Supplemental Digital Content is available in the text.
Keywords: heart failure, myocardial infarction, proteomics, transcriptome
Abstract
Background:
Heart failure (HF) is the most common long-term complication of acute myocardial infarction (MI). Understanding plasma proteins associated with post-MI HF and their gene expression may identify new candidates for biomarker and drug target discovery.
Methods:
We used aptamer-based affinity-capture plasma proteomics to measure 1305 plasma proteins at 1 month post-MI in a New Zealand cohort (CDCS [Coronary Disease Cohort Study]) including 181 patients post-MI who were subsequently hospitalized for HF in comparison with 250 patients post-MI who remained event free over a median follow-up of 4.9 years. We then correlated plasma proteins with left ventricular ejection fraction measured at 4 months post-MI and identified proteins potentially coregulated in post-MI HF using weighted gene co-expression network analysis. A Singapore cohort (IMMACULATE [Improving Outcomes in Myocardial Infarction through Reversal of Cardiac Remodelling]) of 223 patients post-MI, of which 33 patients were hospitalized for HF (median follow-up, 2.0 years), was used for further candidate enrichment of plasma proteins by using Fisher meta-analysis, resampling-based statistical testing, and machine learning. We then cross-referenced differentially expressed proteins with their differentially expressed genes from single-cell transcriptomes of nonmyocyte cardiac cells isolated from a murine MI model, and single-cell and single-nucleus transcriptomes of cardiac myocytes from murine HF models and human patients with HF.
Results:
In the CDCS cohort, 212 differentially expressed plasma proteins were significantly associated with subsequent HF events. Of these, 96 correlated with left ventricular ejection fraction measured at 4 months post-MI. Weighted gene co-expression network analysis prioritized 63 of the 212 proteins that demonstrated significantly higher correlations among patients who developed post-MI HF in comparison with event-free controls (data set 1). Cross-cohort meta-analysis of the IMMACULATE cohort identified 36 plasma proteins associated with post-MI HF (data set 2), whereas single-cell transcriptomes identified 15 gene-protein candidates (data set 3). The majority of prioritized proteins were of matricellular origin. The 6 most highly enriched proteins that were common to all 3 data sets included well-established biomarkers of post-MI HF: N-terminal B-type natriuretic peptide and troponin T, and newly emergent biomarkers, angiopoietin-2, thrombospondin-2, latent transforming growth factor-β binding protein-4, and follistatin-related protein-3, as well.
Conclusions:
Large-scale human plasma proteomics, cross-referenced to unbiased cardiac transcriptomics at single-cell resolution, prioritized protein candidates associated with post-MI HF for further mechanistic and clinical validation.
Clinical Perspective.
What Is New?
We combined 2 powerful unbiased discovery tools, aptamer-based proteomics and single-cell transcriptomics, to prioritize 83 post–myocardial infarction heart failure candidates using human plasma from 2 different acute myocardial infarction patient cohorts and 4 mouse and human single–cardiac cell transcriptomic studies.
Six top candidates were consistently associated with the development of post–myocardial infarction heart failure in both patient cohorts and validated in the single-cell data sets: NT-proBNP/BNP-32 (N-terminal pro B-type natriuretic peptide/brain natriuretic peptide-32 [NPPB gene]), TNNT2 (troponin T), ANGPT2 (angiopoietin-2), THBS2 (thrombospondin-2), LTBP4 (latent transforming growth factor beta binding protein 4), and FSTL3 (follistatin-related protein 3).
Besides the 6 top candidates, our bioinformatics approach identified an additional 19 intermediate-priority and 58 lower-priority proteins that may be useful for future investigation of post–myocardial infarction heart failure biomarkers and drug targets.
What Are the Clinical Implications?
The emergence of high-throughput unbiased screening tools is rapidly changing the way we approach biomarker and drug-target discovery.
These discovery tools have their limitations; therefore, prioritized candidates need further validation in future studies.
Alongside B-type natriuretic peptide and cardiac troponin, angiopoietin-2 and thrombospondin-2 are emerging as important biomarkers in ischemic cardiomyopathy.
Editorial, see p 1422
Acute myocardial infarction (MI) commonly precedes heart failure (HF).1 Few biomarkers associated with HF after MI have gained widespread acceptance in mainstream clinical testing despite the discovery of many potential candidates.2 Of greater concern, there have only been a limited number of new treatments to prevent HF after MI in recent years.3 These unmet needs with respect to biomarker and drug discovery necessitate a prioritization of post-MI HF candidates for the research community.
We performed large-scale proteomic profiling of plasma obtained at 30 days after an index MI to identify circulating proteins associated with incident HF occurring beyond 30 days. Network analysis revealed top-ranked circulating protein candidates, for which association with HF events was then validated in an independent post-MI cohort. Differentially expressed proteins were further cross-referenced with single-cell and single-nucleus transcriptomes of cardiac myocyte (CM) and non-CM cardiac cells to identify candidates presenting consistent associations in murine models of MI and HF, and in patients with dilated cardiomyopathy (DCM). Last, the expression patterns of top-priority candidates were confirmed in human and mouse primary cardiac cell culture models of disease. The overarching goal was to provide a curated series of protein candidates for focused biomarker discovery and possible drug targeting.
Methods
Detailed methods are available in the Data Supplement. The data, analytical methods, and study materials for the purposes of reproducing the results or replicating procedures are available online at https://github.com/ArisStefanosSn/HFproteomics.
Study Design
A flow diagram summarizing the entire study design is provided in Figure 1.
Figure 1.
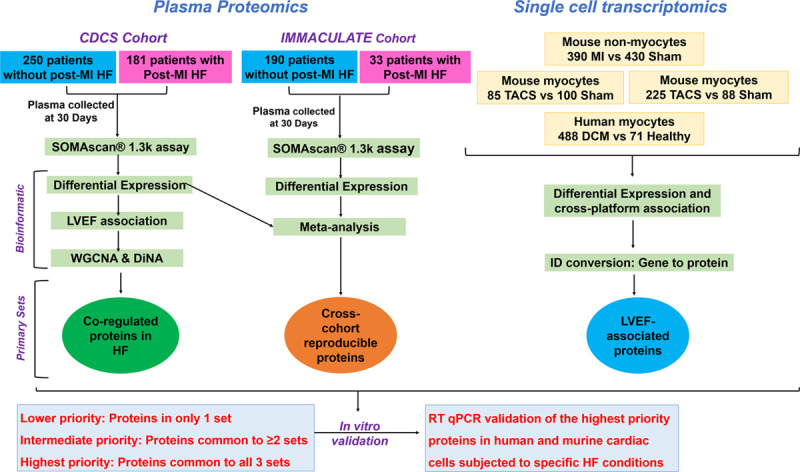
Study design, proteomics, and transcriptomics workflow.
Patients with recent MI were sampled at 30-day posthospitalization in both cohorts, CDCS and IMMACULATE. We performed an aptamer-based proteomics that measures 1305 proteins in a total of 654 patients post-MI and identified candidate proteins through differential expression and network analyses. We also analyzed 4 single-cell data sets to identify candidate genes that show consistent associations in murine and human MI and HF models. Candidates that were common to CDCS, IMMACULATE, and a single-cell data set were further investigated in murine and human cardiac cells subjected to specific HF conditions. CDCS indicates Coronary Artery Disease Cohort Study; DCM, dilated cardiomyopathy; DiNA, differential network analysis; HF, heart failure; IMMACULATE, Improving Outcomes in Myocardial Infarction through Reversal of Cardiac Remodelling; LVEF, left ventricular ejection fraction; MI, myocardial infarction; RT qPCR, real-time quantitative polymerase chain reaction; TAC, transverse aortic constriction; and WGCNA, weighted gene coexpression network analysis.
