Abstract
The non-synonymous mutations of SARS-CoV-2 isolated from across the world have been identified during the last few months. The surface glycoprotein spike of SARS-CoV-2 forms the most important hotspot for amino acid alterations followed by the ORF1a/ORF1ab poly-proteins. It is evident that the D614G mutation in spike glycoprotein and P4715L in RdRp is the important determinant of SARS-CoV-2 evolution since its emergence. P4715L in RdRp, G251V in ORF3a and S1498F of Nsp3 is associated with the epitope loss that may influence pathogenesis caused by antibody escape variants. The phylogenomics distinguished the ancestral viral samples from China and most part of Asia, isolated since the initial outbreak and the later evolved variants isolated from Europe and Americas. The evolved variants have been found to predominant globally with the loss of epitopes from its proteins. These have implications for SARS-CoV-2 transmission, pathogenesis and immune interventions.
Keywords: SARS-CoV-2, COVID-19, Non-synonymous mutation, Epitope loss, Phylogenomics
The current outbreak of COVID-19 caused by SARS-CoV-2 enforced the greatest global health and a socio-economic threat to mankind [1]. It has been first reported in late December 2019 from Wuhan, China [2,3], become an epidemic and rapidly spread across the globe to become a pandemic with devastating impact [4]. At the end of January 2020 India reported its first case of COVID-19 from the state of Kerala. Two months since its outbreak by the middle of March 2020, Europe and North America have become the new epicenters to the pandemic with remarkable expansion of this disease with a huge number of fatalities. A total of 3435894 positive cases and 239604 deaths worldwide and 42,533 confirmed incidence and 1373 deaths from India has been reported by the WHO situation report, 4th May 2020.
SARS-CoV-2 is a novel member of the beta-coronavirus genus with single-stranded positive-sense RNA. They have similarities with the severe acute respiratory syndrome coronavirus (SARS-CoV) and to several bat coronaviruses [5]. SARS-CoV-2 comprises of around 29,903 nucleotides organized into specific genes characteristics within its genome. In the 5′ region more than two-thirds of the genome comprises a set of non-structural proteins (Nsp) produced as cleavage products of the ORF1a and ORF1ab viral polyproteins [6] assemble to facilitate viral replication and transcription. RNA-dependent RNA polymerase (RdRp, also known as Nsp12), is the key component that regulates the synthesis of viral RNA with the assistance of Nsp7 and Nsp8 as co-factors [7]. The 3′ region consists of genes encoding structural proteins including surface (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. Surface glycoprotein spike is involved in the interaction with the host’s receptor, Ace2, further ingress, and forms one of the vital factors for rapid human-to-human transmission [8]. Additionally, 6 accessory proteins are encoded by ORF3a, ORF6, ORF7a, ORF7b, and ORF8 genes (Fig. 1 a). Compared to SARS-CoV emerged in 2002 and MERS-CoV in 2012, SARS-CoV-2 exhibits faster human transmission [9] and leads to the declaration of a worldwide public health emergency by WHO [4]. Three factors that make SARS-CoV-2 associated disease more infectious include higher transmissibility, high mortality, and humans have no prior immunological history against it.
Fig. 1.
(a) Structure of the SARS-CoV-2 genome. (b) The relative occurrence of non-synonymous mutation across the genome of SARS-CoV-2. Black indicates neutral mutation and red are deleterious mutations.
In general RNA viruses are vulnerable to a high rate of mutations [10,11] which may be correlated with the geographical region-specific virulence of virus variants. The rapid global spread of SARS-CoV-2 provides an ample opportunity for natural selection to act upon rare but favorable mutations. Deleterious mutations are indeed thought to always out-number beneficial mutations [12]. Mutation rates are ever evolving in accordance with the environmental changes. In a constant environment, almost no mutations are likely to occur and in a perfectly adapted environment, the rate of beneficial mutation will be zero [13]. On the other hand, if an organism is allowed in a completely new environment, the potential rate of beneficial mutation will be non-zero [13]. From the above fact it can be derived that as SARS-CoV-2 is adapted to the environment in the last 6 months and hence appears to show more number of deleterious and neutral mutations than that of beneficial mutation. Thus, in the present study the non-synonymous mutational function due to neutral and harmful mutations are considered.
