Abstract
Efficiently copying mixed-sequence oligonucleotide templates nonenzymatically is a long-standing problem both with respect to the origin of life, and with regard to bottom up efforts to synthesize artificial living systems. Here we report an efficient and sequence-general nonenzymatic process in which RNA templates direct the synthesis of a complementary strand composed of N3′→P5′ phosphoramidate DNA (3′-NP-DNA) using 3′-amino-2′,3′-dideoxyribonucleotides activated with 2-aminoimidazole. Using only the four canonical nucleobases (A, G, C, and T) of modern DNA, we demonstrate the chemical copying of a variety of mixed-sequence RNA templates, both in solution and within model protocells, into complementary 3′-NP-DNA strands. Templates up to 25 nucleotides long were chemically transcribed with an average stepwise yield of 96–97%. The nonenzymatic template-directed generation of primer extension products long enough to encode active ribozymes and/or aptamers inside model protocells suggests possible routes to the synthesis of evolving cellular systems.
Introduction
RNA is a logical candidate for the primordial genetic polymer because in modern biology it both catalyzes critical chemical reactions such as protein synthesis, and acts as a genetic information carrier.1 Prior to the emergence of ribozyme-catalyzed RNA replication, chemical replication of the genetic material would have been necessary in order to initiate Darwinian evolution.2 This requirement has inspired over 5 decades of research into nonenzymatic template-directed RNA copying chemistry. We recently showed that short activated oligoribonucleotides that bind to an RNA template downstream of an activated ribonucleotide can catalyze the reaction of the monomer with an upstream RNA primer.3 The efficiency of the primer extension reaction is further enhanced through the use of 2-aminoimidazole (2AI) as a nucleotide activating group.4 Combined, these two advances enable the nonenzymatic copying of short mixed-sequence RNA templates in one-pot reactions.4 However, the extent to which an RNA template can be efficiently copied into a complementary RNA strand is currently limited to 7 nucleotides in solution and 5 nucleotides within membrane vesicles.4,5
Clearly, additional factors must come into play in order to improve the extent of nonenzymatic template copying to the point that sequences long enough to encode useful catalytic functions could be copied.
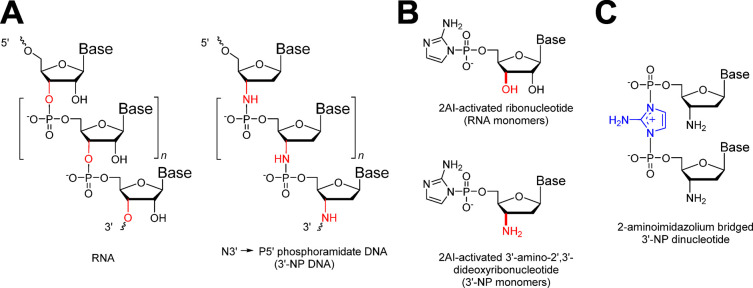
Exploring alternative genetic materials may provide insight into how longer RNA sequences could be propagated nonenzymatically. One such material is N3′→P5′ phosphoramidate DNA (3′-NP-DNA, Figure 1A), a genetic polymer with a phosphoramidate backbone6−9 that can be assembled by the polymerization of activated 3′-amino-2′,3′-dideoxyribo-nucleotide monomers (3′-NH2-2AIpddN, Figure 1B). Of the nonbiological genetic systems studied to date, 3′-NP-DNA is particularly attractive given that the 3′-NP-DNA duplex bears a striking geometric resemblance to the classical RNA A-type duplex structure, which is thought to be optimal for efficient nonenzymatic template-directed nucleotide polymerization.10,11 Moreover, 3′-NP-DNA forms stable duplexes with complementary RNA or DNA strands.12,13 Although no prebiotically plausible synthesis of 3′-amino-2′,3′-dideoxyribonucleotides has been demonstrated, 3′-NP-DNA could potentially form the genetic basis of artificial synthetic life forms that are biochemically distinct from modern biological systems. We have previously studied the template-directed synthesis of 3′-NP-DNA on short mixed-sequence RNA or 3′-NP-DNA templates using 2-methylimidazole (2MI) activated 3′-amino mononucleotides in the presence of N-hydroxyethyl-imidazole (HEI) as an organocatalyst.6,7 In these experiments, HEI is thought to act by displacing the 2-methylimidazole leaving group to generate highly reactive but labile HEI-activated monomers, which may then react directly with the 3′-aminonucleotide terminated primer to generate extended products. Our earlier results showed that the fidelity of template copying under these conditions is poor with the canonical nucleobase monomers, due to the formation of G:T and A:C mismatches.7 This problem was significantly ameliorated by replacing T with 2-thio-T, which we showed significantly decreases the frequency of wobble-pairing errors.7 The Richert group recently reported the copying of mixed-sequence DNA templates containing either two letters (G and C) or all four letters using oxyazabenzotriazole-activated 3′-amino-protected mononucleotides, which must react directly with the primer as monomeric units. Using downstream helper oligonucleotides to improve monomer binding, and repeated cycles of single nucleotide primer extension followed by 3′-amine deprotection, the copying of templates containing only G and C was shown to be feasible, whereas the chemical copying of templates containing all four letters was shown to be so error-prone that sequence information was lost over two rounds of template copying.14
Figure 1.
(A) Chemical structure of RNA and N3′→P5′ phosphoramidate DNA (3′-NP-DNA). The phosphodiester linkages of RNA and the phosphoramidate linkages of 3′-NP-DNA are highlighted in red. (B) Chemical structure of ribonucleotides and 3′-amino-2′,3′-dideoxyribonucleotides (3′-NP) activated with 2-aminoimidazole (2AI) (3′-NH2-2AIpddN, where N signifies the identity of the nucleobase). (C) Chemical structure of 2-aminoimidazolium-bridged 3′-amino-2′,3′-dideoxy-dinucleotide.
