Abstract
Key message
Two major QTLs associated with low seed coat deficiency of soybean seeds were identified in two biparental populations, and three SNP markers were validated to assist low-SCD natto soybean breeding selection.
Abstract
Soybean seed coat deficiency (SCD), known as seed coat cracking during soaking in the natto production process, is problematic because split or broken beans clog production lines and increases production costs. Development of natto soybean cultivars with low SCD is crucial to support the growth of the natto industry. Unfortunately, information on the genetic control of SCD in soybean, which is desperately needed to facilitate breeding selection, remains sparse. In this study, two F2 populations derived from V11-0883 × V12-1626 (Pop 1) and V11-0883 × V12-1885 (Pop 2) were developed and genotyped with BARCSoySNP6K Beadchips and F2-derived lines were evaluated for SCD in three consecutive years (2016–2018) in order to identify quantitative trait loci (QTLs) associated with low SCD in soybean. A total of 17 QTLs underlying SCD were identified in two populations. Among these, two major and stable QTLs, qSCD15 on chromosome 15 and qSCD20 on chromosome 20, were detected across multiple years. These QTLs explained up to 30.3% of the phenotypic variation for SCD in Pop 1 and 6.1% in Pop 2 across years. Three SNP markers associated with the qSCD20 were validated in additional four biparental populations. The average selection efficiency of low-SCD soybean was 77% based on two tightly linked markers, Gm20_34626867 and Gm20_34942502, and 64% based on the marker Gm20_35625615. The novel and stable QTLs identified in this study will facilitate elucidation of the genetic mechanism controlling SCD in soybean, and the markers will significantly accelerate breeding for low-SCD soybean through marker-assisted selection.
Electronic supplementary material
The online version of this article (10.1007/s00122-020-03662-5) contains supplementary material, which is available to authorized users.
Introduction
Because of its high content of protein, fiber, amino acids, and isoflavones, soybean (Glycine max L. Merr.) has become increasingly appealing for human consumption as a nutritional and functional food (Gibbs et al. 2004; Ikeda et al. 2006; Sanjukta and Rai 2016). Natto, fermented whole soybeans, is a popular soyfood product in Japan and is well known for its nutrition, unique flavor and stickiness (Hu et al. 2010; Wei and Chang 2004). The USA has been the largest exporter of natto-type soybeans to Japan since the early 1990s. Utilization of US soybeans for natto production in Japan has been predicated on the ability of domestic growers to continuously meet the needs of natto manufacturers by supplying them with value-added, food-grade soybean seeds that result in high quality natto products (Ikeda et al. 2006; Yoshikawa et al. 2014). Soybean seeds must meet several quality standards for natto production: seeds must be small (< 9 g/100 seeds), have high water absorption capacity (Cook and Rainey 2010), be uniform in size, have minimum change in constituents during storage, be round in shape, and have clear hilum and yellow and smooth seed coat (Cui et al. 2004; Geater et al. 2000; Hosoi and Kiuchi 2003). Seeds with cracked seed coats or seed discoloration, or excessively flat seeds, were unqualified for natto production (Montague Farms, Inc., Center Cross, VA, personal communication). Thus, the quality of natto products is mainly determined by soybean cultivars, processing conditions (soaking, cooking and fermentation), and bacteria strains (Wei and Chang 2004). The initial water absorption in the natto producing process, where seeds are softened and soluble sugars are released, is an important step before natto fermentation (Cook and Rainey 2010). During this process, the seed coat regulates water absorption and prevents the destruction of seed tissue at the beginning of seed imbibition (Copeland and McDonald 2001; Koizumi et al. 2008). It is ideal for natto soybean to keep the seed coat intact despite weakening and expanding during absorption. However, inferior seed coat integrity has been frequently observed in USA, which was mainly caused by the seed coat cracking during water absorption and defined as seed coat deficiency (SCD) (Fig. 1S) (Cook and Rainey 2010 and personal communications). SCD is an undesirable trait for natto soybeans because it affects final natto appearance and clogs production lines which increases production costs and reduces profit (Yasui et al. 2017). The phenotypic data of SCD indicated that breeding selection is feasible to efficiently reduce SCD (Cook and Rainey 2010), but it is challenging due to the time-consuming, subjective, and laborious nature of phenotyping and the poorly understood influence of environment (Escamilla et al. 2019). Marker-assisted selection (MAS) has been incorporated into most soybean breeding programs (He et al. 2004); however, it has not been implanted in the low-SCD soybean selection due to limited genetic control information of this SCD trait. A recent study identified eight QTLs located on chromosomes 4, 6 and 8 that are associated with seed coat cracking after soaking and cooking (Yasui et al. 2017), but none of these QTLs were validated for MAS in breeding selection. SoyBase (http://www.soybase.org/ accessed Aug. 26, 2019) also reported 14 QTLs related to seed coat cracking in dry seeds; however, the phenotype of those QTLs was obtained by determining physical seed coat cracking after harvest, which was physiologically different compared to seed coat deficiency in natto soybean production. Currently, there is a widespread lack of available molecular tools that can be used for MAS of the low-SCD trait in soybean.
Understanding of the genetic control of SCD is essential for development of molecular tools that can be used to improve breeding selection for low-SCD natto soybean cultivars. Therefore, the objectives of this study were to (1) identify QTLs associated with seed coat deficiency in soybean using two populations across multiple environments, and (2) validate these QTLs through Kompetitive Allele Specific PCR (KASP) assays using 86 breeding lines from four validation populations.
Materials and methods
Population development and experiment design
Two populations were developed by crossing small-seeded soybean breeding lines V11-0883 × V12-1626 (Pop 1) and V11-0883 × V12-1885 (Pop 2) (Pedigree was shown in Fig. 2S). The female parent V11-0883 produces the high-SCD phenotype, while the two male parents, V12-1626 and V12-1885, produce the low-SCD phenotype. Crosses were made in Blacksburg, VA in 2014, and the F1 generation was planted at the same location the following year. Six SSR markers (Satt449, Satt197, Satt281, Satt268, Satt431 and Satt345), which were polymorphic between parents, were used to verify true hybrids. A total of 240 and 153 F2 individuals from Pop 1 and Pop 2, respectively, were advanced to F3 at a winter nursery during the winter of 2015. In 2016, the F2:3 lines from two populations were separately spaced planted (0.03 m) in single, 3.05-mm-long rows with 0.76 m row spacing (with a seeding rate of 70,542 plants per hectare) arranged in a complete randomized design with three replications in Blacksburg, VA. F2:4 and F2:5 lines were harvested and replanted in subsequent years at the same growing location using the same experimental design.