There might have complex interplay between amino acids that can confer immune resistance to the virus and the fitness landscape of the particular variant. Mutation of single amino acid within an antigenic determinant or epitope can potentially overcome the antibody recognition. The ability to identify epitope is vital to combat the infection through antibody-mediated immunity and also essential in several biomedical applications like rational vaccine design, disease diagnostic, and immune-therapeutics [14,15]. As a growing need for the development of suitable therapeutics against SARS-CoV-2 for effective disease management, the diagnostic assays based on peptides have become indispensable for their advantages over conventional methods [16,17]. Identification of appropriate epitopes within a particular protein antigen can elicit an immune response and could have been used for the synthesis of an immunogenic peptide. Most of the studies have been focused to predict conserved immunodominant epitopes from the surface glycoprotein of SARS-CoV-2 [18,19]. Some studies have mapped the B-cell epitopes of membrane glycoprotein and nucleo-capsid phospho-protein [20]. However, very little is known about the potential epitopes from the longest chunk of non-structural proteins of ORF1ab and 6 other accessory proteins encoded by ORF3a, ORF6, ORF7a, ORF7b, and ORF8. In the present work, we have tried to locate the loss of epitope due to non-synonymous mutations using a comparative genomics approach. Non-synonymous mutation leading to the loss of epitope allows escaping antibody immunity. Antibody escape due to epitope loss may be responsible for re-infections among individuals. The nature and locations of epitope losses from the various proteins of SARS-CoV-2 due to non-synonymous mutations are the main focus of the present study. The immediate and continuous release of complete genome sequences of SARS-CoV-2 from samples of diverse geographical regions have helped scientists to monitor the rapid evolution of the SARS-CoV-2 to gain insights into the pattern and dynamics global spread of COVID-19. We have performed a comparative genome analysis of the viral samples from a varied geographical location with special reference to India. The mutation of SARS-CoV-2 genome responsible for epitope loss shows a specific pattern of emergence outside China as the virus have migrated through different epicenters of the world. Based on such non-synonymous mutations in the SARS-CoV-2 genome, the initial viral isolates are under different clusters than the later evolved ones. For example, the Indian isolates of SARS-CoV-2 clearly distributed and depicted that the initial samples from Kerala are having similarity with the Wuhan isolates and the rest are predominantly found within isolates originated from patients having travel history from Italy. These findings have important implications to understand the impact of new emerging mutations in the pathogenesis and immune evasion of SARS-CoV-2.
2. Materials and methods
List of tools used in the present study is described in Table S2 of SI.
2.1. Genome analysis
586 SARS-CoV-2 high-quality complete genome sequences from known patient status sequenced across the world and reported from January to April 2020 were accessed from GISAID Epicov [21] platform. The infection load and geographical locations from various continents viz. Asia, Africa, Europe, North, Central, and South Americas, and Oceania were considered for complete genome selections. The redundant sequences with 100% identity were removed. To reduce the number of false-positive variants, we have excluded sequences with more than 50 ambiguous bases. One representative sequence from each country (except India and China) has been considered in the present analysis. All the Indian isolates available till the middle of April 2020 were taken for genome analysis to unravel the nature of SARS-CoV-2. As the outbreak occurred from China, we took multiple sequences to focus the mutation and thus evolution of the virus outside China. Thus a total of 87 SARS-CoV-2 genome sequences representative of 586 genome sequences available till April, 2020 had been utilized in the present analysis (Table S1). All sequences were uniformly annotated using the RAST server [22]. Protein domain analysis was conducted with Pfam [23].
2.2. Multiple sequence alignment
Clustal omega [24] has been employed to align ORF1a, ORF1ab, spike, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8 and N proteins of SARS-CoV-2 sequences originated from all affected countries. Thus the non-synonymous mutations were identified from the multiple sequence alignment.
2.3. Effect of amino acid alterations
PROVEAN algorithm was used to predict the functional effect of non-synonymous mutations [25]. PROVEAN utilizes delta scores to analyze amino acid alterations. In the present study, the cut-off for PROVEAN scores was set to −2.5 for high balanced accuracy. Low delta scores are interpreted as amino acid variations leading to a deleterious effect on protein function, while high delta scores are interpreted as variations with a neutral effect on protein function. IEDB-AR has been utilized for B-cell epitope prediction using Bepipred Linear Epitope Prediction based on a Random Forest Regression (RF) algorithm [26] with a 5-fold cross-validation approach considering computed volume, hydrophobicity, polarity, together with the relative surface accessibility and secondary structure. B-cell epitopes are located using a combination of a hidden Markov model and a propensity scale method. The residues with scores above the threshold (default value 0.35) are predicted to be a part of an epitope and colored in yellow on the graph (Y-axes:residue scores; X-axes residue position in the sequence). Prediction of the change of secondary structure of the respective proteins due to such amino acid alterations have been done with PSIPRED [27].
2.4. Structural changes due to mutations
Vibrational entropy changes and binding conformational enthalpy change due to mutation has been implemented using normal mode analysis (NMA) by Dynamut web interface [28] (http://biosig.unimelb.edu.au/dynamut/). NMA has been applied to the study which utilizes harmonic motions in a system, providing insights into its dynamics and accessible conformations due to mutation. It has been widely used for studies of protein dynamics as an alternative to more computationally intensive molecular dynamics approaches. While molecular dynamics approaches provide motion trajectories for a given molecule over time, conformational fluctuations can be evaluated by NMA via superposition of normal modes (Eigenvectors) and their associated frequencies (Eigenvalues) [29]. NMA can also use simplified representations of the protein structure, such as modeling the amino acids using their Cα atoms, reducing computational cost. The performance of DynaMut tool outperforms alternative algorithms that also provide measurements of effects of single-point mutations on protein stability. Energy minimization of the wildtype and mutated structures has been done using GROMOS96 53a6 force-field [30] until the maximum force of the system became < 100 kJ/mol/nm to relieve steric clashes and inappropriate geometry. The minimized structures are superimposed in order to calculate RMSD using Visual Molecular Dynamics(VMD) (http://www.ks.uiuc.edu/) developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana–Champaign. Visual analysis of the structure has been done with PYMOL molecular graphics system.
2.5. Docking studies
Docking studies have been carried out using HADDOCK [31]. HADDOCK is an information-driven flexible docking for the site-directed approach. HADDOCK recognizes itself from ab initio docking methods with the fact that it encodes information from identified or predicted protein interfaces in ambiguous interaction restraints (AIRs) to manage the docking process. These AIR files have information about active residues of the macromolecule. The result with the lowest HADDOCK score and Z-Score is considered as the best interaction between the molecules [31]. Prodigy [32] is used to calculate ΔG and Kd to predict the binding affinity at 25 °C.
