Abstract
Diabetes is a major threat to people’s health and has become a burden worldwide. Current drugs for diabetes have limitations, such as different drug responses among individuals, failure to achieve glycemic control, and adverse effects. Exploring more effective therapeutic strategies for patients with diabetes is crucial. Currently pharmacogenomics has provided potential for individualized drug therapy based on genetic and genomic information of patients, and has made precision medicine possible. Responses and adverse effects to antidiabetic drugs are significantly associated with gene polymorphisms in patients. Many new targets for diabetes also have been discovered and developed, and even entered clinical trial phases. This review summarizes pharmacogenomic evidence of some current antidiabetic agents applied in clinical settings, and highlights potential drugs with new targets for diabetes, which represent a more effective treatment in the future.
Keywords: Diabetes, New targets, Pharmacogenomics, Precision medicine
Key Summary Points
| The number of patients with diabetes is increasing and diabetes has become a threat to people’s health, but current drugs for diabetes have limitations. |
| It is crucial to explore more effective therapeutic strategies for diabetes. |
| Pharmacogenomics of current antidiabetic agents has provided potential for individualized drug therapy and drugs with new targets for diabetes have potential advantages, hopefully leading to an effective treatment of diabetes. |
| Investigating pharmacogenomics of current antidiabetic drugs in greater depth, accumulating definite clinical evidence, and developing more drugs according to new targets for diabetes are necessary in the future. |
Digital Features
This article is published with digital features to facilitate understanding of the article. You can access the digital features on the article’s associated Figshare page. To view digital features for this article go to 10.6084/m9.figshare.12854816.
Introduction
Diabetes is a chronic, metabolic disease caused by insufficient insulin secretion, insulin resistance, or both. It can be divided into six types: type 1 diabetes (T1D), type 2 diabetes (T2D), hybrid forms of diabetes, other specific types, unclassified diabetes, and hyperglycemia first detected during pregnancy [1]. People with diabetes are at a higher risk of clinical complications such as cardiovascular disease, retinopathy, neuropathy, and kidney disease due to high blood glucose levels [1, 2]. According to the International Diabetes Federation (IDF), in 2019 there are 463 million adult patients with diabetes worldwide, which is about 9.3% of adults aged 20–79 years, and in particular there were over 116 million adult Chinese with diabetes; and the number of patients with diabetes is still increasing [3]. Diabetes has become a threat to people’s health and is a major global health and social concern.
Treatment of diabetes is a timely clinical issue. In addition to diet modulation and adequate exercise, antidiabetic drugs are vital approaches in the clinical management of diabetes. Currently, several hypoglycemic agents are used in the treatment of diabetes, including insulin and insulin analogues, sulfonylureas, biguanides, glinides, thiazolidinediones, alpha-glucosidase inhibitors, glucagon-like peptide 1 (GLP-1) receptor agonists, dipeptidyl peptidase 4 (DPP4) inhibitors, and sodium-glucose cotransporter 2 (SGLT2) inhibitors [4]. However, almost half of patients with diabetes cannot achieve treatment goals, including glycemic control, even over 10 years [5–7]. As a result of the multiple antihyperglycemic drugs available and the fact that the same antidiabetic agent can lead to various therapeutic responses, and sometimes even serious adverse effects in each individual, there is growing uncertainty regarding the proper selection and screening of antidiabetic agents. Therefore the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) recommend applications of individualized treatment and precision medicine for diabetes [8, 9]. One way to achieve precision medicine and to guide proper use of antidiabetic agents is to provide medicine based on genetic information of individuals through pharmacogenomic approaches [10, 11]. In addition, potential new drugs for diabetes continue to emerge; in particular, those with new targets are expected to offer more effective treatment for diabetes.
Hence, this review will summarizes the pharmacogenomics of major antidiabetic drugs and some potential new targets for treating patients with diabetes, and explore other more effective treatments for diabetes. This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Diabetes Drugs and Their Therapeutic Responses
Current pharmacologic therapy for diabetes includes oral hypoglycemic agents and insulin. Oral hypoglycemic drugs are divided into eight categories: sulfonylureas, biguanides, glinides, thiazolidinediones, alpha-glucosidase inhibitors, GLP-1 receptor agonists, DPP4 inhibitors, and SGLT2 inhibitors [4]. Although these drugs have relevant therapeutic effects on diabetes, their long-term maintenance of the effect has not been achieved, and their responses in individuals also show differences [12, 13]. Moreover, some agents produce adverse effects, such as hypoglycemia, weight gain, gastrointestinal discomfort, urogenital infections, discomfort at the injection site, and heart failure [4, 9].
