Abstract
The chemical replication of RNA inside fatty acid vesicles is a plausible step in the emergence of cellular life. On the primitive Earth, simple protocells with the ability to import nucleotides and short oligomers from their environment could potentially have replicated and retained larger genomic RNA oligonucleotides within a spatially defined compartment. We have previously shown that short 5′-phosphoroimidazolide-activated “helper” RNA oligomers enable the nonenzymatic copying of mixed-sequence templates in solution, using 5′-phosphoroimidazolide-activated mononucleotides. Here, we report that citrate-chelated Mg2+, a catalyst of nonenzymatic primer extension, enhances fatty acid membrane permeability to such short RNA oligomers up to the size of tetramers, without disrupting vesicle membranes. In addition, selective permeability of short, but not long, oligomers can be further enhanced by elevating the temperature. The ability to increase the permeability of fatty acid membranes to short oligonucleotides allows for the nonenzymatic copying of RNA templates containing all four nucleotides inside vesicles, bringing us one step closer to the goal of building a protocell capable of Darwinian evolution.
Introduction
Compartmentalization was an essential step toward the origin of cellular life.1,2 Primitive cells may have consisted of membrane-bound compartments, possibly assembled from simple monoacyl lipids,3 which encapsulated genetic materials such as RNA.4,5 A balance between RNA retention and membrane permeability could have been a critical feature of primitive heterotrophic protocells able to carry out internal RNA copying chemistry. Prior to the evolution of biosynthetic pathways, model protocells would have had to acquire nutrients from their environment while releasing reaction byproducts as waste in order to provide a favorable chemical environment for replicative reactions.6−8 However, the ability of a protocell to retain larger genomic RNA oligomers would also have been necessary in order for protocells to evolve functional RNAs and to distribute replicated strands to daughter protocells.9,10 Genomic RNA strands are expected to have acquired mutations during replication, and mutations that led to the emergence of functional RNA sequences11 could then have imparted superior fitness to their host protocell. The barrier to the free diffusion of long RNA molecules imposed by the protocell membrane would have ensured that oligomers related by descent and therefore closely related in sequence were kept together over time and could therefore contribute to their further optimization through Darwinian evolution.12−14 Similar arguments would apply to protocells that contained nucleic acid based genetic materials other than RNA, but might not be relevant to radically different genetic systems.
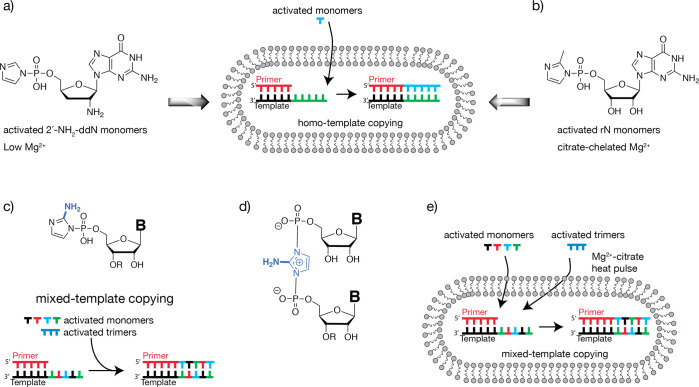
Over the past decade, our laboratory has investigated a variety of model systems, with the goal of developing simple artificial cell-like systems capable of Darwinian evolution. Our studies have also been motivated by the desire to test the plausibility of different potential pathways leading to the origin of cellular life on the early Earth. In particular, we have sought to demonstrate the replication of arbitrary RNA templates within membrane compartments, and we have gradually approached this goal through a step-by-step series of experimental advances. We first demonstrated the efficient chemical copying of homopolymeric oligo-C templates within fatty acid vesicles using 5′-phosphoroimidazoyl-activated 2′-amino-2′-3′-dideoxynucleotides (Figure 1A).15 These activated mononucleotides undergo rapid, template-directed polymerization, in the presence of 1-(2-hydroxyethyl)imidazole as an organocatalyst, generating an N2′–P5′-linked phosphoramidate genetic polymer.15,16 The amine nucleophile enables efficient RNA copying in the absence of Mg2+, thus allowing copying to proceed within fatty acid vesicles. The presence of low concentrations of Mg2+ was subsequently shown to increase nucleotide permeability through fatty acid membranes,17 but levels of Mg2+ above 4 mM destabilize such membranes. Because efficient RNA template copying using activated ribonucleotides requires very high Mg2+ concentrations, the low Mg2+ threshold for fatty acid vesicle stability results in very poor template-directed RNA synthesis within vesicles, leading to a paradoxical issue for the RNA system. In 2013, our group showed that this problem could be circumvented through the use of citrate-chelated Mg2+, which protects fatty acid vesicles from Mg2+-induced disruption, while simultaneously allowing the RNA copying reaction to proceed (Figure 1B).18 Despite these advances, only homopolymers have been efficiently copied inside vesicles. However, in order for functional RNA sequences to replicate and evolve, a way for mixed-sequence RNAs to be copied within vesicles must be identified.
Figure 1.
