subjected to further pharmacodynamic evaluation SPR and ACE2 inhibition assays. Results show that 8 components, including neochlorogenic acid, amygdalin, prunasin, forsythoside I, rutin, forsythoside A, glycyrrhizin and rhein, exhibited binding affinities to ACE2 with KD values ranging from 0.221 to 82.40 μmol/L. The ACE2 Inhibitor Screening Kit further verified the activity of the selected constituents. Finally, rhein, forsythoside A, forsythoside I, neochlorogenic acid and its isomers showed ACE2 inhibitory effects. Together, these active LHQW components circulating in human may play synergistic roles therapeutically with reduced side effect. It is worth mentioning that these constituents not only showed good affinity to ACE2 but also could effectively bind to the contact surface of ACE2 and spike complex, which was confirmed by computer-aided docking results. To the best of our knowledge, this was the first report of comprehensive study on human exposure to LHQW components. More importantly, it was found that several LHQW components exposed to human might play potential roles in inhibiting SARS-CoV-2 by significantly affecting the binding between ACE2 and S protein, which is an important route of preventing virus infection. This study provided direct chemical and biochemical evidences associated with molecular mechanisms of clinical use of LHQW for prevention and treatment of COVID-19. In addition, this study demonstrates the utility of human exposure-based approach to identifying pharmacologically active components in a herbal medicine product with approved therapeutic effects.
Acknowledgments
The authors would like to thank Prof. Chuan Li in Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Shanghai, China) to provide biological samples and technical guidance. This research was supported by Natural Science Foundation of China, China, (Grant Nos. 81773688, U1903119, 81973291, and 81973275); Zhejiang University Special Scientific Research Fund for COVID-19 Prevention and Control, China; “Phospherus” Project of Shanghai Science and Technology Committee, China, (Grant Nos. 19QA1411500); National Major Scientific and Technological Special Project for "Significant New Drugs Development", China, (Grant No. 2020ZX09201005).
Author contributions
Caisheng Wu, Xiaofei Chen, Yifeng Chai, Mingshe Zhu and Yunlong Wu: conceptualization, methodology, software, review & editing. Xiaofei Chen, Chunyan Zhu, Chun Chen, Yanqiu Gu, Shuping Wang and Jiayun Chen: investigation, validation, data analysis, visualization. Lei Zhang, Lei Lv, Guoqing Zhang and Yongfang Yuan: resources, methodology and supervision. All authors interpreted the results and critically revised the manuscript for scientific content. All authors approved the final version of the article.
Conflicts of interest
All authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.10.002.
Contributor Information
Yifeng Chai, Email: yfchai@smmu.edu.cn.
Mingshe Zhu, Email: mingshe.zhu@yahoo.com.
Caisheng Wu, Email: wucsh@xmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wu C.R., Liu Y., Yang Y.Y., Zhang P., Zhong W., Wang Y.L. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pant S., Singh M., Ravichandiran V., Murty U.S.N., Srivastava H.K. Peptide-like and small-molecule inhibitors against Covid-19. J Biomol Struct Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren J.L., Zhang A.H., Wang X.J. Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res. 2020;155:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Islam M.S., Wang J., Li Y., Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci. 2020;16:1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R.F., Hou Y.L., Huang J.C., Pan W.Q., Ma Q.H., Shi Y.X. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu K., Guan W.J., Bi Y., Zhang W., Li L.J., Zhang B.L. Efficacy and safety of lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine. 2020:153242. doi: 10.1016/j.phymed.2020.153242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding Y.W., Zeng L.J., Li R.F., Chen Q.Y., Zhou B.X., Chen Q.L. The Chinese prescription lianhuaqingwen capsule exerts anti-influenza activity through the inhibition of viral propagation and impacts immune function. BMC Compl Altern Med. 2017;17:130. doi: 10.1186/s12906-017-1585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan Z.P., Jia Z.H., Zhang J., Liu S., Chen Y., Liang L.C. Natural herbal medicine lianhuaqingwen capsule anti-influenza A (H1N1) trial: a randomized, double blind, positive controlled clinical trial. Chin Med J. 2011;124:2925–2933. [PubMed] [Google Scholar]