Pharmacogenomics for Diabetes Mellitus
Both genetic factors and nongenetic factors contribute to different drug responses in individuals. Pharmacogenetics and pharmacogenomics study the association between different drug responses and genetic factors in each individual. Gene polymorphisms determine expression levels of drug receptors, transporters, metabolic enzymes, and others, which might change drug pharmacokinetics and pharmacodynamics. Pharmacogenetics is an approach in which molecular biomarkers such as single nucleotide polymorphisms (SNPs) are considered in diagnostics and therapeutic decisions [14]. Pharmacogenomics considers genetic factors of individuals, and guides medication selection and more efficient use of medication according to genomic and other “omic” information, which can improve the efficacy of pharmacotherapy and reduce risks of adverse effects [15]. Thus, precision medicine is the practice of providing tailored treatments for patients on the basis of individual characteristics, including genetic, environmental, and lifestyle factors, and it is beneficial for overcoming existing limitations of therapeutic interventions [14, 16]. Pharmacogenomics, which studies almost all current antidiabetic agents, is becoming a more effective approach to prevent, classify, manage, and treat diabetes [11].
Biguanides
Metformin is a representative drug of biguanides, and it is used as the first-line treatment for T2D as well as an optimal monotherapy option. Currently, the hypoglycemic mechanism of metformin is still unclear, but it is generally believed that metformin reduces glycogen output in the liver and increases glucose uptake by peripheral tissues such as muscle, and in turn lowers the blood glucose through regulating AMP-activated protein kinase (AMPK) [17]. Moreover, increasing evidence indicates that the gut likely contributes to the antidiabetic action of metformin, as metformin has been found to inhibit glucose absorption in the small intestinal, increase intestinal bile acid, and modulate the composition of the gut microbiota, thereby promoting secretion of GLP-1 and reducing blood glucose [18, 19]. Metformin is excreted directly through bile and urine unchanged instead of undergoing enzyme metabolism. Absorption and excretion of metformin are accomplished through many transporters [20]. Therefore, current studies of the pharmacokinetics of metformin mostly focus on transporters that are responsible for its uptake and excretion, including organic cation transporters (OCTs), multidrug and toxin extrusion transporters (MATEs), and plasma membrane monoamine transporters (PMATs).
Metformin and Transporter Genes
After taking metformin, it is first absorbed into intestinal epithelial cells by PMAT and OCT3, and then transported into the blood by OCT1. Subsequently, metformin binds to corresponding receptors and exerts its effects on various target cells through blood circulation. Metformin is transported to the liver by OCT1 and OCT3, and then transferred into bile by the MATE1 protein. Alternatively, metformin is eliminated through the urine pathway, in which metformin is absorbed by OCT2 in the kidney and transported into the urine by MATE1 and MATE2 (Fig. 1).
Fig. 1.