2.6. Phylogenomic analysis
Phylogenetic analysis of whole-genome sequences have been inferred with REALPHY 1.12 [33]. REALPHY utilizes a reference whole-genome sequence data to align with the query sequences via bowtie2, an ultrafast and memory-efficient tool for aligning sequencing reads to long reference sequences [34] from which the phylogenetic trees are further constructed using PhyML. Maximum likelihood tree is thus generated using GTR (generalized time reversible) nucleotide substitution model. The resulted tree has been visualized and annotated using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results
3.1. Spike glycoprotein is the most important hotspot of amino acid substitution in SARS-CoV-2
The study have shown 47 non-synonymous mutations from all the genome sequences of SARS-CoV-2 isolated from more than fifty representative countries of five continents. The total number of neutral mutations have been almost equal to that of the deleterious mutations (Table 1 ). Spike glycoprotein has been found to be the most vital hotspot of amino acid alterations among proteins of SARS-CoV-2. The surface protein spike mediates receptor recognition through its RBD (receptor binding domain) region with human Ace2. 13 different mutations of spike protein are observed of which D614G is most notable and exist among 37 samples, especially among isolates from the European countries. However, the total number of neutral mutations in spike glycoprotein is higher than the deleterious mutations. However, there has been no amino acid alteration observed in the RBD region of the spike. This might be just because all the samples originated from the patient, which means the virus has gone through successful pathogenesis following the interaction to the host receptor through the spike. Thus there is a high chance that the virus might undergo mutation in RBD of spike protein and become non-infective or negligible in number to be highlighted for such mutation mapping. After spike glycoprotein, it is the orf1a polyprotein that forms the major site of amino acid alterations. Orf1a expresses the viral replicase and protease enzyme. The dominance of deleterious mutation is found high in this region. S1498F is the most frequent mutation at the multi-domain essential replication/transcription complex- Nsp3 of ORF1a and reflected among multiple Indian isolates with travel history to Italy. Nucleocapsid N protein shows 7 non-synonymous mutations of which 4 are neutral and 3 are deleterious. An incident of the concurrence of R203K and G204R is found in Belgium, Brazil, Peru, Mexico, Nigeria, Switzerland, and Vietnam from the present analysis. Similar mutations also have been spotted in 3 samples of Indian isolates having a travel connection to Italy. ORF1ab consist of an equal number of neutral and deleterious mutation effects. P4715L variant of ORF1ab is remarkable to occur among 37 samples in the RNA dependent RNA polymerase (RdRp) site. This prime spot for mutation is especially seen among the European samples (Table S1). The Indian isolates from patients with European travel history have revealed such kind of variation deviating from the ancestral samples from China. ORF3a has shown a more deleterious effect of amino acid substitutions. The incidence of occurrence of G251V is high among various samples irrespective of geographical locations. The change of amino acid L84S in ORF8 has been observed among the viral samples of more than 12 countries. Such a mutation can be observed in various affected countries from different geographical locations. Primary samples isolated in India from Kerala in January 2020 bear such mutation. V74F is the deleterious mutation found in ORF7b among an isolate from Kuwait and five Indian isolates sampled at Iran in March 2020. It can be called that V74F variation in ORF7b is primarily restricted to the Asian continent. Membrane glycoprotein consists of a neutral and deleterious mutation not having any amino acid alteration on Indian samples. Amino acid alterations in envelope protein and two other accessory proteins ORF6 and ORF7a have not been found among the isolates chosen for the present study. The relative occurrence of non-synonymous mutations among gene products of SARS-CoV-2 has been shown in Fig. 1b.
Table 1.
The non-synonymous substitutions and their effects among various global isolates of SARS-CoV-2 included in this study. The mutation hotspots that demarcate clades (G, V, S, GH and GR) available at the public database of the Global Initiative on Sharing All Influenza Data (GISAID) is highlighted.