In all years, fertilizer was applied according to soil test recommendations and pre-emergent herbicide (Dual Magnum) was applied at rate of 2 L ha−1 to reduce weed pressure. No irrigation or insecticides was applied. Rows were inspected for flower and pubescence color each year in order to control population purity and avoid contamination. Seeds were harvested 5–10 days after 95% of the plants in a row reached R8 maturity.
Determination of seed coat deficiency
Seeds were stored in the seed storage room until seed moisture stabilized between 10 and 12.2% (Cook and Rainey 2010). Seeds with cracked seed coat or seed discoloration, or excessively flat seeds, were removed. One hundred intact soybean seeds of each plot were subsampled for SCD determination. A modified method from protocols described in previous studies (Cook and Rainey 2010; Rodda et al. 1973) was used for easy observation. Briefly, a 100-seed sample was placed in a plastic container with 50 ml of 1% commercial bleach solution for ten minutes; after soaking, the samples were drained and scored for SCD. Seeds that showed cracking were severely blistered around the hilum, or whose seed coats had detached from the hull were considered seed coat deficient (Fig. 1S) (Cook and Rainey 2010). The percentage of seed coat deficient seeds was used to score SCD.
Statistical data analysis
The SPSS statistical version 20.0 (SPSS Inc., Chicago, USA) was used to summarize the descriptive statistics of the SCD for each population. Normality assumption was assessed by the Shapiro–Wilk test and normal probability plots. Analysis of variance (ANOVA) was used to evaluate variation of SCD within and between each environment in each population. Histograms of SCD distributions were elaborated by R function “hist()”. The variance components were calculated and used to estimate the broad-sense heritability using the following equation:
where H2 is heritability, is genotypic variance, is genotype × environment interaction variance, s2 is error variance, r is the number of replications, and e is the number of environments (Nyquist and Baker 1991).
DNA extraction and genotyping
Leaf tissue samples from F2 individuals and parental lines were collected from the field in centrifuge tubes and stored at −80 °C until extraction. For genomic DNA extraction, leaf tissue samples were freeze-dried at − 0.220 mbar with the collector temperature set at − 56 °C (FreeZone 6 Dryer system, Labconco, Kansas City, MO, USA). Lyophilized tissues were ground in liquid nitrogen using glass stirring rods. Total genomic DNA of each sample was isolated following a modified CTAB method (Saghai-Maroof et al. 1984).
Fifty nanograms of genomic DNA for each genotype were sent to USDA–ARS Soybean Genomics and Improvement Laboratory (Beltsville, MD) for genotyping using the Illumina 6000-SNP BARCSoySNP6K Beadchip, selected from the SoySNP50K (Song et al. 2013). The SNP allele calling was conducted in GenomeStudio Module v2.0.3 (Illumina, Inc.). The low seed coat deficiency parents were scored as A, and high seed coat deficiency parents were scored as B. SNPs with no call and the monomorphic SNPs between parents were discarded. SNPs with low minor allele frequency (MAF) (< 10%) and high missing data ratio (< 5%), as well as severe segregation distortion, were filtered for quality control.
Linkage map construction and QTL analysis
Linkage maps were constructed by Joinmap 4.0 (Van Ooijen 2006) using a regression approach with a minimum logarithm of odds (LOD) threshold of 3. Recombination frequencies were converted to centimorgan (cM) using Kosambi mapping function (Kosambi 1943). To ensure consistency, QTL analyses were performed by single-marker analysis (SMA), interval mapping (IM) and composite interval mapping (CIM) implemented in ICiMapping v 4.1 (Wang et al. 2016). For SMA, p < 0.0001 was used as the experiment wide threshold for significant markers. In the CIM and IM, the experiment wide threshold was determined by 1000 permutation at significance level of 0.05 with a walk speed of 1 cM. MapChart (Voorrips 2002) was used to create the LOD plots based on JoinMap 4.0 and ICiMapping v 4.1 results.
KASP marker development
The SNPs tightly linked to major QTL identified in the mapping populations were converted into Kompetitive Allele Specific PCR (KASP) SNP genotyping assays (LGC, Middlesex, UK) with the flanking sequences obtained from the G. max genome Glyma.Wm82.a1 (Schmutz et al. 2010). The KASP oligos were synthesized by Integrated DNA Technologies (IDT, Iowa, USA), with primers carrying FAM tail (5′-GAAGGTGACCAAGTTCATGCT-3′) or VIC tail (5′-GAAGGTCGGAGTCAACGGATT-3′), and the target SNP in the 3′ end. Primer mix and PCR reaction were set up following LGC Genomics recommendation (46 µL distilled water, 30 µL common primer [100 µM], and 12 µL of each tailed primer [100 µM]). Thermocycling conditions consisted of the initial hot-start step at 95 °C for 15 min, followed by 10 cycles of touchdown PCR (annealing 65 °C to 57 °C, decreasing 0.8 °C per cycle), then 35 cycles of 20 s at 94 °C and 60 s at 57 °C. PCR and fluorescent endpoint reading were performed in FLUOstar Omega microplate reader (BMG LABTECH).
KASP marker validation
A total of 86 breeding lines from four biparental populations (MFS-561 × V09-0579, MFS-561 × V09-3876, V05-5973W × V09-3876, V05-5973W × V09-3984) were planted in Warsaw, VA for marker validation. The female parents, MFS-561 and V05-5973W produce the low-SCD phenotype, while all male lines produce the high-SCD phenotype. SCD was determined by the same method as in the mapping populations. Breeding lines with SCD less than or equal to that of either male parent were considered low-SCD lines. Selection efficiency (SE) of the selected markers linked to low SCD was calculated as follows: SE = (NC/NS) × 100, where NC is the number of low-SCD lines selected correctly by marker and NS is the total number of lines selected as low SCD by marker.