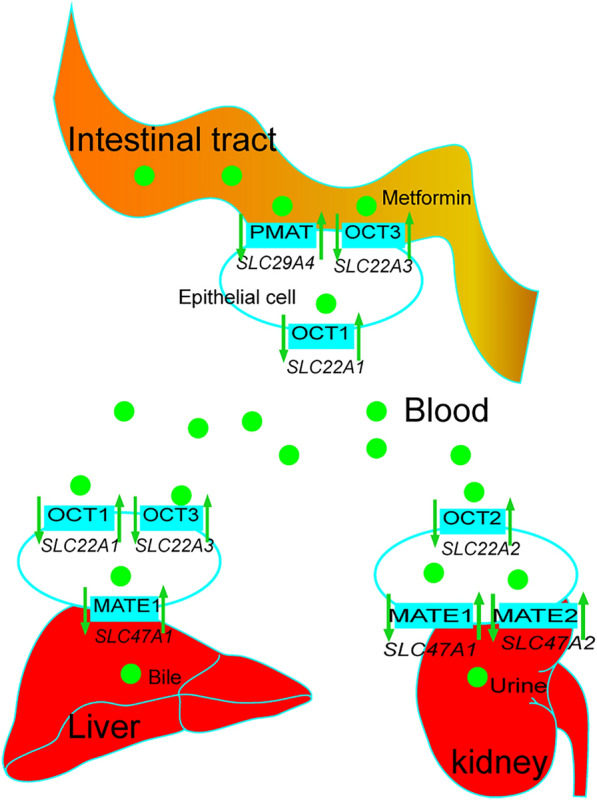
The pharmacokinetic process of metformin and the potential effect of its pharmacogenomic genes. After entering the gastrointestinal tract, metformin in intestinal endothelium is absorbed into intestinal epithelial cells by PMAT (encoded by SLC29A4) and OCT3 (encoded by SLC22A3), and transported into blood by OCT1 (encoded by SLC22A1). When metformin gets to the liver, OCT1 and OCT3 in membranes of hepatocytes take it up, and MATE1 (encoded by SLC47A1) transports it into the liver where it is ultimately eliminated with bile. Metformin also is taken up into epithelial cells of the kidney by OCT2 (encoded by SLC22A2). MATE1 and MATE2 (encoded by SLC47A2) are responsible for excretion of metformin into urine
OCT1, OCT2, and OCT3 are encoded by SLC22A1, SLC22A2, and SLC22A3 genes, respectively, and these proteins are responsible for transfer and uptake of metformin. OCT1 is involved in metformin absorption in enterocytes and hepatic uptake. The effect of SLC22A1 polymorphisms on metformin treatment has been extensively studied with inconsistent outcomes, likely due to the different races of the individuals. SLC22A1 rs622342 polymorphism has been reported to be associated with therapeutic efficacy in South Indian patients with T2D [21]. SLC22A1 and SLC47A1 variants, rs594709 and rs2289669, respectively, may simultaneously affect metformin efficacy in Chinese patients with T2D [22]. Nonsynonymous variants of OCT1 in Chinese and Japanese populations may also lead to differential responses to metformin [23, 24]. On the other hand, no significant association has been found between the rs628031 variant of SLC22A1 and the effect of metformin treatment in Iranian patients with diabetes [25].
In addition, SLC22A1 variants of R61C (rs12208357) and M420del (rs72552763) have been shown to be associated with higher odds of common metformin-induced gastrointestinal adverse effect [26, 27]. SLC22A1 rs622342, rs628031, and rs594709 polymorphisms are significantly related to an increase in hemoglobin A1c (HbA1c) levels in Mexican but not in Jordanian patients with diabetes undergoing metformin treatment [28, 29]. A study applying novel imaging techniques has shown reduced hepatic distribution in patients carrying M420del (rs72552763) and R61C (rs12208357) variants in SLC22A1 [30].
OCT2 (SLC22A2) is involved in transport of metformin from the bloodstream into the kidney, and mutations of OCT2 may affect clearance of metformin from the blood. SLC22A2 808G>T variant has been observed to improve glucose-lowering efficiency of metformin in Chinese patients with T2D through delaying transport of metformin into the kidney [31]. However, the effect of OCT2 gene polymorphisms on renal clearance of metformin has not been confirmed in the Caucasian population [32]. SLC22A2 rs201919874 polymorphism is associated with therapeutic efficacy of metformin with respect to HbA1c, random blood glucose, and fasting plasma glucose (FPG) levels in Pakistani patients [33]. Furthermore, SLC22A2 rs316019 increases average changes in HbA1c levels in dominant and co-dominant models in South Indian patients, but not in Mexican and Caucasian patients with T2D [29, 34, 35], while SLC22A2 rs10755577 and other SNPs have no significant glycemic control effects in Jordanian patients with T2D [28].
OCT3 (SLC22A3) is responsible for uptake of metformin in the intestine and liver. SLC22A3 rs3088442G>A variant has shown a strong correlation with metformin response in Pakistani patients with T2D [36]. SLC22A3 rs12194182 SNP is associated with a lower mean HbA1c level in Jordanian patients with T2D, but not in male Caucasians [28, 32].