- 9.Ma Y.J., Zhang Z.Y., Wei L.B., He S., Deng X., Ji A.M. Efficacy and safety of Reduqing granules in the treatment of common cold with wind-heat syndrome: a randomized, double-blind, double-dummy, positive-controlled trial. J Tradit Chin Med. 2017;37:185–192. doi: 10.1016/S0254-6272(17)30043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao P., Yang H.Z., Lv H.Y., Wei Z.M. Efficacy of lianhuaqingwen capsule compared with oseltamivir for influenza A virus infection: a meta-analysis of randomized, controlled trials. Altern Ther Health Med. 2014;20:25–30. [PubMed] [Google Scholar]
- 11.Wang C.H., Zhong Y., Zhang Y., Liu J.P., Wang Y.F., Jia W.N. A network analysis of the Chinese medicine Lianhua-Qingwen formula to identify its main effective components. Mol Biosyst. 2016;12:606–613. doi: 10.1039/c5mb00448a. [DOI] [PubMed] [Google Scholar]
- 12.You W.L., Wan J.E., Gao H.R., Liu P.L., Zhang L.Z., Wang D.D. Network pharmacological approach to explore the mechanisms of lianhua qingwen capsule in coronavirus disease 2019. Precis Med Res. 2020;2:67–77. [Google Scholar]
- 13.Cheng C.R., Yang M., Yu K., Guan S.H., Wu X.H., Wu W.Y. Metabolite identification of crude extract from Ganoderma lucidum in rats using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;941:90–99. doi: 10.1016/j.jchromb.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Wang X.J., Ren J.L., Zhang A.H., Sun H., Yan G.L., Han Y. Novel applications of mass spectrometry-based metabolomics in herbal medicines and its active ingredients: current evidence. Mass Spectrom Rev. 2019;38:380–402. doi: 10.1002/mas.21589. [DOI] [PubMed] [Google Scholar]
- 15.Wang X.J., Zhang A.H., Zhou X.H., Liu Q., Nan Y., Guan Y. An integrated chinmedomics strategy for discovery of effective constituents from traditional herbal medicine. Sci Rep. 2016;6:18997. doi: 10.1038/srep18997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M.G., Jiang M., Ying X.H., Cui Q.X., Han Y.Q., Hou Y.Y. Identification and comparison of anti-inflammatory ingredients from different organs of Lotus nelumbo by UPLC/Q-TOF and PCA coupled with a NF-κB reporter gene assay. PloS One. 2013;8 doi: 10.1371/journal.pone.0081971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia W.N., Wang C.H., Wang Y.F., Pan G.X., Jiang M.M., Li Z. Qualitative and quantitative analysis of the major constituents in Chinese medical preparation lianhua-qingwen capsule by UPLC–DAD-QTOF-MS. Sci World J. 2015;2015:731765. doi: 10.1155/2015/731765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang Z.P., Wang F., Dai S.Y., Lu J.Q., Wu X.D., Zhang J.Y. Profiling and identification of (–)-epicatechin metabolites in rats using ultra-high performance liquid chromatography coupled with linear trap-Orbitrap mass spectrometer. Drug Test Anal. 2017;9:1224–1235. doi: 10.1002/dta.2155. [DOI] [PubMed] [Google Scholar]
- 19.Wu C.S., Zhang H.Y., Wang C.H., Qin H.L., Zhu M.S., Zhang J.L. An integrated approach for studying exposure, metabolism, and disposition of multiple component herbal medicines using high-resolution mass spectrometry and multiple data processing tools. Drug Metab Dispos. 2016;44:800–808. doi: 10.1124/dmd.115.068189. [DOI] [PubMed] [Google Scholar]
- 20.Zhu C.Y., Cai T.T., Jin Y., Chen J.Y., Liu G.Q., Xu N.S. Artificial intelligence and network pharmacology based investigation of pharmacological mechanism and substance basis of Xiaokewan in treating diabetes. Pharmacol Res. 2020;159:104935. doi: 10.1016/j.phrs.2020.104935. [DOI] [PubMed] [Google Scholar]