Study Populations
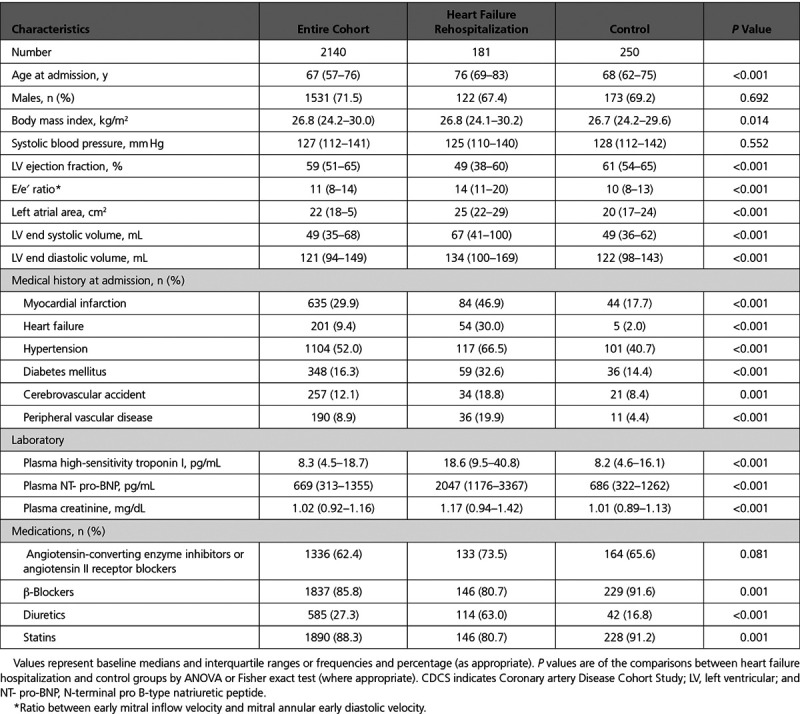
The primary cohort was selected from CDCS (Coronary Disease Cohort Study, ACTRN 12605000431628), which consisted of 2140 patients hospitalized for an acute coronary syndrome in 2 tertiary hospitals in New Zealand from 2002 to 2009 and followed up for a median of 5.1 years (interquartile range, 3.7–6.8 years; maximum 9.5 years).4 Inclusion criteria were ischemic chest discomfort plus ≥1 of the following: electrocardiographic changes (ST-segment depression or elevation ≥0.5 mm, T-wave inversion of ≥3 mm in ≥3 leads, or left bundle-branch block) and elevated cardiac markers. Patients were excluded if they had a severe comorbidity that reduced their life expectancy to <3 years. Clinical data, blood samples, and echocardiographic measurements were obtained at ≈30 days, 4 months, and 12 months after hospital admission. Subsequent clinical events and mortality were obtained from the New Zealand National Health Information System. The New Zealand Multi-region Ethics Committee approved the study (CTY/02/02/018), and all participants gave written informed consent before study participation. For the current study, we selected nested cases of 181 patients post-MI who had readmission for HF (HF group) and another 250 patients post-MI who remained free of HF hospitalization or death from a cardiovascular cause during follow-up (control group). Controls were age- and sex-matched to patients post-MI with HF by using the MatchIT package in R.5 In the HF group, the median time to HF hospitalization was 1.1 years (interquartile range, 99 days to 3.2 years; maximum, 8.8 years). Controls remained free of HF hospitalization for a median of 4.9 years (interquartile range, 3.8–6.5 years; maximum, 9.4 years).
The external cohort was selected from the IMMACULATE registry (Improving Outcomes in Myocardial Infarction through Reversal of Cardiac Remodelling), which consisted of 859 patients hospitalized for MI at 3 hospitals in Singapore from 2013 to 2017 and followed up for a median of 2.0 years (interquartile range, 1.9–2.1 years; maximum, 6.2 years). Inclusion criteria were as follows: clinically diagnosed ST-segment–elevation MI or non–ST-segment–elevation MI with typical history of ischemic chest pain or angina-equivalent symptoms with a typical rise and fall of cardiac marker concentrations (cardiac troponin value exceeding the 99th percentile (in ng/L or pg/mL) and angiographic findings of >50% occlusion of ≥1coronary arteries. The exclusion criteria were severe renal impairment (estimated glomerular filtration rate <15 mL·min–1·m–2); anemia; hemoglobin <8 g/dL (men) and 7 g/dL (women); cardiogenic shock unable to be weaned off inotropes or intra-aortic balloon pump; history of malignancy or other conditions limiting life expectancy diagnosed within the last 12 months. Clinical data, blood samples, and echocardiographic measurements were obtained within 24 to 72 hours of admission, 30 days, 6, 12, and 24 months after hospital admission. We selected a cohort of the first 223 consecutively enrolled patients who had 30-day plasma samples available and therefore had the longest follow-up period at the time of study completion. Of these 223 patients, 33 were hospitalized for HF during follow-up. The institutional review board and the ethics committee at Singapore’s National Healthcare Group Domain Specific Review Board (DSRB 2013/00248 and 2013/00635) approved the study protocol, and all patients gave written informed consent before participation.
For both cohorts, comparisons between groups (HF and controls) and baseline characteristics were tested with analysis of variance, χ2, and log-rank tests, as appropriate. Clinical data are presented as n (%) or medians and interquartile ranges, unless stated otherwise.
Clinical End Points
The primary clinical end point was hospitalization for HF, defined as clinically diagnosed acute HF requiring hospitalization for >24 hours or treatment with diuretics if duration of stay was <24 hours.
Proteomic Analysis
Plasma samples (50 µL) collected 30 days after the index event were analyzed using a Slow Off-rate Modified Aptamer (SOMAmer)–based capture array called SOMAscan (somaLogic, Inc). We measured 1305 human proteins (47% secreted proteins, 28% extracellular domains, and 25% intracellular proteins). The experimental processes and normalization methods have been previously described6,7 (Methods in the Data Supplement). In brief, fluorescently labeled SOMAmers were quantified as relative fluorescence units that correlate with the protein concentration in the original plasma sample. Quality control was performed at the sample and SOMAmer level. The former involved the use of hybridization controls to monitor sample-by-sample variability, whereas the latter used control SOMAmers for data normalization and calibration samples to control interassay and intra-assay variabilities. The sample data were first normalized to remove within-run hybridization variation followed by median normalization across all samples and finally calibrated to eliminate interplate and interrun differences. The acceptance criteria for normalization were 0.4 to 2.5 and calibration scale factor for the SOMAmer within ±0.4 of the median.
Statistical Analysis
The raw SOMAscan expression profiles of the 1128 proteins passing the quality control in CDCS cohort samples were log2 transformed and adjusted for a set of confounding factors. For each protein, we used linear modeling to assess any independent effects of the following confounders: sex, age, body mass index, smoking status, clinical history (diabetes mellitus, hypertension, hyperlipidemia, culprit vessel, and renal disease), and medication status (β-blockers and angiotensin-converting enzyme inhibitors/angiotensin receptor blockers) on protein abundance. The variables with a false discovery rate (FDR) <5% were fitted in a multiple regression linear model to minimize confounding effects (Methods in the Data Supplement). Differential expression analysis was performed on the adjusted protein levels with the LIMMA model.8 We estimated the log2 fold changes of HF versus control groups and the FDR for each cohort separately. We then tested associations between protein abundance (at 30 days) with left ventricular ejection fraction (LVEF) at 4 months post-MI for each of the 1128 proteins by Spearman correlation. The rationale for correlating protein abundance with LVEF measured 4 months after the index MI was to enrich for proteins correlating with cardiac function as a quantitative phenotype during post-MI cardiac remodeling.
To further prioritize protein candidates for validation, we used a combination of network analysis and functional annotation tools to enrich for protein candidates with the potential to influence progression from MI to HF (Methods in the Data Supplement). We hypothesized that proteins with highly correlated plasma concentrations may be coregulated, or functionally related, and that some proteins differentially coregulated in the HF group may promote progression from MI to HF. In brief, we estimated the protein coexpression network of HF and control sets separately by using weighted gene co-expression network analysis (WGCNA)9 on all 1128 proteins of each set. Within each network, we performed unsupervised hierarchical clustering with dynamic branch cutting to identify HF-specific and control-specific clusters of highly correlated proteins (functional modules): the stronger the correlations between proteins in a module, the greater the chance the module represents a network of coregulated, functionally related proteins10 (Methods in the Data Supplement). The biological functions of each module were estimated by Gene Set Enrichment Analysis.11 Last, we used comparative correlation network analysis to estimate statistically significant module differences between the HF and control networks. Our approach, based on Differential Network Analysis (DiNA),12 quantified the network differences in terms of protein correlation patterns. In combination with the evidence from the differential expression analysis, it pointed to large HF-upregulated protein hubs associated with unique functions and pathways potentially influencing the biological processes that underlie progression from MI to HF (Methods in the Data Supplement). Cytoscape13 was used to visualize the significant modules and hubs of interest.