Schematic representation of recent advances in nonenzymatic primer extension. (a) Homopolymeric RNA templates copied using activated 2′-amino-2′-3′-dideoxynucleotides within fatty acid vesicles at low Mg2+ concentration, generating an N2′–P5′-linked phosphoramidate polymer. (b) Homopolymeric RNA templates copied using activated ribonucleotides within fatty acid vesicles in the presence of citrate-chelated Mg2+, generating an RNA polymer. (c) Chemical structure of ribonucleotides activated with 2-aminoimidazole. Activated trinucleotides catalyze primer extension and allow for the one-pot copying of RNA templates containing all four nucleotides. R signifies either hydrogen for activated mononucleotides, or dinucleotides for activated trimers. (d) Imidazolium-bridged intermediate responsible for efficient primer extension. R is either a hydrogen or a dinucleotide. (e) Mixed-sequence RNA templates being copied using activated ribonucleotides and trinucleotides within fatty acid vesicles in the presence of citrate-chelated Mg2+.
It has been known for more than two decades that the presence of an imidazoyl-activated mononucleotide that could bind to the template in the primer+2 position, i.e., downstream of the monomer that is poised to react with the primer, greatly enhances the efficiency of primer extension.19 While reinvestigating this phenomenon, our group discovered that 5′-phosphoro-2-methylimidazoyl-activated trinucleotide downstream binders were optimal for the catalysis of template-directed primer extension and enabled the copying of templates containing all four nucleotides.20 This was particularly helpful for copying the traditionally slower reacting mononucleotides adenosine (A) and uridine (U). Efforts to elucidate the mechanism of the primer extension reaction led to the discovery of an imidazolium-bridged dinucleotide intermediate that is formed by the reaction of two activated monomers, which is responsible for the bulk of primer extension using activated monomers (Figure 1C).21 In addition, we have recently discovered a superior leaving group, 2-aminoimidazole, which significantly accelerates the copying of a variety of short RNA templates (Figure 1D).22 In conjunction with the use of downstream-activated trinucleotide helpers, the novel 5′-phosphoro-2-aminoimidazoyl-activated mononucleotides allow efficient copying of various RNA templates with mixed sequences (Figure 1D). The presence of the 2-amino functional group of the imidazole increases the stability of the bridged intermediate and accounts for the more efficient RNA copying.23 Furthermore, a prebiotically plausible synthetic route to 2-aminoimidazole has been discovered, and the resulting activated mononucleotides primarily generate the natural 3′–5′ phosphodiester linkages during template copying.24−26
A major hurdle in envisioning trimer-assisted RNA copying inside model protocells arises from the low permeability of fatty acid membranes to short oligomers.17 Because the sequence-general copying of RNA within vesicles is likely a necessary step in the origin of cellular life, we were interested in finding conditions where short activated oligonucleotides could diffuse into vesicles and participate in the nonenzymatic copying of RNA templates with mixed sequences. Several factors influence fatty acid membrane permeability including membrane composition, solute size and polarity, the presence of divalent cations such as Mg2+, and temperature.15,17,27−30 Here we show that the rate of passive diffusion of short RNA oligomers through model fatty acid membranes is significantly stimulated by the presence of citrate-chelated Mg2+, thereby enabling the trimer-assisted copying of mixed-sequence RNA templates. We describe, for the first time, the copying of mixed-sequence RNA templates inside vesicles with two different fatty acid membrane compositions, using 2-aminoimidazoyl-activated mononucleotides, 2-aminoimidazoyl-activated helper trimers, and citrate-chelated Mg2+ (Figure 1E). In addition, we show that a 90 °C heat pulse further increases the primer extension efficiency. Our demonstration of the copying of short mixed-sequence RNA templates within fatty acid vesicles brings us one step closer to the laboratory assembly of model protocells capable of replication and evolution.
Results and Discussion
Effect of Mg2+ on Fatty Acid Permeability
We assessed the permeability of fatty acid membranes to mono- and oligo-ribonucleotides using vesicles prepared from a 2:1 mixture of myristoleic acid (MA)/monomyristolein (GMM) since these membrane compositions are known to be more tolerant to ionic solutes31,32 and high temperatures17 than other monoacyl lipids. Although these specific lipids are prebiotically unlikely, they are a useful model system, and similar saturated lipids have been synthesized via Fischer–Tropsch-type reactions in potentially prebiotic conditions.33 In addition, short-chain fatty acids have been identified in meteorites.34 Radiolabeled RNAs ranging from mononucleotides to pentanucleotides were encapsulated within MA/GMM vesicles, and leakage was monitored over time at room temperature (SI Figure 1). To examine the effect of chelated Mg2+ on the permeability of MA/GMM membranes, we monitored the leakage of the above series of RNAs in the presence and absence of citrate-chelated Mg2+ (50 mM Mg2+ mixed with 200 mM citrate, Figure 2a). In the absence of Mg2+ cations, the unmodified nucleotide monophosphate AMP was unable to cross the vesicle membranes, and more than 90% of the nucleotide remained encapsulated after 24 h. Short 5′-phosphorylated oligonucleotides from dimers to pentamers were similarly unable to cross the membrane. In the presence of Mg2+-citrate, however, the MA/GMM membranes became more permeable, with approximately 91% of a mononucleotide, 63% of a dinucleotide, and 46% of a trinucleotide diffusing across the vesicle membrane after 24 h (Figure 2a).
Figure 2.
