Abstract
Early protocells are likely to have arisen from the self-assembly of RNA, peptide, and lipid molecules that were generated and concentrated within geologically favorable environments on the early Earth. The reactivity of these components in a prebiotic environment that supplied sources of chemical energy could have produced additional species with properties favorable to the emergence of protocells. The geochemically plausible activation of amino acids by carbonyl sulfide has been shown to generate short peptides via the formation of cyclic amino acid N-carboxyanhydrides (NCAs). Here, we show that the polymerization of valine-NCA in the presence of fatty acids yields acylated amino acids and peptides via a mixed anhydride intermediate. Notably, Nα-oleoylarginine, a product of the reaction between arginine and oleic acid in the presence of valine-NCA, partitions spontaneously into vesicle membranes and mediates the association of RNA with the vesicles. Our results suggest a potential mechanism by which activated amino acids could diversify the chemical functionality of fatty acid membranes and colocalize RNA with vesicles during the formation of early protocells.
Introduction
Modern cell membranes consist of a phospholipid bilayer with protein channels that control the uptake of nutrients and the export of waste and metabolic products. These structurally complex lipid membranes are hypothesized to have evolved from the membranes of protocells,1 the ancestors of modern biological cells. Protocell membranes are thought to have assembled from prebiotically available amphiphiles that form bilayer membranes similar to but simpler than those in modern cells. In particular, fatty acids are attractive protocell membrane components because they can self-assemble to form bilayer membrane vesicles2 and are relatively permeable to both ions (e.g., Na+ or K+)3 and small organic molecules (e.g., 5′-imidazole activated nucleotides) useful for the replication of encapsulated genetic materials such as RNA4 or alternative information-coding polymers.5 In addition, fatty acids can be synthesized via prebiotically plausible routes, in particular by Fischer–Tropsch-type processes,6,7 and short-chain fatty acids have also been found in carbonaceous meteorites.8
Understanding how simple membranes may enhance metabolic reactions has been a major aim in studying the origin of the first cells. Beyond encapsulating nucleic acids and other reactive solutes, our group and others have begun to uncover routes through which fatty acid membranes can directly influence chemical transformations by providing a unique chemical and physical environment.9−12 At the same time, the mutual compatibility of laboratory-developed prebiotic chemistries is an important logical validation of proposed reactions. Previous studies suggest that geochemical conditions relevant to the early Earth could support the formation of amino acids13 and subsequently peptides.14−19 One prebiotically plausible route to peptide synthesis is the decarboxylative ring-opening polymerization of α-amino acid carboxyanhydrides (NCAs).20,21 However, the chemistry of this polypeptide-forming reaction in the presence of protocellular membranes, particularly those formed from a simple fatty acid, has not been previously investigated.
To explore chemically compatible and potentially synergistic interactions among the constituents of a model protocellular system, here we describe the prebiotic synthesis of fatty acylated amino acids (AAs) and peptides in the presence of fatty acid monomers, micelles, and vesicles. Specifically, we show that in the presence of oleic acid vesicles the N-carboxyanhydride of valine, a prebiotically available AA,22,13 can generate oleoylated AAs and peptides. Notably, Nα-oleoylarginine (Ol-Arg), a product observed when arginine is mixed with Val-NCA and oleic acid, incorporates spontaneously into vesicle membranes and mediates RNA–vesicle association. Localization of RNAs to the membrane may have significant implications for model protocells,23 as it increases the local concentration of otherwise dilute functional RNAs, potentially facilitating RNA-substrate binding and catalysis. Our study sheds light on potential chemical pathways leading to the formation of lipophilically modified AAs and peptides and suggests a novel mechanism by which prebiotically plausible activated AAs may have played a role in the early coevolution of cell membranes and RNA.
Results
Peptide Formation in the Presence of Fatty Acids
To facilitate peptide synthesis, we used Val-NCA (1) (Figure 1a), which we obtained in large quantity via conventional synthesis.24 We treated 1 (a suspension of 0.04 mmol in a 2 mL reaction volume) with a buffered [sodium salt of 300 mM 4-(2-hydroxyethyl)-1-piperazine-propanesulfonic acid (EPPS)] mixture of a free AA substrate (20 mM, see the Methods and Table S1 for the list of AAs) and either oleic acid [C18:1(cis)9] or octanoic acid (C8) (20 mM) at 20 °C and pH ca. 8.5. At this pH, oleic acid, with an observed pKa (pKa-obs) of ca. 8.5,25 forms bilayer membranes, whereas octanoic acid exists only as deprotonated monomers.26,27
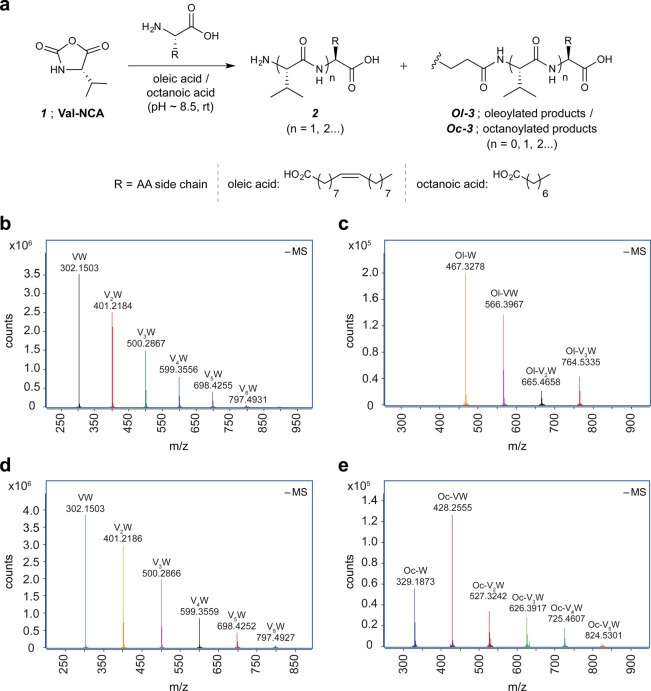
Figure 1.