| Variant | Infected country | Mutation effect | |
|---|---|---|---|
| ORF1a polyprotein | D58E | New Zealand | Deleterious |
| L952P | China(hCoV−19/Shandong/LY003/2020) | Neutral | |
| E955K | China(hCoV−19/Tianmen/HBCDC−HB−07/2020) | Deleterious | |
| S1498F | Multiple Indian isolates | Deleterious | |
| N1559T | Russia | Neutral | |
| A3203V | USA | Neutral | |
| G4227R | Russia | Deleterious | |
| A4297G | Mexico | Deleterious | |
| F4304L | Sweden | Deleterious | |
| ORF1ab polyprotein | P4715L | Multiple countries isolatesa | Neutral |
| Y232C | Australia,USA | Deleterious | |
| F1657L | New Zealand | Neutral | |
| A1906V | Canada | Deleterious | |
| V1973L | New Zealand | Neutral | |
| G2374R | France | Deleterious | |
| Spike protein | Y145del | India(hCoV−19/India/1-27/2020) | Neutral |
| N354D | China(hCoV−19/Shenzhen/SZTH−004/2020) | Neutral | |
| D364Y | China(hCoV−19/Shenzhen/SZTH−004/2020) | Deleterious | |
| R416I | India(hCoV−19/India/1-27/2020) | Neutral | |
| S438F | India(hCoV−19/India/763/2020,777/2020) | Neutral | |
| Y508H | France | Neutral | |
| D614GG | Multiple countries isolatesa | Neutral | |
| Q675H | Scotland | Neutral | |
| T791I | Taiwan | Neutral | |
| F797C | Sweden | Deleterious | |
| A930V | India(hCoV−19/India/1-31/2020) | Deleterious | |
| I1216T | China(hCoV−19/Shanghai/SH0067/2020) | Deleterious | |
| P1263L | England | Neutral | |
| ORF3a | A31T | Hungary | Neutral |
| Q57HGH | Russia, Congo, Saudi Arabia | Deleterious | |
| V88L | Cambodia | Neutral | |
| H93Y | Wales | Deleterious | |
| G196V | Chile, Georgia | Deleterious | |
| G251VV | France, Sweden, Australia, China(hCoV−19/HongKong/CUHK1/2020) | Deleterious | |
| Membrane glycoprotein | D3G | Finland | Neutral |
| T175M | Belgium, Brazil | Deleterious | |
| ORF7a | V74F | Kuwait, India(hCoV−19/India/1073/2020,1093/2020,1100,2020,1115/2020,1063/2020) | Deleterious |
| S81L | New Zealand | Deleterious | |
| ORF8 | V62L | New Zealand | Neutral |
| L84SS | Multiple countries isolatesb | Neutral | |
| Nucleocapsid protein | L121H | China(hCoV−19/Shandong/LY003/2020) | Deleterious |
| T148I | China(hCoV−19/Shenzhen/SZTH−004/2020) | Deleterious | |
| S193I | Wales | Deleterious | |
| S197L | Spain, Chile, Georgia | Neutral | |
| R203KGR | Multiple countries isolatesc | Neutral | |
| G204RGR | Multiple countries isolatesc | Neutral | |
| I292T | Switzerland | Neutral |
Refer Supplementary table1.
Australia, Spain, Chile, USA, S. Korea, Georgia, China (hCoV-19/Hangzhou/ZJU-08/2020, hCoV-19/Beijing/235/2020, hCoV-19/Guangzhou/GZMU0047/2020, hCoV-19/Shandong/LY003/2020), India(hCoV-19/India/1–31/2020), Georgia, New Zealand.
Belgium, Brazil, Peru, Mexico, Nigeria, Vietnam, Switzerland, India(hCoV-19/India/c32/2020, hCoV-19/India/2020c32/2020, hCoV-19/India/c31/2020).
3.2. Loss of epitope for non-synonymous mutation
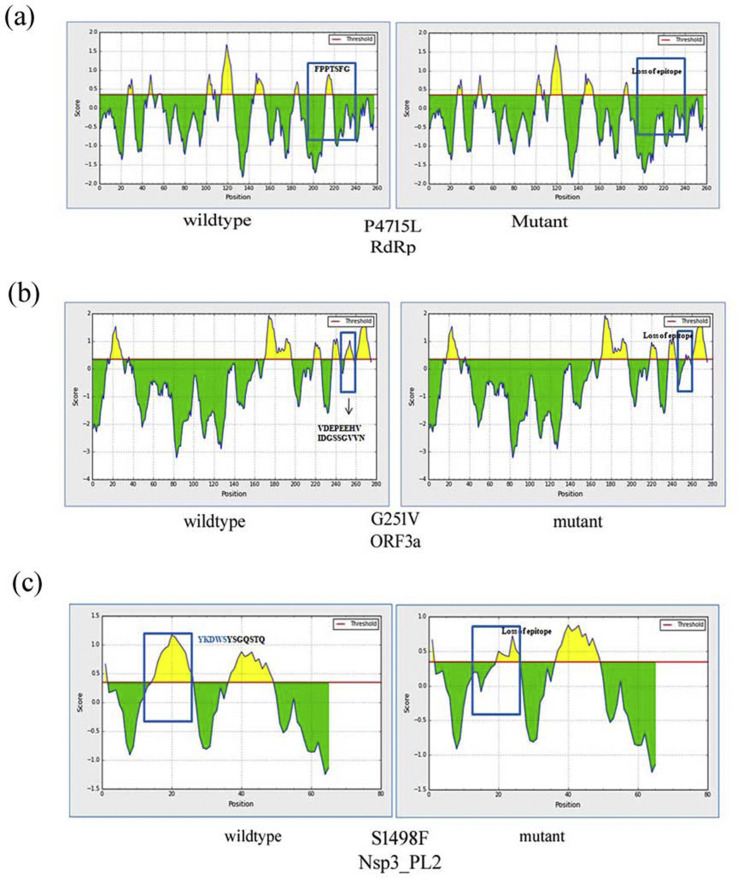
The present study reports three cases of epitope loss due to amino acid alterations. Screening of all B-cell epitopes is presented in table S3. P4715L substitution in RdRp protein from the ORF1ab region is linked with ‘FPPTSFG’ epitope loss from the site (Fig. 2 a). Wild type RdRp consists of seven epitope regions that are reduced to six due to the amino acid substitutions Despite P to L mutational data suggesting a neutral substitution, it has been found to elicit the change from a “turn” to “helix” in RdRp adjacent secondary structure and loss of an epitope (Fig. S1). The second epitope loss is associated with G251V in ORF3a (Fig. 2b). Six putative epitopes in the wild-type ORF3a are replaced by five in the mutant variants. The third loss of epitope S1498F is found in non-structural protein Nsp3 of ORF1a among Indian isolates. In this case, it is observed that the epitope loss is partial. S1498F mutation of Nsp3 is responsible for the loss of linear-epitope consisting of five residues ‘YKDWS’ from the region (Fig. 2c). Such loss of epitope might allow the new variant to escape interaction with host antibodies and influence disease profile by evading the antibody-mediated neutralization.
