Abstract
A defining step in the biogenesis of a membrane protein is the insertion of its hydrophobic transmembrane helices into the lipid bilayer. The nine-subunit ER membrane protein complex (EMC) is a conserved co- and post-translational insertase at the endoplasmic reticulum. We determined the structure of the human EMC in a lipid nanodisc to an overall resolution of 3.4 Å by cryo-electron microscopy, permitting building of a nearly complete atomic model. We used structure-guided mutagenesis to demonstrate that substrate insertion requires a methionine-rich cytosolic loop and occurs via an enclosed hydrophilic vestibule formed by the subunits EMC3 and EMC6 within the membrane. We propose that the EMC uses local membrane thinning and a positively charged patch to decrease the energetic barrier for insertion into the bilayer.
One sentence summary:
The structure of the human ER membrane protein complex provides insights into the mechanism of membrane protein biogenesis at the endoplasmic reticulum.
Main:
The human genome encodes over 5000 integral membrane proteins, all of which contain hydrophobic transmembrane helices (TMs) that must be inserted into the lipid bilayer (1). At the endoplasmic reticulum (ER), multiple insertion pathways operate in parallel to accommodate the enormous topological and biophysical diversity of these substrates (2-7). The ER membrane protein complex (EMC) is a ubiquitously expressed and widely conserved membrane protein insertase (3, 8-10), which both post-translationally inserts tail-anchored proteins and co-translationally inserts some multipass membrane proteins (3, 11). One of the membrane-spanning subunits of the EMC, EMC3, belongs to the Oxa1 superfamily of insertases, which includes the bacterial YidC, the archaeal Ylp1, and the eukaryotic WRB (12, 13). An atomic model of the EMC would thus both provide insight into substrate insertion at the ER, and further define the general principles of membrane protein biogenesis across all domains of life.
Using cells stably expressing GFP-tagged EMC2, the human EMC was affinity purified using an immobilized, protease-cleavable, GFP-nanobody and reconstituted into lipid nanodiscs (fig. S1) (14). Using cryo-electron microscopy (cryo-EM), we generated a reconstruction of the resulting nine-subunit EMC to an overall resolution of 3.4 Å; correction of inter-domain flexibility resulted in reconstructions of the cytosolic and lumenal regions to 3.6 and 3.2 Å resolution, respectively (Fig. 1; figs. S2, S3; Table S1).
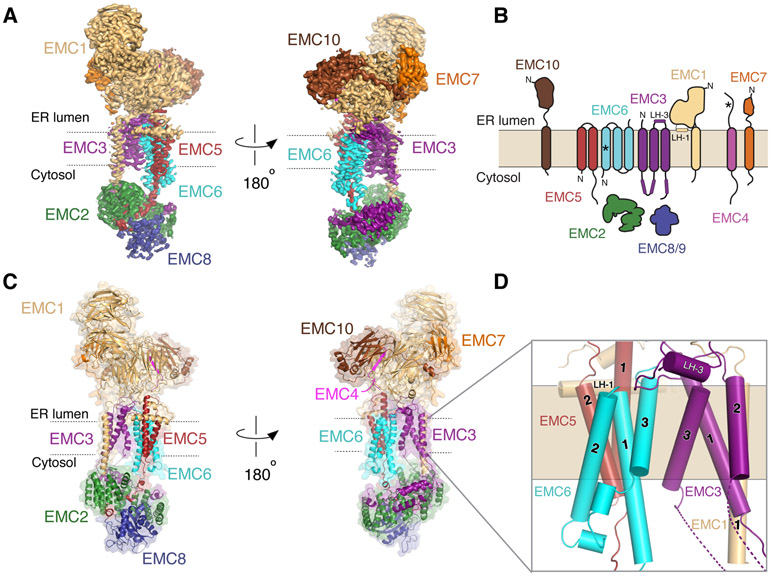
Figure 1. The structure of the human EMC.