MATE1 and MATE2 are multidrug and toxin extrusion transporters encoded by SLC47A1 and SLC47A2 genes, respectively. MATE1 is located in the kidney and liver, and is responsible for transporting metformin into urine and bile, and for eliminating metformin. Population-based cohort studies of patients with T2D have shown that SLC47A1 rs2289669 polymorphism is related to metformin responses, and carriers of the AG or AA genotype have a maximal HbA1c reduction, likely through delaying renal elimination of metformin or increasing basal GLP-1 [35, 37–40]. The AA genotype of both SLC47A1 (rs2289669) and SLC22A1 (rs594709) may cooperate to increase efficacy of metformin, and have a maximal glucose-lowering effect in Chinese patients with T2D [22]. However, no association between the response to metformin and the SLC47A1 (rs2289669) polymorphism has been described in Indian and Mexican populations with T2D [29, 41].
MATE2, which is present in kidney tubules, plays a role in metformin clearance by transporting metformin into urine. Patients with T2D and SLC47A2 rs12943590 polymorphism GA genotype, especially those also carrying SLC22A2 rs316019 polymorphism GG genotype, show significantly increased average changes in HbA1c levels and decreased failure of metformin treatment [34]. A study in patients with newly diagnosed T2D from ethnically diverse American populations has shown that poorer glycemic responses to metformin are significantly correlated with homozygous common 5′-untranslated region (5′-UTR) variant in MATE2-K (an isoform of MATE2) [42].
PMAT is expressed in enterocytes, is encoded by the SLC29A4 gene, and functions in the intestinal absorption of metformin. Higher odds of gastrointestinal intolerance have been demonstrated in a significant proportion of patients with T2D carrying the G allele in SLC29A4 rs3889348 after metformin treatment [26]. PMAT is also located in renal tubules and is likely involved in renal tubular reabsorption of metformin, as c.883-144A>G SNP (rs3889348) in the SLC29A4 gene has shown significantly increased renal clearance of metformin compared to the wild-type gene in the Korean population [43].
Metformin and Other Associated Genes
SLC2A2 encodes a glucose transporter 2 (GLUT2), which is responsible for glucose release and may be a target of metformin to reduce hepatic glucose output [20]. One C allele of SLC2A2 rs8192675 SNP in patients with newly diagnosed T2D has shown a large reduction in fasting glucose levels, and is related to greater reduction of HbA1c in 10,577 participants of European ancestry undergoing metformin treatment [44, 45].
In summary, a better understanding of polymorphisms of transporter genes for metformin in different races should assist more efficient use of metformin and increase success of diabetes treatment.
Sulfonylureas
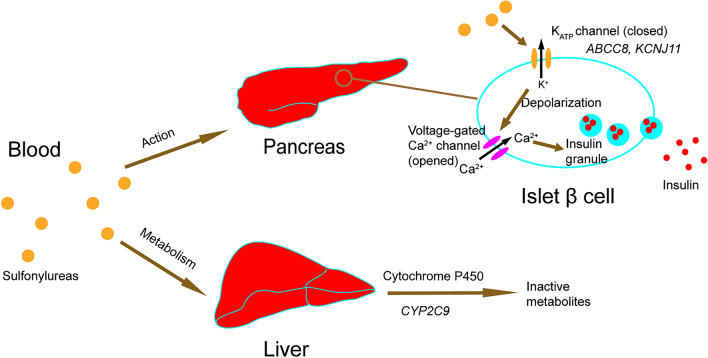
Sulfonylureas play a role in glucose control by promoting insulin secretion. Sulfonylureas function through combining and closing pancreatic β-cell ATP-sensitive potassium channel (KATP channel), which consists of sulfonylurea receptor 1 (SUR1) and inward-rectifier potassium ion channel (Kir6.2), and in turn leads to depolarization of β-cell membranes as well as opening of voltage-gated calcium channels that are essential in insulin secretion. Sulfonylureas include gliclazide, glibenclamide, glimepiride, and glipizide. Most sulfonylureas are metabolized by cytochrome P450 (CYP), a metabolic enzyme in the liver. Mutations of genes encoding for target receptors or metabolic enzymes can affect responses to sulfonylureas in patients with T2D (Fig. 2).
Fig. 2.
