Supplemental Digital Content is available in the text.
Keywords: Colorectal cancer, Inflammatory bowel disease, Organoids, Personalized medicine
Abstract
BACKGROUND:
Colorectal cancer and IBD account for a large portion of the practice of colorectal surgery. Historical research models have provided insights into the underlying causes of these diseases but come with many limitations.
OBJECTIVE:
The aim of this study was to systematically review the literature regarding the advantage of organoid models in modeling benign and malignant colorectal pathology.
DATA SOURCES:
Sources included PubMed, Ovid-Medline, and Ovid Embase
STUDY SELECTION:
Two reviewers completed a systematic review of the literature between January 2006 and January of 2020 for studies related to colon and intestinal organoids. Reviews, commentaries, protocols, and studies not performed in humans or mice were excluded.
RESULTS:
A total of 73 articles were included. Organoid models of colorectal disease have been rising in popularity to further elucidate the genetic, transcriptomic, and treatment response of these diseases at the individual level. Increasingly complex models utilizing coculture techniques are being rapidly developed that allow in vitro recapitulation of the disease microenvironment.
LIMITATIONS:
This review is only qualitative, and the lack of well utilized nomenclature in the organoid community may have resulted in the exclusion of articles.
CONCLUSIONS:
Historical disease models including cell lines, patient-derived tumor xenografts, and animal models have created a strong foundation for our understanding of colorectal pathology. Recent advances in 3-dimensional cell cultures, in the form of patient-derived epithelial organoids and induced human intestinal organoids have opened a new avenue for high-resolution analysis of pathology at the level of an individual patient. Recent research has shown the potential of organoids as a tool for personalized medicine with their ability to retain patient characteristics, including treatment response.
Of the diseases that colorectal surgeons treat, both colorectal cancer (CRC) and IBD account for a dominant portion of our practice. Colorectal cancer continues to grow in prevalence worldwide; it now accounts for about 1 in every 10 cancers,1 constituting the second most common malignancy in the world and the third most common malignancy in the United States.2 Despite numerous scientific advances and an increasing number of therapeutics, more than 700,000 deaths occur annually in the world.3 Inflammatory bowel disease primarily refers to 2 types of chronic relapsing-remitting inflammatory disorders of the bowel, Crohn’s disease and ulcerative colitis. An estimated 3.0 million Americans currently have IBD, resulting in significant morbidity and financial burden to patients and the health care system4 despite the advances in medical therapies that have decreased the need for surgical treatment over the past 6 decades.5
Historically, IBD research has used mouse models of disease that are derived through chemical and genetic transformation and bulk tissue analysis.6 Since the discovery that intestinal stem cells could be cultured, there has been a large shift toward the derivation and use of patient samples to advance research into the treatment and pathogenesis of IBD. For both CRC and IBD, because disease heterogeneity is now recognized at the cellular and molecular levels, clinical and basic science investigators are seeking patient-derived models that 1) more closely mimic disease pathology and can be analyzed at the cellular, molecular, and functional levels and 2) are used as avatars or representations of the human tissue from which it was derived to identify and test patient biomarkers and potential individual patient responses to therapy (Fig. 1). Development of the 3-dimensional intestinal organoid has advanced to the stage that it has the potential to meet these challenges. Herein, we summarize the history of intestinal models used for these purposes, then introduce the organoid as a new model that will obviate many of the challenges to analyzing the biology of both CRC and IBD. Therefore, to allow for a thorough appraisal of the literature, we will use “Are patient-derived organoids an improvement over current technologies for recapitulating patient disease and treatment response” as the overarching question driving this review.
FIGURE 1.
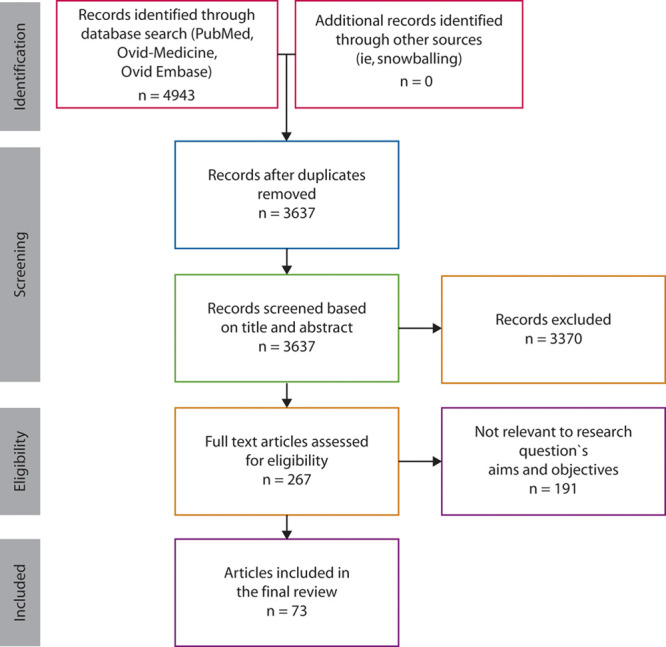
PRISMA flow diagram. 4943 articles were identified using the terms organoid, colonoid, enteroid, or spheroid, as well as colon, intestine, and intestinal (Supplemental Figure 1 http://links.lww.com/DCR/B337). Following deduplication, 3637 unique articles were identified for further review. 3370 articles were screened by reviewing the title and abstract, resulting in 267 full-text research articles that were assessed. Of these, 73 articles were found to be appropriate for inclusion in our review.
MATERIALS AND METHODS
This review was designed and performed in accordance with the Preferred Reporting Items for Systematic Review and Meta-Analyses.7
Search Strategy and Eligibility
A search was constructed using PubMed, Ovid-MEDLINE, and Ovid Embase from January 1, 2006 to January 15, 2020 to identify research articles related to colon or intestinal organoids. The search terms utilized were organoid, spheroid, colonoid, intestine and colon. For PubMed and Ovid-Medline, a combination of MeSH terms and keywords were used, whereas, for Embase, subject headings were utilized along with keywords. For all databases the explode function was utilized where appropriate. Our search strategy is outlined in Supplemental Figure 1 http://links.lww.com/DCR/B337. The references of the included articles were also queried for additional articles that may not have been discovered in the initial search.