Validation in External Cohort
We combined the differential expression estimates of the CDCS and IMMACULATE cohorts to identify the reproducible up- and downregulated proteins in HF. To alleviate the low power for detecting differential expression in the IMMACULATE cohort attributable to its smaller number of HF cases than in the CDCS cohort (33 HF cases in IMMACULATE versus 181 HF cases in CDCS), we performed cross-cohort meta-analysis using the Fisher-based P value combination method from the metaRNASeq R package14 (Methods in the Data Supplement). We considered proteins to be reproducible if they had the same logFC direction across the 2 cohorts with FDR≤5% in CDCS, P≤5% in IMMACULATE and meta-analysis FDR≤5%. We then used supervised random forests to quantify and compare the accuracy of the reproducible proteins in discriminating the patients with HF from the control patients in both cohorts. In addition, we ran a 2-dimensional principal components analysis on the 36 reproducible proteins to determine if the difference in 2-dimensional means of the 36 selected proteins comparing the HF and control groups was statistically larger than their respective differences in 10 000 random 36-protein data sets (generated from the initial 212 differentially expressed proteins). Equality of the 2-dimensional means would imply that the principal components clusters overlap completely and the proteins did not separate the 2 patient groups adequately. The significance of the test was estimated from 10 000 random protein sets of the same size as  (Methods in the Data Supplement).
(Methods in the Data Supplement).
Murine and Human Single-Cell RNA-Sequencing Analysis
In parallel, we prioritized protein candidates for validation by identifying those with consistent associations in murine models of MI and HF, and human patients with DCM, versus controls (Methods in the Data Supplement). In brief, we cross-referenced protein candidates with single-cell transcriptomic data from 4 data sets: (1) an unpublished, in-house single-cell data set from a mouse MI model (permanent left anterior descending coronary artery ligation); (2) our published mouse HF model (transverse aortic constriction [TAC]) single-nucleus data set15; (3) a different published single-cell data set, also from a mouse TAC HF model16; and (4) a published single-cell data set from human patients with DCM.16 Single-cell isolation for data set 1 was performed as previously described.17 All animal experiments were approved by the Institutional Animal Care and Use Committee of the National University of Singapore and performed in accordance with Singapore National Advisory Committee for Laboratory Animal Research guidelines.18
Priority Ranking of Proteins
These analytic steps are expected to yield 3 enriched protein data sets: CDCS plasma proteomic analysis (data set 1), IMMACULATE plasma proteomic analysis (data set 2), and single-cell transcriptomic analysis (data set 3). We then ranked proteins according to 3 priorities: lower priority referring to proteins observed in only 1 of 3 data sets, intermediate priority referring to proteins observed in 2 of 3 data sets, and high priority referring to proteins observed in all 3 data sets. A final enrichment step was then performed by examining which of the prioritized proteins were directly linked with proteins in the DiNA analysis, thus forming strong network hubs.
Targeted Gene Expression Analysis
Expression of top-ranked candidates were measured by quantitative polymerase chain reaction in diverse cell populations, including CM, cardiac fibroblasts (CFs), smooth muscle cells (SMCs), and endothelial cells, that were exposed to prohypertrophic (phenylephrine, isoproterenol, and ET-1), profibrotic (transforming growth factor B1), and proinflammatory (interleukin-1β) stimuli intended to mimic post-MI HF conditions. Gene expression levels of high-priority candidates in different human cell types were compared by using ANOVA with Bonferroni correction.
Results
Cohort Characteristics and Outcomes
In the CDCS cohort, patients in the HF and control groups were well matched for sex and ethnicity, but less so for age and body mass index (Table 1). In the IMMACULATE cohort, there were no differences in sex, ethnicity, age, or body mass index between the different groups (Table I in the Data Supplement). In comparison with the IMMACULATE cohort, the CDCS cohort was older (67 years versus 55 years, P<0.001) with a lower proportion of men (72% versus 93%, P<0.001). Approximately one-third of the patients in CDCS had experienced a previous MI and close to a tenth had previous HF, cerebral vascular accident, and peripheral vascular disease. In comparison, one-tenth of the patients in IMMACULATE had previous MI and <1% had previous HF.
Table 1.
CDCS Baseline Characteristics
Proteins Associated With Post-MI HF
Differential expression analysis of the 1128 SOMAscan proteins identified 212 differentially expressed proteins associated with post-MI HF events at FDR ≤ 5% (Figure 2A and Table II in the Data Supplement. Of these, concentrations of 128 proteins were higher and concentrations of 84 proteins were lower in patients who developed HF in comparison with control patients. The top 20 included not only proteins of established prognostic significance in HF, such as NT-proBNP (N-terminal pro B-type natriuretic peptide), TNNT2 (cardiac troponin T), and TNNI3 (troponin I), but also many emerging candidates, including TFF3 (trefoil factor 3), FSTL3 (follistatin-related protein 3), THBS2 (thrombospondin-2), and ANGPT2 (angiopoietin-2; Table II in the Data Supplement). NT-proBNP was the top-ranked differentially expressed protein, regardless of whether the proteins were ranked on fold change or statistical significance (1.8-fold higher in HF than in control, 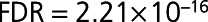 ).
).
Figure 2.
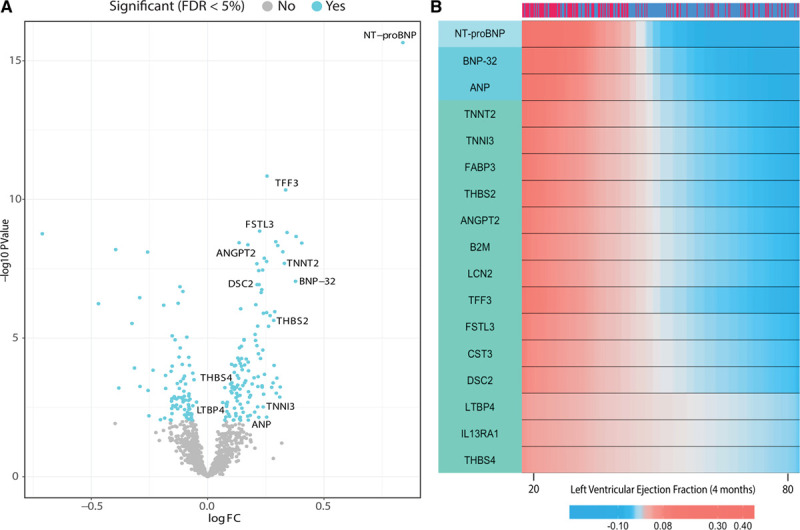
Plasma proteins associated with post–myocardial infarction heart failure and 4-month post–myocardial infarction left ventricular ejection fraction.
A, Volcano plot of the 1128 proteins measured in CDCS and their protein differential expression estimates by Limma. Colored dots represent significantly associated proteins at FDR ≤ 5%. B, Heat map of the protein expression levels vs left ventricular ejection fraction at 4 months (x axis). The strength of the correlation between protein expression and LVEF is indicated by the red and blue gradients of the heat map; a deeper shade of red indicates that higher protein levels (overexpression) correlate more strongly with a particular LVEF value, whereas a deeper shade of blue indicates that lower protein levels (underexpression) correlate more strongly with a particular LVEF value. The 17 proteins all show a negative correlation with LVEF such that high protein levels (deeper red) is observed with lower LVEF values. Patient group is indicated as HF in dark red and control in dark blue (top bar). A subset of the 96 significant proteins with the most highly correlated coefficients are shown (FDR ≤ 5%). The left bar shows the unsupervised hierarchical protein clusters. Protein expression levels have been smoothed by a nonparametric regression model. CDCS indicates Coronary Artery Disease Cohort Study; FC, fold change; FDR, false discovery rate; HF, heart failure; LVEF, left ventricular ejection fraction; and NT-proBNP, N-terminal pro B-type natriuretic peptide.