Given the problems with fidelity in copying templates nonenzymatically using highly activated canonical monomers that react directly with the primer, we decided to explore an alternative approach based on activated monomers that react with the primer primarily through the formation of imidazolium-bridged dinucleotide intermediates (Figure 1C).15 We have previously shown that these highly preorganized covalent intermediates lead to 3′-5′ regioselective RNA primer extension, suggesting that they may also lead to enhanced fidelity in template copying.16 Here, we show that this approach does indeed lead to significantly enhanced fidelity compared to our previous studies of primer extension with nucleotides that must react directly with the primer. We also show that primer extension using 3′-amino-2′,3′-dideoxyribonucleotides activated with 2AI enable the efficient one-pot nonenzymatic copying of mixed-sequence RNA templates up to 25 nucleotides long into a complementary 3′-NP-DNA strand. Finally, we show that this chemical copying of RNA to generate a complementary 3′-NP-DNA strand can occur within fatty acid vesicles, bringing us one step closer to generating an artificial cellular system capable of Darwinian evolution.
Results and Discussion
Given that 2AI(vs 2MI)-activated ribonucleotides exhibit significantly accelerated primer extension reactions on RNA templates,4 we asked whether 2AI activation would also enhance the RNA-template-directed polymerization of 3′-amino-2′,3′-dideoxyribonucleotides into 3′-NP-DNA (Figure 1B). We carried out these reactions in the absence of HEI, so as to favor primer extension via the highly preorganized structure of the imidazolium-bridged dinucleotide intermediate.15 After developing an efficient method for preparing 2AI activated 3′-aminomononucleotides (see SI for synthetic details), we evaluated the degradation profiles of 3′-NH2-2AIpddT as a representative example of a reactive mononucleotide. Using real-time 31P nuclear magnetic resonance (NMR) monitoring, we observed 3′-5′ cyclization as the main mode of monomer degradation, as previously reported.6 We calculated the half-life of 3′-NH2-2AIpddT as approximately 23.5 h under the conditions we used for template-directed synthesis reactions (Figure S1, see Supporting Information for details), both at low (3 mM) and elevated (50 mM) MgCl2 concentrations. We then asked whether these novel monomers could mediate the copying of short RNA homotemplates N4 (N = A, G, C or U) (SI Figure S2) by the primer extension synthesis of 3′-NP-DNA. Copying of the G4 and C4 RNA templates was rapid and efficient in the presence of 3′-NH2-2AIpddC and 3′-NH2-2AIpddG, respectively (>80% primer extension to the +3 product in 10 min). The slower addition of the fourth nucleotide was consistent with primer extension occurring through the covalent imidazolium-bridged dinucleotide intermediate, as previously shown for 2AI-activated ribonucleotides.17 For example, since the G-2AI-G intermediate can bind to the template by two Watson–Crick base-pairs at each of the first three positions on a C4 template, the first three G residues can be added rapidly; however, at the last position, the intermediate can only bind to the template by one Watson–Crick base-pair, leading to slower primer extension.
In contrast to the fast primer extension seen on G4 and C4 templates, primer extension on A4 and U4 templates was much slower. Complete conversion of the primer to the +3 and +4 products took 7 h on the U4 template in the presence of 3′-NH2-2AIpddA, while copying of the A4 template in the presence of 3′-NH2-2AIpddT took 48 h. In summary, all four canonical nucleobases were able to participate in homotemplate copying, but copying was more rapid for G and C monomers than for A and T monomers.
To show more directly that the primer extension reaction mechanism proceeds via the covalent imidazolium-bridged dinucleotide intermediate, we made use of the RNA template overhang 3′-GCCC-5′. When only 3′-NH2-2AIpddC was added, the addition of one C residue opposite the G in the template was very slow (t1/2 = 16 min). The simultaneous addition of 3′-NH2-2AIpddG led to a large rate enhancement (t1/2 < 0.5 min). In contrast, the addition of unactivated 3′-NH2-pddG had a small detrimental effect (t1/2 = 26 min) (Figure 2). The simplest explanation for this result is that primer extension with 2AI-activated 3′-amino-2′,3′-dideoxyribonucleotides occurs via formation of a 5′-5′ imidazolium-bridged dinucleotide C-2AI-G, which can only form from the reaction of the two activated nucleotides.15
Figure 2.