Study Selection
Reviews, commentaries, protocols, and studies not performed in humans or mice were excluded. Two reviewers (R.K.D. and E.H.H.) screened PubMed, Ovid-MEDLINE and Embase records based on titles with subsequent abstract review following removal of those articles not applicable to the above search. The full search strategy is included in Supplemental Figure 1 http://links.lww.com/DCR/B337. The remaining full-text articles were reviewed, and articles not focused on organoids or organoid models of colorectal pathology were removed (Fig. 1).
Historical Models
Conventionally, investigation of CRC pathogenesis relies on tissue analysis of the stages of the adenoma-to-carcinoma transition during which normal epithelium evolves to adenoma and eventually to carcinoma through a sequence of acquired genetic alterations in the APC,1 K-RAS, DCC, and TP53 genes.8 Throughout the 1990s and early 2000s, much of the experimental work regarding CRC was performed using 3 models: 1) animal cancer models, 2) cancer cell lines, or 3) patient-derived tumor xenografts (PDTXs). Animal cancer models have served the cancer research community well, with numerous genetic (genetically engineered mice) and chemical models of CRC that have allowed us to delineate many of the basic alterations that occur between benign and malignant tissue. Unfortunately, these models are costly and time consuming, and they fail to recapitulate the complex pathologic processes seen in human patients.9,10
One of the most frequently used tools in cancer research is cancer cell lines. These are derived from primary patient tissue and adapted to 2-dimensional culture for use worldwide. Over the course of decades, these have advanced our understanding of cancer genetics, epigenetics, molecular, and cell biology. However, they have numerous limitations, including the loss of tissue heterogeneity from the primary tumor and an absence of matched healthy control lines. The most frequently used and well-characterized colon cancer cell lines are HCT-116, HT-29, and SW480, but many others including cell lines derived from lymph nodes and metastatic disease exist.11 The PDTX model is generated by transplanting a fresh patient-derived tissue sample either subcutaneously or orthotopically in an immunocompromised mouse. Patient-derived tumor xenografts models allow for in vivo preclinical testing of novel cancer therapies. Despite the advantages provided by in vivo implantation, the recapitulation of the normal tumor microenvironment is limited, because immunocompromised mice must be used. As with other mouse models, the PDTX model is also limited by expense, technical challenges, and resource consumption, as well as the presence of mouse-specific tissue changes.12
Inflammatory bowel disease research primarily has used animal models, as no current standard cell lines reflect any stage of IBD. Broadly, these animal models are either genetic or chemical models of inflammation. Genetic models of IBD include the interleukin-10 (IL-10) null mouse, the Winnie mouse (MUC –/–), and the Samp/Yit mouse. Interleukin-10 is required to maintain immune homeostasis throughout the GI tract, and knocking out the IL-10 gene leads to induction of spontaneous colitis and increased sensitivity to infectious, drug-induced, or autoimmune colitis.13 N-Ethyl-N-nitrosourea-induced mutagenesis screening produces a phenotype of watery diarrhea and rectal bleeding arising from a MUC2 mutation. This mouse strain was named the Winnie mouse and shows a phenotype of mild spontaneous ulcerative colitis.14 Unlike the IL-10 null and Muc2 mouse that have specific single-gene mutations, the Samp/Yit mouse arose from the accumulation of mutations during breeding that has led to a phenotype of chronic intestinal inflammation with skip lesions localizing to the ileum and cecum.15 Further breeding of this strain led to the development of the SAMP1/YitFc mouse that has a phenotype even more similar to Crohn’s disease, including perianal fistulizing disease and intestinal structuring.16 Recently, increased efficacy and decreased off-target effects have been achieved in genetically engineered mouse models of IBD via the use of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)- associated protein 9 (Cas9).17 Chemical mouse models of colitis vary, with the most widely used being dextran sulfate sodium (DSS) added to animals’ drinking water.18 Dextran sulfate sodium induces an acute chemical colitis in mice that leads to diarrhea, hematochezia, and weight loss within 6 to 10 days with concomitant histopathologic changes, including erosions and crypt abscesses. With recurrent DSS administration, colitis-associated cancer will develop in about 10% of mice.19 Building on this work, azoxymethane, a procarcinogen that becomes an alkylating agent in the host, induces colitis-associated cancer in rodents receiving DSS over several weeks.20 Although these models have provided insight into the underlying genetics of IBD, they remain extremely limited because they do not recapitulate either the human immune system or the intestinal microbiome.
In Vitro Models of Intestinal Stem Cells and the Rise of Organoids
In the early 1990s, Evans and colleagues21 developed one of the first methods to grow rat intestinal epithelial cells 2-dimensionally in culture. Unfortunately, these cells were only stable in culture for about 14 days before they became senescent. Significant progress was made in the following years regarding the physiologic signaling pathways, primarily Wnt, in the proliferation and maintenance of intestinal epithelia.22,23 Several other studies outlined the roles of other various factors in the maintenance and differentiation of epithelial cell lineages including noggin, bone morphogenetic protein, and other growth factors.24,25
In 2007, Barker et al26 discovered Lgr5 as a marker of intestinal stem cells and showed that these were capable of differentiating into the cell types that make up the intestinal villus. Building on this work, a method for isolating and maintaining Lgr5+ intestinal stem cells was developed that has led to an explosion of novel research.27 Further characterization of Lgr5 stem cells revealed that that they are found at the bottom of crypts between terminally differentiated Paneth cells producing lysozyme and defensins. These characteristics held true for both mouse and human small intestine, as well as in colonic adenomas with Lgr5+ cells associating with CD24(+) cells, which represents the colonic functional equivalent of small intestinal Paneth cells and thus setting the stage for the development of organoid models (Fig. 2).28 Along with normal tissue, several potential cancer stem cell markers have been identified, including CD133 (PROM1), CD44, or aldehyde dehydrogenase, which demonstrate the hallmark stem cell characteristic of self-renewal (Fig. 3).29–33
FIGURE 2.