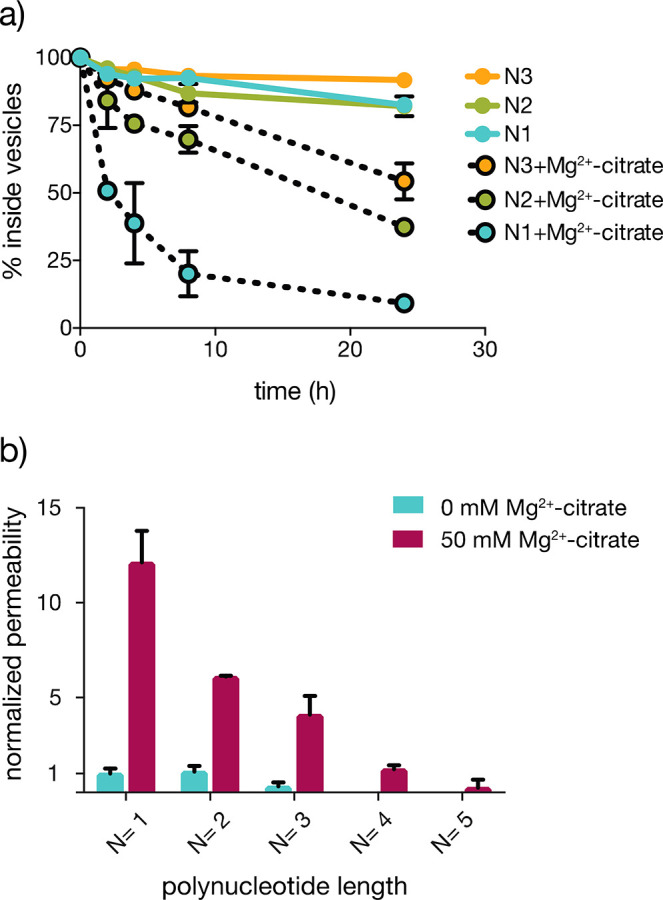
Mg2+-citrate enhances RNA permeability through MA/GMM membranes. (a) The retention of mono-, di-, and trinucleotides within MA/GMM vesicles as a function of chain length (N), in the presence or absence of 50 mM Mg2+ chelated with 200 mM citrate, at 22 °C. (b) Permeability coefficients for various RNAs in the absence (blue) and presence (maroon) of Mg2+-citrate, normalized to the permeability of AMP in the absence of Mg2+-citrate. The sequences of RNA used to represent oligomers of different lengths were nonactivated AMP (N = 1), GA (N = 2), AGC (N = 3), AAAA (N = 4), and AAAAA (N = 5). All oligonucleotides were 5′-phosphorylated; permeability studies were conducted in 0.2 M Na+-bicine, pH 8.5. n = 3; error bars represent standard deviation.
To quantitatively compare permeabilities as a function of RNA size, we calculated the permeability coefficient according to a classical diffusion mechanism using Fick’s law35 (SI Table 1). We then expressed relative permeability coefficients as the ratio with respect to the permeability of AMP through MA/GMM vesicles in the absence of Mg2+-citrate (Figure 2b). The presence of citrate-chelated Mg2+ increased permeability 12-fold for the mononucleotide AMP. Dimers and trimers experienced similar increases in permeability (6- and 12-fold, respectively) in the presence of citrate-chelated Mg2+ (SI Table 1). As expected, RNA permeability decreased monotonically with increasing length. While a tetramer exhibited measurable permeability in the presence of Mg2+-citrate, a pentamer was essentially impermeable.
The enhanced membrane permeability to oligonucleotides in the presence of Mg2+-citrate could, in principle, be due to interactions with either the RNA or the membrane. Previous work by Mansy et al. showed that free Mg2+ likely improved nucleotide permeability by neutralizing the negative charge density on nucleotide diphosphates.17 However, the Mg2+-citrate complex is much larger than free Mg2+ and may retain a net negative charge, suggesting that a direct interaction with RNA is unlikely to be the cause of increased permeability. Instead, changes to the membrane itself may be responsible. Previous anisotropy studies showed that citrate-chelated Mg2+ increases membrane fluidity, indicating that chelated Mg2+ interacts with the membrane sufficiently to change its physical properties.18 We used dynamic light scattering (DLS) measurements and observed that increasing Mg2+-citrate concentration increased the average apparent size of MA/GMM vesicles (Figure 3a), most likely due to aggregation,27 further supporting the hypothesis that chelated Mg2+ interacts with MA/GMM membranes. To test the hypothesis that citrate-chelated Mg2+ and not free, unchelated Mg2+ was responsible for the measured increase in RNA permeability, we incubated MA/GMM vesicles with 4 mM MgCl2, the maximum concentration of MgCl2 tolerated by these membranes and far more than the estimated <1 mM residual free Mg2+ in a solution of 50 mM Mg2+/200 mM citrate.18 While 4 mM MgCl2 increased AMP permeability by approximately 3-fold relative to the absence of Mg2+ (Figure 3b), this effect was much smaller than the 12-fold increase observed with 50 mM citrate-chelated Mg2+, indicating that chelated and not free Mg2+ was largely responsible for the increased nucleotide permeability of MA/GMM membranes.
Figure 3.