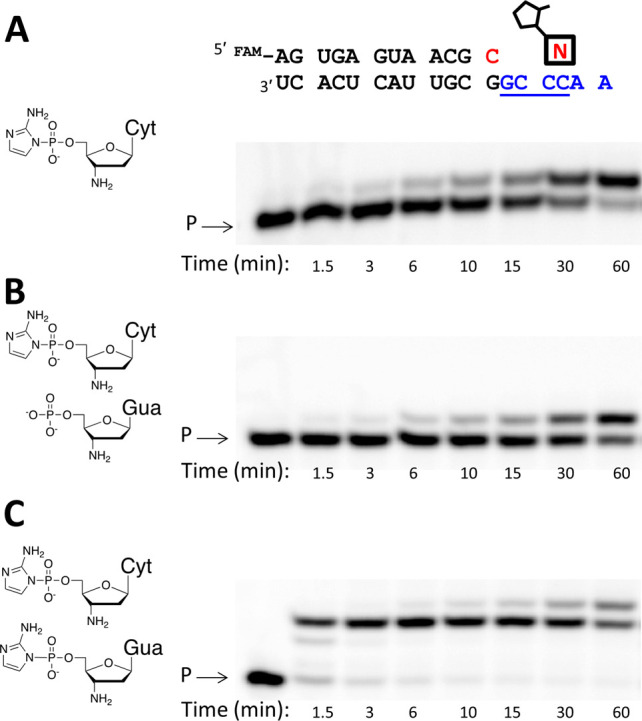
Chemical copying of RNA template 3′-GCCC into 3′-NP-DNA using (A) 3′-NH2-2AIpddC alone, (B) 3′-NH2-2AIpddC and 3′-NH2-5′-PO3-ddG, and (C) 3′-NH2-2AIpddC and 3′-NH2-2AIpddG. The nucleotide highlighted in red indicates the 3′-NH2-ddC at the end of the primer. All primer extension reactions were conducted at pH 8.5, 25 °C, and 200 mM Na+-bicine with 10 mM of each mononucleotide and 50 mM Mg2+. Reaction products were analyzed by polyacrylamide gel electrophoresis (PAGE). The RNA templating region is indicated in blue. The slow primer extension observed in the presence of 3′-NH2-2AIpddC alone (t1/2 = 16 ± 1 min), or 3′-NH2-2AIpddC plus 3′-NH2-ddG-5′-monophosphate (t1/2 = 26 ± 2 min) relative to 3′-NH2-2AIpddC plus 3′-NH2-2AIpddG (t1/2 < 0.5 min) suggests that the polymerization reaction proceeds via the imidazolium-bridged dinucleotide intermediate C-2AI-G. P, primer. Cyt, cytosine. Gua, guanine.
31P NMR analysis of the 3′-NH2-2AIpddC monomer alone in Na+-bicine, pH 8.5, in the presence of Mg2+ (50 mM) showed the presence of a major signal corresponding to the monomer (−10.7 ppm), and a second more shielded signal (−12.3 ppm) corresponding to the 5′-5′ imidazolium-bridged dinucleotide (SI Figure S3).15 Moreover, MS analysis revealed m/z signals consistent with both the monomer and dinucleotide species (SI Figure S3).
To provide further evidence for the role of the imidazolium-bridged dinucleotide intermediate in primer extension, we examined the copying of RNA template sequences 3′-ACCC-5′ and 3′-UCCC-5′ (SI Figure S4). In both cases, the primer extension reactions proceeded efficiently when carried out in the presence of the two complementary monomers, showing that single T and A residues can be added to a primer in high yield and faster than on homopolymeric A4 and T4 templates respectively, presumably through tighter binding of the T-2AI-G and A-2AI-G intermediates to the template, relative to T-2AI-T and A-2AI-A, respectively.
As an initial test of fidelity, we examined the extent of primer extension by single mismatched bases using the template sequences 3′-NCCC-5′ (N = A, G, or U) with 3′-NH2-2AIpddG as the sole activated monomer (SI Figure S4). We observed a low yield (<10%) of primer extension products on all three templates even at times of 60–75 min (see SI for details), showing that G is incorporated very poorly across from A, G or U in the template. This is expected if the imidazolium- bridged dinucleotide must bind to the template via two Watson–Crick base-pairs in order to set up the appropriate geometry for efficient reaction with the primer. We assessed the fidelity of the chemical synthesis of 3′-NP-DNA on mixed-sequence RNA templates by liquid chromatography mass spectrometry (LC-MS) analysis of the primer extension products. In previous work from our laboratory, using HEI as an organocatalyst, low fidelity was observed for the chemical copying of the template 3′-GAGAC-5′ using 2MI activated 3′-NH2-ddC and 3′-NH2-ddT, and for copying of the template 3′-CUCUA-5′ using 2MI activated 3′-NH2-ddG and 3′-NH2-ddA.7 The high frequency of errors, which ranged from 15 to 50%, was due to both C:A and G:T pair mismatches. In contrast, when we performed template-directed polymerization on similar templates using 2AI-activated mononucleotides in the absence of HEI, we observed rapid (complete reaction in <4 h) and accurate primer extension in both cases (Figure 3). The LC-MS analysis of fully extended primer +5 products revealed excellent fidelity for the chemical copying of template 3′-GAGAG-5′, with only the desired primer + C3T2 species detectable (Figure 3A). With template 3′-CUCUC-5′, a small amount (<5%, from MS and PAGE analysis, Figure S5) of G–T mismatch product, primer + G4A1, was detected along with the fully extended correct product primer + G3A2 (Figure 3B).
Figure 3.