Val-NCA-mediated acylation of amino acids and peptides. (a) Schematic of Val-NCA-mediated peptide acylation in the presence of either oleic acid or octanoic acid. (b–e) Overlay of extracted high-resolution mass spectrometry (HRMS) spectra ([M – H]−1) for peptide or acyl peptide products observed in 24 h incubated reaction mixtures starting from Val-NCA (20 mM), tryptophan (W) (20 mM), EPPS (300 mM), and either oleic acid (20 mM) (b and c) or octanoic acid (20 mM) (d and e). For similar HRMS analyses of the oligo-valine products (Vn) [R2 = CH(CH3)2], see Figure S1, SI). HRMS experiments were carried out in negative mode. Panels (b) and (d) highlight the W-containing native peptides observed in the presence of oleic acid and octanoic acid, respectively. Panels (c) and (e) highlight the W-containing oleoylated (Ol-) and octanoylated (Oc-) products, respectively. See the SI for tabulation of mass errors and an HPLC-extracted ion chromatogram showing the product distribution observed in the case of oleic acid (b, c).
Qualitative analysis of reaction aliquots by electrospray ionization/high-resolution mass spectrometry (ESI-HRMS) indicated the presence of a complex mixture of peptides 2 (up to 6 AA residues or longer) composed of both (Val)n-AA and (Val)n peptides both in reactions containing oleic acid and in reactions containing octanoic acid (Figure 1a; for HRMS analyses of oligo-Val products see Figure S1). Surprisingly, these analyses also revealed fatty acylated products of both the newly formed peptides and the free AA substrates in both fatty acid systems. As a representative example, Figure 1b–e shows HRMS analyses of the polymerization of 1 in the presence of Trp and either oleic acid (Figure 1b,c) or octanoic acid (Figure 1d,e). In order to measure the yields of the major acylated and unacylated peptides accurately, we specifically relied on the UV absorption of the products containing tryptophan, which has a distinct UV absorbance at 280 nm. Therefore, we determined the overall yield of Trp-containing products by UV absorption spectroscopy of purified fractions (see the SI for details). Starting from a 20 mL suspension of 0.4 mmol of substrates (1, Trp, and either oleic or octanoic acid), Trp-containing, unacylated peptides (2) were obtained in 23% and 30% overall yields, respectively. Of the Trp-containing products, 2–3% were acylated (3). In the absence of Val-NCA (1), we observed neither peptides nor acylated species, suggesting that 1 was necessary for the formation of peptides, and that an acylating agent was indeed generated during the course of the reactions when both 1 and fatty acids were present.
In the case of the reactions starting from a 5-fold higher concentration of octanoic acid (100 mM), the fraction of Trp-containing products that were octanoylated increased significantly, from ca. 2% to 10%. Furthermore, when the initial concentrations of each substrate (1, Trp and octanoic acid) were increased by 5-fold (2 mmol in 20 mL reaction volume), both the acylation and the peptide formation efficiency improved further, with ca. 18% of the Trp-containing products being octanoylated. This particular reaction mixture was slightly turbid and required a larger concentration of EPPS buffer (600 mM) to maintain the pH at around 8.5. In the presence of oleic acid vesicles, ca. 5% of the Trp-containing products were oleoylated.28 Importantly, the formation of acylated amino acids could be achieved in the absence of EPPS (vide infra).
Mechanistic insights were gained for the acylation of amino acids and peptides formed in situ.
In addition to ring-opening polymerization, NCAs have been reported to form isocyanato carboxylates by N-H deprotonation above pH 930 or acyclic phospho-carboxy anhydrides by reaction with phosphate nucleophiles.31,32 With regard to the former pathway, we did not observe peptidyl urea products of Val (e.g., N,N-divalylurea or N,N-valyl-AA-urea), which would result from trapping Val-isocyanato carboxylate by either free Val, generated by the hydrolysis of 1, or the AA substrate initially introduced into the reaction mixtures. The latter reaction mechanism, which involves oxygen nucleophiles, is more directly relevant to our experimental observations. In the context of prebiotic phosphorylation of alcohols, Biron et al. showed31 that 1 and HPO42– form a carboxylic acid–phosphoric acid mixed anhydride, which selectively phosphorylates, instead of acylating, methanol, affording methyl phosphate. Subsequently, Leman et al. demonstrated32 the formation of aminoacyl adenylates through the addition of adenosine 5′-monophosphate to NCAs. These reports suggest that delocalized oxyanions such as phosphate are sufficiently nucleophilic to attack NCAs and induce ring opening. Similarly, here we hypothesize (Figure 2) that at a pH near or above the pKa of the fatty acid the carboxylate anion attacks 1 at its more electrophilic carbonyl center, C-5,33 generating a putative mixed anhydride 4 (in equilibrium with its protonated form) that can either undergo intramolecular rearrangement to give Nα-acylvaline (path i, Figure 2), induce peptide bond formation (path ii),34 or serve as an intermolecular acylating agent (path iii).29
Figure 2.
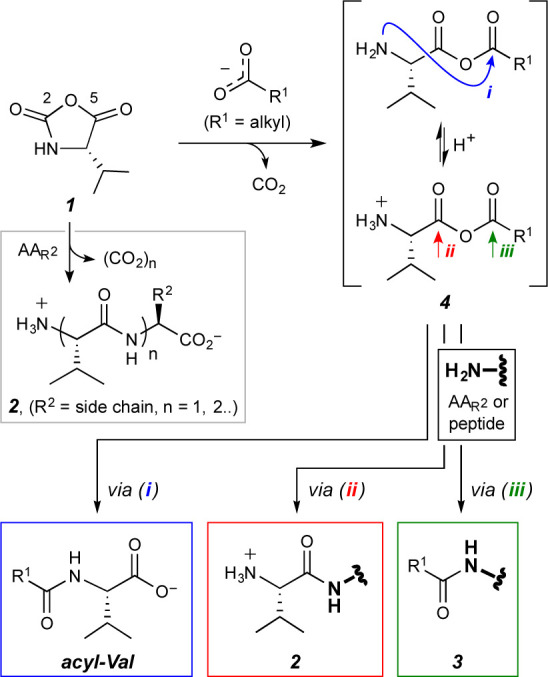
Possible pathways of a reaction between Val-NCA (1) and a carboxylate nucleophile.29 Product 3 also represents acyl-Val since free Val (from hydrolysis of 1) reacts with 4 via path iii.