Results
Phenotypic analysis of seed coat deficiency
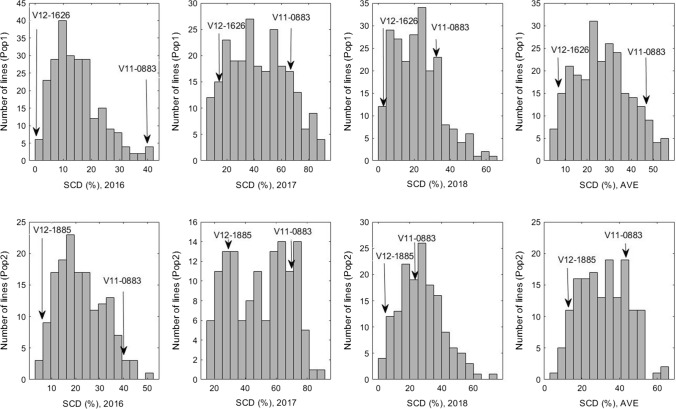
Both populations and their parental lines were scored for SCD during 2016–2018. In both populations, the SCD of the female parent V11-0883 (45.6%) exceeded that of both male parents (V12-1626, 7.3% and V12-1885, 13.7%) in all 3 years (Table 1). Among eight tests (two populations in 3 years plus mean across 3 years), three (SCD in 2017 and mean for Pop1; SCD in 2018 for Pop2) showed normal distribution (K–S test, p >0.2), while the rest showed continuous distribution (Table 1, Fig. 1), suggesting SCD was inherited as a quantitative trait. Large SCD variation was observed among individuals in both populations across all 3 years. For both populations, a larger degree of variation was observed in 2017 than in 2016 and 2018. The skewness was positive for SCD in all years and populations with the exception of Pop 2 in 2017, which displayed asymmetry toward the low-SCD end of the curve. Significant differences (p ≤ 0.0001) were detected when comparing SCD among genotypes, years, and the interaction between genotype and year in both populations (Table 1). The broad-sense heritability (H2) of SCD was 0.67 and 0.83 for Pop 1 and Pop 2, respectively, with an overall mean of 0.75.
Table 1.
Descriptive statistics for seed coat deficiency (SCD) of two populations in 2016, 2017 and 2018
| Populationa | Year | Max (%)b | Min (%)c | Mean (%) | SEd | Skewness | Kurtosis | p value | P1e | P2f | P3g | H2h | K–S testi |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pop 1 | 2016* | 48.0 | 0 | 14.8 | 0.39 | 0.88 | 0.52 | < 0.0001 | 40.0 | 0.5 | – | – | p < 0.001 |
| 2017* | 94.0 | 3.0 | 43.3 | 1.02 | 0.18 | − 0.83 | < 0.0001 | 68.0 | 18.5 | – | – | p >0.2 | |
| 2018* | 70.0 | 0 | 21.7 | 0.66 | 0.71 | 0.31 | < 0.0001 | 33.0 | 3.0 | – | – | p = 0.004 | |
| 2016/2017/2018** | 55.8 | 4.5 | 26.8 | 0.46 | 0.24 | − 0.69 | < 0.0001 | 47.0 | 7.3 | – | 0.67 | p >0.2 | |
| Pop 2 | 2016* | 59.0 | 2.0 | 22.0 | 0.78 | 0.43 | − 0.36 | 0.0133 | 40.0 | – | 6.5 | – | p = 0.017 |
| 2017* | 96.0 | 6.0 | 49.3 | 1.98 | − 0.07 | − 1.23 | < 0.0001 | 68.0 | – | 29.0 | – | p = 0.001 | |
| 2018* | 75.0 | 2.0 | 27.0 | 1.10 | 0.56 | 0.24 | 0.0132 | 24.5 | – | 5.5 | – | p >0.2 | |
| 2016/2017/2018** | 65.0 | 5.3 | 31.6 | 1.02 | 0.11 | − 0.72 | 0.0111 | 44.2 | – | 13.7 | 0.83 | p = 0.039 |
aPop 1 and Pop 2 were developed from the crosses of V11-0883 × V12-1626 and V11-0883 × V12-1885, respectively
bMax: maximum
cMin: minimum
dSE: standard error
eP1: V11-0883
fP2: V12-1626
gP3: V12-1885
hH2: Broad-sense heritability in combined environments (2016,2017, and 2018)
iK–S test: Kolmogorov–Smirnov test for normality distribution, probability was shown
*Significant difference among genotypes at the p < 0.0001
**Significant difference among genotypes (G), years (Y) and G × Y interaction at the p < 0.0001
Fig. 1.
Distribution of seed coat deficiency (SCD) in 3 years (2016, 2017 and 2018) and their mean overall years (AVE) of Pop 1 (V11-0883 × V12-1626) and Pop 2 (V11-0883 × V12-1885)
QTL associated with SCD
Out of 6000 SNPs, 1318 and 1637 SNPs were polymorphic between the parental lines and subsequently selected after date filtration for linkage map construction in Pop 1 and Pop 2, respectively. A total of 1258 SNPs were mapped to 20 chromosomes (Chr.) in Pop 1 with a total genetic distance of 1826 cM ranging from 44.9 cM (Chr. 16) to 128.3 cM (Chr. 3) with an average length of 91.3 cM (Table 1S). The average distance between adjacent markers in Pop 1 was 1.6 cM. For Pop 2, a total of 1604 SNPs were mapped to 20 chromosomes (Table 2S). The total genetic length of LGs for Pop 2 was 1189 cM, with Chr. 10 being the longest (90.5 cM) and Chr. 16 being the shortest (33.3 cM). The average length of LG was 59.3 cM with an average marker interval of 0.8 cM.
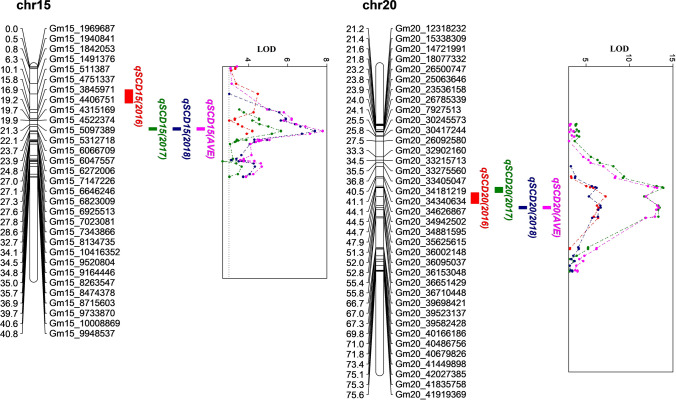
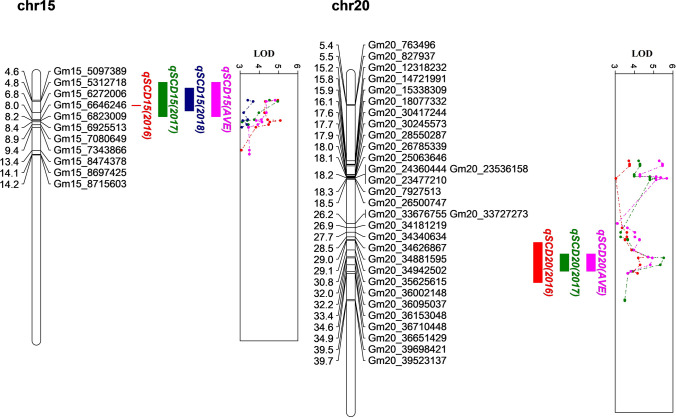
Genome-wide QTL analysis was performed based on the genetic map and phenotypic data of each population using composite interval mapping (CIM) and interval mapping (IM) in each year, as well as across 3 years. In Pop 1, two major QTLs were detected on Chr. 15 and Chr. 20 (Table 2, Fig. 2), while six other QTLs were detected on Chr. 3, 5, 6, 7, 10 and 14. The two stable QTLs, namely qSCD15 and qSCD20, were detected in each year and across 3 years, explaining 5.3–23.8% phenotypic variation. Meanwhile, qSCD15 and qSCD20 were also repeatedly detected in Pop 2 (Table 2, Fig. 3), along with seven other QTLs distributed on Chr. 1, 13, 16, 18 and 19. Among the two stable QTLs detected in Pop 2, qSCD15 was detected in individual and across years, explaining 3.1–10.4% phenotypic variation, while qSCD20 were detected in 2016, 2017 and across years, explaining 3.0–13.8% phenotypic variation. The additive effect of all detected QTLs was negative, indicating the male parents (V12-1626 and V12-1885) contributed negative allele decreasing SCD, while the female parent (V11-0883) contributed positive allele increasing SCD.