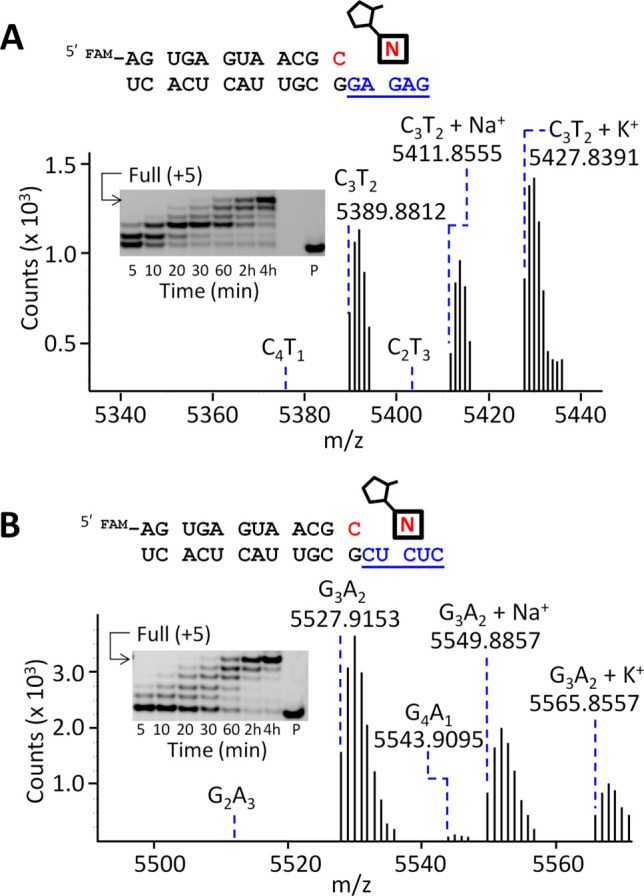
Low level of mismatches during copying of RNA templates (A) 3′-GAGAG and (B) 3′-CUCUC into 3′-NP-DNA. (A) The 3′-GAGAG template was copied by primer extension in the presence of 10 mM 3′-NH2-2AIpddC and 3′-NH2-2AIpddT. (B) The 3′-CUCUC template was copied by primer extension in the presence of 10 mM 3′-NH2-2AIpddA and 3′-NH2-2AIpddG. The nucleotide highlighted in red indicates the 3′-NH2-ddC at the end of the primer. Reactions were conducted in 200 mM Na+-HEPES pH 8.0, 25 °C, 50 mM Mg2+. Plots show MS analysis of the full-length extension products (+5) from the 4 h time point. Inset: PAGE analysis of the products of the primer extension reaction. P, primer. See SI Figure S5 for Sanger-type sequencing results.
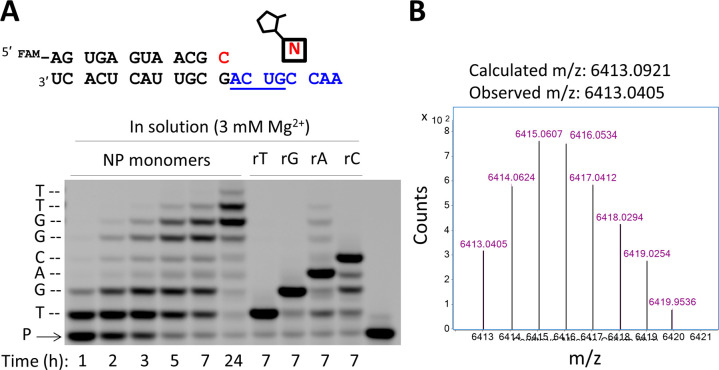
Encouraged by the yields and fidelities obtained during the copying of the RNA templates containing two letters, we proceeded to examine primer extension on an RNA template containing all four letters (3′-ACUGCCAA-5′), again generating a 3′-NP-DNA product (Figure 4). Conversion of the primer into primer +4 (and longer) products was observed within 3 h, with essentially complete conversion to primer +7/8 by 24 h (SI Figure S7). This copying of a mixed-sequence template occurred using only the four 2AI-activated 3′-amino mononucleotides, in the absence of the downstream activated oligonucleotides that are required for the sequence-general copying of RNA templates with 2AI-activated ribonucleotides.3,4 In addition, the polymerization reaction was carried out as a one-pot process, without requiring stepwise protection and deprotection of the nucleophilic 3′-amino functionality as used in the Richert group’s approach to copying DNA templates.14,18
Figure 4.
Nonenzymatic primer extension on a mixed-sequence RNA template using all four 2AI activated 3′-amino nucleotides. (A) Chemical copying of an RNA template into 3′-NP-DNA. The nucleotide highlighted in red indicates the 3′-NH2-ddC at the end of the primer. The six lanes at left show the time course of the reaction. The four lanes at right show an assessment of the fidelity of 3′-NP-DNA synthesis. Each 3′-NH2-nucleotide was replaced, one at a time, with a ribonucleotide, which at 3 mM Mg2+ results in chain termination (see SI Figure S6 for full time-course). P, primer. (B) Deconvoluted monoisotopic mass distribution for the full-length primer extension product (+8) showing correct composition: calc. mass, 6413.0921 Da; obs. mass, 6413.0405 Da; error, 8.0 ppm. Primer extension reactions were carried out using 10 mM activated NP monomers (3′-NH2-2AIpddA, 3′-NH2-2AIpddG, 3′-NH2-2AIpddC, 3′-NH2-2AIpddT), 200 mM Na+-HEPES pH 8.0, 25 °C and (A) 3 mM MgCl2, (B) 50 mM MgCl2. See SI for full description of methods. The RNA templating region is indicated in blue.
To gain insight into the fidelity with which a template containing all four nucleotides was copied, we carried out an assay adapted from Sanger sequencing.19 We repeated the primer extension reaction on the 3′-ACUG-5′ template at a Mg2+ concentration (3 mM) sufficiently low that template-directed polymerization of activated ribonucleotides is greatly reduced. By successively replacing each of the activated 3′-amino mononucleotides with the corresponding 2AI-activated ribonucleotides, we generated primer extension products that chain-terminated after the addition of each ribonucleotide (Figure 4A). A clear ladder corresponding to the sequential synthesis of the expected products was obtained, suggesting that at each step of primer extension the major product was correct. To confirm the identity of the predominant full-length primer extension product, we carried out LC-MS analysis of the primer extension reaction; the mass of the fully extended product was consistent with the expected composition (Figure 4B). Taken together, these results suggest that the 2AI-activated 3′-amino-2′,3′-dideoxyribonucleotides are excellent substrates for the fast and accurate copying of RNA templates into complementary NP-DNA products. A more complete analysis of the fidelity of this copying chemistry will require the development of single molecule sequencing methods for NP-DNA, which is underway in our group.