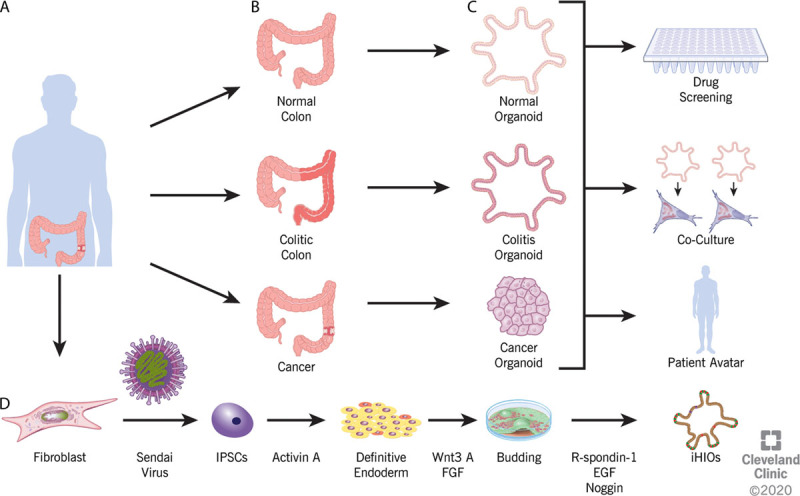
Organoid generation. A, B, Specimens are obtained from patients via either surgical resection or endoscopic biopsy. Specimens as small as 1 mm3 have been used to obtain viable organoids. C, The organoids generated will recapitulate the phenotype of the tissue sampled, be it normal, colitic, or malignant. The organoids can be used for various downstream analyses, including drug screening, genomic analysis, transcriptional analysis, or to derive a patient avatar that allows the testing of varying treatment strategies for either IBD or malignancy on patient-specific samples. D, Induced human intestinal organoids can be generated from colon fibroblast, skin fibroblast, or blood. Sendai virus containing the Yamanaka factors is used to reprogram the differentiated cell back to a pluripotent state. Through treatment with a series of various factors, induced pluripotent stem cells (IPSCs) undergo directed differentiation to definitive endoderm, then buds, and finally progress to an induced human intestinal organoid. EGF = epidermal growth factor; FGF = fibroblast growth factor; iHIO = induced human intestinal organoid.
FIGURE 3.
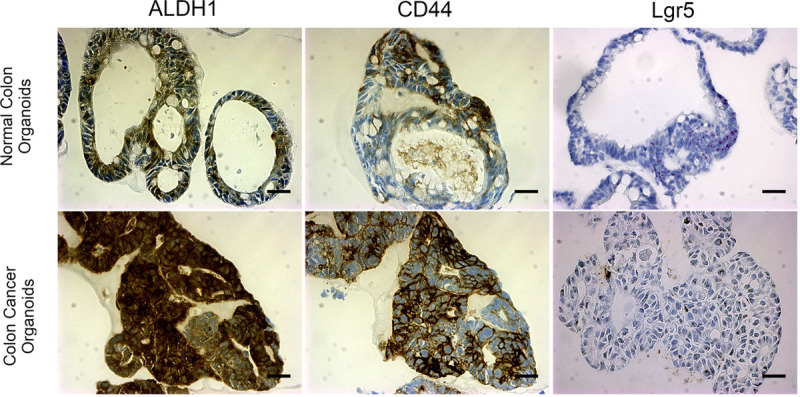
Representative immunohistochemistry and in situ hybridization of normal and colon cancer organoids. A–C, Normal and cancer organoids with immunohistochemistry staining for stem cell markers ALDH1, CD44, and in situ hybrization for Lgr5. ALDH1 is less specific, in this case, whereas CD44 marks a more specific cell population. Lgr5 (RNAScope) marks cells with high specificity at the base of crypts. D–F, Stem cell markers show increased enrichment throughout colon cancer organoids especially for ALDH1 and CD44 (p < 0.005 for ALDH1, p < 0.05 for CD44, p < 0.02 for Lgr5). n ≥ 3 organoids; scale bar = 100 μm.
Organoids are defined as a part of a given organ that is able to replicate in 3-dimensional culture a structure similar in structure and function to the tissue from which it is derived.34–36 Culturing technique has become more refined since the landmark study of Barker et al,26 but the primary components of organoid culture, Matrigel, a solubilized basement membrane protein derived from Engelbreth-Holm-Swarm mouse sarcoma that serves as an extracellular matrix, and the growth factors Wnt3A, R-Spondin, and Noggin, remain integral to the successful prolonged culture of intestinal epithelial cells.37–40 As shown in Figure 2, the models generated by Barker et al are limited to the expression of intestinal epithelia.41 These epithelial organoids can be used to study biology, including signaling pathways and molecular inhibitors (Fig. 2). Some researchers have also utilized an air-liquid interface system that increases cell viability due to improved oxygenation.42 This culturing system was recently shown to recapitulate “homeostasis-injury-regeneration” cycles of epithelial alterations that occur in vivo through hypoxia and endoplasmic reticulum stress like those seen in IBD.43 Using other novel technologies, some groups have combined the use of decellularized human colon with epithelial organoids derived from normal human tissue to assess genetic drivers of CRC invasion and progression.44
Their genomic and phenotypic stability along with the ability to undergo prolonged culture allows organoids to be used to investigate cancer biology in ways that previously were both technologically and financially nearly impossible. The adenoma-to-carcinoma pathway has been well documented, but CRISPR-Cas9 editing of epithelial organoids has led to new insights. CRISPR-Cas9 has been used to generate mutant organoids harboring the most common modifications to APC, p53, KRAS, and SMAD4.45–48 When xenografted into mice, these quadruple mutant epithelial organoids develop into invasive carcinomas.45 Similarly, cancer organoids can be used to recapitulate the accumulated mutations seen in mismatch repair-deficient CRC.49 Similar methodology has been applied to analyze the development of tumor metastasis. CRISPR-Cas9-modified organoids implanted into mice revealed that the metastatic potential of CRC required mutations in each of the genes p53, KRAS, APC, and SMAD4.50
Concurrent with the derivation of epithelial organoids based on intestinal stem cells, several other groups focused on using induced pluripotent stem cells to derive intestinal and colonic organoids (induced human intestinal organoids) (Fig. 2D). Unlike epithelial cell organoids, these have the advantage of possessing all 3 germ layers with mesoderm and nonfunctioning ectoderm arising during the generation of definitive endoderm in the traditional model (Figs. 2D and 4).51,52 Recently, the absence of an enteric nervous system has been overcome by successfully coculturing human pluripotent stem cell-derived neural crest cells and induced human intestinal organoids, opening a new frontier to further study the physiologic function of the GI tract.53 Along with study of the enteric nervous system, induced human intestinal organoids have been leveraged to further elucidate the role of the mesoderm, in particular, myofibroblasts, in the intestinal fibrosis that may occur in IBD.54
FIGURE 4.