Correlation Between Plasma Proteins and 4-Month Post-MI LVEF
Global Spearman nonparametric correlation coefficient showed that 96 of the 212 differentially expressed proteins correlated with LVEF measured at 4 months post-MI (Figure 2B and Table III in the Data Supplement). The top 5 proteins showing negative correlation with LVEF were NT-proBNP (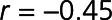 ,
, 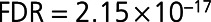 ), BNP-32 (brain natriuretic peptide-32), VEGF-D (vascular endothelial growth factor D), TFF3, and FSTL3 (all
), BNP-32 (brain natriuretic peptide-32), VEGF-D (vascular endothelial growth factor D), TFF3, and FSTL3 (all 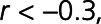
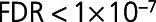 ). Seventeen proteins positively correlated with LVEF including 6-phosphogluconate dehydrogenase (
). Seventeen proteins positively correlated with LVEF including 6-phosphogluconate dehydrogenase (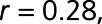
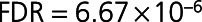 ), S100A6 (S100 calcium-binding protein A6;
), S100A6 (S100 calcium-binding protein A6; 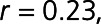
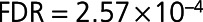 ), and ENTPD3 (ectonucleoside triphosphate diphosphohydrolase-3;
), and ENTPD3 (ectonucleoside triphosphate diphosphohydrolase-3; 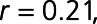
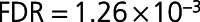 ).
).
Bioinformatic Enrichment Through Network Analysis
To identify proteins with highly correlated plasma concentrations that may be coregulated and influence progression from MI to HF, WGCNA was applied to all 1128 SOMAscan proteins in the HF and control groups of the CDCS cohort. Unsupervised hierarchical clustering revealed 3 modules in the HF group (Figure 3A and Figures I and II in the Data Supplement), designated HF1 (153 proteins), HF2 (662 proteins), and HF3 (313 proteins), and 2 modules in the control group (Figures I and II in the Data Supplement), designated Ctrl1 (824 proteins) and Ctrl2 (304 proteins).
Figure 3.
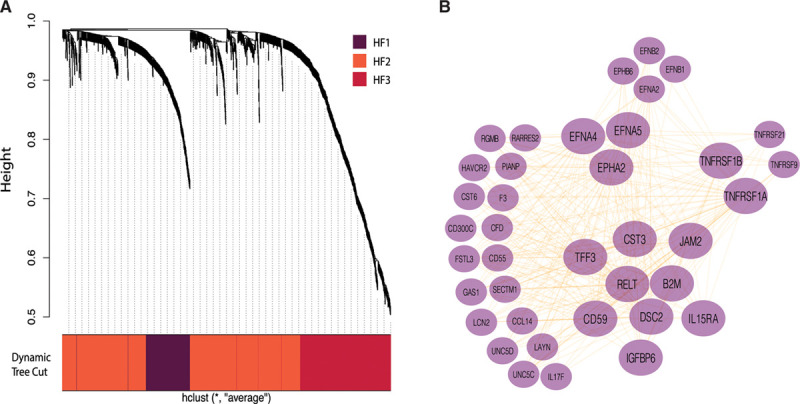
Network analysis of plasma proteins.
A, Hierarchical clustering highlighting the estimated, color-coded WGCNA modules in heart failure. B, The WGCNA Heart Failure network of the HF1 module proteins highlighting the significant coexpression hubs. In large font are the significant proteins of the Differential Network Analysis model. Only the connections with weighted correlations aii > 0.2 are shown. HF indicates heart failure; and WGCNA, weighted gene co-expression network analysis.
Because WGCNA modules may represent networks of coregulated, functionally related proteins,9 we explored the potential biological function of the modules associated with post-MI HF using Gene Set Enrichment Analysis with a customized background protein set (Methods in the Data Supplement). The proteins of the HF1 module were uniquely associated with the regulation of actin filament process, ephrin receptor signaling, and regulation of muscle system processes (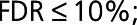 Figure IIIA in the Data Supplement). The enrichment of the ephrin-signaling pathway was independently validated by REACTOME GO analysis using g:Profiler.19
Figure IIIA in the Data Supplement). The enrichment of the ephrin-signaling pathway was independently validated by REACTOME GO analysis using g:Profiler.19
Next, we compared the HF and control correlation networks by DiNA to highlight their key differences in protein coregulation and associate them with specific biological functions and pathways (Methods in the Data Supplement). DiNA identified a set of 15 proteins up-regulated in HF at 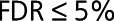 . Of these, 14 proteins belonged to the HF1 module (enrichment Fisher test
. Of these, 14 proteins belonged to the HF1 module (enrichment Fisher test 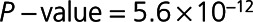 ) and had statistically stronger correlations to other proteins in HF than in control at resampling-based
) and had statistically stronger correlations to other proteins in HF than in control at resampling-based 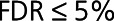 (Figure IIIB in the Data Supplement and Table IV in the Data Supplement). These 14 proteins were TFF3, B2M (β2 microglobulin), CST3 (Cystatin C), EPHA2 (Ephrin-A2), EFNA4 (Ephrin-A4), EFNA5 (ephrin-A5), TNFRSF1α, TNFRSF1β, and RELT (tumor necrosis factor receptor superfamily members 1α, 1β, and 19 L), DSC2 (desmocollin-2), IL15RA (interleukin-15 receptor subunit A), CD59 glycoprotein, JAM2 (junctional adhesion molecule B), and IGFBP6 (insulin-like growth factor binding protein 6). Cytoscape13 (Figure 3B) illustrated the top hubs formed by these proteins in HF and control, implying that the HF1 module contained strong protein hubs associated with unique biological functions differentiating the HF group from the control group (Figure IIIC in the Data Supplement). We found that 63 of the differentially expressed proteins of HF1 were also correlated with 4-month post-MI LVEF as a quantitative phenotype (Table IV in the Data Supplement).
(Figure IIIB in the Data Supplement and Table IV in the Data Supplement). These 14 proteins were TFF3, B2M (β2 microglobulin), CST3 (Cystatin C), EPHA2 (Ephrin-A2), EFNA4 (Ephrin-A4), EFNA5 (ephrin-A5), TNFRSF1α, TNFRSF1β, and RELT (tumor necrosis factor receptor superfamily members 1α, 1β, and 19 L), DSC2 (desmocollin-2), IL15RA (interleukin-15 receptor subunit A), CD59 glycoprotein, JAM2 (junctional adhesion molecule B), and IGFBP6 (insulin-like growth factor binding protein 6). Cytoscape13 (Figure 3B) illustrated the top hubs formed by these proteins in HF and control, implying that the HF1 module contained strong protein hubs associated with unique biological functions differentiating the HF group from the control group (Figure IIIC in the Data Supplement). We found that 63 of the differentially expressed proteins of HF1 were also correlated with 4-month post-MI LVEF as a quantitative phenotype (Table IV in the Data Supplement).