Fig. 2.
Epitope loss linked with non-synonymous mutations. The predicted score above the threshold level is the Yellow region showing epitope. (a) Effect of P4715L mutation in RdRp. (a) B-cell epitope in non-mutated RdRp (left) and P4715L mutant (right). It is linked with such epitope loss FPPTSFG from the site. (b) Epitope loss linked with ORF3a G251V. The B-cell epitope of wildtype ORF3a (left). G251V mutant (right) causes loss of DGSSGVV(250 …. 256aa). (c) Epitope loss linked with S1498F in the papain-like protease domain of NSP3 in the ORF1a region. The B-cell epitope of wildtype sample (left), S1498F mutation causes the loss YKDWS (right).
3.3. Structural impact due to P4715L in RdRp and D614G in spike protein
To study the effect of the mutation on the tertiary structure of RdRp and spike protein PDB ID: 6M71, chain A, PDB ID: 6VSB, chain A have been used as wild type structures, respectively. P4715L in RdRp accounts for stable conformation change confirmed by binding conformational enthalpy change (ΔΔG = 1.540 kcal/mol). The vibrational entropy changes (ΔΔSVib ENCoM = −4.074 kcal mol−1 K−1) for this mutation signify decrease in molecular flexibility (Fig. S2). In both the conformations of RdRp of Pro4715 and Leu4715 can form intramolecular hydrophobic interactions with Phe4788. The mutation P4715L led to the formation of an intramolecular hydrogen bonding between Leu4715 and Phe4718. The oxygen atom of Leu4715 acting as hydrogen bond acceptor is meant here to interact with hydrogen atom of Phe4718 (hydrogen bond donar). The distance of such h-bond interaction is 2.1892 Å. Energy minimization of wildtype and mutant variant of RdRp shows RMSD 0.169 nm. In spike glycoprotein, D614G is favored by the attainment of molecular stability (ΔΔG = 1.128 kcal/mol) and vibrational entropy changes (ΔΔSVib ENCoM = −4.531 kcal mol−1 K−1) leading to decrease in molecular flexibility (Fig. S2). The inter-atomic interactions with the neighboring residues remain the same after D614G mutation. Energy minimization of wild-type and mutant variant of spike shows RMSD 0.405 nm. However, careful structural studies indicate that - the sidechain of D614 potentially can form a hydrogen bond with T859 and electrostatic bond with K854 of the neighboring protomer (Table 2 ). G614 is unable to participate in such inter protomer interactions. D614G mutation of spike protein does not effect human Ace2 binding. The binding free energy and dissociation constant between the docked complex of wild-type and mutant spike protein bound with human Ace2 (Table S4) does not show much difference in binding affinity.
Table 2.
Spike SARS-CoV-2 interactions with neighboring protomer through D614 side chains, examined from cryoEM structure [42].
| Bond type | Interacting residues and atoms | Distance (Å) |
|---|---|---|
| H-bond | Thr859:OG1-Asp614:OD2 | 2.74 |
| Electrostatic | Lys854:NZ-Asp614:OD2 | 5.20 |
3.4. Coexistence of P4715L in RdRp and D614G in spike protein among European and American samples
It is interesting to find that L4715 RdRp and D614G spike variants show co-occurrence (Fig. 3 ). These variants from two completely different proteins are highly correlated. Viral sequences consistently contain both mutations regardless of when the sequence is obtained. Such modifications in spike and RdRp are adopted more in SARS-CoV-2 isolated from patients of Europe and Americas than those isolated from Asia. However, the initial isolates from China lack such variation completely. The pandemic burden of COVID-19 in India consists of both types of SARS-CoV-2. Indian patients with travel history from Europe and Americas are mainly affected by such variants. In Africa only two samples are considered and both of them showed such a new variant of spike and RdRp. The global infection fatality rate (deaths/cases) as per WHO situation report, 4th May 2020 (the time when the present study was analyzed) has found to be much higher in Europe and America than the rest of world. The P4715L mutation of RdRp and D614G mutation in spike protein, predominant in viral strains of Europe and America, has a major role in outrageous infection evident from the high fatality rate (Fig. S3).
Fig. 3.
P4715L RdRp and D614G spike variants show co-occurrence. (a) The incidence (%) of occurrence of P4715 and L4715 of RdRp in various geographical locations. (b) The incidence (%) of occurrence of D614 and G614 of spike protein in various geographical locations. (c) The overlap in the mutation across the world is depicted from the combined plot. The distribution of these two unique mutant sites of vital SARS-CoV-2 proteins can be completely superimposed.