- 21.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X.Y., Hirano M., Werner R.A., Decker M., Higuchi T. Novel 18F-labeled PET imaging agent FV45 targeting the renin-angiotensin system. ACS Omega. 2018;3:10460–10470. doi: 10.1021/acsomega.8b01885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 virus targeting the cns: tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y., Zhao Z.X., Wang Y.J., Zhou Y.Q., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X.T., Chen P., Wang J.F., Feng J.N., Zhou H., Li X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y.Q., Chen X., Wang Y., Liu Y., Zheng L.Y., Li X.Q. Development of 3-mercaptopropyltrimethoxysilane (MPTS)-modified bone marrow mononuclear cell membrane chromatography for screening anti-osteoporosis components from Scutellariae Radix. Acta Pharm Sin B. 2020;10:1856–1865. doi: 10.1016/j.apsb.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X.F., Cao Y., Zhang H., Zhu Z.Y., Liu M., Liu H.B. Comparative normal/failing rat myocardium cell membrane chromatographic analysis system for screening specific components that counteract doxorubicin-induced heart failure from Acontium carmichaeli. Anal Chem. 2014;86:4748–4757. doi: 10.1021/ac500287e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X.F., Cao Y., Lv D.Y., Zhu Z.Y., Zhang J.P., Chai Y.F. Comprehensive two-dimensional HepG2/cell membrane chromatography/monolithic column/time-of-flight mass spectrometry system for screening anti-tumor components from herbal medicines. J Chromatogr A. 2012;1242:67–74. doi: 10.1016/j.chroma.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 29.Ruan C.H., Liu L.S., Lu Y.F., Zhang Y., He X., Chen X.L. Substance P-modified human serum albumin nanoparticles loaded with paclitaxel for targeted therapy of glioma. Acta Pharm Sin B. 2018;8:85–96. doi: 10.1016/j.apsb.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo P., Liu D., Li J. Pharmacological perspective: glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int J Antimicrob Agents. 2020;55:105995. doi: 10.1016/j.ijantimicag.2020.105995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L.D., Lv D.Y., Chen X.F., Liu M.D., Wang D.Y., Liu Y. Biosensor-based active ingredients recognition system for screening STAT3 ligands from medical herbs. Anal Chem. 2018;90:8936–8945. doi: 10.1021/acs.analchem.8b01103. [DOI] [PubMed] [Google Scholar]
- 32.Lan J., Ge J.W., Yu J.F., Shan S.S., Zhou H., Fan S.L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 33.Yan R.H., Zhang Y.Y., Li Y.N., Xia L., Guo Y.Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guy J.L., Jackson R.M., Jensen H.A., Hooper N.M., Turner A.J. Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis. FEBS J. 2005;272:3512–3520. doi: 10.1111/j.1742-4658.2005.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rushworth C.A., Guy J.L., Turner A.J. Residues affecting the chloride regulation and substrate selectivity of the angiotensin-converting enzymes (ACE and ACE2) identified by site-directed mutagenesis. FEBS J. 2008;275:6033–6042. doi: 10.1111/j.1742-4658.2008.06733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han Y.X., Kral P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C.H., Wu C.S., Qin H.L., Zhang J.L. Rapid discovery and identification of 68 compounds in the active fraction from Xiao-Xu-Ming decoction (XXMD) by HPLC–HRMS and MTSF technique. Chin Chem Lett. 2014;25:1648–1652. [Google Scholar]
- 39.Chen C., Fan Z.Q., Xu H., Tan X.J., Zhu M.S. Metabolomics-based parallel discovery of xenobiotics and induced endogenous metabolic dysregulation in clinical toxicology. Biomed Chromatogr. 2019;33:e4413. doi: 10.1002/bmc.4413. [DOI] [PubMed] [Google Scholar]
- 40.Li Q., Yin J., Ran Q.S., Yang Q., Liu L., Zhao Z. Efficacy and mechanism of lianhua qingwen capsules (LHQW) on chemotaxis of macrophages in acute lung injury (ALI) animal model. China J Chin Mater Med. 2019;44:2317–2323. doi: 10.19540/j.cnki.cjcmm.20190210.001. [DOI] [PubMed] [Google Scholar]
- 41.Dong L., Xia J.W., Gong Y., Chen Z., Yang H.H., Zhang J. Effect of lianhuaqingwen capsules on airway inflammation in patients with acute exacerbation of chronic obstructive pulmonary disease. Evid Based Complement Alternat Med. 2014;2014:637969. doi: 10.1155/2014/637969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C., Zhou Q.Q., Li Y.Z., Garner L.V., Watkins S.P., Carter L.J. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen S.F., Ho Y.C. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130:2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.