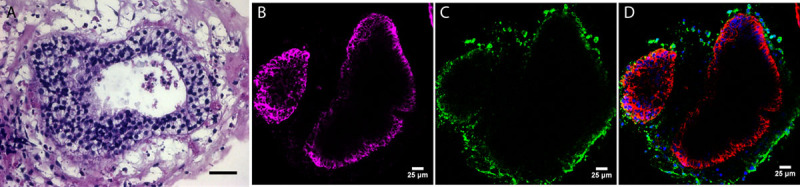
Representative H&E and immunofluorescence staining of induced pluripotent stem cell-derived colitic organoids. A, H&E of iPSC colitis-derived organoid. Note multilayered nature of epithelia. B, Green fluorescence marks vimentin, a fibroblast marker in the mesenchyme. C, Purple staining for CK19 highlights colonic organoid epithelial cells. D, Both CK19 (purple) and vimentin (green) show the distinct epithelial and mesenchymal populations. n = 3 organoids; scale bar = 25 μm. H&E = hematoxylin and eosin; iPSC, induced pluripotent stem cell.
Organoid Bio-Banking and the Future of Personalized Medicine
A principal advantage of organoid technology is that both the physiologic and pathologic features of the tissue of origin are maintained despite prolonged culturing.55 Despite there being only roughly 12 known signaling pathways that drive CRC tumorigenesis, every tumor within a patient has distinct genetic alterations as well as intratumoral heterogeneity that defines both pathology and treatment response.56,57 Consequently, intestinal organoids present the opportunity to better tailor treatment to the individual patient with CRC. Individual patient tumors are cultured to generate organoids that are then used to screen drugs to determine the best chemotherapeutic strategy for that patient. Ashley et al58 performed a small proof-of-concept study using organoids cultured with and without Matrigel and cell death enumeration to elucidate the effects of staurosporine, an apoptosis inhibitor, on CRC organoids. They showed that organoids were able to recapitulate the phenotype of an individual patient’s malignancy and were responsive to drug therapy.58 In another proof-of-concept study, Van de Wetering and colleagues59 derived organoids from tumors obtained from 20 patients with CRC and tested treatment response to 25 drugs showing that the genetic mutations present in the original tumor were recapitulated within the organoid and had varying responses to therapy based on these mutations. Since the advent of organoid technology, rapid advances in culturing capabilities have allowed organoid lines to be derived from nearly all types of colorectal cancer.60 Patient-derived organoids from treatment-naïve metastatic colorectal and gastroesophageal tumors, when used as in vivo and orthotropic mouse tumor xenografts, recapitulated patient response to chemotherapy.61 Various other studies have tested patient-derived and induced pluripotent stem cell-derived CRC organoids alongside normal colonic organoids to assess both treatment response and off-target toxicity to normal colonic epithelium with various chemotherapeutic agents.62–64 One study showed that clinically used mitogen-activated protein kinase kinase inhibitors inadvertently induced stem cell plasticity and gene signatures consistent with cancer recurrence.65 Two studies used epithelial organoids derived from patients with rectal cancer to investigate clinical guideline-based chemoradiation treatment.66,67 Both groups concluded that patient-derived organoids reliably predicted the primary tumor response, providing hope that organoids will provide a novel way to better individualize a patient’s treatment regimen.