Table 2.
Quantitative trait loci for seed coat deficiency (SCD) in the mapping populations
| Popa | Envb | QTL namec | Position (cM) | Chrd | Flanking markerse | GI (cM)f | Wm82.a2.v1 Physical interval (bp)g | LODh | PVE(%)i | Addj | Methodk |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pop 1 | 2016 | qSCD14 | 28 | 14 | Gm14_6024101-Gm14_4956317 | 26.5–29.5 | 6,135,158–5,040,727 | 3.4 | 4.6 | − 1.9 | CIM |
| qSCD15 | 12 | 15 | Gm15_511387-Gm15_4751337 | 8.5–13.5 | 511,864–4,770,814 | 4.5 − 6.1 | 6.6 − 10.1 | − 2.8 to − 2.9 | CIM, IM, SMA | ||
| qSCD20 | 48 | 20 | Gm20_35625615-Gm20_36002148 | 46.5–50.5 | 36,720,824–37,097,315 | 7.3 − 8.4 | 7.5 − 11.3 | − 3.0 to − 3.1 | CIM, IM, SMA | ||
| 2017 | qSCD3 | 4 | 3 | Gm03_241447-Gm03_715393 | 0–8.5 | 239,949–714,448 | 4.5 − 7.2 | 4.2 − 5.3 | − 4.4 to − 6.1 | CIM, IM, SMA | |
| qSCD10 | 25 | 10 | Gm10_11518881-Gm10_26188429 | 22.5–25.5 | 11,710,604–16,707,334 | 3.4 − 4.5 | 4.1 − 5.0 | − 4.3 to − 6.1 | CIM, IM, SMA | ||
| qSCD15 | 23 | 15 | Gm15_5312718-Gm15_6066709 | 22.5–23.5 | 5,331,364–6,085,794 | 5.0 − 5.7 | 5.3 − 7.0 | − 5.0 to − 6.9 | CIM, IM, SMA | ||
| qSCD20 | 45 | 20 | Gm20_34881595-Gm20_35625615 | 44.5–46.5 | 36,021,058–36,720,824 | 14.1 − 15.2 | 12.2 − 23.8 | − 9.3 to − 10.4 | CIM, IM, SMA | ||
| 2018 | qSCD5 | 98 | 5 | Gm05_36671535-Gm05_41740936 | 95.5–98 | 36,962,030–38,597,425 | 3.1 | 4.9 | − 2.7 | CIM | |
| qSCD6 | 51 | 6 | Gm06_17258654-Gm06_38499465 | 46.5–56.5 | 17,303,937–39,188,086 | 5.5 − 5.9 | 9.3 − 9.5 | − 3.6 to − 4.6 | CIM, IM, SMA | ||
| qSCD7 | 72 | 7 | Gm07_17030454-Gm07_16979586 | 71.5–72.5 | 17,116,283–17,065,562 | 3.1 | 4.8 | − 2.6 | CIM | ||
| qSCD15 | 23 | 15 | Gm15_5312718-Gm15_6066709 | 22.5–23.5 | 5,331,364–6,085,794 | 6.3 − 7.4 | 10.2 − 10.4 | − 3.7 to − 4.9 | CIM, IM, SMA | ||
| qSCD20 | 52 | 20 | Gm20_36002148-Gm20_36095037 | 51.5–52.5 | 37,097,315–37,190,252 | 6.1 − 6.7 | 9.4 − 9.7 | − 3.7 to − 4.7 | CIM, IM, SMA | ||
| AVE | qSCD6 | 51 | 6 | Gm06_17258654-Gm06_38499465 | 46.5–56.5 | 17,303,937–39,188,086 | 5.2 − 7.3 | 7.3 − 7.8 | − 3.0 to − 4.7 | CIM, IM, SMA | |
| qSCD10 | 26 | 10 | Gm10_23967467-Gm10_12080275 | 22.5–27.5 | 20,275,514–12,272,454 | 3.1 − 3.8 | 3.8 | − 3.3 | IM, SMA | ||
| qSCD14 | 30 | 14 | Gm14_5603904-Gm14_5655487 | 29.5–30.5 | 5,714,996–5,766,579 | 2.9 | 3.5 | − 2.1 | CIM | ||
| qSCD15 | 23 | 15 | Gm15_5312718-Gm15_6066709 | 22.5–23.5 | 5,331,364–6,085,794 | 7.3 − 7.8 | 7.5 − 9.5 | − 3.5 to − 4.6 | CIM, IM, SMA | ||
| qSCD20 | 52 | 20 | Gm20_36002148-Gm20_36095037 | 51.5–52.5 | 37,097,315–37,190,252 | 13.7 − 15.1 | 12.9 − 20.8 | − 5.1 to − 6.0 | CIM, IM, SMA | ||
| Pop 2 | 2016 | qSCD1_1 | 15 | 1 | Gm01_1887205-Gm01_49322760 | 13.5–16.5 | 1,887,609–50,206,347 | 4.6 | 5.3 | − 4.3 | IM, SMA |
| qSCD1_2 | 25 | 1 | Gm01_1653315-Gm01_50262496 | 24.5–26.5 | 1,653,600–51,147,675 | 5.4 − 7.2 | 6.8 − 17.4 | − 3.7 to − 5.2 | CIM, IM, SMA | ||
| qSCD1_3 | 50 | 1 | Gm01_31906055-Gm01_7052156 | 46.5–55.5 | 33,203,133–7,071,117 | 4.9 | 4.1 | − 3.9 | IM, SMA | ||
| qSCD13 | 21 | 13 | Gm13_29677928-Gm13_28206014 | 20.37–22.37 | 30,875,555–29,399,384 | 5.5 | 4.3 | − 3.8 | IM, SMA | ||
| qSCD15 | 6 | 15 | Gm15_5312718-Gm15_6272006 | 5.5–6.5 | 5,331,364–6,291,081 | 4.8 | 3.9 | − 4.1 | IM, SMA | ||
| qSCD18_1 | 21 | 18 | Gm18_15324039-Gm18_7348168 | 20.5–21.5 | 15,066,204–7,370,390 | 4.1 | 3.3 | − 3.5 | IM, SMA | ||
| qSCD20 | 32 | 20 | Gm20_35625615-Gm20_36002148 | 29.5–36.5 | 36,720,824–37,097,315 | 4.7 | 3.6 | − 3.7 | IM, SMA | ||
| 2017 | qSCD1_3 | 51 | 1 | Gm01_31906055-Gm01_7052156 | 48.5–55.5 | 33,203,133–7,071,117 | 4.0 − 5.5 | 4.8 − 14.0 | − 6.7 to − 8.6 | CIM, IM, SMA | |
| qSCD15 | 5 | 15 | Gm15_5312718-Gm15_6272006 | 1.5–7.5 | 5,331,364–6,291,081 | 4.9 | 3.9 | − 8.3 | IM, SMA | ||
| qSCD16 | 2 | 16 | Gm16_103052-Gm16_1755870 | 0–7.5 | 102,882–1,772,720 | 4.2 | 3.6 | − 7.5 | IM, SMA | ||