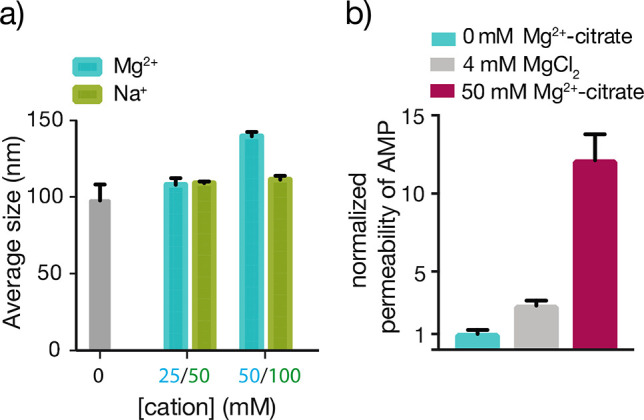
Effects of Mg2+-citrate on MA/GMM vesicles. (a) The presence of 50 mM Mg2+/200 mM citrate induces an increase in average vesicle size, determined by DLS. Replacing the Mg2+ cation with Na+ (100 mM) does not increase average particle size. n = 3; error bars represent standard deviation. (b) Permeability coefficients of AMP through MA/GMM vesicles in the presence of 4 mM MgCl2 (gray) or 50 mM Mg2+-citrate (maroon) normalized to AMP permeability in the absence of Mg2+ (blue).
Effect of High Temperature on Fatty Acid Permeability
We considered whether other environmental factors, especially factors important for RNA replication, could further increase membrane permeability. Transient exposure to high temperature is a simple way of driving RNA strand separation and has previously been shown to enhance fatty acid membrane permeability to short oligomers.17 We therefore decided to investigate the effect of elevated temperature combined with Mg2+-citrate on RNA permeability. In the absence of Mg2+-citrate, incubating vesicles at 90 °C significantly increased RNA mono- and dinucleotide permeability (Figure 4a). However, a trimer diffused across MA/GMM membranes slowly, with only 15% crossing the membrane in 1 h even at 90 °C in the absence of Mg2+. Strikingly, the combination of chelated Mg2+ and elevated temperature significantly increased trimer permeability (Figure 4a, red). After 1 h of continuous exposure to 90 °C or 1.3 h of thermal cycling (40 min at 90 °C), more than half of the trimer had crossed the vesicle membrane. Thermal cycling has previously been shown to effectively separate annealed strands inside fatty acid vesicles and also provides multiple opportunities to rearrange secondary structures.17 Interestingly, the total trimer leakage is approximately proportional to the total time at 90 °C, suggesting that the cycling itself had little effect.
Figure 4.
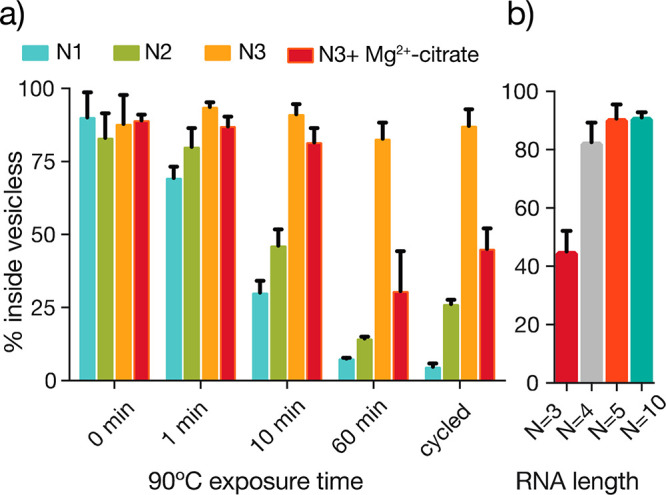
Incubation at 90° C increases Mg2+-citrate-enhanced RNA permeability of MA/GMM vesicles. (a) Oligomer permeability of nucleotide monophosphate (AMP), dinucleotide (NAD), and trinucleotide (pAGC) RNA through 2:1 MA/GMM membranes at 90 °C in the absence (blue, green, orange) and presence (red) of Mg2+-citrate (50 mM Mg2+/200 mM citrate). The cycled condition consists of 2 min at 25 °C and 2 min at 90 °C, repeated 20 times. (b) Retention of RNA oligomers in MA/GMM vesicles as a function of RNA size after 24 h of incubation with Mg2+-citrate at RT and an initial exposure to 90 °C for 1 h. n = 3; error bars represent standard deviation.
Because the ultimate goal is to copy encapsulated RNA templates following the entry of externally supplied short oligonucleotide substrates and catalysts, the environmental conditions used to enable permeability to short oligonucleotides must not lead to leakage or loss of longer genomic RNAs. We therefore assessed the critical length of RNA consistent with membrane permeation. Vesicles containing tri-, tetra-, penta-, or decamer oligonucleotides were exposed to 50 mM Mg2+-citrate at room temperature (RT) for 24 h and 90 °C for 1 h, conditions that yielded high membrane permeability to short RNA fragments. The tetramer was the longest oligonucleotide to exhibit measurable permeability (Figure 4b), while less than 10% of pentamer or decamer leakage was detected after 24 h of vesicle exposure to Mg2+-citrate and 1 h at 90 °C. We also examined the stability and permeability of a short-chain, prebiotically plausible membrane made from decanoic acid (DA)/decanol (DOH):/glycerol monodecanoate (GDM) (4:1:1) in the presence of Mg2+-citrate.