The action mechanism and metabolism of sulfonylureas and their potential pharmacogenomic genes. Sulfonylureas combine and close ATP-sensitive potassium channel (KATP channel, encoded by ABCC8 gene and KCNJ11 gene) in the membrane of pancreatic β-cells, which leads to depolarization of membranes, and then opens voltage-gated calcium channel. Ca2+ influx stimulates exocytosis of insulin granules. Sulfonylureas are metabolized by cytochrome P450 (CYP2C9 gene) in the liver
Sulfonylureas and Target Genes
SUR1, encoded by the ABCC8 gene, is a protein constituting the KATP channel. A significant association between ABCC8 Ser1369Ala polymorphisms and response to sulfonylureas has been found by a novel association screening method based on global probabilities in Mexican patients [46]. The Ser1369Ala variant in the ABCC8 gene can influence the therapeutic efficacy of gliclazide [47]. ABCC8 polymorphisms in exon 16 and exon 31 are related to concentrations of HbA1c and triglyceride in patients with T2D treated with sulfonylurea therapy [48]. On the other hand, some studies indicate that the Ser1369Ala (rs757110) variant in ABCC8 is not associated with severe hypoglycemia response to sulfonylurea treatment in patients with T2D [49], and it is not related to favorable glycemic control in patients treated with glibenclamide [50].
KCNJ11 is located next to the ABCC8 gene and encodes protein Kir6.2, which is another subunit of the KATP channel. The most studied KCNJ11 SNPs are variants rs5219 (E23K) and rs5215. A study among Caucasian patients has shown that carriers of the K allele of KCNJ11 rs5219 polymorphism exhibit higher HbA1c reduction after gliclazide treatment [51]. An association between the KCNJ11 rs5219 variant and effect of gliclazide treatment is also reported in the Chinese population [52]. One study has shown a significant association between the KCNJ11 rs5219 polymorphism and Hb1Ac reduction after sulfonylurea treatment in Mexicans, whereas another study shows no association between KCNJ11 rs5219 variant and glycemic control [46, 50]. However, KCNJ11 rs5219 and rs5215 polymorphisms have not been found to alter risk of hypoglycemia among patients with T2D treated with sulfonylurea [49, 53]. One study has shown a lower risk of severe hypoglycemia in patients with T2D and KCNJ11 E23K polymorphism, and this variant locus can reduce the response to sulfonylurea therapy, which results in increased HbA1c [54]. The KCNJ11 rs5219 variant is associated with an increased risk of sulfonylurea treatment failure [55]. The KCNJ11 rs5219 variant is not associated with sulfonylurea therapy efficacy in Caucasian and Korean patients [48, 56].
Sulfonylureas and Other Associated Genes
The enzyme encoded by the CYP2C9 gene belongs to the CYP superfamily and is involved in metabolism of sulfonylureas. Some studies have shown that CYP2C9 loss-of-function alleles (CYP2C9*2/*3 variants) are associated with higher sulfonylurea levels and decreased risk of therapy failure [57]. Recently, the CYP2C9*3 polymorphism was found to be associated with favorable glycemic control in patients with T2D treated with glibenclamide [50]. Although the aforementioned results have shown that CYP2C9 polymorphisms are associated with response to sulfonylureas, some results have also demonstrated no significant difference among patients with CYP2C9 variants on sulfonylurea treatment [49].
KCNQ1 is expressed in a wide variety of tissues and encodes a pore-forming subunit of a voltage-gated K+ channel (KvLQT1) that plays a crucial role in repolarization of the cardiac action potential, and water and salt transport in epithelial tissues. Genome-wide association studies have reported that KCNQ1 is associated with T2D. The KCNQ1 rs163184 variant was found to markedly reduce FPG in patients with diabetes on sulfonylurea treatment [58]. Two studies from China revealed that KCNQ1 polymorphisms are associated with response to sulfonylurea therapy in patients with newly diagnosed T2D [59, 60].
IRS1 encodes insulin receptor substrate 1 that plays a key role in the insulin receptor signaling pathway. Some studies have found that Caucasian patients with T2D with the IRS1 Arg972 (rs1801278) polymorphism have a significantly increased risk of secondary failure to sulfonylurea [61, 62]; IRS1 Arg972 carriers also have significantly higher HbA1c levels [62].