| qSCD18_2 | 5 | 18 | Gm18_122382-Gm18_1308443 | 0–5.5 | 122,380–1,308,798 | 5.5 | 4.9 | − 8.5 | IM, SMA | ||
| qSCD19 | 67 | 19 | Gm19_41918030-Gm19_50486916 | 65.5–68.5 | 42,120,908–50,607,336 | 5.3 | 5.8 | − 9.5 | IM, SMA | ||
| qSCD20_2 | 18 | 20 | Gm20_26785339-Gm20_25063646 | 15.5–21.5 | 27,947,431–26,214,468 | 4.8 | 3.8 | − 8.0 | IM, SMA | ||
| qSCD20 | 33 | 20 | Gm20_36095037-Gm20_36153048 | 31.5–34.5 | 37,190,252–37,248,263 | 4.1 − 5.4 | 4.3 − 13.8 | − 6.8 to − 8.4 | CIM, IM, SMA | ||
| 2018 | qSCD15 | 5 | 15 | Gm15_5312718-Gm15_6272006 | 2.5–6.5 | 5,331,364–6,291,081 | 3.8 | 10.4 | − 5.1 | CIM, IM, SMA | |
| AVE | qSCD1_2 | 25 | 1 | Gm01_1653315-Gm01_50262496 | 24.5–26.5 | 1,653,600–51,147,675 | 3.6 | 3.1 | − 5.0 | IM, SMA | |
| qSCD1_3 | 50 | 1 | Gm01_31906055-Gm01_7052156 | 46.5–55.5 | 33,203,133–7,071,117 | 4.2 − 6.0 | 4.1 − 15.5 | − 4.5 to − 5.6 | CIM, IM, SMA | ||
| qSCD15 | 5 | 15 | Gm15_5312718-Gm15_6272006 | 2.5–6.5 | 5,331,364–6,291,081 | 4.9 | 3.1 | − 5.3 | IM, SMA | ||
| qSCD20_2 | 12 | 20 | Gm20_827937-Gm20_12318232 | 7.5–14.5 | 824,049–8,318,718 | 5.7 | 4.3 | − 5.8 | IM, SMA | ||
| qSCD20 | 33 | 20 | Gm20_36095037-Gm20_36153048 | 31.5–34.5 | 37,190,252–37,248,263 | 4.9 | 3.0 | − 5.0 | IM, SMA |
Bold letters or values are QTLs names or the least or the maximum percentage of variance explained by corresponding QTL, respectively, mentioned in the text
aPop 1 and Pop 2 were developed from the crosses of V11-0883 × V12-1626 and V11-0883 × V12-1885, respectively
bEnv: the individual years and across years (AVE) in which the QTL was detected
cThe name of each QTL is a composite of SCD followed by the chromosome number, QTLs detected in different methods with the same or overlapping maker interval were designated as on QTL
dChr: chromosome
eFlanking markers: the markers flanking the QTL
fGI: genetic interval
gPhysical interval according to genome position at Wm82.a2.v
hLOD: logarithm of odds, data showed the range of the values detected across methods
iPVE: percentage of variance explained, data showed the range of the values detected across methods
jAdd: additive effect, data showed the range of the values detected across methods
kMethods: by which the QTL was detected, data presented were the range of different methods
Fig. 2.
Mapping for seed coat deficiency (SCD) in the Pop 1 (V11-0883 × V12-1626) in individual and across years (AVE). QTL nomenclature is in the form of qTraitChr. Colored intervals refer to the mapped QTLs detected by CIM and/or IM in different environments, the curves indicate the physical position of markers against LOD score detected on chromosomes, and lines with different colors represent different environments
Fig. 3.
Mapping for seed coat deficiency (SCD) in the Pop 2 (V11-0883 × V12-1885) in individual and across years (AVE). QTL nomenclature is in the form of qTraitChr. Colored intervals refer to the mapped QTLs detected by CIM and/or IM in different environments; the curves indicate the physical position of markers against LOD score detected on chromosomes, and lines with different colors represent different environments
The two stable QTLs, qSCD15 and qSCD20, were identified in both populations across multiple years and could together explain 21.4–30.8% and 6.3% of phenotypic variation for Pop 1 and Pop 2, respectively. The high stability and explained phenotypic variation percentage suggest that qSCD15 and qSCD20 are major QTLs for SCD in soybeans. Furthermore, the single-marker analysis (SMA) confirmed the significant associations between SCD and eleven markers within the two major QTL regions identified in Pop 1 (Table 3). Two markers within the region of qSCD15 (Gm15_5312718 and Gm15_6272006) were detected in 2018 and across years, and nine markers from the qSCD20 region were detected in 2017 and across years.
Table 3.