Novel drug discovery can also leverage organoid technology to rapidly assess the efficacy and toxicity of drugs before they are tested in costly and expensive prolonged clinical trials. Plocabulin, a novel microtubule-disrupting compound originally isolated from a marine sponge, had a strong cytotoxic effect when it was used to treat CRC organoids from 3 patients.68 A study by Fiore and colleagues69 showed the versatility of the organoid models to assess the effects of chemotherapeutic agents, in this case rimonabant, to induce cytotoxicity in cancer organoids while minimizing off-target effects on normal organoids. Genetic factors that predict response to drugs can be elucidated as well.70–74 Tissue-derived organoids can be cocultured with local tumor-infiltrating immune cells to assess the response to chemotherapeutic agents and radiation.75 Along with the ability to assess the effects of chemotherapeutic agents and radiation directly on patient tumor samples, organoids also allow us to more deeply analyze the transcriptional and proteomic profiles of individual patients. Using normal and cancer organoids derived from the same patient, Cristobal and coworkers76 determined a CRC-specific proteomic profile that showed a significant enrichment in Wnt signaling as would be expected, along with unique tumor proteomic signatures. Organoid technology allows analysis at the single-cell transcriptome level; thus, intratumor heterogeneity that gives rise to marked differences in response to chemotherapeutic agents can be analyzed.77,78 Air-liquid interface culturing techniques permit coculture of patient-derived organoids and native immune cells, including macrophages and T, B, and NK cells, with the goal of enabling precision immune-oncology investigations and personalized immunotherapy testing.79 Chimeric antigen receptor-engineered lymphocytes have been tested using CRC organoids with a chimeric antigen receptor strategy directed against FRIZZLED receptors with some success in vitro.80 Other treatment strategies including CRISPR-Cas9 gene correction have also been performed successfully in organoids to correct mutations in the CFTR (cystic fibrosis) gene in intestinal stem cells.81 At least 1 institution has begun clinical trials integrating patient whole-exome sequencing with high-throughput tumor organoid drug screening to identify the ideal drug combinations for individual patients.82
Organoids and New Frontiers in IBD
Organoids have been utilized with good success to further elucidate the mechanisms behind current biologic therapies for IBD. Bradford and associates83 sought to better understand this mechanism by using organoids and genetic mouse models. Organoid technology has also improved our understanding of the mechanism of colitis-associated cancer development in patients with long-standing ulcerative colitis. Unlike spontaneous CRC, which is associated with late mutations in p53, colitis-associated cancer has been shown to have frequent early mutations in p53.84 Organoid technology combined with azoxymethane/DSS models of colitis revealed that p53 promoted continuous inflammation through nuclear factor κB (NF-κB) activation with associated tissue damage.84 One long-standing theory of IBD is that, even in pathologic and endoscopic remission, the previously inflamed colonic epithelia remain dysregulated. Biopsy specimens from histologically normal colonic tissue from patients with IBD and healthy controls exhibited an aberrant transcriptional profile showing increased expression of genes seen in small intestinal Paneth cells and gastric epithelia even in the setting of histologically normal epithelia, a finding consistent with a chronic underlying genetic change.85–87 In mouse intestinal organoids, chronic inflammation was generated by treating the organoids for 60 weeks with a combination of cytokines (tumor necrosis factor-α, IL-1β and IL-6) and bacterial components (lipopolysaccharide and flagellin).88 After treatment, significant induction of target genes in the NF-κB pathway was found, and these genes remained induced despite withdrawal of treatment for up to 60 weeks, again suggesting that changes likely occur in the progenitor and stem cell compartments that are “remembered” even after the inflammatory stimuli are removed. Single-cell analysis of small-bowel organoids derived from patients with Crohn’s disease show similar findings, with factors in the inflammatory microenvironment modifying intestinal stem cells.89 Coculturing of macrophages and organoids have produced physiologic models recapitulating the epithelial-to-mesenchymal transition that occurs with fibrosis in Crohn’s disease and leukocyte migration.90,91 Other groups have used epithelial organoids to determine epithelial barrier responses to interferon-γ-induced permeability changes.92 Similar studies have been performed by other groups using mouse intestinal organoids to show novel genes involved in intestinal epithelial integrity and tight junction formation.93 Williamson and colleagues94 developed a novel microinjection platform that allows for injection of luminal microbiota into patient-derived organoids to assess the effects of patient-specific fecal microbiota on epithelial integrity and physiology. Induced human intestinal organoids provide a more advanced tool as myofibroblasts are present in the mesoderm-derived stroma of the cells and respond to profibrotic stimuli with a response consistent with isolated human myofibroblasts.54
As with cancer, patient-derived organoids can be used to identify epigenetic and transcriptional changes present in the development of other intestinal diseases.95–98 For example, despite the fact that NF-κB signaling pathway mutations are significantly enriched throughout the colon in patients with ulcerative colitis, these pathways are exceedingly rare in areas of the colon with dysplasia or colitis-associated cancer.99 A recent study comparing epithelial organoids derived from 71 patients, 16 without colitis, 29 with colitis lacking dysplasia, and 26 from patients with colitis-associated cancer, showed mutations in the IL-17 signaling pathway that confer resistance to an IL-17A-induced apoptotic response.100 Like novel chemotherapeutic testing for cancer, ulcerative colitis and normal colonic organoids can be treated with cytokines and bacterial components to recapitulate the colitis phenotype, and then treated with novel agents to assess suppression of the immune response and regeneration of normal crypts.101–103 One of these studies showed that naltrexone alleviated endoplasmic reticulum stress in organoids of patients with IBD after treatment with inflammatory stimuli.104 Interleukin-22 has shown promise as a promising target for ulcerative colitis therapy due to its role in epithelial homeostasis and repair, which lead Patnaude and colleagues105–107 to trial treatment of inflammation-stimulated organoids with IL-22 that showed improved epithelial regeneration and production of membrane mucus. Han and colleagues108 took this idea a step further and microinjected patient-derived fecal supernatants with the probiotic Lactobacillus rhamnosus. Their group showed that fecal supernatants could induce epithelial barrier dysfunction which improved in the presence of L rhamnosus.
CONCLUSION
As cellular and molecular understanding of colorectal cancer and IBD have advanced, so too have the models for in vitro and in vivo investigation. Patient-derived organoid models, reproducing disease heterogeneity and bearing not only colonic epithelia but also the mesenchyme, will increase our scientific and clinical understanding and may permit high-throughput analyses of chemotherapeutic strategies. Furthermore, genetic manipulation of organoids is a strategy to increase therapeutic options by allowing for the examination of novel targets. For cancer, organoids provide a strategy to interrogate DNA damage and repair mechanisms in response to various treatments, including radiation. These strategies continue to advance our ability to provide personalized medicine for both CRC and IBD and will promote high-throughput treatment screening to determine the most efficacious therapy for an individual patient. To date, numerous studies have been performed showing the utility of organoid models (Supplemental Table 1 http://links.lww.com/DCR/B339). In the near term, organoid studies from both CRC and IBD can be combined with pharmacologic treatments and next-generation sequencing techniques to better elucidate underlying genetic, transcriptomic, proteomic, and epigenetic signatures that predict treatment success and failure. Organoid models can also be leveraged to help provide novel insights into the carcinoma-to-metastasis pathway through matched analysis of primary and metastatic tissues. Along with disease-specific insights, they will likely play a large role in drug discovery because intestinal organoids have been shown to recapitulate cell death and survival of intestinal epithelial cells, one of the most common side effects of chemotherapy, radiotherapy, and drug-mediated toxicity.109 The field of organoid research still has numerous limitations, including various culture and derivation techniques that may induce unknown changes to the tissue, inability to recapitulate the complete human tumor immune environment, being epithelial only in nature for patient-derived organoids without associated stroma, and reliance on Matrigel as the extracellular matrix. Future studies will be required to validate the accuracy of intestinal organoids to fully discriminate the heterogeneity of human disease to parallel in vivo clinical treatment and responses.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML and PDF versions of this article on the journal’s Web site (www.dcrjournal.com).