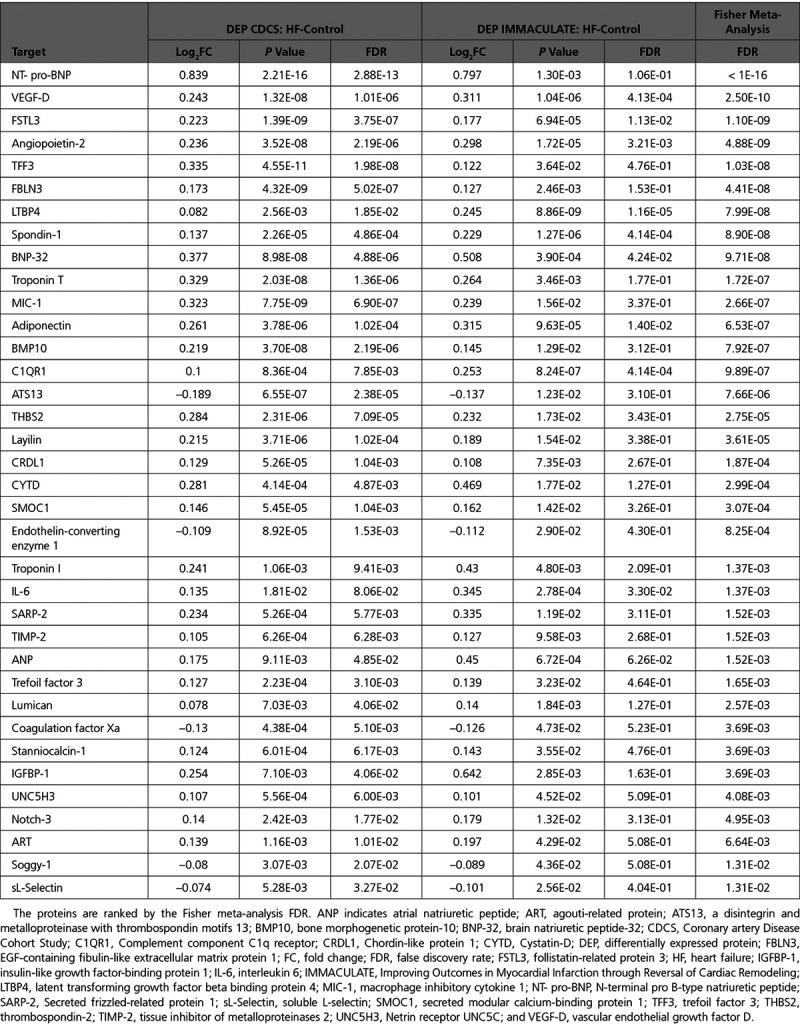
Cross-Cohort Validation of Plasma Proteins
The cross-cohort meta-analysis of the CDCS and IMMACULATE cohorts identified 36 reproducible proteins with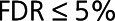 in CDCS,
in CDCS,  in IMMACULATE, and meta-analysis
in IMMACULATE, and meta-analysis 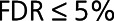 (Table 2). The difference in 2-dimensional means of the HF and control principal components clusters was significantly >10 000 random 36-protein sets generated from the initial 212 differentially expressed proteins (resampling-based t test
(Table 2). The difference in 2-dimensional means of the HF and control principal components clusters was significantly >10 000 random 36-protein sets generated from the initial 212 differentially expressed proteins (resampling-based t test 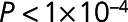 ) indicating that the separation of the HF and control groups was more significant than random (Figure IVA in the Data Supplement). Supervised random forests indicated that, when compared with other targeted protein sets, the 36 reproducible proteins exhibited the highest prediction accuracy (minimal estimation errors) and excellent trade-off between the number of reproducible proteins and predictive accuracy (Methods in the Data Supplement and Figure IVB in the Data Supplement).
) indicating that the separation of the HF and control groups was more significant than random (Figure IVA in the Data Supplement). Supervised random forests indicated that, when compared with other targeted protein sets, the 36 reproducible proteins exhibited the highest prediction accuracy (minimal estimation errors) and excellent trade-off between the number of reproducible proteins and predictive accuracy (Methods in the Data Supplement and Figure IVB in the Data Supplement).
Table 2.
The 36 Reproduced Plasma Proteins in the CDCS and IMMACULATE Cohorts
Cross-Referencing Plasma Proteins With Cardiac Single-Cell Transcriptomes
To identify candidates presenting consistent associations in murine models of MI and HF, and patients with DCM, as well, we cross-referenced them with single-cell transcriptomic data from mouse MI and HF disease models and human patients with DCM.
The first data set consisted of 2031 single nonmyocyte cells from 6 mice 7 days after we had performed coronary ligation–induced MI (versus sham) surgery (1074 cells from 3 MI mice and 957 cells from 3 sham mice). Forty-three stable clusters (Figure VA in the Data Supplement) were identified and grouped into 5 major cell populations (Figure VB in the Data Supplement). Of those, the 830 CFs (390 MI and 440 sham) showed the highest degree of corroboration with the human plasma protein results (Figure VC in the Data Supplement) with 39 CF-enriched genes corresponding to proteins detected in the SOMAscan array (Table V in the Data Supplement). This consistency adds validity and emphasizes the key contributory roles of nonmyocyte cell types such as CFs in post-MI pathology. Six genes were identified with significant differential expression directionally consistent with the corresponding clinical plasma protein results: THBS2, ANGPT2, EIF5 (eukaryotic translation initiation factor-5), ECE1 (endothelin-converting enzyme 1), SFRP1 (secreted frizzled-related protein-1), and IL13RA1 (interleukin-13 receptor subunit α-1; Figure VIA in the Data Supplement).
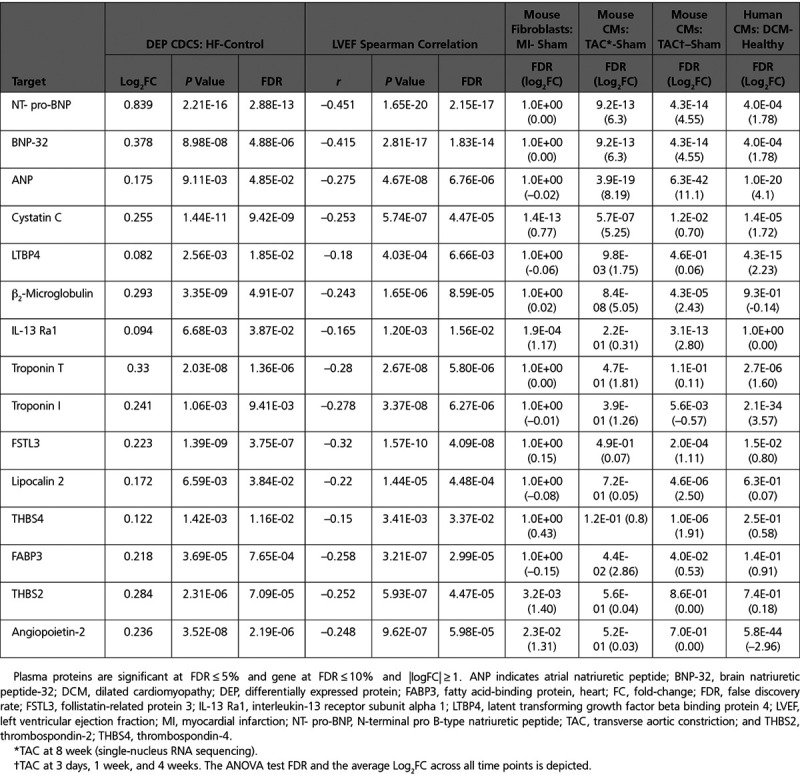
As a second comparative data set, we analyzed our published single-nucleus RNA-sequencing analysis of mouse CMs isolated 8 weeks after we had performed TAC (versus sham) surgery, a technique commonly used to induce a HF phenotype in mice.15 A total of 302 differentially expressed genes were identified, 282 of which were upregulated in the TAC group. Forty of these genes corresponded to proteins detected on the SOMAscan array (Table VI in the Data Supplement), implicating CMs as a major source of plasma protein biomarker candidates in post-MI HF. For 13 of these genes, differential expression was both statistically significant and directionally consistent with corresponding plasma protein results: ANP (atrial natriuretic peptide; NPPA gene), NT-proBNP/BNP-32 (NPPB gene), CST3, B2M, HINT1 (histidine triad nucleotide-binding protein 1), LDHB (L-lactate dehydrogenase B chain), FABP3 (fatty acid-binding protein, heart), ATP50 (ATP synthase subunit O, mitochondrial), LUM (lumican), EIF5, IGFBP5 (insulin-like growth factor-binding protein 5), ANXA1 (annexin A1), and LTBP4 (latent transforming growth factor beta binding protein 4; Figure VIB in the Data Supplement).