3.5. The more evolved form of SARS-CoV-2 is indicated from phylogenomic analysis
The adaptive evolution of this novel pathogen is not biased with geographic location but is related to the non-synonymous mutation located in spike glycoprotein and RdRp of ORF1ab. P4715L RdRp and D614G spike is the primary determining factor to cluster the different SARS-CoV-2 isolates into different clades. The phylogenetic tree using whole-genome sequence elicits two types of clades-the green represented by the ancestral type from China and red with the evolved variant in spike and RdRp (Fig. 4 ). Thus the red group is the mutant form showing the loss of epitope among various infected countries. The green group is more prominent in China and other parts of Asia and the red group mainly belongs to Europe and America. From the Indian perspective, it is very clear that Indian samples isolated from patients with the contact or travel history linked to Wuhan are green variants and the rest linked with other than Asia, especially Europe and America, are the red variants. The red variants seem to be more infectious than the green and the mutations with epitope loss may have a definite role in this.
Fig. 4.
Phylogenomics with different SARS-CoV-2 viral samples across the world. The tree is distinctly divided into 2 clades: green shows the original (ancestral) form of the virus isolated from Dec, 2019 to Feb, 2020 and red clades are the evolved variant isolated after Feb, 2020 to April, 2020. Loss of epitope is found in the evolved variant due to non-synonymous mutation. Indian isolates are highlighted in purple belonging to both the green and red clades. The samples from Kerala, India are Wuhan representative (within green clade) and the rest are the variants from Italy (within red clade).
4. Discussion
The impact of epitope loss due to non-synonymous mutation is the biggest and unique concern highlighted in this study. It could potentially be linked to immune evasion and thus higher viral spread and pathogenesis. SARS-CoV-2 is recognized by the “pattern recognition receptors” (PRR) of the immune cells that induce cytokines release activating more immune cells to produce a large number of pro-inflammatory cytokines, tissue factors and vasoactive peptides [35]. Cytokine storm syndrome related to hypercytokinaemia with multiorgan failure can be observed [36]. In patients infected by SARS-CoV-2 increase in T-helper 2 (TH2) cytokines (IL-4 and IL10) are reported in addition to the T-helper 1 (TH1) cytokines (IL1B, IFNγ, IP10, and MCP1) previously detected in other coronavirus infections [37]. Patients with more severe cases had higher leukocyte and neutrophil count, lower lymphocyte count and higher neutrophil-to-lymphocyte ratio (NLR) [38]. Humoral immune responses have a substantial role in COVID-19 infections. It has been found that nucleocapsid protein (NP)-specific antibody response, and anti-spike RBD antibody atleast 3 days of post symptoms onset [39]. It has been reported that IgM peaked at day 9 after disease onset and then switched to IgG by week 2 [40]. Immunoglobulin class switching from IgM to IgG can be observed within 1 week after the first virus exposure [40]. Concomitantly with the decrease of IgM, IgG levels have raised gradually from week 3 to week 7, indicating the activation of the humoral immune response against the virus [41]. The real-time evolution of SARS-CoV-2 has been tracked from the available whole genome sequences around the globe through the phylogenomic analysis. It has been found that there are many point mutations incorporated in the genome of SARS-CoV-2 from the variants associated with the first outbreak in China to the present variants isolated in Europe. SARS-CoV-2 genome sequences available at the public database of the Global Initiative on Sharing All Influenza Data (GISAID) till 02-10-2020 classifies the variants into several clades like (i) L-original lineage, (ii) G-variant of spike protein causing D614S mutation, (iii) S- variant ORF8 responsible for L84S mutation, (iv) V- variant of the ORF3a coding protein N3S responsible for G251V mutation, (v) GH- a G derivative characterized by ORF3a: Q57H mutation, (vi) GR-nucleocapsid gene mutations- R203K and G204R, (vii) O- other combinations that do match from the rest [42]. In present analyses we have included all these variants (Table 1) along with the loss of epitope due to such mutation in ‘V’ variant. The second most common mutation is among these variants is P4715L of RdRp [42]. All the emerged SARS-CoV-2 genomes from new epicenters belong to either of these clades. The present study identified all the popular mutations along with some other mutations and reported the phenomenon of epitope loss that may influence pathogenesis caused by antibody escape variants. Hence, the result obtained from the 586 sequences is a brief reflection of >82,000 sequences present in GISAID database. It is further interesting to note that all the samples isolated after the middle of March 2020 to 30th April 2020 show the major prevalence of epitope loss due to such amino acid alterations. Mutations that appeared at multiple times include D614G in spike glycoprotein and P4715L in RdRp. The bulletin from WHO on variant analysis of SARS-CoV-2 reported on June, 2020 [43] described P4715L mutation in ORF1ab from 6319 samples. D614G mutation in spike is the dominant pandemic form that may indicate a fitness advantage [44] and related to severe reduced antigenic specificity [45]. Such mutations have occurred primarily as the virus started perpetuating outside China during the second sharp infection and morbidity in Italy, Spain, and other parts of Europe. The present analysis clearly justifies the concurrence of these two mutation sites in the same viral sample. Yin, 2020 also reported the frequent mutation of spike protein and RdRp in SARS-CoV-2 and their co-occurrence [46]. Increased transmission of such mutations might have a selective advantage for positive selection. In the present study, apart from the phenomenon of co-occurrence, we have focused into linear B-cell epitope loss due to such mutation in RdRp that may promote antibody escape in the western world variant than that of the eastern world. The phylogenomics study prominently have represented the evolution of SARS-CoV-2 since its outbreak in Dec, 2019. The later variants of the virus especially that are found in Europe and America lacks the epitope in certain proteins than that of the original SARS-CoV-2 strains from China. The study delineates the change in traits in SARS-CoV-2 as it came outside China to rest of the world. Conformational changes in SARS-CoV-2 protein structure due to much non-synonymous mutation and epitope loss could be immensely interesting both for plasma mediated therapy or serological detection of COVID-19. However, it can be anticipated that the replacement of negatively charged Asp into non-polar