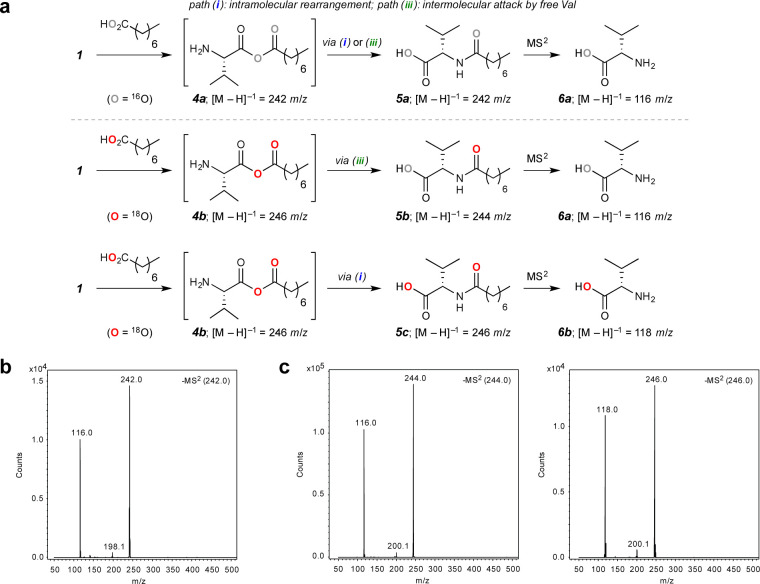
Our initial efforts to observe the protonated form of 4 by mass spectrometry were not successful in the case of either oleic or octanoic acid. However, if such an intermediate indeed forms, at least one of the chemically equivalent oxygen atoms of the carboxylate nucleophile must be incorporated into the acylated product after either the intramolecular rearrangement of 4 (path i) or nucleophilic attack at the fatty acyl carbonyl center (path iii). Therefore, we sought to explore the acylation mechanism by using an isotopically labeled carboxylic acid and then detecting the change in mass of the resulting acylated Val species. To accomplish this, we performed tandem mass spectrometry (Figure 3) of reaction mixtures obtained by treating 1 (40 mM) with a wet mixture of triethylamine (TEA) (1 M) and either octanoic acid (1 M), as the control, or 18O-enriched octanoic acid (1 M, 87% isotopic enrichment). See Figure S5 for a similar investigation carried out under more dilute conditions. The volatile base TEA was used to set the initial reaction pH in a manner compatible with direct injection ESI-MS. In the case of natural octanoic acid, we did not observe a m/z 244 signal in positive ionization mode, which would correspond to the protonated mixed anhydride species [M + H]+1, presumably because this species is quite transient. However, we did detect the [M – H]−1m/z 242 signal in negative ionization mode, suggesting that an intramolecular rearrangement or an intermolecular acylation of free Val may have occurred to generate the isomeric and negatively ionizable Nα-octanoyl-Val (5a; the 1H NMR spectrum of the crude reaction mixture matched that of the product isolated from a reaction of Val and octanoyl chloride; see the SI for details). Isolation and fragmentation of this m/z 242 ion gave rise to a major m/z 116 fragment (Figure 3b), as expected, for the monoisotopic mass of natural abundance Val (6a). In the case of oleic acid, we also observed this m/z 116 signal as a result of fragmenting the m/z 380 precursor, which corresponds to Nα-oleoylvaline formed from 1 and oleic acid (for details see Figure S4). Notably, under anhydrous and anaerobic conditions, 1 underwent nearly full conversion to the corresponding Nα-fatty acylvaline, presumably through path i, in the presence of ca. 50 equiv fatty acid and TEA.
Figure 3.
Mechanistic investigation of the formation of a mixed anhydride from Val-NCA 1 and octanoic acid. (a) (a) Schematic of the reaction between 1 and either 16O-enriched or 18O-enriched octanoic acid with observable [M – H]–1 ions. (b) MS/MS product ion spectra highlighting the isolated 16O-enriched species and its fragmentation product. (c) MS/MS product ion spectra highlighting the isolated 18O-enriched species and their fragmentation products.
Similar experiments carried out with 18O-enriched octanoic acid provided both the [M – H]−1m/z 244 and [M – H]−1m/z 246 signals in negative ionization mode. The presence of the former species suggests that an intermolecular displacement via path iii had occurred with the natural abundance Val to yield a mono-18O-containing octanoylvaline 5b, while the m/z 246 signal is consistent with formation of a di-18O-containing counterpart 5c by intramolecular rearrangement (path i), such that one 18O is on the valine carboxylate and one on the amide carbonyl. Isolation and fragmentation of the m/z 244 and 246 precursors gave rise to m/z 116 and 118 species, respectively (Figure 3c), which we attribute to the natural abundance 6a and mono-18O-containing fragmentation product 6b. We also observed the decarboxylation products resulting from fragmentation of the precursor ions. While the natural abundance m/z 242 ion gave rise to a m/z 198 signal, both the mono- and di-18O-containing m/z 244 and 246 ions fragmented into a m/z 200 species, as expected from the mono-18O-containing decarboxylated amido products. These observations are consistent with the formation of a transient mixed anhydride intermediate 4, which we propose is the major component that drives fatty acylation of Val (via path i) and of free AA substrates and the short peptides generated in situ (via path iii).29 This hypothesis is in line with the recent mechanistic investigations of Murillo-Sánchez et al.12 on the formation of mixed anhydrides from 5(4H)-oxazolones and carboxylic acids, as well as the earlier postulate of Miva and Stahmann that the benzoate anion could attack NCAs to form a carboxylic–benzoic mixed anhydride.35
Insertion of Nα-Oleoylarginine into Phospholipid and Oleic Acid Membranes
As fatty acylated amphiphiles have been shown to preferentially localize to lipid membranes,36,37 we investigated whether an Nα-oleoylated AA with a cationic side chain could also localize to lipid membranes. Because short arginine-containing peptides have been shown to localize RNA effectively to fatty acid membranes,38 we focused on Nα-oleoylarginine (Ol-Arg), one of the acylation byproducts formed when Arg and oleic acid are mixed with 1 at pH ca. 8.5 (Figure S3). We tested the incorporation of Ol-Arg (see Methods) into preformed oleic acid and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) vesicles. To monitor the membrane growth that would occur upon the insertion of Ol-Arg into the membrane, we used a Förster resonance energy transfer (FRET) assay37 (Figure S8) by labeling the phospholipid and oleic acid membranes with a FRET donor–acceptor fluorescent dye pair [N-(7-nitro-2,1,3-benzoxadiazol-4-yl) (NBD) and rhodamine]. This assay reports changes in membrane surface area that occur when vesicles absorb material into their membrane. The increase in membrane surface area affects the distance between FRET dyes, leading to a decrease in FRET efficiency. We prepared standard curves for both types of vesicles (Figure S8) by assembling vesicles containing increasing concentrations of FRET dyes and measuring the FRET efficiency in the membrane as a function of dye concentration. Using these standard curves, we examined the change in the surface area of oleic acid vesicles (Figure 4a) and POPC vesicles (Figure 4b) upon the addition of Ol-Arg by measuring the resulting change in FRET efficiency. As a positive control, we added oleate micelles (OA micelles), which have previously been shown to induce membrane growth in both phospholipid and fatty acid membranes.39,40 We observed that both Ol-Arg and OA micelles incorporated into preformed vesicles of both compositions (Figure 4). The addition of just the HCl or NaOH used to dissolve these amphiphiles (see the Methods) did not change vesicle surface area, indicating that membrane growth is due to the incorporation of OA or Ol-Arg.