SNPs highly significant associated with seed coat deficiency (SCD) detected by single-marker analysis (SMA) in Pop1 (V11-0883 × V12-1626) within two stable QTL regions
| QTLa | Marker ID | Position (cM) | LODb | |||
|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | AVE | |||
| qSCD15 | Gm15_5312718 | 22.1 | – | – | 6.7* | 5.9* |
| Gm15_6272006 | 24.8 | – | – | 6.7* | 6.0* | |
| qSCD20 | Gm20_34626867 | 44.1 | – | 13.1* | – | 12.1* |
| Gm20_34942502 | 44.5 | – | 13.8* | – | 12.8* | |
| Gm20_34881595 | 44.7 | – | 13.9* | – | 13.2* | |
| Gm20_35625615 | 47.9 | 7.3* | 11.7* | – | 11.8* | |
| Gm20_36002148 | 51.3 | 6.3* | 13.3* | 6.7* | 12.9* | |
| Gm20_36095037 | 52.0 | 6.4* | 13.5* | 6.7* | 13.1* | |
| Gm20_36153048 | 52.8 | 6.3* | 13.3* | 6.4* | 13.1* | |
| Gm20_36651429 | 55.4 | – | 13.3* | 6.4* | 12.9* | |
| Gm20_36710448 | 55.8 | 6.4* | 11.8* | – | 11.9* | |
aQTL: the QTL detected by CIM and/or IM, see in Table 2
bLOD: logarithm of odds
*SNPs significantly associated with seed coat deficiency at p < 0.0001
SNP validation via KASP genotyping assay
Eleven SNPs from the two stable QTLs (qSCD15 and qSCD20) that showed tight linkage to SCD (p < 0.001) were selected for marker validation (Table 3). None of the SNPs derived from the QTLs located on Chr. 15 segregated in the parental lines of the validation populations. In addition, three SNPs (Gm20_34626867, Gm20_34942502 and Gm20_35625615) located on Chr. 20 were found to be polymorphic between the parental lines of the validation populations. Gm20_34626867 and Gm20_34942502, located only 0.39 cM apart, had high selection efficiency (83% and 100%, respectively) in MFS-561-derived populations, and low selection efficiency (25% and 50%) in V05-5973 W-derived populations (Table 4). The other marker, Gm20_35625615, located 3.4 cM away from Gm20_34942502, showed variable selection efficiency across all four populations (57%, 92%, 33% or 50%). The combined average selection efficiency of marker Gm20_34626867 and Gm20_34942502 across all four validation populations was 77%, which exceeded that of marker Gm20_35625615 (64%) by just over 12%.
Table 4.
Markers selection efficiency (SE) in four validation populations developed in crosses using one low-SCD soybean in each combination and three SNPs significantly associated with SCD
| Validation population | Line no. | Gm20_34626867 | Gm20_34942502 | Gm20_35625615 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NCa | NSb | SE (%) | NCa | NSb | SE (%) | NCa | NSb | SE (%) | ||
| MFS-561 × V09-0579 | 34 | 5 | 6 | 83 | 5 | 6 | 83 | 12 | 21 | 57 |
| MFS-561 × V09-3876 | 23 | 12 | 12 | 100 | 12 | 12 | 100 | 11 | 12 | 92 |
| V05-5973W × V09-3876 | 16 | 1 | 4 | 25 | 1 | 4 | 25 | 1 | 3 | 33 |
| V05-5973W × V09-3984 | 13 | 2 | 4 | 50 | 2 | 4 | 50 | 3 | 6 | 50 |
| Total | 86 | 20 | 26 | 77 | 20 | 26 | 77 | 27 | 42 | 64 |
aNC: number of lines correctly selected as low SCD by marker
bNS: number of lines selected as low SCD by marker
Discussion
The quality of natto is largely cultivar dependent. Given its ramifications for consumer acceptance and production costs, developing natto soybean cultivars with lower incidence of SCD is of considerable interest to the natto industry. However, screening lines for SCD soybean is time-consuming and an effective selection tool is needed. In an effort to identify QTL and markers associated with low SCD, two sets of biparental populations and their parents were evaluated for SCD, QTLs associated with SCD were identified on Chr. 15 and 20. SNP markers derived from the two stable QTLs were developed and validated to select low-SCD natto soybean.
Large SCD variance was observed among individuals in Pop 1 (0–94%) and Pop 2 (2–96%), consistent with previously reported SCD variation (Cook and Rainey 2010; Yasui et al. 2017). Significant variation in SCD was observed among different years in both populations. SCD was much more severe in 2017 than in 2016 and 2018 for both populations, suggesting that expression of this trait may be particularly influenced by environmental conditions (Table 1). Interestingly, the average temperature and total precipitation during pod development stage (August and September) were much lower in 2017 than in 2016 and 2018 (Table 3S). Low temperature can directly jeopardize soybean seed quality, as chilling temperatures during flowering and low moisture content are both conducive to seed coat cracking (Koizumi et al. 2008). Despite the differences in environmental conditions, continuous distributions were still observed in both populations in individual and across years. This suggests that while environmental influence holds potential to exacerbate absolute SCD incidence, recovery of genotypes that show consistently lower relative SCD is possible through targeted breeding.
Although SCD is widely recognized as an important trait for natto cultivar development, few studies have investigated its genetic control. To address this need, we utilized two biparental mapping populations and identified seventeen QTLs associated with SCD (Table 2). Two stable QTLs were detected across multiple years and populations. A similar QTL study was conducted to study seed coat cracking during natto producing processes (soaking and cooking) in 126 recombinant inbred lines (Yasui et al. 2017). They found that QTL regions on Chr. 4, 6, and 8 were likely responsible for seed coat cracking; however no stable QTL was identified. The reliability of QTL mapping depends on the phenotyping methods, population size, linkage map density, and diverse environment factors. In order to increase the likelihood of identifying stable QTLs, we evaluated larger mapping populations over 3 years (environments) while observing greater marker density per chromosome. Thus, the QTLs identified in this study hold greater potential for future studies regarding the molecular mechanism and genetic basis of SCD in soybean.
The QTL qSCD15, flanked by Gm15_5312718 and Gm15_6272006, included two significant SNPs. Gm15_5312718_C_T and Gm15_6272006_T_C were located at 5,331,364 bp and 6,291,081 bp (respectively) on Chr. 15 of the Wm82.a2.v1 reference genome. Even though no QTL associated with soybean seed traits was reported within this region, a previous study identified nearby QTLs associated with soybean seed coat hardness (Kuroda et al. 2013). Though further correlation studies must be conducted, the evidence from both our study and the study by Kuroda et al. (2013) suggest that seed coat deficiency and seed coat hardness might be closely related and that Chr. 15 may be particularly important for modification of seed coat traits in soybean.