The transcription factor 7 like 2 (TCF7L2) gene is considered the strongest risk factor for T2D. The gene encodes a transcription factor that is involved in the WNT signaling pathway, which is associated with pancreatic islet β-cells. An association has been reported between its common variants (rs7903146 and rs12255372) and sulfonylurea treatment. TCF7L2 variants influence response to sulfonylureas in patients with T2D by reducing HbA1c and FPG levels, which establishes that genetic variation could predict response to therapy in T2D [63]. In contrast, no significant differences are observed in patients with T2D with TCF7L2 rs7903146 and rs12255372 variants receiving sulfonylurea treatment [50, 56].
Glinides
Glinides, a type of insulin secretagogue consisting of repaglinide, nateglinide, and mitiglinide, act on the KATP channel on the pancreatic islet β-cell membrane to stimulate insulin secretion and to reduce blood glucose levels. The structure of the glinides class is different from that of sulfonylureas. Glinides exert faster effects but with shorter duration than sulfonylureas, even though they work by a similar mechanism. Pharmacogenomics studies on glinides have mainly focused on repaglinide and has mostly been conducted in the Chinese population.
Glinides and Target Genes
Glinides absorbed into the blood bind to the KATP channel, which consists of SUR1 (encoded by the ABCC8 gene) and Kir6.2 (encoded by the KCNJ11 gene). Patients carrying the CT genotype of the ABCC8 rs1801261 SNP exhibit significant reduction in FPG and HbA1c compared with those with the CC genotype [64]. Patients with the homozygous CC genotype in the exon 16–3 T/C variant (rs1799854) of the ABCC8 gene show reduced homeostasis model assessment of insulin resistance (HOMA-IR), suggesting that repaglinide improves insulin sensitivity in these patients [65]. Moreover, patients with the K allele of the KCNJ11 E23K (rs5219) variant show significantly reduced HbA1c and postprandial plasma glucose (PPG) levels compared with those with the homozygous EE genotype, which indicates that E/K and K/K genotypes are associated with more favorable responses to repaglinide in stimulating insulin secretion [65]. Analogously, patients with at least one A allele of the KCNJ11 rs5219 polymorphism have significantly decreased PPG, FPG, and HbA1c levels [66].
Glinides and Transporter Gene
Before glinides are metabolized in the liver, they are transported into hepatocytes by transporter organic anion-transporting polypeptide 1B1 (OATP1B1) transporter, which is an organic anion-transporting polypeptide encoded by the solute carrier organic anion transporter family member 1 (SLCO1B1) gene. SLCO1B1 c.521T>C polymorphism is associated with increased Cmax (peak plasma concentration) of repaglinide in healthy Chinese and Caucasian volunteers [67, 68]. The effect of SLCO1B1 c.521T>C polymorphism on increased Cmax and area under the curve of plasma concentration (AUC) of repaglinide persists throughout relevant dose ranges [69]. SLCO1B1*1B/*1B genotype shows a strong correlation with lower plasma concentrations of repaglinide and higher clearance of repaglinide in Finnish and Chinese individuals [70, 71]. Moreover, the SLCO1B1 polymorphism affects the extent of interactions between OATP1B1 inhibitors and repaglinide in Chinese individuals [68].
Glinides and Other Associated Genes
KCNQ1, also expressed in pancreatic islet β-cells, encodes a voltage-gated K+ channel and is responsible for repolarization of the action potential in cardiac muscle. Patients with the rs2237892 T allele and rs2237895 C allele of the KCNQ1 gene are more likely to have a favorable response to repaglinide [72]. CT and TT genotypes of the KCNQ1 rs2237892 polymorphism are significantly associated with reduction in FPG, PPG, HOMA-IR, HbA1c, and low-density lipoprotein cholesterol (LDL-c), and with increase in postprandial serum insulin (PINS) compared with the homozygous CC genotype [73]. Compared with other genotypes, lower 2-h postprandial glucose levels are found in patients with the TT genotype of the rs2237892 polymorphism [74].
TCF7L2 is a T2D susceptibility gene and is associated with function of pancreatic β-cells. Chinese patients with the TT genotype of the TCF7L2 rs290487(C/T) polymorphism show a more favorable response in terms of fasting serum insulin, triglyceride, and LDL-c levels, which may indicate that these polymorphisms are more sensitive to repaglinide [66].