To determine the fraction of possible RNA sequences that could potentially be copied by activated 3′-amino-2′,3′-dideoxyribo-nucleotides, we examined the one-pot chemical copying of a heterogeneous population of RNA templates. A pool of 256 template sequences (randomized region 3′-NNNNUU-5′, N = A, G, C or U) were simultaneously copied by primer extension in a one-pot reaction containing all four activated mononucleotides. Approximately 74% of the initial primer was converted into fully extended products (primer +4 or more) in solution after 72 h (Figure 5), suggesting that most but not all RNA sequences can be copied by activated monomers.
Figure 5.
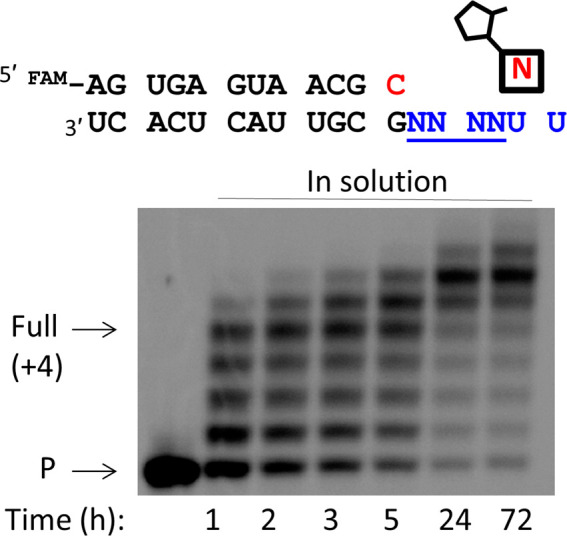
Simultaneous chemical copying of a randomized pool of RNA templates into 3′-NP-DNA using 3′-NH2-2AIpddNs in solution, where N = A, G, C, or U. The nucleotide highlighted in red indicates the 3′-NH2-ddC at the end of the primer. Primer extension reactions were carried out using 10 mM activated NP monomers (3′-NH2-2AIpddA, 3′-NH2-2AIpddG, 3′-NH2-2AIpddC, 3′-NH2-2AIpddT), 50 mM MgCl2, 200 mM Na+-HEPES pH 8.0, 25 °C. P, primer.
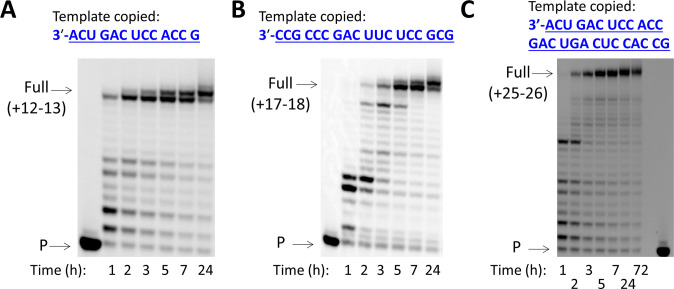
A long-standing goal in nonenzymatic nucleic acid replication has been to copy mixed-sequence templates long enough to encode useful functions such as molecular recognition or catalysis. The replication of longer sequences, at a sufficiently low mutation rate, could enable the evolution of selectively advantageous sequences in a population of replicating protocells. In addition, alleviating the need for specific auxiliaries such as activated downstream oligonucleotides3,4,14,20 would enable a much simpler means of replicating these longer oligomeric templates. We asked whether we could chemically copy a series of RNA templates of increasing length, to generate 3′-NP-DNA products in which the primer had been extended by at least 12, 17, or 25 nucleotides (Figure 6). Strikingly, the copying of RNA into 3′-NP-DNA was efficient in all three cases, with average single-nucleotide extension yields of 96–97% and full length yields of 67 ± 2, 63 ± 1 and 49 ± 1%. Thus, the copying all four letters in RNAs longer than 12 nts into 3′-NP-DNA is feasible using 2AI-activated 3′-amino mononucleotides. To our knowledge, this is the first demonstration of the nonenzymatic copying of templates that are potentially of sufficient length to encode catalytic activity,21,22 if sequence length requirements for catalysis are similar for 3′-NP-DNA and DNA or RNA sequences. Our findings suggest that the nonenzymatic propagation of genetic information containing four-letter mixed-sequences may be possible.
Figure 6.
Efficient chemical copying of long mixed-sequence RNA templates into 3′-NP-DNA using 3′-NH2-2AIpddNs, with templates (A) 13 nucleotides, (B) 18 nucleotides, and (C) 26 nucleotides in length. All primer extension reactions were carried out in solution using 10 mM activated NP monomers (3′-NH2-2AIpddA, 3′-NH2-2AIpddG, 3′-NH2-2AIpddC, 3′-NH2-2AIpddT), 50 mM MgCl2, 200 mM Na+-HEPES pH 8.0, 25 °C. P, primer.