Funding/Support: Drs Huang and Sarvestani are supported by NIH R01 CA237304, U01 CA214300. Dr DeHaan is supported by the Cleveland Clinic Foundation Crile Fellowship award. This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland, 4UL1TR002548-01 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Financial Disclosures: None reported.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020; 70:7–30 [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012; 380:2095–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kappelman MD, Rifas-Shiman SL, Kleinman K, et al. The prevalence and geographic distribution of Crohn’s disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol. 2007; 5:1424–1429 [DOI] [PubMed] [Google Scholar]
- 5.Frolkis AD, Dykeman J, Negrón ME, et al. Risk of surgery for inflammatory bowel diseases has decreased over time: a systematic review and meta-analysis of population-based studies. Gastroenterology. 2013; 145:996–1006 [DOI] [PubMed] [Google Scholar]
- 6.Liu TC, Stappenbeck TS. Genetics and pathogenesis of inflammatory bowel disease. Annu Rev Pathol. 2016; 11:127–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009; 151:264–9, W64 [DOI] [PubMed] [Google Scholar]
- 8.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990; 61:759–767 [DOI] [PubMed] [Google Scholar]
- 9.Kamb A. What’s wrong with our cancer models?. Nat Rev Drug Discov. 2005; 4:161–165 [DOI] [PubMed] [Google Scholar]
- 10.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011; 6:479–507 [DOI] [PubMed] [Google Scholar]
- 11.Ahmed D, Eide PW, Eilertsen IA, et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013; 2:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-David U, Ha G, Tseng YY, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017; 49:1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993; 75:263–274 [DOI] [PubMed] [Google Scholar]
- 14.Heazlewood CK, Cook MC, Eri R, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008; 5:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto S, Okabe Y, Setoyama H, et al. Inflammatory bowel disease-like enteritis and caecitis in a senescence accelerated mouse P1/Yit strain. Gut. 1998; 43:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivera-Nieves J, Bamias G, Vidrich A, et al. Emergence of perianal fistulizing disease in the SAMP1/YitFc mouse, a spontaneous model of chronic ileitis. Gastroenterology. 2003; 124:972–982 [DOI] [PubMed] [Google Scholar]
- 17.Platt RJ, Chen S, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014; 159:440–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990; 98:694–702 [DOI] [PubMed] [Google Scholar]
- 19.Okayasu I, Yamada M, Mikami T, Yoshida T, Kanno J, Ohkusa T. Dysplasia and carcinoma development in a repeated dextran sulfate sodium-induced colitis model. J Gastroenterol Hepatol. 2002; 17:1078–1083 [DOI] [PubMed] [Google Scholar]
- 20.Okayasu I, Ohkusa T, Kajiura K, Kanno J, Sakamoto S. Promotion of colorectal neoplasia in experimental murine ulcerative colitis. Gut. 1996; 39:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans GS, Flint N, Somers AS, Eyden B, Potten CS. The development of a method for the preparation of rat intestinal epithelial cell primary cultures. J Cell Sci. 1992; 101pt 1219–231 [DOI] [PubMed] [Google Scholar]
- 22.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003; 17:1709–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhnert F, Davis CR, Wang HT, et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004; 101:266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dignass AU, Sturm A. Peptide growth factors in the intestine. Eur J Gastroenterol Hepatol. 2001; 13:763–770 [DOI] [PubMed] [Google Scholar]
- 25.Haramis AP, Begthel H, van den Born M, et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004; 303:1684–1686 [DOI] [PubMed] [Google Scholar]
- 26.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007; 449:1003–1007 [DOI] [PubMed] [Google Scholar]
- 27.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009; 459:262–265 [DOI] [PubMed] [Google Scholar]
- 28.Sato T, van Es JH, Snippert HJ, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011; 469:415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007; 445:106–110 [DOI] [PubMed] [Google Scholar]
- 30.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007; 445:111–115 [DOI] [PubMed] [Google Scholar]
- 31.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007; 104:10158–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpentino JE, Hynes MJ, Appelman HD, et al. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009; 69:8208–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shenoy AK, Fisher RC, Butterworth EA, et al. Transition from colitis to cancer: high Wnt activity sustains the tumor-initiating potential of colon cancer stem cell precursors. Cancer Res. 2012; 72:5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaushik G, Ponnusamy MP, Batra SK. Concise review: current status of three-dimensional organoids as preclinical models. Stem Cells. 2018; 36:1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol. 2017; 216:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spence JR. Taming the Wild West of organoids, enteroids, and mini-guts. Cell Mol Gastroenterol Hepatol. 2018; 5:159–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011; 141:1762–1772 [DOI] [PubMed] [Google Scholar]
- 38.Hofmann C, Obermeier F, Artinger M, et al. Cell-cell contacts prevent anoikis in primary human colonic epithelial cells. Gastroenterology. 2007; 132:587–600 [DOI] [PubMed] [Google Scholar]
- 39.Ray S, Langan RC, Mullinax JE, et al. Establishment of human ultra-low passage colorectal cancer cell lines using spheroids from fresh surgical specimens suitable for in vitro and in vivo studies. J Cancer. 2012; 3:196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VanDussen KL, Marinshaw JM, Shaikh N, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015; 64:911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finkbeiner SR, Spence JR. A gutsy task: generating intestinal tissue from human pluripotent stem cells. Dig Dis Sci. 2013; 58:1176–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ootani A, Li X, Sangiorgi E, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009; 15:701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Chiang IL, Ohara TE, et al. Long-term culture captures injury-repair cycles of colonic stem cells. Cell. 2019; 179:1144–1159.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen HJ, Wei Z, Sun J, et al. A recellularized human colon model identifies cancer driver genes. Nat Biotechnol. 2016; 34:845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drost J, van Jaarsveld RH, Ponsioen B, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015; 521:43–47 [DOI] [PubMed] [Google Scholar]