(A) Two views of the sharpened ‘overall’ density map (fig. S3) from the perspective of the two intramembrane sides of the EMC colored by subunit. (B) Schematic representation of the topology of the nine EMC subunits as determined by the structure. EMC8 and 9 are functional paralogs, and their binding to EMC2 is mutually exclusive. For simplicity, we refer only to EMC8 throughout the text, though most observations will apply to both EMC8 and 9. Helices of EMC1 and 3 that are positioned in the lumenal plane of the membrane are labeled LH-1 and LH-3. Asterisks indicate newly determined topologies based on the structure and experimental data. Note, we cannot unambiguously define the topology of EMC4, but structural data is most consistent with it containing a single TM (fig. S7). (C) Atomic model of the EMC, in the same orientation as the density map in (A). (D) Close-up of the nine core TMs of the EMC and their subunit assignment.
The EMC extends ~200 x 70 x 100 Å and has a tripartite organization: (i) the membrane spanning region, composed of twelve TMs, nine of which form the central ordered core; (ii) a basket-shaped cytosolic region anchored by EMC2 and 8; and (iii) an L-shaped lumenal region comprised of EMC1, 4, 7, and 10. The cryo-EM density maps were sufficient to unambiguously assign and build nearly complete atomic models for EMC1, 2, 3, 5, 6, and 8, and the lumenal domains of EMC4, 7, and 10 (Fig. 1B, C; figs. S3-5, Table S2; movie S1). In doing so, we found that EMC6 unexpectedly contained three TMs. We demonstrated that the poorly hydrophobic TM1 of EMC6 (ΔG=3.8; (15)) inserted only upon assembly with EMC5, a conserved strategy for stabilizing poor TMs at subunit interfaces (fig. S6) (16, 17). We also observed weak density for three putative TMs extending from the lumenal domains of EMC4, 7, and 10 (fig. S7; movie S2). The flexible nature of these TMs is consistent with the limited roles of EMC4, 7, and 10 in EMC stability (18).
The membrane-spanning region of the EMC is pseudo-symmetric: three TMs of EMC6 abut the three TMs of EMC3 at the complex’s midline. On either side, EMC5 and 1 anchor the cytosolic and lumenal regions, respectively. In addition, the EMC contains at least two helices within the lumenal plane of the bilayer contributed by EMC1 and 3 (Fig. 1D). Similar to the amphipathic EH-1 helix of YidC, these helices may position the complex within the membrane and locally remodel the bilayer (19, 20).
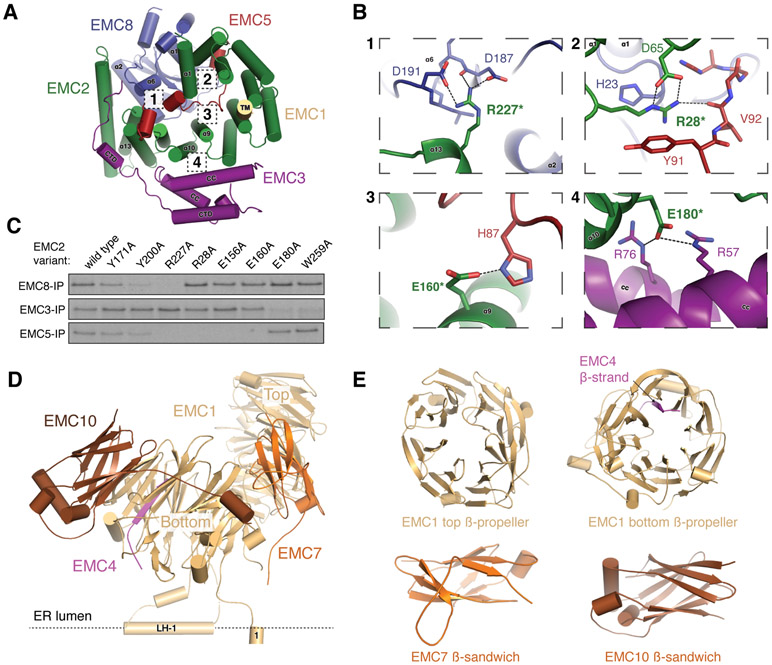
In the cytosol, EMC2 acts as an architectural scaffold for EMC8 and the cytosolic regions of EMC3, 5, and 1, consistent with its essential role in stability of the EMC (Fig. 2A) (18). EMC2 forms an α-solenoid that binds the three-helix bundle formed by the coiled coil and C-terminus of EMC3 (Fig. 2A; fig. S8). The C-terminus of EMC1 forms π-stacking interactions within a cleft of EMC2, and EMC2 clamps around EMC8 through an extensive hydrophobic surface (Table S3). Together, EMC2 and 8 form a composite interface with the C-terminal tail of EMC5, which traverses through the center of EMC2 to the cytosolic face of the complex. Mutations at the interfaces between EMC2, 3, 5, and 8 disrupted subunit binding in vitro, verifying the atomic model (Fig. 2B, C; fig. S8).