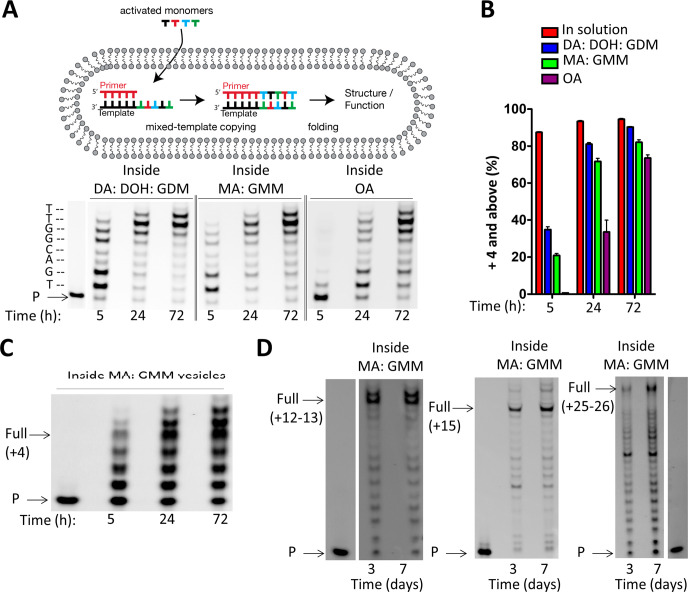
Finally, we asked whether the efficient copying of RNA templates into 3′-NP-DNA containing all four letters was compatible with compartmentalization. We repeated the RNA copying reactions within model protocell vesicles prepared with three different membrane compositions (decanoate:decanol:glycerol monodecanoate 4:1:1 (DA:DOH:GDM), myristoleate:glycerol monomyristoleate 2:1 (MA:GMM), and oleate (OA)), all of which generate robust vesicles that are permeable to polar solutes.23,24 In terms of prebiotic relevance, similar fatty acids have been synthesized using Fischer–Tropsch-type reactions25 in potentially prebiotic conditions, by iterated cycles of aldol condensation and reduction,26 and short-chain fatty acids have been identified in meteorites.27 The primer-template duplex was encapsulated within vesicles, and unencapsulated material removed by size exclusion chromatography. Activated mononucleotides were then added to the outside of the vesicles to initiate the reaction, in order to simulate the uptake of nutrients, by a heterotrophic protocell, such as nucleotides from the surrounding environment. As observed previously, the template-directed synthesis was slower within vesicles than in solution, owing in part to mononucleotides having to cross the membranes in order to initiate primer extension (Figure 7A).5 In addition, the reaction was slower with increasing fatty acid chain length, suggesting that entry of the activated monomers into the vesicle interior was rate limiting. Nonetheless, a high yield of full-length primer extension products was observed within membrane vesicles of all three compositions after 72 h (Figure 7A,B). Similar results were observed for primer extension on the randomized template (3′-NNNN-5′, Figure 7C), as well as on longer RNA templates leading to primer extension by 12–13, 17–18, and 25–26 nucleotides (Figure 7D). The latter primer extension reactions were conducted in MA:GMM vesicles only, given that reaction within these membranes was of intermediate efficiency (slower than within DA:DOH:GDM, but faster than within OA). This is a significant improvement over previously reported mixed-sequence RNA-template-directed polymerization within membrane compartments, which has been limited to primer extension by at most 5 nucleotides.5,24
Figure 7.
Chemical copying of mixed-sequence RNA templates into 3′-NP-DNA within model protocells. (A) Schematic representation of RNA copying leading to products long enough to exhibit structure and/or function. Chemical copying of RNA template (3′-AC UGC CAA-5′) into 3′-NP-DNA within vesicles of three different membrane compositions. (B) Comparison of time course reactions in solution (SI Figure S7) and inside vesicles using 3′-NH2-2AIpddNs. Data points are reported as the mean ± s.d. from triplicate experiments. (C) Chemical copying of a randomized pool of RNA templates (Figure 3, 3′-NNNN UU-5′) into 3′-NP-DNA using 3′-NH2-2AIpddNs within MA: GMM vesicles, where N = A, G, C, or U in the template. D) Chemical copying of long mixed-sequence RNA template into 3′-NP-DNA using 3′-NH2-2AIpddNs, with templates greater than 10 (3′-ACU GAC UCC ACC G), 15 (3′-CCG CCC GAC UUC UCC GCG), or 25 (3′-ACU GAC UCC ACC GAC UGA CUC CAC CG) nucleotides in length within MA:GMM vesicles (as shown in Figure 6). It should be noted that the lanes containing the primer species only were from the same PAGE experiment as the primer extension reaction. All primer extension reactions were carried out using 10 mM activated NP monomers (3′-NH2-2AIpddA, 3′-NH2-2AIpddG, 3′-NH2-2AIpddC, 3′-NH2-2AIpddT), 50 mM MgCl2, 200 mM sodium citrate, 200 mM Na+-bicine pH 8.5, 25 °C. P, primer. Control reactions outside of vesicles are reported in the SI.
Our results suggest that the accurate nonenzymatic copying of RNA templates is feasible with the four canonical nucleobases of the modern genetic alphabet, if the corresponding nucleotides are activated with the prebiotically plausible activating compound, 2AI.28 In contrast, previous studies using monomers that are highly activated, and which react directly as monomers with the growing primer terminus, have exhibited comparatively poor fidelity.7 The copying of RNA with activated 2AI substrates, which has been shown to proceed through a covalent dinucleotide intermediate,17 may exhibit higher fidelity as a result of the geometric preorganization of the imidazolium-bridged dinucleotide. Further studies will be required to measure the context dependent fidelity of template copying in these systems accurately, and to elucidate the reasons behind the different fidelities exhibited as a consequence of these distinct reaction pathways fully. The combination of the imidazolium-bridged intermediate pathway with the use of 2-thio-U/T in place of U/T may lead to further enhanced fidelity, which would enable the propagation of additional functionally important genetic information.