Figure 4.
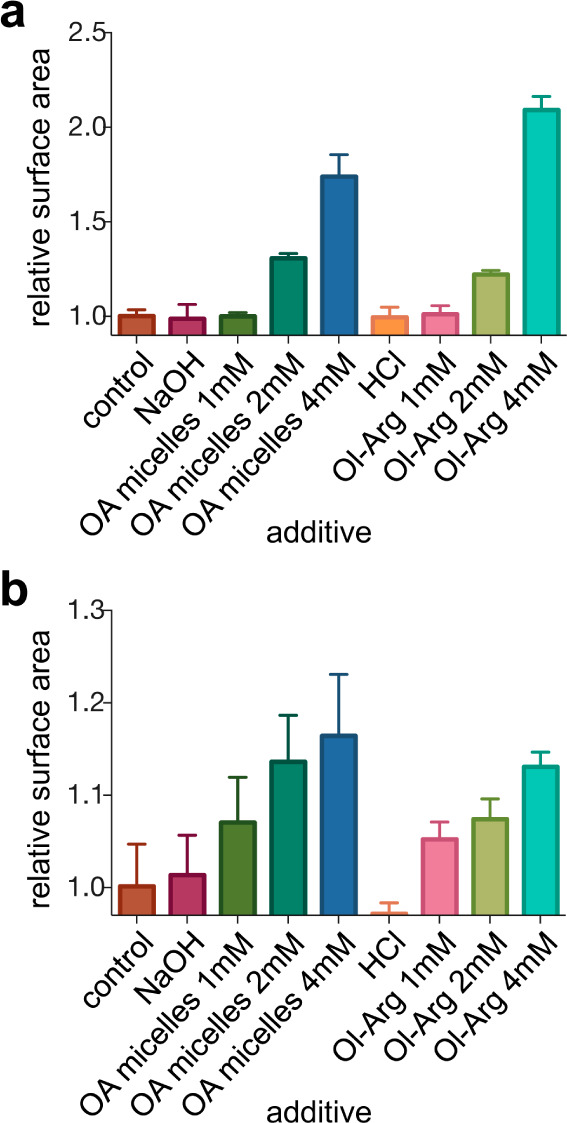
Oleic acid membrane growth (a) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane growth (b) upon oleate (OA) micelle and Ol-arginine (Ol-Arg) addition. The change in membrane surface area is reported upon the addition of various additives to 7.5 mM oleate or POPC vesicles in 200 mM bicine buffer, pH 8.5. OA micelles or Ol-Arg were dissolved by addition of either 4 mM NaOH or 4 mM HCl, respectively, and control additions of either the base or acid alone are indicated. n = 3; error bars represent standard deviation.
RNA Localization to Vesicles Containing Ol-Arg
We next sought to determine whether the presence of Ol-Arg in preformed vesicles could induce RNA localization to vesicle membranes (Figure 5). In principle, localization of RNA to vesicle membranes provides a potential route to concentrating functional RNA sequences which might otherwise exist at significantly lower concentrations and enhance intermolecular chemical transformations. We recently observed that cationic and membrane-associating peptides can localize short RNA oligomers (15-mer) to the surface of phospholipid and fatty acid membranes.38 Based on our observations, fatty acylated amino acids might have formed naturally on the prebiotic earth and could have similarly served to mediate the interaction of RNA with protocell membranes. To investigate this possibility, we prepared giant vesicles (GVs) that contained Ol-Arg in their membranes from either oleic acid or POPC. We then added a fluorescently tagged 15-mer RNA strand and incubated each sample for 30 min at room temperature (see the Methods for details). Membranes without Ol-Arg did not localize RNA with either vesicle composition (top rows, Figure 5a,b). However, both OA and POPC membranes containing Ol-Arg localized RNA to the vesicles. Interestingly, the predominant form of RNA association with OA vesicles that contained Ol-Arg was encapsulation inside of the vesicles (bottom row, Figure 5a). In POPC vesicle samples containing Ol-Arg (Figure 5b, bottom row), we observed that RNA clearly binds to the surface of the POPC membrane but does not penetrate the outer membrane; we also observed the appearance of RNA aggregates bound to the surface of vesicles.
Figure 5.