Figure 2. Architecture of the cytosolic and lumenal regions of the EMC.
(A) View from the membrane of the cytosolic region of the EMC. (B) Close-up of the primary interfaces between the cytosolic subunits of the EMC indicated in (A). Dashed lines represent polar interactions, and asterisks indicate mutations that disrupt complex assembly (fig. S8). (C) 35S-methionine labeled wild type EMC2 or the indicated point mutants were translated in rabbit reticulocyte lysate (RRL) and tested for binding to FLAG-tagged EMC8, EMC3 or EMC5 by co-immunoprecipitation using anti-FLAG resin. (D) Side view of the EMC lumenal region. (E) Cartoon model of the globular N-termini of EMC1, 7 and 10. EMC1 and EMC4 together form one of the four-stranded blades of the bottom ß-propeller.
Finally, the lumenal region is composed of the N-termini of EMC1, 7, 10, and the tail of EMC4 (Fig. 2D, E; fig. S8). EMC1 contains two eight-bladed β-propellers, and stabilizes the entire complex, contacting six of the remaining eight subunits. EMC7 and 10 form β-sandwiches that are anchored to EMC1 via primarily hydrophobic contacts (Table S3).
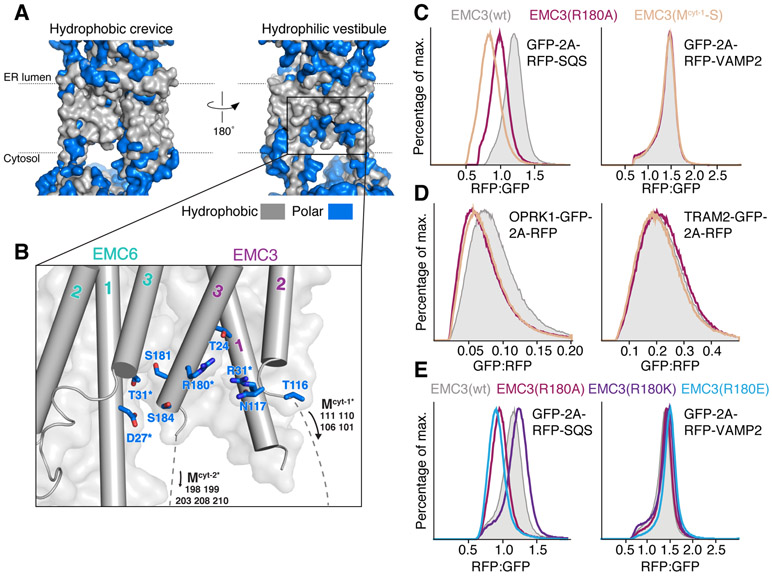
This atomic model permitted detailed interrogation of how EMC facilitates substrate insertion. We first used site-specific crosslinking to establish EMC3 as the primary interaction partner of a tail-anchored substrate with purified EMC (fig. S9). However, the EMC contains two intramembrane surfaces that could be involved in insertion and are consistent with an EMC3-substrate crosslink (Fig. 3A). On one side is a hydrophobic crevice that runs perpendicular to the plane of the membrane and could accommodate a TM (fig. S10). On the opposite side is a lipid-exposed cytosolic vestibule composed of EMC3 and 6, which is partially enclosed by the TMs of EMC4, 7, and 10, and sealed by EMC3’s lumenal helix (Fig. 3B; fig. S7; movie S2). Within this vestibule, EMC3 contains a positive patch in the bilayer that is surrounded by hydrophobic residues (Fig. 3B; fig. S11). We postulated that this hydrophilic vestibule formed the insertase based on analogy to YidC: the bacterial homolog of EMC3 relies on positively charged residues in the membrane for insertion (fig. S12) (19). Consistent with this model, the sequence conservation of the hydrophilic vestibule is significantly higher than that of the hydrophobic crevice (fig. S11). In particular, R31 of EMC3 is a positive charge in all eukaryotes.
Figure 3. Substrate insertion by the EMC requires a positive patch in the bilayer and a flexible methionine-rich loop.