Thiazolidinediones
Thiazolidinediones, a class of peripheral insulin sensitizer that consists of pioglitazone and rosiglitazone, cooperate with nuclear peroxisome proliferator-activated receptor γ (PPARγ) in the peripheral tissue, and mediates transcription of genes related to glucose metabolism, and in turn enhances peripheral tissue sensitivity to insulin to lower blood glucose [75]. Thiazolidinediones modulate glucose but do not target pancreatic islet β-cells.
Thiazolidinediones and Transporter Gene
SLCO1B1 encodes the hepatic drug transporter OATP1B1, which may participate in transporting thiazolidinediones from the blood into the liver, and plays a role in thiazolidinedione metabolism. The SLCO1B1 521T>C variant shows a substantial association with an enhanced glycemic response in rosiglitazone-treated patients with T2D, but no statistically significant difference has been found in the pharmacokinetics of thiazolidinediones in healthy volunteers [76–78].
Thiazolidinediones and Target Gene
PPARγ is a nuclear hormone receptor that is encoded by the peroxisome proliferator-activated receptor γ gene (PPARG). The Pro12Ala (rs1801282) polymorphism is the most common variation in the PPARG and also an associated risk factor for T2D. A potential association between the Pro12Ala polymorphism and better therapeutic response to pioglitazone has been consistently reported in different populations, including those from South Indian, China, Iran, and menopausal women in Mexico [79–82]. However, some studies demonstrate statistically non-significant differences between the response to thiazolidinedione and the Pro12Ala polymorphism [83, 84].
Other Antidiabetic Agents and Associated Genes
Some other antidiabetic agents including α-glucosidase inhibitors, GLP-1 receptor agonists, DPP4 inhibitors, and SGLT2 inhibitors have not shown strong pharmacogenomic evidence.
α-Glucosidase inhibitors, working as a hypoglycemic drug for patients with T2D, act in the intestinal tract and delay absorption of complex carbohydrates by inhibiting α-glucosidases in the upper gastrointestinal tract [85]. The STOP-NIDDM trial assessed the association between treatment with acarbose, an α-glucosidase inhibitor, and PPAR-alpha (PPARA), PPAR-gamma2 (PPARG2), PPAR-gamma coactivator 1alpha (PPARGC1A), and hepatic nuclear factor 4alpha (HNF4A) SNPs without tests in clinic [86].
GLP-1, an incretin hormone that can be degraded by the DPP4 enzyme, stimulates insulin secretion, inhibits glucagon secretion in a glucose-dependent way, and delays gastric emptying (GE) [87]. GLP-1 receptor is expressed in a number of organs, such as the pancreas. GLP-1 receptor agonists, including exenatide, liraglutide, albiglutide, dulaglutide, lixisenatide, and semaglutide, are resistant to DPP4-mediated inactivation and are applied to the treatment of diabetes. The drug target gene GLP1R is the main focus in pharmacogenetic studies of GLP-1 receptor agonists. Some studies have shown that the carriers of the A allele in GLP1R rs6923761 have a propensity for a greater decrease in weight or a greater delay in GE after treatment with GLP-1 receptor agonists [88, 89]. On the other hand, glycemic and weight response to exenatide have been consistently confirmed as an association with the GLP1R rs6923761 variant in overweight patients with T2D from China [90]. However, another study indicates that GLP1R gene polymorphisms have no significant correlations with the response of treatment with GLP-1 analogue in patients with T2D [91]. In addition, the gene TCF7L2 rs7903146 variant also shows inconsistent results in pharmacogenetic studies of GLP-1 receptor agonists [88, 92].
The incretin hormones including GLP-1 and glucose-dependent insulinotropic polypeptide (GIP) are rapidly degraded by DPP4. DPP4 inhibitors including alogliptin, sitagliptin, linagliptin, and vildagliptin promote insulin secretion by preventing inactivation of the incretin hormones. There are a few gene variants correlated to DPP4 inhibitor treatment. Two studies among Caucasian patients have shown that the GLP1R rs6923761 (Gly168Ser) variant is associated with a smaller reduction in HbA1c after gliptin therapy [93, 94]. Genetic variation of GLP1R rs3765467 is associated with DPP4 inhibitor efficacy in Korean patients with T2D [95]. Moreover, some other gene variants are also reported to be associated with the response to DPP4 inhibitors, including KCNJ11 rs2285676, KCNQ1 rs163184, and TCF7L2 variants [96–98].