Gly in D614G mutation of spike protein consequences from the replacement of hydrophilic to the hydrophobic residue. Furthermore the flexibility has also been compromised due to such mutation as reflected from the vibrational entropy changes. Thus the loss of acidic residue and impairment in structural flexibility might play a role in altered recognition towards human Ace2 receptor recognition and fusion of viral membrane. Structural analysis has indicated that the D to G mutation at 614th position in spike protein does not cause significant variation as both of them attain sheet-like conformation in their secondary structure. Spike D614G mutation site is not proximally situated to the RBD-Ace2 binding interface and unable to differentiate much in Ace2 interactions. However, careful examination of the cryoEM structure [47] predicts that the interactions with the neighbouring protomer due to D614G mutation is disrupted in absence of negatively charged/hydrogen bond accepting group from the side chain of non-polar glycine. The interaction between the two protomers due to Asp614 is very critical as it brings together the S1 unit of one protomer to the S2 unit of the other protomer. These two sites play vital role in furin and S2 cleavage [48]. Thus, it may be hypothecated that D614G mutation diminishes the interaction between the S1 and S2 units, facilitating the shedding of S1 from viral-membrane-bound S2. This process may trigger the binding of S1 subunit to host cell receptors leading to enhanced pathogenicity due to D614G mutation. In the tertiary structure, it has appeared that such mutation stabilized the structure better however decreased the structural flexibility as accounted from binding conformational enthalpy change. The phenomenon of epitope loss due to P4715L in ORF1ab polyprotein is supported because Leu (L) is the hydrophobic residue which prefers normally to buried within the protein rather than to expose outside to act as an epitope for immunological interactions [49,50]. The change of secondary structure from loop to helix linked to P4715L supports epitope loss as epitope antibody residues are enriched by loops and depleted of strands and helixes [49]. This is logical due to the substitution of helix breaker –Pro (P) with a most stabilizing residue in alpha helix, Leu (L) might cause structural stabilization. The ellipse-like shape of the epitope [51] ‘FPPTSFG’ becomes distorted due to amino acid alteration leading to the loss of epitope. Thus the change of shape of the epitope also supports the cause behind the loss of antigenic determinant. Structural changes in RdRp might influence viral replication as it is one of the key players in viral replications. It can be hypothesized, that the decrease in flexibility of the overall structure of spike and RdRp due to mutations and epitope loss has made them less accessible to antibody causing less responsive immunity by the host. Such stabilizing mutation might thus account for positive selection and adaptive advantage as when introduced to new variant sit rapidly becomes the dominant form. The detailed structural dynamics associated with epitope loss on paratope interaction at epitope:paratope interface shall be interesting to focus in future. The epitope loss linked with G251V in ORF3a might cause a change in flexibility due to the substitution of small hydrogen moiety of Gly (G) to the bulky branched side chain in Val (V). The tendency for epitopes to be depleted of small hydrophobic amino acids like Val [49,50] supports the epitope loss associated with G251V. Such changes in the ORF3a-viroporin domain require further experimental outcomes to precisely understand the effect exerted. While trying to unravel the role of non-synonymous mutation from the Indian perspective, it has been found that the Indian SARS-CoV-2 samples isolated from the patient of Kerala state with travel history from Wuhan on January lack D614G mutation in their spike glycoprotein but additionally shows a neutral mutation R416I and an alteration of A930V with deleterious effect. Deletion is also found at Y145 in the initial viral sample hCoV-19/India/1–27/2020 during the report of onset of infection in India. The D to G mutation at 614th position among Indian samples has been found with European travel connections. Increased infectivity might be consistent with rapid spread, and also the association of higher viral load with G614 that we observed in more infected countries. For P4715L mutation in RdRp, a mixed outcome has been noticed. P4715 with the epitope ‘FPPTSFG’ remains conserved to all the samples isolated from patients without travel history outside Asia. L4715 with the loss of epitope is found in the samples with travel history from Italy. Indian isolates from European origin show a new mutation S1498F in the papain-like protease domain of Nsp3 in the ORF1a region. It is responsible for releasing Nsp1, Nsp2, and Nsp3 from the N-terminal region of polyproteins 1a/1 ab [52]. Ser (S) to Phe (F) mutation led to a shift from polar hydrophilic to the non-polar hydrophobic group. The residues in epitope are enriched by polar amino acids and depleted of hydrophobic amino acid compared to residues of non-epitope residues [[49], [50], [51], [52]] supports the observation of epitope loss from this site. As the properties of these two amino acids are different, the mutations might presumably modify the replication/transcription function of Nsp3. V74F mutation appears in the ORF7a accessory protein among isolates from the COVID-19 patients of India who migrated from Iran. This accessory protein is suggestive to have a potential role in binding with the IL-1 receptor of the host cell [53]. Such a mutation from V to F is deleterious and may have an impact as the side chain isopropyl group is substituted by a phenyl group. R203K and G204R in nucleocapsid protein are found in SARS-CoV-2 of three Indian patients who traveled from Italy in April 2020. Membrane glycoprotein and ORF3a is uniform among all the Indian isolates. Multiple emergent variants of viral genomes both from Europe and China are circulating in the Indian population. Sequences submitted from Kerala are similar to the original Wuhan virus while the rest are similar to Italy. There can be a better understanding through large-scale global sequencing studies which clears the most perpetuating SARS-CoV-2 in the Indian population. Meanwhile, understanding how mutations could lead to loss of epitope and how this in turn related to probable immune evasion would be an important topic of research to combat the pandemic. Biological impact of epitope loss of RdRp, ORF3a and Nsp3 on immune recognisation has not been reported yet. The loss of antigenic determinant impairs immune recognisation is an established fact in other viral disease models [54] which can be followed to know the antigenic variation and immune escape by SARS-CoV-2 in near future.