(A) Surface filling representation of the membrane-spanning region of the EMC colored with hydrophobic residues in grey and polar residues in blue. Displayed are the two sides of the complex: the ‘hydrophobic crevice’ (left) and the ‘hydrophilic vestibule’ (right) as in Fig. 1A and C. (B) Close-up view of the hydrophilic vestibule formed by EMC3 and 6, with polar residues shown in blue and displayed as sticks. Residues that were mutated in functional assays are highlighted with asterisks (fig. S13). (C) HEK293 cells were generated that stably expressed exogenous wild type or mutant EMC3, as well as the tail-anchored substrates RFP-squalene synthase (SQS; EMC-dependent) or RFP-VAMP2 (EMC-independent) (3). The relative RFP fluorescence, normalized to an internal expression control (GFP), is plotted as a histogram. (D) As in (C) but with the co-translational substrates Opioid Receptor Kappa 1 (OPRK1)-GFP (EMC-dependent) and TRAM2-GFP (EMC-independent). (E) As in (C), analysis of the role of positive charge in the hydrophilic vestibule.
We introduced mutations to residues of EMC3 and 6 that line the hydrophilic vestibule but do not affect complex assembly, and tested their effect on substrate biogenesis using an established assay for EMC insertion (Fig. 3C-E; fig. S13) (3, 11). The mutations R31A and R180A in EMC3 destabilized representative post- and co-translational EMC-dependent substrates (SQS and OPRK1), but had no effect on the matched EMC-independent controls (VAMP2 and TRAM2). Furthermore, a positive charge at these positions is required for EMC insertion: R31E and R180E caused an insertion defect that was rescued by R31K/R180K for some substrates (Fig. 3E; fig. S13). Mutations to the polar residues of EMC6 had a modest, but detectable, effect on EMC-dependent insertion (fig. S13). We therefore concluded that the hydrophilic vestibule of the EMC is required for insertion of both post- and co-translational substrates.
It was recently noted that Oxa1 superfamily insertases contain methionine-rich cytosolic loops that were proposed to interact with substrates, in analogy to SRP54 and Get3 (21, 22). In the structure of the EMC, these loops were flexible. Nevertheless, mutation of methionines adjacent to TM2 in EMC3 specifically disrupted biogenesis of co- and post-translational EMC substrates (Fig. 3C, D; fig. S13). These methionines are positioned in the cytosol just below the insertase vestibule, and could transiently orient a substrate on its path into the membrane. We do not exclude a direct role for the cytosolic region in substrate binding; however, we could not identify a suitable hydrophobic surface or groove in the structure.