The greater nucleophilicity of the 3′-amine of activated 3′-amino-2′,3′-dideoxyribonucleotides allows for faster and more extensive primer extension than previously observed with identically activated ribonucleotides. This raises the intriguing question of whether RNA copying with ribonucleotides could be improved by making the 3′-hydroxyl of ribonucleotides more nucleophilic. One possibility would be to replace the catalytic metal ion Mg2+ with a different divalent cation with a lower pKa. We have recently shown that replacing Mg2+ with Fe2+ results in faster primer extension at neutral pH; however, Fe2+ catalyzes very rapid monomer hydrolysis, and also forms insoluble oxyhydroxide complexes at pH values above 7.5, which strongly inhibit the primer extension reaction.29 Another possibility, currently under investigation in our laboratory, would be to employ a metal chelating ligand, either noncovalently or covalently attached to the RNA, to help to bind and precisely position the catalytic metal ion so as to lead to enhanced catalysis. Enhanced metal ion catalysis combined with other approaches, such as the continuous reactivation of hydrolyzed monomers,30 and the use of activated downstream helper oligonucleotides, may ultimately lead to the identification of a prebiotically realistic means for the effective copying of RNA templates.
The heterotrophic protocell model posits the emergence of functional nucleic acid structures in a complex and dynamic environment, where nutrients could be assimilated from the external environment. We have now shown that activated mononucleotides alone can efficiently participate in the template-directed synthesis of a genetic polymer to produce mixed-sequence products long enough to encode advantageous functions, all within prebiotically plausible model protocell vesicles. This advance raises the possibility that 3′-NP-DNA could potentially form the genetic basis for an artificial form of cellular life with a biochemistry distinct from that of extant biological life. However, several obstacles must be overcome before this possibility can be realized. First, the efficiency of template directed 3′-NP-DNA synthesis must be further increased (e.g., from stepwise yields of >96% to >99%). The accumulation of stalled incomplete copying products currently limits the yield of full-length copies, but the reasons for the accumulation of these partial copies are unclear. Stalling could be due to misincorporation, chain-terminating chemical amine modifications, or the presence of template bound oligomers that block continued primer extension; further work is required to determine whether some or all of these possibilities lead to stalled primer extension products. In addition, template regions may have varying degrees of secondary structure, which may impact the efficiency of nonenzymatic primer extension chemistry. Another issue with the use of activated 3′-amino-2′,3′-dideoxyribonucleotides as substrates for template copying is that these monomers decay predominantly through 5′-3′ cyclization. At present there is no known pathway by which such cyclic nucleotides can be recycled into activated monomers, suggesting that a continuous flow system to bring in new activated monomers and remove cyclized dead-end products might be required to achieve continuous template copying in the 3′-NP-DNA system. Finally, the problem of duplex strand separation or strand displacement synthesis must be solved to allow for multiple generations of genetic replication. If the above roadblocks can be overcome, the synthesis of a replicating, evolving cellular system may be possible.
Acknowledgments
The authors are indebted to Mr. Constantin Giurgiu, Dr. Anna Wang, and Dr. Tom Wright for helpful discussions, and to Dr. Victor Lelyveld for help with the LC-MS experiments. The authors thank the Szostak group for helpful feedback. J.W.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by a grant (290363) from the Simons Foundation to J.W.S. and by a grant from the NSF (CHE-1607034) to J.W.S. D.K.O. is a recipient of a Postdoctoral Research Scholarship (B3) from the Fonds de Recherche du Quebec–Nature et Technologies (FRQNT), Quebec, Canada, and a Postdoctoral Fellowship from Canadian Institutes of Health Research (CIHR) from Canada.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b04858.
Supplementary methods, Figures S1–S7, and Table S1 (PDF)
Author Contributions
∇ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Szostak J. W.; Bartel D. P.; Luisi P. L. Synthesizing Life. Nature 2001, 409, 387–390. 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Szostak J. W. The Narrow Road to the Deep Past: In Search of the Chemistry of the Origin of Life. Angew. Chem., Int. Ed. 2017, 56, 11037–11043. 10.1002/anie.201704048. [DOI] [PubMed] [Google Scholar]
- Prywes N.; Blain J. C.; Del Frate F.; Szostak J. W. Nonenzymatic Copying of RNA Templates Containing All Four Letters Is Catalyzed by Activated Oligonucleotides. eLife 2016, 5, e17756 10.7554/eLife.17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Prywes N.; Tam C. P.; O'Flaherty D. K.; Lelyveld V. S.; Izgu E. C.; Pal A.; Szostak J. W. Enhanced Nonenzymatic RNA Copying with 2-Aminoimidazole Activated Nucleotides. J. Am. Chem. Soc. 2017, 139, 1810–1813. 10.1021/jacs.6b13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Flaherty D. K.; Kamat N. P.; Mirza F. N.; Li L.; Prywes N.; Szostak J. W. Copying of Mixed-Sequence RNA Templates inside Model Protocells. J. Am. Chem. Soc. 2018, 140, 5171–5178. 10.1021/jacs.8b00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Zhang N.; Blain J. C.; Szostak J. W. Synthesis of N3′-P5′-Linked Phosphoramidate DNA by Nonenzymatic Template-Directed Primer Extension. J. Am. Chem. Soc. 2013, 135, 924–932. 10.1021/ja311164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Blain J. C.; Zielinska D.; Gryaznov S. M.; Szostak J. W. Fast and Accurate Nonenzymatic Copying of an RNA-like Synthetic Genetic Polymer. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 17732–17737. 10.1073/pnas.1312329110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas Stütz J. A.; Richert C. A Steroid Cap Adjusts the Selectivity and Accelerates the Rates of Nonenzymatic Single Nucleotide Extensions of an Oligonucleotide. J. Am. Chem. Soc. 2001, 123, 12718–12719. 10.1021/ja011448i. [DOI] [PubMed] [Google Scholar]