While these vesicles were stable in morphology in the presence of citrate-chelated Mg2+ for several hours, we observed substantial leakage of oligomers 10 or 30 nucleotides in length (SI Figure 2a) over the course of 24 h. However, DA/DOH/GDM membranes also exhibited higher permeability to short oligomers compared to the longer chain MA/GMM membranes; for example, when incubated with 50 mM Mg2+-citrate, DA/DOH/GDM vesicles released 80% of encapsulated RNA trimers (SI Figure 2b) after a few hours while retaining longer oligomers. As result, we expect RNA copying within the shorter chain DA/DOH/GDM vesicles to be faster than in MA/GMM vesicles.
Nonenzymatic RNA Template Copying in Vesicles
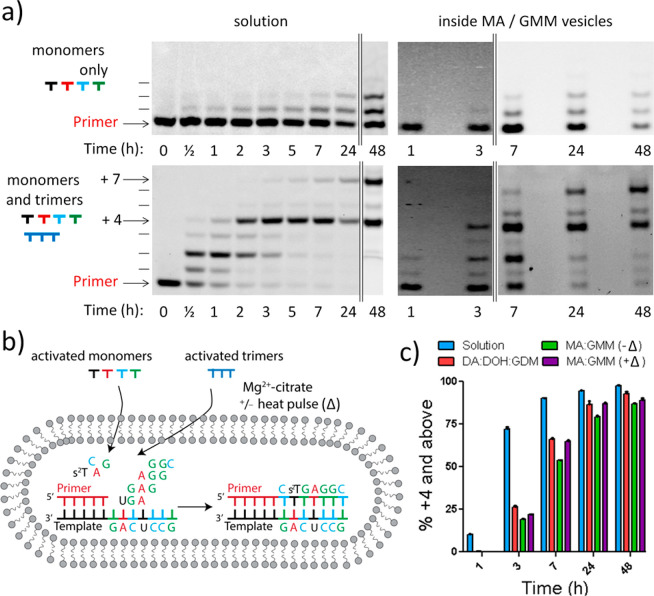
Having determined conditions that enable trinucleotide permeability while maintaining membrane stability, we asked whether such RNA oligomers, when added to the outside of vesicles, could diffuse to the interior and participate in template-copying reactions. We first used a template sequence similar to a previously studied primer extension, in which the template sequence 3′-GACUCCG-5′ was being copied, to examine the effect of replacing free Mg2+ with citrate-chelated Mg2+ (Figure 5). In this reaction, the activated trimers act as catalysts to enable the efficient and sequence-general nonenzymatic copying of RNA templates with 2-aminoimidazole-activated monomers; the activated trimers are thought to promote primer extension via the formation of an imidazolium-bridged monomer–trimer intermediate (Figure 1d, where R is a dinucleotide).20,21 We used 2-thio-ribothymidine as an activated monomer in place of uridine, as we have previously shown that this improves both the fidelity and rate of primer extension.36 As expected, the 5′-activated trimers are required for efficient primer extension on a mixed sequence template in solution (Figure 5a, left-hand side).20,22 The copying reaction proceeds almost quantitatively to +4 by trimer-assisted monomer addition; in addition, a +7 product appears through the slower ligation of the last activated trimer.
Figure 5.
Nonenzymatic primer extension on a mixed template inside vesicles. All reactions were conducted at pH 8.5, 22 °C, 200 mM Na+-bicine, 50 mM Mg2+–200 mM citrate, 10 mM 2AI-activated monomers, and 0.5 mM 2AI-activated trimers. (a) PAGE analysis of the products of the primer extension reaction in solution and inside MA/GMM vesicles using RNA monomers and the four downstream helper trimers activated with 2-aminoimidazole. Arrows indicate the +4 and +7 (full-length) products. (b) Schematic diagram of a primer extension reaction incorporating all four RNA mononucleotides, with four respective downstream helper trimers as conducted in panel (a). (c) Product formation summary of trimer-assisted time course reactions in solution and inside vesicles using 2AI-activated nucleotides and trimers. +Δ represents a 1 min pulse at 90 °C. n = 3; error bars represent standard deviation. See SI Figures 3–6 for PAGE. See SI Figure 7 for primer extension decay plot. Full sequences of primer and template complexes can be found in the SI.
Although the reaction proceeds to a very high yield in the presence of citrate-chelated Mg2+, it is somewhat slower than seen previously with free Mg2+.18,22 To assess the efficiency of this reaction in vesicles, we first encapsulated the RNA primer/template duplex inside DA/DOH/GDM or MA/GMM vesicles. We then added 2-aminoimidazole-activated mononucleotides, A, 2-thioribothymidine (s2T), G, and C, as well as four activated trimers, UGA, GAG, AGG, and GGC, to the outside of the vesicles (Figure 5a, right-hand side). We observed the formation of full-length primer extension products inside vesicles when both monomers and trimers were added, but as expected observed negligible yields of fully extended products if only activated mononucleotides were added. The reaction proceeded more rapidly in free solution than inside either DA/DOH/GDM or MA/GMM vesicles, suggesting that entry of the activated trimers into the vesicle interior was rate limiting (Figure 5c). Nevertheless, the RNA monomers and trimers were capable of accessing the vesicle interior and were able to completely copy the encapsulated mixed-sequence template. Primer extension was more efficient within DA/DOH/GDM vesicles than within MA/GMM membranes, as expected from their higher permeability to mononucleotides and trinucleotides.