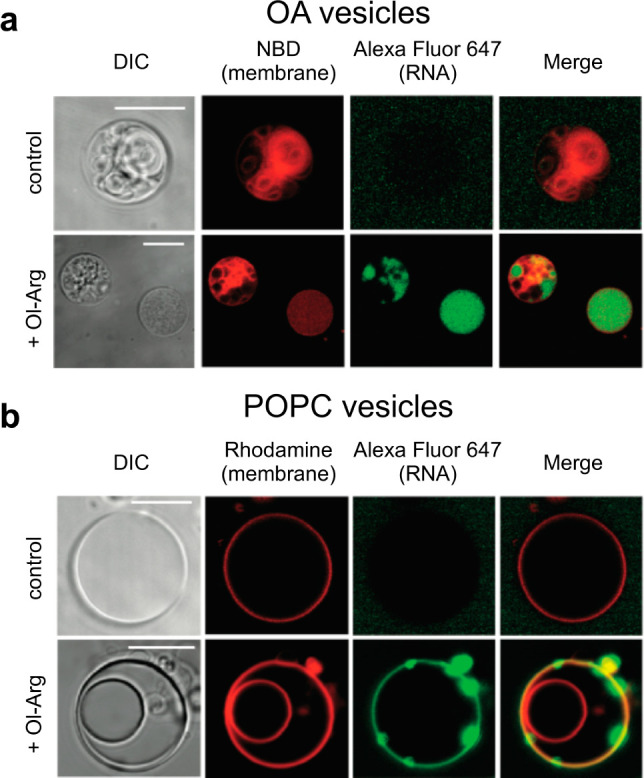
Microscopy of RNA-membrane localization with Ol-Arg. Confocal images of a 5′ Alexa Fluor-647 labeled 15-mer RNA oligonucleotide (green) association with giant vesicles composed of (a) 100% oleic acid (OA) (b) 100% POPC vesicles. An NBD-PE dye (red, panel a) and Lissamine Rhodamine PE dye (red, panel b) was used to label OA and POPC vesicle membranes, respectively. (Top rows, panels a and b) In the absence of Ol-Arg (control), RNA does not associate with either OA or POPC vesicles. (Bottom rows, panels a and b) Upon incubation with RNA, OA containing 25 mol % Ol-Arg and POPC vesicles containing 50 mol % Ol-Arg internalize or localize RNA. Increased regions of RNA fluorescence indicate aggregates of RNA that have bound the membrane. Differential interference contrast (DIC) images (gray) and fluorescence images (NBD or Rhodamine (red), AlexaFluor-647 (green), and a merge of the Rhodamine and Alexa Fluor-647 channels) are shown for each field of view to show the location of RNA with respect to the vesicle membranes. Images are contrast adjusted. Scale bars represent 10 μm.
Discussion
We have shown that Val-NCA (1) can mediate the condensation of unactivated amino acids and peptides with fatty acids to generate Nα-acylated products. Our hypothesis for the mechanism of this fatty acylation process is that the nucleophilic addition of the fatty acid carboxylate to carbonyl C-5 of 1 generates a mixed anhydride intermediate 4, which can serve as a prebiotic fatty acyl transfer agent. In the case of octanoic acid, the efficiency of this acyl transfer mechanism is proportional to the concentration of octanoic acid. The observation that free AAs and peptides are octanoylated at pH ca. 8.5 in the presence of octanoic acid, which does not form vesicles, suggests that vesicle formation, which occurs with the longer chain oleic acid, is not necessary for the fatty acylation chemistry to take place. The relative flux of the reaction through these competing pathways is likely to be influenced by the abundance of competing free AA and peptide nucleophiles, and by whether the reactions are taking place in aqueous solution or in a drying mixture. It is also notable that intermediate 4 is subject to hydrolysis, particularly in a pH regime where the Nα of AAs and peptides is largely unprotonated and therefore has adequate nucleophilic character (ca. pH ≥ 8). We have measured the observed rate constant of hydrolysis of 1 at pH ca. 8.5 as 0.018 s–1 (see Figure S7 for details). Although this hydrolysis competes with the fatty acylation pathway, a constant supply of geochemical energy (e.g., in the form of carbonyl sulfide) could regenerate NCAs and lead to the continued formation of both peptides and acylated peptides. In addition, once acylated, AAs and peptides should be quite stable under moderate pHs and temperatures because amide bonds have half-lives on the order of 100–500 years at room temperature41 and could potentially accumulate within protocells over time by selectively partitioning into fatty acid vesicles.
The microscopy experiments carried out with POPC and OA vesicles containing Ol-Arg indicate a significant increase in the local concentration of a 15-mer RNA. In many POPC + Ol-Arg vesicles, RNA was present both uniformly on the membrane and in larger aggregates. We occasionally observed smaller POPC + Ol-Arg vesicles encapsulated within a larger vesicle; in such cases, only the outermost membrane showed RNA labeling, suggesting that this particular vesicle membrane composition did not allow the associated RNA to transit the membrane. The appearance of RNA aggregates on the surface of POPC membranes that contain Ol-Arg may be related to previous observations of the interaction of RNA with arginine rich peptides, which form condensed phases in aqueous solution.42 In contrast, the predominant form of RNA association with OA vesicles that contained Ol-Arg was encapsulation inside of the membrane vesicles. In some large, apparently unilamellar OA + Ol-Arg vesicles, RNA had clearly become concentrated in the internal lumen of the vesicle. Although the mechanism behind this surprising effect is unclear, one possible pathway would involve initial binding of RNA to Ol-Arg on the outer surface of the membrane, followed by translocation of the RNA to the inner surface and equilibration with the internal volume of the vesicle; alternatively, Ol-Arg-induced vesicle fusion may allow RNA to become internalized within larger vesicles. However, in other more complex and multilamellar OA + Ol-Arg vesicles, RNA appeared internalized inside smaller membrane compartments or localized between internal membranes, suggesting that significant membrane reorganization must occur to explain the observed distribution of RNA within OA + Ol-Arg vesicles. We suggest that strong intermembrane adhesion resulting from the charge–charge interaction of oleate and Ol-Arg may lead to the engulfment of smaller vesicles by larger vesicles. This mechanism of RNA internalization and membrane reorganization in the presence of Ol-Arg is currently the subject of ongoing study in our laboratory.
Conclusion
We have presented a mechanism for fatty acylation of AAs and in situ formed peptides from prebiotically plausible substrates. Motivated by our observation of prebiotic fatty acylation, we then investigated the effect of Ol-Arg, an amphiphilic product generated when Val-NCA was mixed with Arg and oleic acid, on fatty acid and POPC vesicles. We showed that Ol-Arg incorporates spontaneously into the membrane of both oleic acid and POPC vesicles. Notably, fatty acid vesicles preformed in the presence of Ol-Arg enabled a 15-mer RNA oligonucleotide to increase its local concentration by being either localized on the membrane exterior or encapsulated inside the vesicle.
As a two-dimensional matrix, the lipid membrane of the cell surface has been demonstrated to promote a variety of chemical transformations.43 Similarly, primordial membranes derivatized with amino acids or peptides could have provided advantages to early cells by localizing functional RNA sequences, including ribozymes, to a surface; functional interactions could have been facilitated by the reduction in translational degrees of freedom.