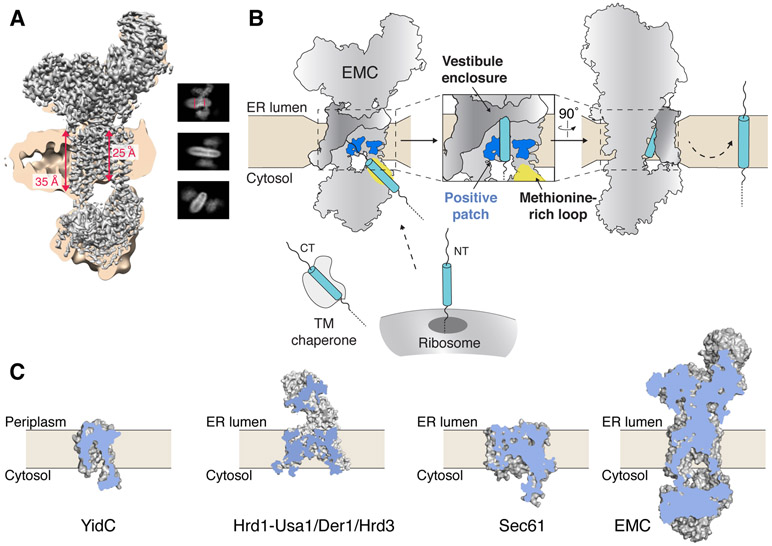
We therefore propose a model for EMC-mediated co- and post-translational substrate insertion (Fig. 4). A substrate would first be captured and guided towards the membrane by the flexible methionine-rich cytosolic loop of EMC3, possibly assisted by those of nearby EMC4 and 7. Prior to substrate engagement, the lipid-exposed hydrophilic vestibule of the EMC is axially sealed by the lumenal helix of EMC3 and laterally partially enclosed by the dynamic TMs of EMC4 and 7. The few contacts between EMC3 and 6 suggest this may be a potential site for subunit rearrangement during insertion.
Figure 4. Model for membrane protein insertion by the EMC.
(A) Unsharpened EM density maps are shown at low (tan) and high contour (grey) to highlight the thickness of the lipid nanodisc. Distances measured within the density are shown in red (fig. S14). Insets are representative 2D class averages that depict the local thinning of the lipid bilayer by the EMC. (B) Post- and co-translational EMC substrates are released from either a TM chaperone (e.g. calmodulin) or the ribosome, respectively. The flexible methionine-rich loop of EMC3 is positioned to capture substrates for insertion through the hydrophilic vestibule along the surface of EMC3 and 6. The EMC decreases the energetic barrier for insertion via local thinning of the membrane and a positively charged patch in the bilayer. The TMs of EMC4, 7, and 10 enclose the cytoplasmic vestibule and facilitate insertion. (C) Cut-away view of the space filling-models for the bacterial YidC (PDB 3WO6), the fungal Hrd1-Usa1/Der1/Hrd3 complex (6VJZ), mammalian Sec61 (3J7Q), and the human EMC. A hydrophilic conduit from the cytosol to the membrane is a general feature of evolutionarily diverse protein conducting channels.
The EMC decreases the energetic cost of insertion in two ways. First, by inducing a local thinning of the membrane by ~10 Å. Similar to other translocases, EMC thus decreases the distance that a substrate’s soluble lumenal domain must travel through the hydrophobic bilayer (Fig. 4A; fig. S14; movie S3) (23-25). Second, the EMC positions polar and positively charged residues within the bilayer, which could provide a way station for the substrate’s lumenal domain, enforce the positive-inside rule, and potentially stabilize the hydrophilic residues that are highly enriched in the TMs of EMC substrates (Fig. 4B) (26, 27).