Next, 169 differentially expressed genes were further identified in a third data set of murine CMs isolated at 3 days (69 cells), 1 week (83 cells), and 4 weeks (73 cells) post-TAC and sham (88 cells) from Nomura et al16 (Table VII in the Data Supplement). Of these, 10 gene candidates: ANP (NPPA gene), NT-proBNP/BNP-32 (NPPB gene), IL13RA1, LCN2 (neutrophil gelatinase-associated lipocalin), B2M, THBS4 (thrombospondin-4), EIF5, PAFAH1β2 (platelet-activating factor acetylhydrolase IB subunit beta), ESD (S-formylglutathione hydrolase), and FSTL3, were statistically significant and directionally consistent with the results of plasma protein measurements in the CDCS clinical samples (Figure 3C and Figure VIC in the Data Supplement).
A fourth data set, from the same publicly available data set of Nomura et al,16 was a human single-cell transcriptome of 559 CM cells from patients with DCM versus controls. Differential expression was significant for 11 gene candidates with the altered expression directionally consistent with that of corresponding translated plasma proteins: ANP (NPPA gene), TNNI3, LTBP4, CFD (complement factor D), LUM, NT-proBNP/BNP-32 (NPPB gene), CST3, SOD3 (extracellular superoxide dismutase [Cu-Zn]), TNNT2, IGFBP6, and KLRF1 (killer cell lectin-like receptor subfamily F member 1; Table VIII in the Data Supplement; Figure 3C; and Figure VID in the Data Supplement).
In summary, in single-cell studies we found a total of 40 genes with altered expression corresponding to the directional shifts of the concentrations of 30 unique proteins significantly associated with HF in the CDCS plasma samples (Figure VII in the Data Supplement). Of these 30 proteins, 15 candidates were also correlated with LVEF measured at 4 months post-MI as a quantitative phenotype: TNNT2, B2M, NT-proBNP, BNP, LCN2, TNNI3, FABP3, CST3, ANGPT2, ANP, IL13RA1, LTBP4, THBS2, THBS4, and FSTL3 (Table 3).
Table 3.
Candidates That Were Statistically Significant and Directionally Consistent at Both Plasma Protein and Transcriptomic Levels
Prioritization and In Vitro Quantitative Polymerase Chain Reaction Validation of Post-MI HF Candidates
We compared the 3 enriched protein data sets from the preceding analysis: the 63 proteins from the HF1 module that were correlated with LVEF in CDCS (data set 1), the 36 proteins from the cross-cohort meta-analysis in IMMACULATE (data set 2), and the protein identities of the prioritized genes from the single-cell transcriptomic data (data set 3). Fifty-eight proteins were found in only 1 data set (lower-priority candidates), 19 candidates were found in 2 data sets (intermediate-priority candidates), and 6 candidates were found in all 3 data sets (highest-priority candidates), making a total of 83 prioritized proteins (Table IX in the Data Supplement). When the 6 highest-priority proteins were mapped back to the 14 DiNA proteins within the densely correlated HF1 module of the WGCNA analysis, TNNT2, ANGPT2, THBS2, LTPB4, and FSTL3 all mapped directly to ≥1 DiNA proteins, but NT-proBNP did not map directly to any DiNA proteins, suggesting closer coregulation of the former 5 proteins than NT-proBNP within the HF1 module of proteins.
To further investigate the source of the 6 top-priority candidates, we obtained and cultured primary human cardiac CFs, SMCs, and endothelial cells, alongside human embryonic stem cell–derived CMs, and exposed these cells to prohypertrophic, profibrotic, or proinflammatory stimuli to mimic conditions that lead to HF after MI. Cell- and condition-specific patterns of candidate gene expression were detected by quantitative polymerase chain reaction (Figure 4). NPPB and TNNT2 showed the highest gene expression in CMs, with NPPB further upregulated by prohypertrophic stimulation. In contrast, FSTL3 and THBS2 showed the highest expression in CFs, with presence also in SMCs. Proinflammatory and profibrotic stimuli increased THBS2 gene expression in nonmyocytes. Of note, profibrotic stimulation markedly increased FSTL3 and LTBP4 expression in CFs and SMCs, whereas prohypertrophic stimulation increased FSTL3 and LTBP4 expression in CMs, therefore suggesting multicellular sources and roles for FSTL3 and LTBP4 in post-MI HF. The robustness of these data is further demonstrated by reproduction in primary cultured murine CMs and CFs (Figure VIII in the Data Supplement). Only ANGPT2 showed discordant interspecies gene expression and was detectable in murine but not human CFs; this was probably because of the presence of some endothelial cells retained in these mouse CF cultures as previously reported.17
Figure 4.
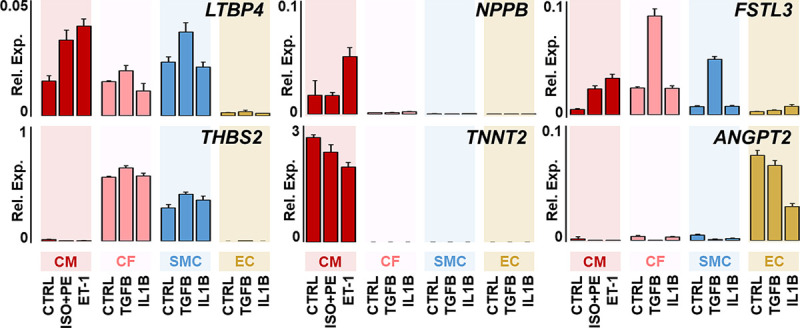
Expression of top-priority candidate genes in human cardiac cell cultures.
Human cardiac cells were cultured and stimulated in serum-free conditions. After stimulation, RNA was collected, and candidate gene expression was detected by quantitative polymerase chain reaction, relative to 18S/10 000 expression. Data show means, and error bars show standard deviation; n=3 biological replicates. Prohypertrophic (phenylephrine, isoproterenol, ET-1), fibrotic (TGFB), or inflammatory (IL1B). ANGPT2 indicates angiopoietin-2; CF, cardiac fibroblast; CM, cardiac myocyte; Ctrl, control; EC, endothelial cell; FSTL3, follistatin-related protein 3; IL1B, interleukin 1β; ISO, isoproterenol; LTBP4, latent transforming growth factor beta binding protein 4; NPPB, NT-proBNP/BNP; PE, phenylephrine; Rel. Exp., relative expression; SMC, smooth muscle cell; TGFB, transforming growth factor-B; THBS2, thrombospondin-2; and TNNT2, troponin T.
Discussion
Large-scale proteomics of plasma obtained 30 days after MI revealed 212 plasma proteins associated with subsequent HF hospitalization. Of these protein candidates, 96 were correlated with LVEF measured at 4 months post-MI. Bioinformatic enrichment further revealed 63 highly correlated proteins of a single module among patients who had post-MI HF but not among event-free patients. Meta-analysis of plasma proteins in an independent post-MI cohort revealed 36 proteins that were associated with post-MI HF. Unbiased single-cell transcriptomics of murine MI and HF model systems or human subjects with DCM further identified 30 candidates of which 15 candidates correlated with 4-month LVEF. Of the 83 prioritized candidates, the 6 highest-priority proteins were common to the plasma proteomic analyses of both patient cohorts and the single-cell transcriptomic analysis; 2 candidates were well-established biomarkers of post-MI HF, NT-proBNP and TNNT, whereas the other 4 are newly emerging biomarkers, ANGPT2, THBS2, LTBP4, and FSTL3 (Figure 5).
Figure 5.