The unique reactivity of NCAs has the potential to enable diverse chemical transformations, enzymatic variants of which are also seen in modern biology. For example, the N-acetylation of proteins is hypothesized to have evolved as a mechanism for protecting against spontaneous degradation in eukaryotic cells.44 In addition, many signaling pathways depend upon N-acylation of proteins for membrane localization.45 Specifically, α-amino lipidation (e.g., Nα-myristoylation; C14) of peptides is a cotranslational or post-translational covalent modification in eukaryotes, which plays an essential role in protein–protein46 and protein–membrane interactions,47 as well as cellular trafficking of proteins between membrane compartments.48 Prebiotically formed N-acylated peptides are therefore of great biological interest as a potential substrate for subsequent evolutionary processes, once cellular life formed. Using the chemical energy harvested in cyclic anhydrides to activate carboxylates could be a generalized approach to prebiotic acylation of diverse nucleophiles, including nonpeptide-based nucleophilic constituents of protocells, such as nucleotides. Studying these reactions will lead us closer to understanding how the interactions among RNA, fatty acids, and peptides could have led to the emergence of the first protocells.
Methods
Materials
LCMS-grade water (Optima) was purchased from Thermo Fisher Scientific and used in all experiments unless otherwise reported. Amino acids (AAs: tryptophan (Trp), arginine (Arg), valine (Val), histidine (His), lysine (Lys), glutamine (Gln), asparagine (Asn)) and 4-(2-hydroxyethyl)-1-piperazinepropane-sulfonic acid (EPPS) were purchased from Sigma-Aldrich (St. Louis, MO). Oleic acid and octanoic acid were purchased from Nu-Chek Prep (Elysian, MN). The phospholipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was purchased from Avanti Polar Lipids. The phospholipids N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (NBD-PE) and 1,2-dioleoyl-sn-glycero-3-phosphoethanol-amine-N-(Lissamine Rhodamine B sulfonyl) (ammonium salt) (Liss Rh-PE) were purchased from Life Technologies (Grand Island, NY). RNA (HPLC purified) was purchased from Integrated DNA Technologies (IDT, Coralville, IA). Nα-Oleoylarginine (Ol-Arg) was prepared following a previously reported protocol.49
Valine-N-carboxyanhydride (Val-NCA)-Mediated Polymerization of AAs in the Presence of Fatty Acids
Into a 4 mL screw thread vial containing Val-NCA24 (30 μmol, 1 equiv) and equipped with a magnetic stir bar was added a mixture (1.5 mL) of a single AA (30 μmol, 1 equiv, e.g., tryptophan, arginine, histidine, lysine, glutamine or asparagine, also see Table S1), the fatty acid (oleic acid or octanoic acid; 30 μmol, 1 equiv), and EPPS buffer (300 mM, pH 8.5–8.8) at room temperature. The reaction vial was sealed, and the content was mixed first by vortexing for 1 min and then by stirring on a stir plate over 24 h. At the end of this mixing period, an aliquot (ca. 10 μL) was taken and diluted 1000-times with HPLC-grade water for electrospray ionization/high-resolution mass spectrometry ESI-HRMS analysis (for Trp as the AA substrate, see Figure 1b–e and Figure S1 and for Arg as the AA substrate, see Figure S3). Typically, the pH of the reaction media decreases by ca. 0.3 units during the course of the experiment, and the product distributions show essentially no variation after 12 h.
Mass Spectrometry
For mechanistic fragmentation analysis, reaction samples were analyzed by direct injection from a syringe pump into a Bruker Esquire 6000 ESI-ion trap mass spectrometer. The mechanistic studies shown in Figure 3 were carried out in the presence of 1 M octanoic acid and 1 M triethylamine. For more dilute conditions, see the SI. Precursor ions were selected for MS/MS fragmentation with an isolation width of m/z 1 and a fragmentation amplitude between 0.2–0.3. For high-resolution LC–MS analyses of peptides and acylated products, reaction samples were separated and analyzed on an Agilent 1200 HPLC coupled to an Agilent 6520 Q-TOF or an Agilent 6230 TOF. The systems were equipped with a solvent degasser, column oven, autosampler, and diode array detector. Samples were analyzed in a multimode source operated in the mixed-mode (atmospheric pressure chemical ionization (APCI) and ESI) configuration using the following settings: drying gas temperature, 250 °C; vaporizer temperature, 200 °C; drying gas flow, 5 L/min; nebulizer pressure, 40 psig; charging electrode, 2000 V; capillary voltage, 2500 V; corona current 1 μA; fragmentor, 140 V; and skimmer, 65 V. Polarity for mass analysis was as indicated in figure legends. The scan range was m/z 100–1200 with a scan rate of 2 spectra/s. Samples were separated on a 75 mm Agilent ZORBAX Eclipse Amino Acid Analysis column (4.6 mm i.d. and 3.5 μm particle size) using a mobile phase of water/methanol with a step gradient (from 2 to 100% methanol at a flow rate of 0.450 mL/min). Extracted spectra for products were obtained using Agilent’s Find by Formula algorithm in Agilent’s MassHunter Qualitative Analysis platform.
Vesicle Preparation
POPC and OA vesicles for the membrane growth assay were prepared by thin-film hydration of the lipid or fatty acid in a chloroform solution. The solution was dried with N2 on the surface of a glass vial, and the solvent was evaporated in a vacuum oven for >12 h. Lipid films were hydrated with BICINE buffer (200 mM, pH 8.5) and tumbled overnight before extrusion through 100 nm membranes. For microscopy, giant unilamellar vesicles were prepared by thin film hydration methods on the bottom of glass vials. Vesicles were prepared from POPC or oleic acid and contained 0.15 mol % Liss Rh-PE or NBD-PE. Vesicle samples with Ol-Arg were prepared by premixing POPC or OA with 50 mol % Ol-Arg and 0.15 mol % Liss Rh-PE or NBD-PE in chloroform before being dried on the surface of glass vials. Solvent was evaporated in a vacuum oven for >12 h, and lipid films were hydrated with sucrose solution (200 mM) for POPC vesicles or sucrose + Tris solution (200 mM sucrose +100 mM Tris-HCl buffer, pH 8.5) for OA vesicles and heated at 65 °C overnight to form giant unilamellar vesicles.