Since MA/GMM membranes are less permeable than DA/DOH/GDM membranes, and MA/GMM vesicles showed correspondingly less efficient internal template copying, we sought to enhance the encapsulated primer extension reaction by using a 1 min 90 °C heat pulse to allow for uptake of the activated trimers. We chose 1 min for the heat pulse since this does not cause significant degradation of 2-aminoimidazole-activated monomers or oligomers. We were pleased to observe a modest but reproducible increase in primer extension efficiency due to the short period of elevated temperature, suggesting that this treatment allowed for a sufficient influx of activated trimers to result in increased catalysis of primer extension.
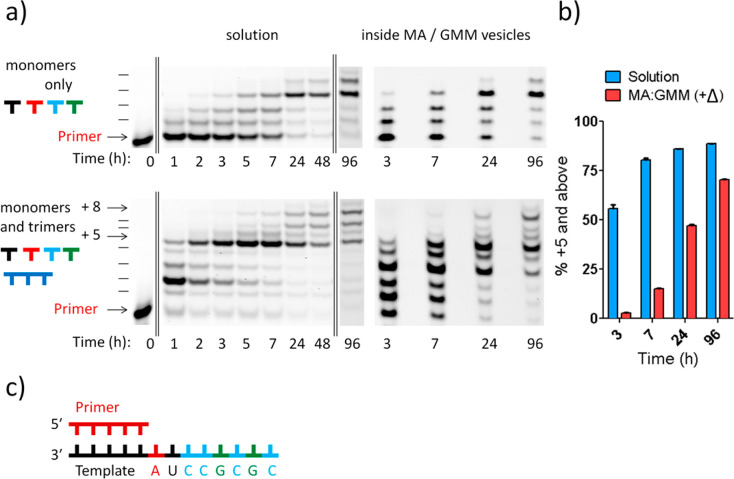
Having demonstrated the one-pot copying of a template containing all four nucleotides inside model protocells, we set out to demonstrate the generalizability of the RNA copying within vesicles. We copied a second mixed-sequence RNA, with the sequence 3′-AUCCGCGC-5′ in the template region (Figure 6), which shared minimal homology to the first template region (3′-GACU-5′). In this reaction, the first five nucleotides of the template, 3′-AUCCG-5′, are copied primarily by trimer-assisted monomer incorporation, with longer products being generated by slower ligation of the final trimer. The solution reaction using activated mononucleotides alone generated minimal formation of products +5 and above even after 96 h (<2%), whereas full extension was observed using trimer assistance after 24 h (>80% of +5 and above). The primer extension reaction was slower for this sequence relative to the first sequence, with approximately 75% product formation after 5 h in solution. We observed similar levels of +5 product (75%) after 96 h within MA/GMM vesicles (Figure 6) using an initial 90 °C heat pulse. The slower primer extension reaction on this template sequence may be due to the presence of a short region of self-complementarity (−CGCG−) within the RNA template. Nevertheless, efficient product formation was observed with the assistance of trimers, suggesting that the short oligomers may have competitively inhibited formation of the undesired secondary structure.
Figure 6.
Nonenzymatic primer extension on a second mixed template inside vesicles. All reactions were conducted at pH 8.5, 22 °C, 200 mM Na+-bicine, 50 mM Mg2+–200 mM citrate, 10 mM 2AI-activated monomers, and 0.5 mM 2AI-activated trimers (except for the 5′-GCG, which was 1 mM given that it was used for two extensions). (a) PAGE analysis of the products of the primer extension reaction in solution and inside MA/GMM vesicles using RNA monomers and the four downstream helper trimers activated with 2-aminoimidazole. Arrows indicate the +5 and +8 (full-length) products. (b) Product formation summary of trimer-assisted time course reactions in solution and inside vesicles using 2AI-activated nucleotides and trimers. +Δ represents a 1 min pulse at 90 °C. n = 3; error bars represent standard deviation. See SI Figures 8 and 9 for PAGE. See SI Figure 10 for primer decay plot. (c) Sequence context of the RNA template being copied. Full sequences of primer and template complexes can be found in the SI.
The nonenzymatic copying of mixed-sequence RNA templates within membrane vesicles represents a significant step toward the goal of generating an artificial life form capable of self-sustained reproduction and Darwinian evolution.
Conclusions
At present it is uncertain whether life could only emerge following one highly specific pathway under highly specific conditions, or whether there exist multiple pathways and sets of environmental conditions that could lead to life. One way to address this overarching question is to try to synthesize simple living cells in the laboratory. Of the many challenges that must be overcome in order to reach this goal, the experimental demonstration of nonenzymatic replication of a genetic polymer and the integration of genetic replication with cellular replication are perhaps the most daunting. Here we have taken a step toward the development of an integrated synthetic cellular system by combining recent advances in the nonenzymatic copying of mixed-sequence RNA templates with the recent discovery of conditions that make RNA copying chemistry compatible with fatty-acid-based model protocell membranes. In the course of combining these advances and thereby demonstrating sequence-general template copying within fatty acid vesicles, we found that both citrate-chelated Mg2+ and elevated temperature enhance the ability of short RNA oligonucleotides to spontaneously cross fatty-acid-based membranes, so that they can reach the interior of model protocells and catalyze the copying of mixed-sequence RNA templates. Nonenzymatic primer extension reactions inside vesicles become faster and more efficient as the membrane permeability of short RNA oligonucleotides increases, and even a brief exposure to 90 °C allows enough activated trimer to enter to enhance primer extension. The synergistic effect of Mg2+-citrate and high temperature on oligonucleotide-membrane permeability represents a very specific combination of environmental factors that are necessary for RNA copying within our current protocell model. If very early protocells were dependent on similarly specific environmental conditions to acquire metabolic building blocks, such as nucleotides and short oligonucleotides, from the external environment, the emergence of life might have been a very low probability event. However, continued research may well reveal much more permissive and widely available environmental conditions and chemical processes, consistent with a more facile origin of life. While our findings support a potential means of generating a synthetic artificial system capable of Darwinian evolution, our approach need not reflect the specific way that life originated on the early Earth.