- 46.Li X, Nadauld L, Ootani A, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014; 20:769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matano M, Date S, Shimokawa M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015; 21:256–262 [DOI] [PubMed] [Google Scholar]
- 48.Roper J, Tammela T, Cetinbas NM, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol. 2017; 35:569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drost J, van Boxtel R, Blokzijl F, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017; 358:234–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fumagalli A, Drost J, Suijkerbuijk SJ, et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci U S A. 2017; 114:E2357–E2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011; 470:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson CL, Mahe MM, Múnera J, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat Med. 2014; 20:1310–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Workman MJ, Mahe MM, Trisno S, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med. 2017; 23:49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodansky ES, Johnson LA, Huang S, Spence JR, Higgins PD. Intestinal organoids: a model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp Mol Pathol. 2015; 98:346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Middendorp S, Schneeberger K, Wiegerinck CL, et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. 2014; 32:1083–1091 [DOI] [PubMed] [Google Scholar]
- 56.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013; 339:1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakayama M, Sakai E, Oshima H, et al. Novel model systems for colon cancer progression by accumulation of multiple driver mutations. 2017: Cancer Science Conference: 76th Annual Meeting of the Japanese Cancer Association, JCA; 109 [Google Scholar]
- 58.Ashley N, Jones M, Ouaret D, Wilding J, Bodmer WF. Rapidly derived colorectal cancer cultures recapitulate parental cancer characteristics and enable personalized therapeutic assays. J Pathol. 2014; 234:34–45 [DOI] [PubMed] [Google Scholar]
- 59.van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015; 161:933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujii M, Shimokawa M, Date S, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016; 18:827–838 [DOI] [PubMed] [Google Scholar]
- 61.Vlachogiannis G, Hedayat S, Vatsiou A, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018; 359:920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costacurta P, Short SP, Thompson J, Washington K, Chen X, Williams CS. Human colorectal cancer organoids: a tractable platform for modeling patient tumors and testing chemotherapeutic efficacy. Gastroenterology. 2016; 150:S139–S140 [Google Scholar]
- 63.Crespo M, Vilar E, Tsai SY, et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med. 2017; 23:878–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dijkmans T, Basten S, Herpers B, et al. Patient-derived 3D tumor cultures for clinical diagnostics and pre-clinical drug development. Cancer Res. 2018; 78suppl (Abstract 4644) [Google Scholar]
- 65.Zhan T, Ambrosi G, Wandmacher AM, et al. MEK inhibitors activate Wnt signalling and induce stem cell plasticity in colorectal cancer. Nat Commun. 2019; 10:2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao Y, Xu X, Yang L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020; 26:17–26.e6 [DOI] [PubMed] [Google Scholar]
- 67.Ganesh K, Wu C, O’Rourke KP, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. 2019; 25:1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costales-Carrera A, Fernández-Barral A, Bustamante-Madrid P, et al. Plocabulin displays strong cytotoxic activity in a personalized colon cancer patient-derived 3d organoid assay. Mar Drugs. 2019; 17:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fiore D, Ramesh P, Proto MC, et al. Rimonabant kills colon cancer stem cells without inducing toxicity in normal colon organoids. Front Pharmacol. 2017; 8:949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koppens MA, Bounova G, Cornelissen-Steijger P, et al. Large variety in a panel of human colon cancer organoids in response to EZH2 inhibition. Oncotarget. 2016; 7:69816–69828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brandt R, Sell T, Lüthen M, et al. Cell type-dependent differential activation of ERK by oncogenic KRAS in colon cancer and intestinal epithelium. Nat Commun. 2019; 10:2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Engel RM, Chan WH, Nickless D, et al. Patient-derived colorectal cancer organoids upregulate revival stem cell marker genes following chemotherapeutic treatment. J Clin Med. 2020; 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu C, Banister CE, Weige CC, et al. PRDM1 silences stem cell-related genes and inhibits proliferation of human colon tumor organoids. Proc Natl Acad Sci U S A. 2018; 115:E5066–E5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsuchiya K, Watanabe S, Shirasaki T, et al. TP53 mutation in human colonic organoids acquires resistance to in vitro long-term inflammation. ECCOJC. 2019; 13suppl 1S023 [Google Scholar]
- 75.Finnberg NK, Gokare P, Lev A, et al. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget. 2017; 8:66747–66757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cristobal A, van den Toorn HWP, van de Wetering M, Clevers H, Heck AJR, Mohammed S. Personalized proteome profiles of healthy and tumor human colon organoids reveal both individual diversity and basic features of colorectal cancer. Cell Rep. 2017; 18:263–274 [DOI] [PubMed] [Google Scholar]
- 77.Roerink SF, Sasaki N, Lee-Six H, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018; 556:457–462 [DOI] [PubMed] [Google Scholar]