Membrane Growth Assay
POPC vesicles were prepared with 0.1 mol % of the FRET dyes NBD PE and Liss Rh-PE. Growth from the addition of OA or Ol-Arg (see the SI for preparation) was assessed by adding each reagent to a vesicle solution (1, 2, and 4 mM) and monitoring the resulting change in FRET signal as previously described.37 Ol-Arg was dissolved in a 100 mM HCl solution, and oleate micelles were prepared by dissolving OA in 500 mM NaOH (20 mM oleate final concentration). FRET was measured using the fluorescence ratio between the donor (λem 517 nm) and the acceptor (λem 590 nm) lipids, with an excitation at 463 nm. The FRET signal was converted into relative surface area through a standard curve correlating mol % of FRET dyes in the membrane to FRET signal (Figure S8). The change in surface area after a 10 h incubation with each reagent was reported.
Microscopy
OA and POPC vesicles were mixed with an equimolar solution of 125 mM Tris-HCl, pH 8.5 and imaged in Lab-Tek II Coverglass (Thermo Fisher Scientific) chambers that were preblocked with a 1% bovine serum albumin (BSA) (Sigma-Aldrich) solution. For OA membranes, vesicles were diluted into a 500 μM solution of oleic acid (200 mOsm) to maintain the concentration of fatty acid above the critical aggregation concentration of oleic acid. Vesicles were imaged on a Nikon (Tokyo, Japan) A1R MP confocal microscope (60X oil objective) and processed in ImageJ. For studies of RNA localization to the outside of vesicle membranes with Ol-Arg, vesicle membranes containing 10, 25, and 50 mol % Ol-Arg were mixed with RNA, so that the final concentration of each component was 100 μM POPC vesicles, 10, 25, or 50 μM Ol-Arg, and 2 μM RNA for studies with POPC vesicles and 10 mM OA vesicles, 1, 2.5, or 5 mM Ol-Arg, and 10 μM RNA for studies with oleic acid vesicles (the oleic acid vesicle concentration was increased to maintain vesicle stability). After a 30 min incubation, 15 μL of the vesicle/peptide/RNA mixture was diluted 20-times into either 125 mM Tris–HCl or 500 μM oleic acid (200 mOsm) and imaged. The RNA used was 5′-(Alexa Fluor-647)-GCG-UAG-ACU-GAC-UGG-3′ (HPLC Purified).
Acknowledgments
We thank Prof. Sheref Mansy, Dr. Lijun Zhou, Dr. Li Li, and Ms. Claudia Bonfio for critical discussions and technical insights. We also thank Dr. Daniel Duzdevich for helpful comments on the manuscript. J.W.S. is an Investigator of the Howard Hughes Medical Institute. A.B. was supported by a fellowship from the Academy of Finland. This work was supported in part by a grant (290363) from the Simons Foundation to J.W.S.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b08801.
Detailed experimental procedures, additional HRMS analyses, FRET assays, synthesis, and spectroscopic characterization of new compounds accompanied by 1H and 13C NMR spectra (PDF)
Author Present Address
⊥ (A.B.) Statens Serum Institut, Artillerivej 5, DK-2300 Copenhagen S, Denmark.
The authors declare no competing financial interest.
Supplementary Material
References
- Chen I. A.; Walde P. Cold Spring Harbor Perspect. Biol. 2010, 2, a002170. 10.1101/cshperspect.a002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel C. L.; Deamer D. W.; Mautner M. N. Biochim. Biophys. Acta, Biomembr. 2002, 1559, 1–9. 10.1016/S0005-2736(01)00400-X. [DOI] [PubMed] [Google Scholar]
- Chen I. A.; Szostak J. W. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 7965–7970. 10.1073/pnas.0308045101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K.; Szostak J. W. Science 2013, 342, 1098–1100. 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy S. S.; Schrum J. P.; Krishnamurthy M.; Tobe S.; Treco D. A.; Szostak J. W. Nature 2008, 454, 122–125. 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom T. M.; Ritter G.; Simoneit B. R. Origins Life Evol. Biospheres 1999, 29, 153–166. 10.1023/A:1006592502746. [DOI] [PubMed] [Google Scholar]
- Rushdi A. I.; Simoneit B. R. Origins Life Evol. Biospheres 2001, 31, 103–118. 10.1023/A:1006702503954. [DOI] [PubMed] [Google Scholar]
- Yuen G. U.; Kvenvolden K. A. Nature 1973, 246, 301–303. 10.1038/246301a0. [DOI] [Google Scholar]
- Blocher M.; Liu D.; Walde P.; Luisi P. L. Macromolecules 1999, 32, 7332–7334. 10.1021/ma990917m. [DOI] [Google Scholar]
- Grochmal A.; Prout L.; Makin-Taylor R.; Prohens R.; Tomas S. J. Am. Chem. Soc. 2015, 137, 12269–12275. 10.1021/jacs.5b06207. [DOI] [PubMed] [Google Scholar]
- Adamala K.; Engelhart A. E.; Szostak J. W. Nat. Commun. 2016, 7, 11041. 10.1038/ncomms11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo-Sánchez S.; Beaufils D.; González Mañas J. M.; Pascal R.; Ruiz-Mirazo K. Chem. Sci. 2016, 7, 3406–3413. 10.1039/C5SC04796J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B. H.; Percivalle C.; Ritson D. J.; Duffy C. D.; Sutherland J. D. Nat. Chem. 2015, 7, 301–307. 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H.; Orgel L. E. J. Mol. Evol. 1975, 6, 185–197. 10.1007/BF01732355. [DOI] [PubMed] [Google Scholar]
- Armstrong D. W.; Seguin R.; McNeal C. J.; Macfarlane R. D.; Fendler J. H. J. Am. Chem. Soc. 1978, 100, 4605–4606. 10.1021/ja00482a053. [DOI] [Google Scholar]
- Huber C.; Wächtershäuser G. Science 1998, 281, 670–672. 10.1126/science.281.5377.670. [DOI] [PubMed] [Google Scholar]