Although short oligonucleotides can enter model protocells, it is clear that in the specific systems we have examined membrane permeation of catalytic trimers is rate limiting for internal nonenzymatic primer extension given the significant difference observed for extra- and intravesicular primer extension rates. This raises the question of whether there is an inevitable conflict between the need for primitive membrane-bound protocells to retain genomic and functional RNA molecules versus their need to access external mono- and oligo-ribonucleotides. One possibility is that other membrane compositions, not yet thoroughly studied, might allow for more ready uptake of oligonucleotides, while retaining overall stability. For example, a small fraction of mono- or diacyl amphiphiles that have longer chains37 or different headgroups38 stabilize membranes formed predominantly from short-chain amphiphiles; such mixed membranes could have advantageous permeability properties. Alternatively, the need for externally synthesized oligonucleotides could be circumvented if short oligonucleotides could be made internally and then retained. Such oligonucleotides might be generated internally either by the nontemplated reaction of activated monomers or by the breakdown of longer genomic or functional RNAs, or as partial replication products. Alternatively, the nonenzymatic assembly of functional sequences could have led to the emergence of catalysts such as nucleotide synthetase ribozymes. In this scenario, protocellular nutrients could have been reduced to nucleosides, rather than the activated mononucleotides and trinucleotides discussed herein.39 In conjunction with a plausible activation pathway, materials required for the copying of mixed-sequence template RNAs could have been assembled in situ.40 In this way, the evolution of internal metabolic reactions could have allowed early life to gradually become independent of strict and limiting environmental conditions. Finally, despite the fitness cost of membranes acting as a partial barrier to needed nutrients, membranes have the potential to colocalize RNA substrates8 and thus to improve reaction rates, which could also benefit the larger protocellular system. The findings described herein advance the goal of generating an artificial life form. In addition, our results add to a growing body of observations suggesting that simple physical principles may have coordinated the initial interactions between genomic building blocks and membranes that ultimately led to the emergence of cellular life.
Acknowledgments
J.W.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by a grant (290363) from the Simons Foundation to J.W.S. and by a grant from the NSF (CHE-1607034) to J.W.S. D.K.O. is a recipient of a Postdoctoral Research Scholarship (B3) from the Fonds de Recherche du Québec–Nature et Technologies (FRQNT), Quebec, Canada, and a Postdoctoral Fellowship from Canadian Institutes of Health Research (CIHR) from Canada. N.P.K. was supported by an appointment to the NASA Postdoctoral Program, administered by Oak Ridge Associated Universities through a contract with NASA, and is supported by the Searle Funds at The Chicago Community Trust. L.L. is a Life Sciences Research Foundation Fellow. The authors thank Lijun Zhou, Constantin Giurgiu, Chun Pong Tam, Sheref Mansy, Lin Jin, and Szostak laboratory members for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b00639.
Supplementary methods and Figures S1–S10 (PDF)
Author Present Address
⊥ F.N.M.: Yale School of Medicine, 333 Cedar Street, New Haven, Connecticut 06510, United States.
Author Present Address
∥ N.P.: Department of Plant and Environmental Sciences, Weizmann Institute, Rehovot, Israel.
Author Contributions
§ D. K. O’Flaherty and N. P. Kamat contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Szostak J. W.; Bartel D. P.; Luisi P. L. Nature 2001, 409, 387–390. 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Chen I. A.; Walde P. Cold Spring Harb. Perspect. Biol. 2010, 2 (7), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves W. R.; Deamer D. W. Biochemistry 1978, 17 (18), 3759–3768. 10.1021/bi00611a014. [DOI] [PubMed] [Google Scholar]
- Orgel L. E. J. Mol. Biol. 1968, 38 (3), 381–393. 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- Pace N. R.; Marsh T. L. Origins Life Evol. Biospheres 1985, 16 (2), 97–116. 10.1007/BF01809465. [DOI] [PubMed] [Google Scholar]
- Grochmal A.; Prout L.; Makin-Taylor R.; Prohens R.; Tomas S. J. Am. Chem. Soc. 2015, 137 (38), 12269–12275. 10.1021/jacs.5b06207. [DOI] [PubMed] [Google Scholar]