Author contributions
AMG curated, analyzed and interpreted the data. JC helped in docking studies. SM supervised the work. All the authors write, review and edited the manuscript.
Declaration of competing interest
The authors wish to declare that they do not have any conflict of interest.
Acknowledgments
We are thankful to the authors, generating and submitting laboratories of the sequences from GISAID’s EpiCoV™ database. AMG is greatful to SNBNCBS for fellowship. Authors are grateful to the anonymous reviewer for all their kind suggestion and recommendation to improve the overall study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2020.10.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Poon L.L.M., Peiris M. Emergence of a novel human coronavirus threatening human health. Nat Med. 2020;26:317–319. doi: 10.1038/s41591-020-0796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziebuhr J. The coronavirus replicase. Curr Top Microbiol Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. 2. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang X., Wu C., Li X., Song Y., Yao X., Wu X. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander H.K., Mayer S.I., Bonhoeffer S. Population heterogeneity in mutation rate increases the frequency of higher-order mutants and reduces long-term mutational load. Mol Biol Evol. 2017;34:419–436. doi: 10.1093/molbev/msw244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loewe L., Hill W.G. The populations of mutations: good, bad and indifferent. Philos Trans R Soc Lond B Biol Sci. 2010;365:1153–1167. doi: 10.1098/rstb.2009.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gershoni J.M., Roitburd-Berman A., Siman-Tov D.D., Tarnovitski Freund N., Weiss Y. Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs. 2007;21:145–156. doi: 10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irving M.B., Pan O., Scott J.K. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr Opin Chem Biol. 2001;5:314–324. doi: 10.1016/S1367-5931(00)00208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., Joshi M.D., Singhania S., Ramsey K.H., Murthy A.K. Peptide vaccine: progress and challenges. Vaccines. 2014;2:515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohanraj U., Chander S., Chavan Y.G. Peptide based viral detection systems for effective diagnosis of common viral infections in India. Curr Protein Pept Sci. 2017;18:939–945. doi: 10.2174/1389203717666160724205226. [DOI] [PubMed] [Google Scholar]
- 18.Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y.P., Chee R.S.L. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee S., Tworowski D., Detroja R., Biswas S.M., Morgenstern M.F. Immunoinformatics and structural analysis for identification of immunodominant epitopes in SARS-CoV-2 as potential vaccine targets. Vaccines (Basel) 2020;8:E290. doi: 10.3390/vaccines8020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., Wang P., Kim Y., Haste-Andersen P., Beaver J., Bourne P.E. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues C.H., Pires D.E. Ascher DB DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46(W1):W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins. 1998;33:417–429. doi: 10.1002/(sici)1097-0134(19981115)33:3<417::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Oostenbrink C., Villa A., Mark A.E., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. Comput Chem. 2004;13:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 31.de Vries S.J., van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 32.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A Mjj. Structural bioinformatics PRODIGY : a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 33.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B., Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L., Liu H.G., Liu W., Liu J., Liu K., Shang J. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jiehe He Huxi Zazhi. 2020;43:203–208. doi: 10.3760/cma.j.issn.1001-0939.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao A.T., Gao C., Zhang S. Profile of specific antibodies to SARS-CoV-2: the first report. J Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S.J., Nguyen V.G., Park Y.H., Park B.K., Chung H.C. A novel synonymous mutation of SARS-CoV-2: is this possible to affect their antigenicity and immunogenicity? Vaccines (Basel) 2020;8:220. doi: 10.3390/vaccines8020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112:3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ofran Y., Schlessinger A., Rost B. Automated identification of complementarity determining regions (CDRs) reveals peculiar characteristics of CDRs and B cell epitopes. J Immunot. 2008;181:6230–6235. doi: 10.4049/jimmunol.181.9.6230. [DOI] [PubMed] [Google Scholar]
- 50.Sun J., Xu T., Wang S., Li G., Wu D., Cao Z. Does difference exist between epitope and non-epitope residues? Analysis of the physicochemical and structural properties on conformational epitopes from B-cell protein antigens. Immunol Res. 2011;7:1–11. [Google Scholar]
- 51.Kringelum J.V., Nielsen M., Padkjær S.B., Lund O. Structural analysis of B-cell epitopes in antibody:protein complexes. Mol Immunol. 2013;53:24–34. doi: 10.1016/j.molimm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hänel K., Stangler T., Stoldt M., Willbold D. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J Biomed Sci. 2006;13:281–293. doi: 10.1007/s11373-005-9043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Francica J.R., Rohena A.V., Medvec A., Plesa G., Riley J.L., Bates P. Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.