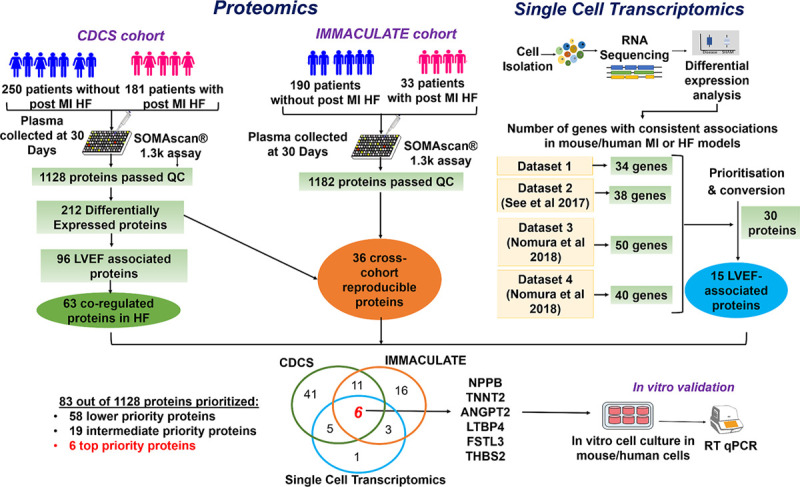
Summary of post–myocardial infarction heart failure candidates.
CDCS indicates Coronary Artery Disease Cohort Study; HF, heart failure; IMMACULATE, Improving Outcomes in Myocardial Infarction through Reversal of Cardiac Remodelling; LVEF, left ventricular ejection fraction; MI, myocardial infarction; QC, quality control; and RT qPCR, real-time quantitative polymerase chain reaction.
Many of the 83 prioritized proteins are matricellular proteins that are typically secretable proteins found most abundantly in non-CMs embedded within the cardiac extracellular matrix.20 Extracellular matrix fibrosis-mediating proteins, including BMP (bone morphogenetic protein), VEGF (vascular endothelial growth factor), and FABP3, and extracellular matrix stress proteins, including GDF15 (growth-differentiating factor 15) CST3, were found in our intermediate-priority list of proteins.21 The 4 newly emergent biomarkers among the highest-priority candidates, ANGPT2, THBS2, LTBP4, and FSTL3, are all proteins with high bioactivity within the cardiac matricellular environment.22 In particular, ANGPT2 and THBS2 have demonstrated strong potential as prognostic biomarkers of cardiac ischemic risk and HF rehospitalization in several other studies.23,24 ANGPT2, which is expressed on endothelial cells in response to hypoxia, destabilizes vascular endothelial integrity and may promote abnormal microvascular remodeling within the myocardium during the chronic postinfarct phase.25 THBS1 (thrombospondin 1) has been previously established as exerting a deleterious effect in ischemic (post-MI) HF,21 but the biological roles of THBS2 in ischemic HF are less clear. THBS2 promotes fibrosis26 and has recently been shown to be associated with incident HF events in large hospital- and community-based cohorts profiled using the same SOMAscan array, in which plasma concentrations of THBS2 declined after successful cardiac transplantation for HF.23 LTPB4 has an established role in activating the profibrotic TGFβ (transforming growth factor beta) pathway, whereas FSTL3 is reportedly secreted by CMs and may regulate CM hypertrophy and activate surrounding CFs.27 We found that exposure to TGFβ stimulation, which would likely be present in the post-MI HF environment, activated FSTL3 gene expression more strongly in CFs and even SMCs than in CMs (Figure 4), a finding recapitulated in primary cultured mouse cardiac cells (Figure VIII in the Data Supplement). Plasma FSTL3 concentrations were higher among patients with HF than among healthy controls in a small cohort study,28 whereas plasma FSTL3 concentrations are also higher in obese individuals with metabolic heart disease and correlate with echocardiographic indices of diastolic dysfunction.29
Our study has several strengths. First, the large scale of the plasma proteomic array and whole-genome approach of the RNA sequencing at single-cell/single-nucleus resolution enabled a relatively unbiased exploration of protein-RNA candidates in post-MI HF. Second, other investigators using the SOMAscan platform to interrogate the plasma proteome have found typically intracellular proteins to be associated with cardiovascular diseases such as pulmonary hypertension, HF, and MI.30–32 Although intracellular plasma proteins may leach into the circulation immediately after cardiac cell necrosis, we deliberately obtained plasma 30 days after the index MI to limit the number of proteins released by cardiac cell necrosis. We were able to detect many proteins in human plasma at 30 days post-MI, indicating the robustness of this approach. Third, our 2 independent cohorts were of White and Asian ethnicity, demonstrating the relevance of our results to post-MI patients of diverse genetic backgrounds.
Our study also has limitations. First, our external cohort (IMMACULATE) was smaller than our primary cohort (CDCS) with fewer post-MI HF events (33 versus 181); as such, the power for detecting differentially expressed proteins in the IMMACULATE cohort is relatively low, a problem alleviated by the meta-analysis pipeline. Second, several candidates identified at both the plasma-protein and gene-expression level did not pass Somalogic quality control checks; examples of these proteins were periostin and tissue inhibitor of metalloproteinase-1, proteins that are considered to play key roles in post-MI cardiac remodeling (Tables V through VIII in the Data Supplement; Figure VIA through VID in the Data Supplement). This raises the concern that the quality control checks used may have been overly conservative. Third, concerns have been raised about aptamer-based protein identification and its ability to unambiguously identify whole proteins.33 This concern is partly allayed by integration and cross-validation of candidates against multiple cardiac single-cell data sets, although these are reliant on transcriptomic rather than direct protein expression analysis. Fourth, the availability of high-resolution CM-specific transcriptomic data sets is currently limited to murine permanent ligation, acute MI, TAC HF, and human DCM, rather than a strictly post-MI HF disease. Cross-referencing of data between different cardiac models and pathologies (MI and TAC), different molecular profiling platforms (protein versus RNA), different compartments (plasma and cardiac cells), different species (murine and human), and different ethnic groups (White and Asian) conceivably risks introducing both false positives and, more likely, false negatives. We postulate that these pathologies share many underlying features including CM stress and accompanying fibrosis, and are likely to exhibit similarities in biomarker profiles, which are readily apparent for multiple identified candidates, such as NPPB and LTBP4. Fifth, a larger SOMAscan protein array is now available with ≈5000 proteins and may identify more candidates than the 1305 array used in our study.
Conclusions
Large-scale plasma proteomics of 2 independent post-MI cohorts, cross-referenced to unbiased transcriptomics of murine MI and TAC HF model systems at single-cell resolution, identified 83 proteins as potential biomarkers and drug discovery targets of post-MI HF. Many of these proteins were secretable matricellular proteins, highlighting the prominence of the extracellular matrix in post-MI HF. ANGPT2, THBS2, LTBP4, and FSTL3, 4 of the 6 most highly enriched proteins, are nascent candidates requiring further validation.
Acknowledgments
The authors thank S.-C. Poh, E. Lim, and F. Ng (National University Heart Center, National University Health System, Singapore) for study coordination, Dr Seneviratna (National Public Health and Epidemiology Unit, Tan Tock Seng Hospital, Singapore) and Dr Carvalho (Universidade Federal de São Paulo, Sao Paolo, Brazil) for clinical oversight of the IMMACULATE Registry patients and Z.-l. Teo for research program management. We also thank the research staff of the Heart Health Research Group (Department of Medicine, University of Auckland) and the Christchurch Heart Institute (Department of Medicine, University of Otago, Christchurch) including L. Skelton who provided oversight of clinical studies coordination and B. Neame who provided expert data management. We thank Dr Sims (Newcastle University, UK) for expert assistance with human embryonic stem cell-cardiomyocyte experiments. We also acknowledge the important work by Nomura et al16 that provided independently corroborative single murine and human cell data to our work.
Sources of Funding
This work was supported by the following grants from Singapore: National Medical Research Council (NMRC)/Clinical Science Award (CSA)-Investigator (INV)/0006/2016 (principal investigator: Dr Chan), NMRC/STaR/0022/2014 (principal investigator: Dr Richards); and from New Zealand: The Coronary Disease Cohort Study was funded by the Health Research Council of New Zealand (Program Grants 02/152, 08/070, 11/1070); National Heart Foundation of New Zealand; New Zealand Lotteries Grant Board; Foundation for Research, Science and Technology, and the Christchurch Heart Institute Trust.
Disclosures
None.
Supplemental Materials
Data Supplement Methods
Data Supplement Tables I–IX
Data Supplement Figures I–VIII
References 34–40
Supplementary Material
Footnotes
Drs Chan, Efthymios, and Tan contributed equally.