- Huber C.; Eisenreich W.; Hecht S.; Wächtershäuser G. Science 2003, 301, 938–940. 10.1126/science.1086501. [DOI] [PubMed] [Google Scholar]
- Leman L.; Orgel L.; Ghadiri M. R. Science 2004, 306, 283–286. 10.1126/science.1102722. [DOI] [PubMed] [Google Scholar]
- Danger G.; Boiteau L.; Cottet H.; Pascal R. J. Am. Chem. Soc. 2006, 128, 7412–7413. 10.1021/ja061339+. [DOI] [PubMed] [Google Scholar]
- Kricheldorf H. R. Angew. Chem., Int. Ed. 2006, 45, 5752–5784. 10.1002/anie.200600693. [DOI] [PubMed] [Google Scholar]
- Danger G.; Plasson R.; Pascal R. Chem. Soc. Rev. 2012, 41, 5416–5429. 10.1039/c2cs35064e. [DOI] [PubMed] [Google Scholar]
- Kvenvolden K.; Lawless J.; Pering K.; Peterson E.; Flores J.; Ponnamperuma C.; Kaplan I. R.; Moore C. Nature 1970, 228, 923–926. 10.1038/228923a0. [DOI] [PubMed] [Google Scholar]
- Strulson C. A.; Molden R. C.; Keating C. D.; Bevilacqua P. C. Nat. Chem. 2012, 4, 941–946. 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]
- Daly W. H.; Poché D. Tetrahedron Lett. 1988, 29, 5859–5862. 10.1016/S0040-4039(00)82209-1. [DOI] [Google Scholar]
- Cistola D. P.; Hamilton J. A.; Jackson D.; Small D. M. Biochemistry 1988, 27, 1881–1888. 10.1021/bi00406a013. [DOI] [PubMed] [Google Scholar]
- Weast R. C., Ed. Handbook of Chemistry and Physics, 51st ed.; The Chemical Rubber Co.: Cleveland, OH, 1970. [Google Scholar]
- Walde P.; Namani T.; Morigaki K.; Hauser H. In Liposome Technology, 3rd ed.; Gregoriadis G., Ed.; Informa Healthcare: New York, 2006; Vol. I, pp 1–19. [Google Scholar]
- We have performed parallel experiments with 100 mM acetate at pH 8.5. We did not observe the formation of any acetylated AAs or peptides by MS, while unacetylated peptides of up to 5-mer in length were observed. This result is consistent with the idea that, under aqueous conditions, the hydrophobic Val-NCA reacts preferentially with hydrophobic fatty acids.
- We have been unable to observe by MS the anions (286 or 424 m/z) that would correspond to the carbamates of Nα-octanoylvaline or Nα-oleoylvaline, even by direct injection of unquenched reactions, suggesting that these intermediates are highly unstable. As a result, we are unable to assess the possibility that slow decarboxylation is what allows the observed intermolecular reactions as shown in paths ii and iii in Figure 2.
- Hirschmann R.; Strachan R. G.; Schwam H.; Schoenewaldt E. F.; Joshua H.; Barkemeyer B.; Veber D. F.; Paleveda W. J.; Jacob T. A.; Beesley T. E.; Denkewalter R. G. J. Org. Chem. 1967, 32, 3415–3425. 10.1021/jo01286a030. [DOI] [PubMed] [Google Scholar]
- Biron J.-P.; Pascal R. J. Am. Chem. Soc. 2004, 126, 9198–9199. 10.1021/ja048189s. [DOI] [PubMed] [Google Scholar]
- Leman L.; Orgel L.; Ghadiri M. R. J. Am. Chem. Soc. 2006, 128, 20–21. 10.1021/ja056036e. [DOI] [PubMed] [Google Scholar]
- Sekiguchi H. Pure Appl. Chem. 1981, 53, 1689–1714. 10.1351/pac198153091689. [DOI] [Google Scholar]
- For a conceptually relevant and complementary novel mechanism to produce peptides and peptide derivatives that could possess useful functions in prebiotic chemistry, see:Forsythe J. G.; Yu S.-S.; Mamajanov I.; Grover M. A.; Krishnamurthy R.; Fernández F. M.; Hud N. V. Angew. Chem., Int. Ed. 2015, 54, 9871–9875. 10.1002/anie.201503792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miva T. K.; Stahmann M. A.. Polyamino Acids, Polypeptides, And Proteins; Stahmann M. A., Ed.; University of Wisconsin Press: Madison, 1962; p 81. [Google Scholar]
- Thomas R. M.; Baici A.; Werder M.; Schulthess G.; Hauser H. Biochemistry 2002, 41, 1591–1601. 10.1021/bi011555p. [DOI] [PubMed] [Google Scholar]
- Chen I. A.; Szostak J. W. Biophys. J. 2004, 87, 988–998. 10.1529/biophysj.104.039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat N. P.; Tobe S.; Hill I. T.; Szostak J. W. Angew. Chem., Int. Ed. 2015, 54, 11735–11739. 10.1002/anie.201505742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin P.; Arrigler V.; Kogej K.; Svetina S.; Walde P. Chem. Phys. Lipids 2009, 159, 67–76. 10.1016/j.chemphyslip.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Zhu T. F.; Szostak J. W. J. Am. Chem. Soc. 2009, 131, 5705–5713. 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzicka A.; Wolfenden R. J. Am. Chem. Soc. 1996, 118, 6105–6109. 10.1021/ja954077c. [DOI] [Google Scholar]
- Jia T. Z.; Fahrenbach A. C.; Kamat N. P.; Adamala K. P.; Szostak J. W. Nat. Chem. 2016, 8, 915–921. 10.1038/nchem.2551. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Walde P.; Umakoshi H.; Stano P.; Mavelli F. Chem. Commun. 2014, 50, 10177–10197. 10.1039/C4CC02812K. [DOI] [PubMed] [Google Scholar]
- Brown J. L.; Roberts W. K. J. Biol. Chem. 1976, 251, 1009–1014. [PubMed] [Google Scholar]
- Resh M. D. Biochim. Biophys. Acta, Mol. Cell Res. 1999, 1451, 1–16. 10.1016/S0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- Zha J.; Weiler S.; Oh K. J.; Wei M. C.; Korsmeyer S. J. Science 2000, 290, 1761–1765. 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- Murray D.; Ben-Tal N.; Honig B.; McLaughlin S. Structure 1997, 5, 985–989. 10.1016/S0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- Resh M. D. Cell. Signalling 1996, 8, 403–412. 10.1016/S0898-6568(96)00088-5. [DOI] [PubMed] [Google Scholar]
- Brady S. F.; Clardy J. Org. Lett. 2005, 7, 3613–3616. 10.1021/ol0509585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.