Once within the membrane, the hydrophobic core of the substrate could interact with the hydrophobic surface of EMC3 above and below these polar residues. The flexible nature of the TMs of EMC4 and 7, which partially enclose the insertase, permit sampling of the lipid bilayer, and may further serve a gating function and/or interact directly with substrate. Finally, the shortened TMs of EMC3 and 6 cannot stably bind a membrane-spanning substrate, favoring its partitioning into the bilayer and dissociation from the complex. The substrate’s soluble lumenal domain would then encounter the β-propellers of EMC1, which may serve as a platform for recruitment of co-factors at the site of nascent protein insertion (26).
We therefore conclude that the Oxa1 superfamily insertases all rely on qualitatively similar mechanisms for insertion, and there is a marked similarity between EMC3/6 and WRB•CAML (17). More broadly, the presence of a hydrophilic cytosolic funnel is a conserved feature of all protein conducting channels including Sec61, Hrd1, YidC, and now EMC (Fig. 4C) (19, 25, 28). However, the significant increase in complexity of the EMC compared to YidC or even Sec61 suggests that its well-defined insertase function represents only a part of its more general role in membrane protein biogenesis and quality control.
Supplementary Material
Acknowledgements
We thank Israel Sanchez-Fernandez, Chi-Min Ho, Naima Sharaf, Chengcheng Fan, Gabe Lander, Andrey Malyutin, and Songye Chen for technical assistance; and Bil Clemons and the entire Voorhees lab for thoughtful discussion. Cryo-EM was performed in the Beckman Institute Center for TEM at Caltech, and data was processed using the Caltech High Performance Cluster, supported by a grant from the Gordon and Betty Moore Foundation. This research used resources of the Advanced Light Source, a DOE Office of Science User Facility under contract no. DE-AC02-05CH11231.
Funding: This work was supported by grants from the Heritage Medical Research Institute, the Kinship Foundation, the Pew-Stewart Foundation, and the NIH’s National Institute of General Medical Sciences (DP2GM137412). T.P. is funded by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Atomic coordinates and cryo-EM maps have been deposited in the Protein Data Bank under accession code 6WW7 and in the Electron Microscopy Data Bank under accession codes EMD-21929 (overall map), 21930 (lumenal map), and 21931 (cytosolic map). Plasmids for the GFP-nanobody purification strategy are available on Addgene, and requests for materials should be addressed to R.M.V.
References and notes:
- 1.The UniProt Consortium, UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviram N et al. , The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 540, 134–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guna A, Volkmar N, Christianson JC, Hegde RS, The ER membrane protein complex is a transmembrane domain insertase. Science 359, 470–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Görlich D, Rapoport TA, Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75, 615–630 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Mariappan M et al. , The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477, 61–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuldiner M et al. , The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefanovic S, Hegde RS, Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147–1159 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Jonikas MC et al. , Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wideman JG, The ubiquitous and ancient ER membrane protein complex (EMC): tether or not? F1000Res 4, 624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chitwood PJ, Hegde RS, The Role of EMC during Membrane Protein Biogenesis. Trends Cell Biol 29, 371–384 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Chitwood PJ, Juszkiewicz S, Guna A, Shao S, Hegde RS, EMC Is Required to Initiate Accurate Membrane Protein Topogenesis. Cell 175, 1507–1519.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennon SW, Soman R, Zhu L, Dalbey RE, YidC/Alb3/Oxa1 Family of Insertases. J Biol Chem 290, 14866–14874 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anghel SA, McGilvray PT, Hegde RS, Keenan RJ, Identification of Oxa1 Homologs Operating in the Eukaryotic Endoplasmic Reticulum. Cell Rep 21, 3708–3716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pleiner T et al. , Nanobodies: site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. eLife 4, e11349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hessa T et al. , Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Carvalho HJF, Del Bondio A, Maltecca F, Colombo SF, Borgese N, The WRB Subunit of the Get3 Receptor is Required for the Correct Integration of its Partner CAML into the ER. Sci Rep 9, 11887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglis AJ, Page KR, Guna A, Voorhees RM, Differential Modes of Orphan Subunit Recognition for the WRB/CAML Complex. Cell Rep 30, 3691–3698.