Nine significant SNPs were identified with the qSCD20 region. This region was flanked by Gm20_34881595 and Gm20_36095037, ranging from 36,021,058 bp to 37,190,252 bp on Chr. 20. QTLs responsible for seed composition (oil and isoflavones), seed set, and hilum color have been previously reported in this region (Fang et al. 2017; Leamy et al. 2017; Meng et al. 2016), all of which are also important for natto soybean breeding selection (Escamilla et al. 2019). Similar to the Chr. 15 QTL, qSCD20 overlapped with a previously identified QTL on Chr. 20 associated with seed coat hardness (Kuroda et al. 2013). Interestingly, qSCD20 also happened to be located approximately 20 Mbp (12.6 cM based on GmConsensus40) away from another previously reported QTL on Chr. 20 associated with seed coat cracking after harvest (Ha et al. 2012). Seed coat cracking is directly related to the strength of the seed coat. Chr. 20 may harbor several genes associated with seed coat strength and hardness, which play an important role or interact with other genes to cause a complex trait as seed coat deficiency.
We identified candidate genes in the intervals that qSCD20 and qSCD15 map using public data on soybase.org. We found 52 genes in the qSCD20 interval and 95 genes in the qSCD15 interval. Among those genes in the qSCD20 interval, we identified one candidate gene, Glyma.20G128600, which is homologous to Arabidopsis CAD4 (AT3G19450.1). This gene is a GroES-like zinc-binding alcohol dehydrogenase family protein involved in lignin biosynthesis. Given that lignin is a key component of the cell wall, it is plausible that alleles governing improved lignin biosynthesis may result in decreased incidence of SCD. We also identified additional candidate genes Glyma.15G078300, Glyma.15G075300, Glyma.15G074700, Glyma.15G074000, and Glyma.15G072300, which are involved in cell wall organization, callose deposition in cell wall, cuticle development, cell wall modification, and secondary cell wall biogenesis (Shao et al. 2007). These candidate genes are targets for future experimental validations of the molecular mechanisms of SCD in soybean. Gene lists underlying both QTLs are provided as a supplementary table with the candidate genes highlighted (Table 4S).
The broad-sense heritability (H2) of SCD in Pop 1 was 0.67, which was also observed for seed coat cracking after soaking (Yasui et al. 2017). Meanwhile, the broad-sense heritability of SCD was higher in Pop 2 (0.83). The reported heritability of other food-grade seed traits such as seed size, protein, sucrose, raffinose, and stachyose concentration range from 0.45 to 0.86 in soybean (Jaureguy et al. 2011). According to Robinson et al. (1949), trait heritability is categorized in three levels: low (0–30%), medium (31–60%), and high (> 60%). Heritability estimates indicate the potential to achieve genetic gain for a trait through breeding selection (Jaureguy et al. 2011). The high broad-sense heritability and large variance in SCD incidence observed in this study suggest that the reduction in SCD in natto soybean cultivars by accumulating favorable alleles through breeding selection is promising. Moreover, the QTLs identified in this study can be used to increase efficiency of breeding selection for the low-SCD trait, which can ultimately permit the development of superior natto soybean cultivars.
Eleven SNP markers were developed from the two stable QTLs located on Chr. 15 and 20 and validated in four diverse biparental populations. None of the markers from Chr. 15 were polymorphic between the parental lines and thus were not suitable for marker-assisted selection in the four populations. Three markers located on Chr. 20 were polymorphic between the parental lines and segregated in the breeding lines of validation populations. Markers Gm20_34626867 and Gm20_34942502 showed the same selection efficiency pattern in the validation populations and both showed higher selection efficiency in populations derived from the low-SCD (2%) parent MFS-561 and relatively lower selection efficiency in populations derived from low-SCD (15%) parent V05-5973W. Marker Gm20_35625615 showed relatively higher selection efficiency in populations derived from MFS-561 than that in populations developed from V05-5973W, which was consistent with the other two markers. The pedigree of MFS-561 shows that its parent MFS-553 is the grand parent of V16-1626 and V12-1885 used for QTL mapping (Fig. 2S). V05-5973W doesn’t share any parents traced back to two generations with V16-1626 and V12-1885. Therefore, the markers explored in this study would be ideally used for SCD selection on populations derived from MFS-553. In addition, the different selection efficiency across populations might indicate the variation of recombination rate between the test markers and low-SCD genes, or the possible involvement or interaction with other QTL. The overall selection efficiency of three markers was 56% in four populations with higher selection efficiency on MFS-561-derived populations and lower selection efficiency on V05-5973W-derived populations, when single-marker selection efficiency was compared. The results indicated that multiple markers should be employed to make better selection decision when using marker-assisted breeding approach.
In summary, two stable QTLs associated with low SCD were identified on chromosomes 15 and 20 by evaluating two biparental populations in three environments. Furthermore, three markers (Gm20_34626867, Gm20_34942502 and Gm20_35625615) were developed based on the stable QTL located on Chr. 20. The markers had a high selection efficiency in two populations with MFS-561 genetic background. The finding of these two major QTLs will shed light on the genetic control of seed coat deficiency, and the markers developed from this study will facilitate molecular marker-assisted selection of low-SCD natto soybean.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was funded by Montague Farms, Inc. (Grant No. 459611) and USDA Hatch Program VA-136296.