Drs Ackers-Johnson, Pilbrow, and Richards contributed equally.
Sources of Funding, see page 1420
The Data Supplement, podcast, and transcript are available as a Data Supplement at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.119.045158.
References
- 1.Gerber Y, Weston SA, Enriquez-Sarano M, Berardi C, Chamberlain AM, Manemann SM, Jiang R, Dunlay SM, Roger VL. Mortality associated with heart failure after myocardial infarction: a contemporary community perspective. Circ Heart Fail. 2016;9:e002460.doi: 10.1161/CIRCHEARTFAILURE.115.002460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibrahim NE, Januzzi JL., Jr. Established and emerging roles of biomarkers in heart failure. Circ Res. 2018;123:614–629. doi: 10.1161/CIRCRESAHA.118.312706 [DOI] [PubMed] [Google Scholar]
- 3.Udelson JE, Stevenson LW. The future of heart failure diagnosis, therapy, and management. Circulation. 2016;133:2671–2686. doi: 10.1161/CIRCULATIONAHA.116.023518 [DOI] [PubMed] [Google Scholar]
- 4.Prickett TC, Doughty RN, Troughton RW, Frampton CM, Whalley GA, Ellis CJ, Espiner EA, Richards AM. C-type natriuretic peptides in coronary disease. Clin Chem. 2017;63:316–324. doi: 10.1373/clinchem.2016.257816 [DOI] [PubMed] [Google Scholar]
- 5.Ho DI, Imai K, King G, Stuart EA. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Political Anal. 2007;15:199–236. doi: 10.1093/pan/mpl013 [Google Scholar]
- 6.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004.doi: 10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, Huang J, Stuccio S, Zingone A, Ryan BM, et al. Assessment of variability in the SOMAscan assay. Sci Rep. 2017;7:14248.doi: 10.1038/s41598-017-14755-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17.doi: 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- 10.Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat. 2010;20:281–300. doi: 10.1080/10543400903572753 [DOI] [PubMed] [Google Scholar]
- 11.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm Genome. 2007;18:463–472. doi: 10.1007/s00335-007-9043-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rau A, Marot G, Jaffrézic F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinformatics. 2014;15:91.doi: 10.1186/1471-2105-15-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.See K, Tan WLW, Lim EH, Tiang Z, Lee LT, Li PYQ, Luu TDA, Ackers-Johnson M, Foo RS. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8:225.doi: 10.1038/s41467-017-00319-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomura S, Satoh M, Fujita T, Higo T, Sumida T, Ko T, Yamaguchi T, Tobita T, Naito AT, Ito M, et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun. 2018;9:4435.doi: 10.1038/s41467-018-06639-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackers-Johnson M, Li PY, Holmes AP, O’Brien SM, Pavlovic D, Foo RS. A simplified, Langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ Res. 2016;119:909–920. doi: 10.1161/CIRCRESAHA.116.309202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess AL, Carayol J, Blædel T, Hager J, Di Cara A, Astrup A, Saris WHM, Larsen LH, Valsesia A. Analysis of circulating angiopoietin-like protein 3 and genetic variants in lipid metabolism and liver health: the DiOGenes study. Genes Nutr. 2018;13:7.doi: 10.1186/s12263-018-0597-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019;47W1W191–W198. doi: 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frangogiannis NG. The extracellular matrix in myocardial injury, repair, and remodeling. J Clin Invest. 2017;127:1600–1612. doi: 10.1172/JCI87491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hausenloy DJ, Garcia-Dorado D, Bøtker HE, Davidson SM, Downey J, Engel FB, Jennings R, Lecour S, Leor J, Madonna R, et al. Novel targets and future strategies for acute cardioprotection: position paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc Res. 2017;113:564–585. doi: 10.1093/cvr/cvx049 [DOI] [PubMed] [Google Scholar]
- 22.Frangogiannis NG. The extracellular matrix in ischemic and nonischemic heart failure. Circ Res. 2019;125:117–146. doi: 10.1161/CIRCRESAHA.119.311148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells QS, Gupta DK, Smith JG, Collins SP, Storrow AB, Ferguson J, Smith ML, Pulley JM, Collier S, Wang X, et al. Accelerating biomarker discovery through electronic health records, automated biobanking, and proteomics. J Am Coll Cardiol. 2019;73:2195–2205. doi: 10.1016/j.jacc.2019.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, Sterling DG, Williams SA. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–2541. doi: 10.1001/jama.2016.5951 [DOI] [PubMed] [Google Scholar]
- 25.Lee SJ, Lee CK, Kang S, Park I, Kim YH, Kim SK, Hong SP, Bae H, He Y, Kubota Y, et al. Angiopoietin-2 exacerbates cardiac hypoxia and inflammation after myocardial infarction. J Clin Invest. 2018;128:5018–5033. doi: 10.1172/JCI99659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinecke H, Robey TE, Mignone JL, Muskheli V, Bornstein P, Murry CE. Lack of thrombospondin-2 reduces fibrosis and increases vascularity around cardiac cell grafts. Cardiovasc Pathol. 2013;22:91–95. doi: 10.1016/j.carpath.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panse KD, Felkin LE, López-Olañeta MM, Gómez-Salinero J, Villalba M, Muñoz L, Nakamura K, Shimano M, Walsh K, Barton PJ, et al. Follistatin-like 3 mediates paracrine fibroblast activation by cardiomyocytes. J Cardiovasc Transl Res. 2012;5:814–826. doi: 10.1007/s12265-012-9400-9 [DOI] [PubMed] [Google Scholar]
- 28.Lara-Pezzi E, Felkin LE, Birks EJ, Sarathchandra P, Panse KD, George R, Hall JL, Yacoub MH, Rosenthal N, Barton PJ. Expression of follistatin-related genes is altered in heart failure. Endocrinology. 2008;149:5822–5827. doi: 10.1210/en.2008-0151 [DOI] [PubMed] [Google Scholar]
- 29.Gopal DM, Ayalon N, Wang YC, Siwik D, Sverdlov A, Donohue C, Perez A, Downing J, Apovian C, Silva V, et al. Galectin-3 is associated with stage B metabolic heart disease and pulmonary hypertension in young obese patients. J Am Heart Assoc. 2019;8:e011100.doi: 10.1161/JAHA.118.011100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob J, Ngo D, Finkel N, Pitts R, Gleim S, Benson MD, Keyes MJ, Farrell LA, Morgan T, Jennings LL, et al. Application of large-scale aptamer-based proteomic profiling to planned myocardial infarctions. Circulation. 2018;137:1270–1277. doi: 10.1161/CIRCULATIONAHA.117.029443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes CJ, Wharton J, Ghataorhe P, Watson G, Girerd B, Howard LS, Gibbs JSR, Condliffe R, Elliot CA, Kiely DG, et al. Plasma proteome analysis in patients with pulmonary arterial hypertension: an observational cohort study. Lancet Respir Med. 2017;5:717–726. doi: 10.1016/S2213-2600(17)30161-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams SA, Murthy AC, DeLisle RK, Hyde C, Malarstig A, Ostroff R, Weiss SJ, Segal MR, Ganz P. Improving assessment of drug safety through proteomics: early detection and mechanistic characterization of the unforeseen harmful effects of torcetrapib. Circulation. 2018;137:999–1010. doi: 10.1161/CIRCULATIONAHA.117.028213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi A, Mayr M. in aptamers they trust: the caveats of the SOMAscan biomarker discovery platform from SomaLogic. Circulation. 2018;138:2482–2485. doi: 10.1161/CIRCULATIONAHA.118.036823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- 35.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800 [DOI] [PubMed] [Google Scholar]
- 36.Scialdone A, Natarajan KN, Saraiva LR, Proserpio V, Teichmann SA, Stegle O, Marioni JC, Buettner F. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021 [DOI] [PubMed] [Google Scholar]
- 37.Xu H, Luo X, Qian J, Pang X, Song J, Qian G, Chen J, Chen S. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One. 2012;7:e52249.doi: 10.1371/journal.pone.0052249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 39.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