- Schrum J. P.; Ricardo A.; Krishnamurthy M.; Blain J. C.; Szostak J. W. Efficient and Rapid Template-Directed Nucleic Acid Copying Using 2′-Amino-2′,3′-Dideoxyribonucleoside–5′-Phosphorimidazolide Monomers. J. Am. Chem. Soc. 2009, 131, 14560–14570. 10.1021/ja906557v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tereshko V.; Gryaznov S.; Egli M. Consequences of Replacing the DNA 3′-Oxygen by an Amino Group: High-Resolution Crystal Structure of a Fully Modified N3′ → P5′ Phosphoramidate DNA Dodecamer Duplex. J. Am. Chem. Soc. 1998, 120, 269–283. 10.1021/ja971962h. [DOI] [Google Scholar]
- Ding D.; Gryaznov S. M.; Wilson W. D. NMR Solution Structure of the N3′ → P5′ Phosphoramidate Duplex d(CGCGAATTCGCG)2 by the Iterative Relaxation Matrix Approach. Biochemistry 1998, 37, 12082–12093. 10.1021/bi980711y. [DOI] [PubMed] [Google Scholar]
- Gryaznov S. M.; Lloyd D. H.; Chen J. K.; Schultz R. G.; DeDionisio L. A.; Ratmeyer L.; Wilson W. D. Oligonucleotide N3′ → P5′ Phosphoramidates. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 5798–5802. 10.1073/pnas.92.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryaznov S. M. Synthesis and Properties of the Oligonucleotide N3′→P5′ Phosphoramidates. Nucleosides Nucleotides 1997, 16, 899–905. 10.1080/07328319708006106. [DOI] [Google Scholar]
- Richert C.; Hänle E. Enzyme-Free Replication with Two or Four Bases. Angew. Chem., Int. Ed. 2018, 57, 8911–8915. 10.1002/anie.201803074. [DOI] [PubMed] [Google Scholar]
- Walton T.; Szostak J. W. A Highly Reactive Imidazolium-Bridged Dinucleotide Intermediate in Nonenzymatic RNA Primer Extension. J. Am. Chem. Soc. 2016, 138, 11996–12002. 10.1021/jacs.6b07977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurgiu C.; Li L.; O’Flaherty D. K.; Tam C. P.; Szostak J. W. A Mechanistic Explanation for the Regioselectivity of Nonenzymatic RNA Primer Extension. J. Am. Chem. Soc. 2017, 139, 16741–16747. 10.1021/jacs.7b08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton T.; Szostak J. W. A Kinetic Model of Nonenzymatic RNA Polymerization by Cytidine-5′-Phosphoro-2-Aminoimidazolide. Biochemistry 2017, 56, 5739–5747. 10.1021/acs.biochem.7b00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser A.; Spies S.; Lommel T.; Richert C. Template-Directed Synthesis in 3′- and 5′-Direction with Reversible Termination. Angew. Chem., Int. Ed. 2012, 51, 8299–8303. 10.1002/anie.201203859. [DOI] [PubMed] [Google Scholar]
- Sanger F.; Nicklen S.; Coulson A. R. DNA Sequencing with Chain-Terminating Inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977, 74, 5463–5467. 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deck C.; Jauker M.; Richert C. Efficient Enzyme-Free Copying of All Four Nucleobases Templated by Immobilized RNA. Nat. Chem. 2011, 3, 603–608. 10.1038/nchem.1086. [DOI] [PubMed] [Google Scholar]
- Ferré-D’Amaré A. R.; Scott W. G. Small Self-Cleaving Ribozymes. Cold Spring Harbor Perspect. Biol. 2010, 2, a003574. 10.1101/cshperspect.a003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W.The Eightfold Path to Non-Enzymatic RNA Replication. J. Syst. Chem. 2012, 3, 2. 10.1186/1759-2208-3-2. [DOI] [Google Scholar]
- Mansy S. S.; Schrum J. P.; Krishnamurthy M.; Tobé S.; Treco D. A.; Szostak J. W. Template-Directed Synthesis of a Genetic Polymer in a Model Protocell. Nature 2008, 454, 122–125. 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K.; Szostak J. W. Nonenzymatic Template-Directed RNA Synthesis Inside Model Protocells. Science 2013, 342, 1098–1100. 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom T. M.; Ritter G.; Simoneit B. R. T. Lipid Synthesis under Hydrothermal Conditions by Fischer–Tropsch Reactions. Origins Life Evol. Biospheres 1999, 29, 153–166. 10.1023/A:1006592502746. [DOI] [PubMed] [Google Scholar]
- Bonfio C.; Caumes C.; Duffy C. D.; Patel B. H.; Percivalle C.; Tsanakopoulou M.; Sutherland J. D. Length-Selective Synthesis of Acylglycerol-Phosphates through Energy-Dissipative Cycling. J. Am. Chem. Soc. 2019, 141, 3934–3939. 10.1021/jacs.8b12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless J. G.; Yuen G. U. Quantification of Monocarboxylic Acids in the Murchison Carbonaceous Meteorite. Nature 1979, 282, 396–398. 10.1038/282396a0. [DOI] [Google Scholar]
- Fahrenbach A. C.; Giurgiu C.; Tam C. P.; Li L.; Hongo Y.; Aono M.; Szostak J. W. Common and Potentially Prebiotic Origin for Precursors of Nucleotide Synthesis and Activation. J. Am. Chem. Soc. 2017, 139, 8780–8783. 10.1021/jacs.7b01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L.; Engelhart A. E.; Zhang W.; Adamala K. P.; Szostak J. W. Catalysis of Template-Directed Nonenzymatic RNA Copying by Iron (II). J. Am. Chem. Soc. 2018, 140, 15016–15021. 10.1021/jacs.8b09617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani A.; Russell D. A.; Javelle T.; Sutherland J. D. A Light-Releasable Potentially Prebiotic Nucleotide Activating Agent. J. Am. Chem. Soc. 2018, 140, 8657–8661. 10.1021/jacs.8b05189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.