- Adamala K. P.; Engelhart A. E.; Szostak J. W. Nat. Commun. 2016, 7, 11041. 10.1038/ncomms11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat N. P.; Tobé S.; Hill I. T.; Szostak J. W. Angew. Chem., Int. Ed. 2015, 54, 11735–11739. 10.1002/anie.201505742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D.; Dworkin J. P.; Sandford S. A.; Bernstein M. P.; Allamandola L. J. Astrobiology 2003, 2, 371–381. 10.1089/153110702762470482. [DOI] [PubMed] [Google Scholar]
- Mansy S. S. Cold Spring Harbor Perspect. Biol. 2010, 2, a002188. 10.1101/cshperspect.a002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce G. F. Nature 1989, 338, 217–224. 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- Chan C. T. Y.; Dyavaiah M.; DeMott M. S.; Taghizadeh K.; Dedon P. C.; Begley T. J. PLoS Genet. 2010, 6, e1001247. 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmáry E.; Demeter L. J. Theor. Biol. 1987, 128 (4), 463–486. 10.1016/S0022-5193(87)80191-1. [DOI] [PubMed] [Google Scholar]
- Robertson M. P.; Joyce G. F.; Noller H. F.; Volpe T.; Martienssen R. A. Cold Spring Harbor Perspect. Biol. 2012, 4, a003608. 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy S. S.; Schrum J. P.; Krishnamurthy M.; Tobé S.; Treco D. a.; Szostak J. W. Nature 2008, 454 (7200), 122–125. 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrum J. P.; Ricardo A.; Krishnamurthy M.; Blain J. C.; Szostak J. W. J. Am. Chem. Soc. 2009, 131, 14560–14570. 10.1021/ja906557v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy S. S.; Szostak J. W. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (36), 13351–13355. 10.1073/pnas.0805086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K.; Szostak J. W. Science 2013, 342, 1098–1100. 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.; Orgel L. E. J. Am. Chem. Soc. 1992, 114 (14), 5496–5501. 10.1021/ja00040a002. [DOI] [PubMed] [Google Scholar]
- Prywes N.; Blain J. C.; Del Frate F.; Szostak J. W. eLife 2016, 5, e17756 10.7554/eLife.17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton T.; Szostak J. W. J. Am. Chem. Soc. 2016, 138, 11996–12002. 10.1021/jacs.6b07977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Prywes N.; Tam C. P.; O’Flaherty D. K.; Lelyveld V. S.; Izgu E. C.; Pal A.; Szostak J. W. J. Am. Chem. Soc. 2017, 139 (5), 1810–1813. 10.1021/jacs.6b13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton T.; Szostak J. W. Biochemistry 2017, 56 (43), 5739–5747. 10.1021/acs.biochem.7b00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurgiu C.; Li L.; O’Flaherty D. K.; Tam C. P.; Szostak J. W. J. Am. Chem. Soc. 2017, 139 (46), 16741–16747. 10.1021/jacs.7b08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenbach A. C.; Giurgiu C.; Tam C. P.; Li L.; Hongo Y.; Aono M.; Szostak J. W. J. Am. Chem. Soc. 2017, 139, 8780–8783. 10.1021/jacs.7b01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R.; Hongo Y.; Fahrenbach A. C. Chem. Commun. 2018, 54, 511–514. 10.1039/C7CC08489G. [DOI] [PubMed] [Google Scholar]
- Paula S.; Volkov A. G.; Van Hoek A. N.; Haines T. H.; Deamer D. W. Biophys. J. 1996, 70 (1), 339–348. 10.1016/S0006-3495(96)79575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treyer M.; Walde P.; Oberholzer T. Langmuir 2002, 18 (4), 1043–1050. 10.1021/la011111u. [DOI] [Google Scholar]
- Boon J. M.; Smith B. D. Curr. Opin. Chem. Biol. 2002, 6 (6), 749–756. 10.1016/S1367-5931(02)00399-X. [DOI] [PubMed] [Google Scholar]
- Davidsen J.; Mouritsen O. G.; Jorgensen K. Biochim. Biophys. Acta, Biomembr. 2002, 1564 (1), 256–262. 10.1016/S0005-2736(02)00461-3. [DOI] [PubMed] [Google Scholar]
- Monnard P.-A.; Apel C. L.; Kanavarioti A.; Deamer D. W. Astrobiology 2002, 2 (2), 139–152. 10.1089/15311070260192237. [DOI] [PubMed] [Google Scholar]
- Chen I. A.; Salehi-Ashtiani K.; Szostak J. W. J. Am. Chem. Soc. 2005, 127 (38), 13213–13219. 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom T. M.; Ritter G.; Simoneit B. R. T. Origins Life Evol. Biospheres 1999, 29, 153–166. 10.1023/A:1006592502746. [DOI] [PubMed] [Google Scholar]
- Lawless J. G.; Yuen G. U. Nature 1979, 282, 396–398. 10.1038/282396a0. [DOI] [Google Scholar]
- Clary L.; Verderone G.; Santaella C.; Vierling P. Biochim. Biophys. Acta, Biomembr. 1997, 1328, 55–64. 10.1016/S0005-2736(97)00076-X. [DOI] [PubMed] [Google Scholar]
- Heuberger B. D.; Pal A.; Del Frate F.; Topkar V. V.; Szostak J. W. J. Am. Chem. Soc. 2015, 137, 2769–2775. 10.1021/jacs.5b00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budin I.; Prwyes N.; Zhang N.; Szostak J. W. Biophys. J. 2014, 107, 1582–1590. 10.1016/j.bpj.2014.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel C. L.; Deamer D. W.; Mautner M. N. Biochim. Biophys. Acta, Biomembr. 2002, 1559, 1–9. 10.1016/S0005-2736(01)00400-X. [DOI] [PubMed] [Google Scholar]
- Ma W.; Yu C.; Zhang W.; Hu J. RNA 2007, 13, 2012–2019. 10.1261/rna.658507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. Life 2017, 7, 49. 10.3390/life7040049. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.