- 78.Tung KL, Chen KY, Negrete M, et al. Integrated chromatin and transcriptomic profiling of patient-derived colon cancer organoids identifies personalized drug targets to overcome oxaliplatin resistance. Genes Dis. https://doi.org/10.1016/j.gendis.2019.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neal JT, Li X, Zhu J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018; 175:1972–1988.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schnalzger TE, de Groot MH, Zhang C, et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019; 38:e100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013; 13:653–658 [DOI] [PubMed] [Google Scholar]
- 82.Pauli C, Hopkins BD, Prandi D, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017; 7:462–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bradford EM, Ryu SH, Singh AP, et al. Epithelial TNF receptor signaling promotes mucosal repair in inflammatory bowel disease. J Immunol. 2017; 199:1886–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cooks T, Pateras IS, Tarcic O, et al. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell. 2013; 23:634–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dotti I, Mora-Buch R, Ferrer-Picón E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017; 66:2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dotti I, Ferrer-Picon E, Planell N, et al. Transcriptional analysis of intestinal epithelial organoid cultures derived from pediatric crohn’s disease patients. J Pediatr Gastr Nutr. 2017; 65suppl 1S5 [Google Scholar]
- 87.Yi J, Bergstrom K, Fu J, et al. Dclk1 in tuft cells promotes inflammation-driven epithelial restitution and mitigates chronic colitis. Cell Death Differ. 2019; 26:1656–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hibiya S, Tsuchiya K, Hayashi R, et al. Long-term inflammation transforms intestinal epithelial cells of colonic organoids. ECCOJC. 2016jjw186. [DOI] [PubMed] [Google Scholar]
- 89.Suzuki K, Murano T, Shimizu H, et al. Single cell analysis of Crohn’s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J Gastroenterol. 2018; 53:1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hahn S, Nam MO, Noh JH, et al. Organoid-based epithelial to mesenchymal transition (OEMT) model: from an intestinal fibrosis perspective. Sci Rep. 2017; 7:2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roh TT, Chen Y, Paul HT, Guo C, Kaplan DL. 3D bioengineered tissue model of the large intestine to study inflammatory bowel disease. Biomaterials. 2019; 225:119517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bardenbacher M, Ruder B, Britzen-Laurent N, et al. Permeability analyses and three dimensional imaging of interferon gamma-induced barrier disintegration in intestinal organoids. Stem Cell Res. 2019; 35:101383. [DOI] [PubMed] [Google Scholar]
- 93.Bayrer JR, Fletterick R, Ingraham H. Targeting intestinal stem cells to promote healing in inflammatory bowel disease. Gastroenterology. 2015; 148:S911–S912 [Google Scholar]
- 94.Williamson IA, Arnold JW, Samsa LA, et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell Mol Gastroenterol Hepatol. 2018; 6:301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Howell KJ, Kraiczy J, Nayak KM, et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018; 154:585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mokry M, Middendorp S, Wiegerinck CL, et al. Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology. 2014; 146:1040–1047 [DOI] [PubMed] [Google Scholar]
- 97.Hibiya S, Tsuchiya K, Nishimura R, et al. Lesion-specific gene expression in the epithelial cells of Crohn’s disease by comparing small intestinal organoids from active and inactive lesion in the same patient. United European Gastroenterol J. 2018; 6supplA425 [Google Scholar]
- 98.Hibiya S, Tsuchiya K, Watanabe M. Establishment of a colonic organoid with Wnt-independent growth by long-term inflammatory stimulation. 2017. Paper presented at: Cancer Science Conference: 76th Annual Meeting of the Japanese Cancer Association, JCA, Yokohama, Japan; 109 [Google Scholar]
- 99.Kakiuchi N, Yoshida K, Uchino M, et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature. 2020; 577:260–265 [DOI] [PubMed] [Google Scholar]
- 100.Nanki K, Fujii M, Shimokawa M, et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature. 2020; 577:254–259 [DOI] [PubMed] [Google Scholar]
- 101.Nishimura R, Shirasaki T, Tsuchiya K, et al. Establishment of a system to evaluate the therapeutic effect and the dynamics of an investigational drug on ulcerative colitis using human colonic organoids. J Gastroenterol. 2019; 54:608–620 [DOI] [PubMed] [Google Scholar]
- 102.Hibiya S, Tsuchiya K, Nishimura R, et al. Establishment of an in vitro system to evaluate the therapeutic effect of the investigational drug on ulcerative colitis using human colonic organoids. Abstracts of the 14th Congress of ECCO – European Crohn’s and Colitis Organisation. J Crohns Colitis. 2019; 13suppl 1S099 [Google Scholar]
- 103.Hibiya S, Tsuchiya K, Watanabe S, et al. Human colonic organoid treated with inflammatory factors might mimic the pathophysiology of epithelial cells in ulcerative colitis. Gastroenterology. 2018; 154suppl 1S1012 [Google Scholar]
- 104.Lie MRKL, van der Giessen J, Fuhler GM, et al. Low dose Naltrexone for induction of remission in inflammatory bowel disease patients. J Transl Med. 2018; 16:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Patnaude L, Mayo M, Mario R, et al. Advancing IL-22-based therapies for inflammatory bowel diseases with human intestinal organoids. Am J Gastroenterol. 2019; 114suppl 1S18 [Google Scholar]
- 106.Patnaude L, Mayo M, Mario R, et al. Leveraging human intestinal organoids to advance Il-22-based therapies for ulcerative colitis. Gastroenterology. 2019; 1566 S1S-37 [Google Scholar]
- 107.Pavlidis P, Tsakmaki A, Niazi U, et al. The interleukin 22 transcriptional programme is activated in human colonic inflammation and associated to anti-TNFalpha primary non-response in Crohn’s. Abstracts of the 14th Congress of ECCO – European Crohn’s and Colitis Organisation. J Crohns Colitis. 2019; 13suppl 1S065 [Google Scholar]
- 108.Han X, Lee A, Huang S, Gao J, Spence JR, Owyang C. Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut Microbes. 2019; 10:59–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grabinger T, Luks L, Kostadinova F, et al. Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death Dis. 2014; 5:e1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