Author contributions
QZ and DE conducted the entire field experiments, constructed the genetic map, conducted the QTL analysis, and prepared the manuscript. XW helped analyze the data and write the manuscript particularly in results and discussion. QS developed the BARCSoySNP6K assay, performed the genotyping analysis of the materials, and provided guidance on data analysis. SL helped on candidate gene identification. NL helped perform the phenotypic assay, and LR helped in all lab activities. FX helped design the experiments. BZ conceived the study, designed the experiment, oversaw the development of the populations, and guided the writing of the entire manuscript. All authors read and revised the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Cook DE, Rainey KM. Seed coat deficiency, trait stability, and other soybean seed quality traits for natto cultivar development. Crop Sci. 2010;50:1244–1249. doi: 10.2135/cropsci2009.06.0300. [DOI] [Google Scholar]
- Copeland LO, McDonald MB. Principles of seed science and technology. New York: Springer; 2001. [Google Scholar]
- Cui Z, James A, Miyazaki S, Wilson R, Carter T. Breeding specialty soybeans for traditional and new soyfoods. In: Liu K, editor. Soybeans as functional foods and ingredients. Urbana: AOCS Press; 2004. pp. 264–322. [Google Scholar]
- Escamilla DM, Rosso ML, Holshouser DL, Chen P, Zhang B. Improvement of soybean cultivars for natto production through the selection of seed morphological and physiological characteristics and seed compositions: a review. Plant Breed. 2019;138:131–139. doi: 10.1111/pbr.12678. [DOI] [Google Scholar]
- Fang C, Ma YM, Wu SW, Liu Z, Wang Z, Yang R, Hu GH, Zhou ZK, Yu H, Zhang M, Pan Y, Zhou GA, Ren HX, Du WG, Yan HR, Wang YP, Han DZ, Shen YT, Liu SL, Liu TF, Zhang JX, Qin H, Yuan J, Yuan XH, Kong FJ, Liu BH, Li JY, Zhang ZW, Wang GD, Zhu BG, Tian ZX. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017;18(1):161. doi: 10.1186/s13059-017-1289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geater CW, Fehr WR, Wilson LA. Association of soybean seed traits with physical properties of natto. Crop Sci. 2000;40:1529–1534. doi: 10.2135/cropsci2000.4061529x. [DOI] [Google Scholar]
- Gibbs BF, Zougman A, Masse R, Mulligan C. Production and characterization of bioactive peptides from soy hydrolysate and soy-fermented food. Food Res Int. 2004;37:123–131. doi: 10.1016/j.foodres.2003.09.010. [DOI] [Google Scholar]
- Ha B-K, Kim H-K, Kang S-T. Mapping QTLs with epistatic effects and QTL-by-environment interactions for seed coat cracking in soybeans. Euphytica. 2012;186:933–942. doi: 10.1007/s10681-012-0719-8. [DOI] [Google Scholar]
- He J, Zhao X, Laroche A, Lu ZX, Liu H, Li Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci. 2004;5:484. doi: 10.3389/fpls.2014.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi T, Kiuchi K. Fermenting cooked soybeans with Bacillus subtilis (natto) In: Farnworth ER, editor. Handbook of fermented functional foods. Boca Raton: CRC Press; 2003. pp. 227–250. [Google Scholar]
- Hu Y, Ge C, Yuan W, Zhu R, Zhang W, Du L, Xue J. Characterization of fermented black soybean natto inoculated with Bacillus natto during fermentation. J Sci Food Agric. 2010;90:1194–1202. doi: 10.1002/jsfa.3947. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Iki M, Morita A, Kajita E, Kagamimori S, Kagawa Y, Yoneshima H. Intake of fermented soybeans, natto, is associated with reduced bone loss in postmenopausal women: Japanese population-based osteoporosis (JPOS) study. J Nutr. 2006;136:1323–1328. doi: 10.1093/jn/136.5.1323. [DOI] [PubMed] [Google Scholar]
- Jaureguy LM, Chen P, Scaboo AM. Heritability and correlations among food-grade traits in soybean. Plant Breed. 2011;130:647–652. doi: 10.1111/j.1439-0523.2011.01887.x. [DOI] [Google Scholar]
- Koizumi M, Kikuchi K, Isobe S, Ishida N, Naito S, Kano H. Role of seed coat in imbibing soybean seeds observed by micro-magnetic resonance imaging. Ann Bot. 2008;102:343–352. doi: 10.1093/aob/mcn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1943;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Yano H, Takada Y, Kato S, Vaughan D. QTL affecting fitness of hybrids between wild and cultivated soybeans in experimental fields. Ecol Evol. 2013;3:2150–2168. doi: 10.1002/ece3.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy LJ, Zhang HY, Li CB, Chen CY, Song BH. A genome-wide association study of seed composition traits in wild soybean (Glycine soja) BMC Genom. 2017;18(1):18. doi: 10.1186/s12864-016-3397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S, He JB, Zhao TJ, Xing GN, Li Y, Yang SP, Lu JJ, Wang YF, Gai JY. Detecting the QTL-allele system of seed isoflavone content in Chinese soybean landrace population for optimal cross design and gene system exploration. Theor Appl Genet. 2016;129:1557–1576. doi: 10.1007/s00122-016-2724-0. [DOI] [PubMed] [Google Scholar]
- Nyquist WE, Baker RJ. Estimation of heritability and prediction of selection response in plant populations. Crit Rev Plant Sci. 1991;10:235–322. doi: 10.1080/07352689109382313. [DOI] [Google Scholar]
- Robinson HF, Comstock RE, Harvey PH. Estimation of heritability and the degree of dominance in corn. Agron J. 1949;41:353–359. doi: 10.2134/agronj1949.00021962004100080005x. [DOI] [Google Scholar]
- Rodda ED, Steinberg MP, Wei LS. Soybean damage detection and evaluation for food use. Trans ASAE. 1973;16:365. doi: 10.13031/2013.37521. [DOI] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjukta S, Rai AK. Production of bioactive peptides during soybean fermentation and their potential health benefits. Trends Food Sci Technol. 2016;50:1–10. doi: 10.1016/j.tifs.2016.01.010. [DOI] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Shao S, Meyer CJ, Ma F, Peterson CA, Bernards MA. The outermost cuticle of soybean seeds: chemical composition and function during imbibition. J Exp Bot. 2007;58(5):1071–1082. doi: 10.1093/jxb/erl268. [DOI] [PubMed] [Google Scholar]
- Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE. 2013;8(1):e54985. doi: 10.1371/journal.pone.0054985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. JoinMap 4®: software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma B. V.; 2006. [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang J, Li H, Zhang L, Meng L. Users’ manual of QTL IciMapping. Beijing and Mexico: Institute of Crop Science, CAAS and Crop Research Informatics Lab; 2016. [Google Scholar]
- Wei Q, Chang SKC. Characteristics of fermented natto products as affected by soybean cultivars. J Food Process Preserv. 2004;28:251–273. doi: 10.1111/j.1745-4549.2004.23047.x. [DOI] [Google Scholar]
- Yasui T, Toda K, Yamada T, Yumoto S, Takahashi K, Takahashi M, Hajika M. Seed coat cracking of soybean (Glycine max [L.] Merr.) after soaking and cooking. Cereal Chem. 2017;94:717–722. doi: 10.1094/CCHEM-12-16-0284-R. [DOI] [Google Scholar]
- Yoshikawa Y, Chen PY, Zhang B, Scaboo A, Orazaly M. Evaluation of seed chemical quality traits and sensory properties of natto soybean. Food Chem. 2014;153:186–192. doi: 10.1016/j.foodchem.2013.12.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.