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volkmar N et al. , The ER membrane protein complex promotes biogenesis of sterol-related enzymes maintaining cholesterol homeostasis. J Cell Sci 132, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumazaki K et al. , Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Drin G, Antonny B, Amphipathic helices and membrane curvature. FEBS Lett 584, 1840–1847 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Borowska MT, Dominik PK, Anghel SA, Kossiakoff AA, Keenan RJ, A YidC-like Protein in the Archaeal Plasma Membrane. Structure 23, 1715–1724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guna A, Hegde RS, Transmembrane Domain Recognition during Membrane Protein Biogenesis and Quality Control. Curr Biol 28, R498–R511 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM, Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci U S A 101, 4083–4088 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y et al. , YidC Insertase of Escherichia coli: Water Accessibility and Membrane Shaping. Structure 25, 1403–1414.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X et al. , Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 368, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shurtleff MJ et al. , The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. eLife 7, e37018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian S et al. , Proteomic Analysis Identifies Membrane Proteins Dependent on the ER Membrane Protein Complex. Cell Rep 28, 2517–2526.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voorhees RM, Fernández IS, Scheres SH, Hegde RS, Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157, 1632–1643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krissinel E, Henrick K, Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM, PDBsum: Structural summaries of PDB entries. Protein Sci 27, 129–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frey S, Görlich D, A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J Chromatogr A 1337, 95–105 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Pleiner T, Bates M, Görlich D, A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J Cell Biol 217, 1143–1154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vera Rodriguez A, Frey S, Görlich D, Engineered SUMO/protease system identifies Pdr6 as a bidirectional nuclear transport receptor. J Cell Biol 218, 2006–2020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariappan M et al. , A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466, 1120–1124 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao S, Rodrigo-Brenni MC, Kivlen MH, Hegde RS, Mechanistic basis for a molecular triage reaction. Science 355, 298–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie TK et al. , Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirchhofer A et al. , Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17, 133–138 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Beckett D, Kovaleva E, Schatz PJ, A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci 8, 921–929 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fairhead M, Howarth M, Site-specific biotinylation of purified proteins using BirA. Methods Mol Biol 1266, 171–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Spurrier J, Butt TR, Strickler JE, Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions. Protein Expr Purif 62, 21–28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma A, Mariappan M, Appathurai S, Hegde RS, In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol 619, 339–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter P, Blobel G, Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol 96, 84–93 (1983). [DOI] [PubMed] [Google Scholar]
- 43.Itakura E et al. , Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol Cell 63, 21–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Felipe P et al. , E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol 24, 68–75 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Zivanov J et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Terwilliger TC, Adams PD, Afonine PV, Sobolev OV, A fully automatic method yielding initial models from high-resolution cryo-electron microscopy maps. Nat Methods 15, 905–908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casañal A, Lohkamp B, Emsley P, Current Developments in Coot for Macromolecular Model Building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci 29, 1069–1078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho CM et al. , Bottom-up structural proteomics: cryoEM of protein complexes enriched from the cellular milieu. Nat Methods 17, 79–85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine PV et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J et al. , Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci U S A 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liebschner D et al. , Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones DT, Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292, 195–202 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Croll TI, ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moriarty NW, Grosse-Kunstleve RW, Adams PD, electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karpenahalli MR, Lupas AN, Söding J, TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics 8, 2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noinaj N, Gumbart JC, Buchanan SK, The β-barrel assembly machinery in motion. Nat Rev Microbiol 15, 197–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harel T et al. , Monoallelic and Biallelic Variants in EMC1 Identified in Individuals with Global Developmental Delay, Hypotonia, Scoliosis, and Cerebellar Atrophy. Am J Hum Genet 98, 562–570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.