As new antidiabetic drugs, SGLT2 inhibitors including dapagliflozin, empagliflozin, ertugliflozin, and canagliflozin target and restrain SGLT2, which is responsible for glucose reabsorption in the proximal renal tubule, and cause loss of glucose through urine and a reduction in serum glucose. SGLT2 is encoded by the SLC5A2 gene, but the SNPs of SLC5A2 have not shown a significant association with response to treatment with empagliflozin in a pharmacogenetic study [99]. Uridine diphosphate glucuronosyltransferase (UGT) isozymes are involved in the metabolic elimination pathway for SGLT2 inhibitors, and an increase in exposure of plasma canagliflozin is observed in carriers with UGT1A9*3 or UGT2B4*2 [100].
In summary, pharmacogenomics studies conducted worldwide have involved almost all current antidiabetic agents but have primarily focused on commonly used drugs such as metformin and sulfonylureas. These findings indicate that some gene polymorphisms are significantly related to response to antidiabetic drugs. Accumulating clinical studies, even though showing variations, have provided new research directions in precision medicine for diabetes in the future.
Potential Therapeutic Drugs with New Targets for Diabetes
It is critical to improve the therapeutic effects of current antidiabetic drugs, decrease the risk of adverse effects, and even fundamentally reverse the development of diabetes through discovering and developing new targets. Currently, many potential antidiabetic agents with new targets are undergoing clinical trials; these agents may become new treatment approaches for diabetes and provide more therapeutic options for patients with diabetes.
Glucokinase Activators
Glucokinase (GK), which is mainly present in pancreatic cells and hepatic cells, is a hexokinase that phosphorylates glucose to glucose-6-phosphate (G6P), which is a limiting step in catalytic transformation. In the pancreas, glucokinase senses and mediates glucose levels. It also promotes insulin release and reduces glucagon secretion when glucose concentration is high. In the liver, glucokinase facilitates glucose uptake and hepatic glycogen synthesis through phosphorylation, thereby subsequently lowering blood glucose. Glucokinase in hepatocytes is inhibited by glucokinase regulatory protein (GKRP). When blood glucose is too high or too low, GKRP will release or bind to glucokinase, which maintains glucose homeostasis [101]. In general, activating glucokinase in the pancreas and liver is beneficial in lowering blood glucose; thus, glucokinase modulation could be a treatment approach for diabetes.
As a potential diabetes treatment approach, research attention has been paid to glucokinase activators (GKAs). However, the existence of adverse effects such as hypoglycemia, increased triglycerides, and diminishing hypoglycemic efficacy has hindered development of GKAs as new drugs for diabetes [102, 103]. A dual-acting full GKA, dorzagliatin (HMS5552), and a liver-selective GKA, TTP399, have overcome these limitations and demonstrated a favorable hypoglycemic effect, adequate tolerance, and improved safety profile as well as no weight gain. Phase II clinical trials of both GKAs have been successfully completed [103, 104–106]. Compared with currently available drugs, GKA is a target in a novel and unique physiological pathway and is expected to supplement current agents.
11-β-Hydroxysteroid Dehydrogenase 1 Inhibitors
11-β-Hydroxysteroid dehydrogenase 1 (11-β-HSD1), an enzyme mainly expressed in the liver and adipose tissue, catalyzes the conversion of inactive cortisone to active cortisol, which can bind and activate the glucocorticoid receptor. Excess systemic glucocorticoid results in visceral obesity, insulin resistance, glucose intolerance, dyslipidemia, hypertension, and increased cardiovascular risk [107]. Numerous studies have reported that 11-β-HSD1 expression and activity in the adipose tissue and liver are associated with obesity, insulin resistance, dyslipidemia, and metabolic syndrome [107, 108]. Therefore, 11-β-HSD1 inhibitors might reduce cortisol and glucocorticoid levels, and improve diabetes management.
INCB013739, MK0916, and selective inhibitors PF-915275 and BI 135585 have entered clinical trials, and have been found to be safe and well tolerated in patients with diabetes [108, 109]. In addition, in a diabetic mouse model, the selective 11-β-HSD1 inhibitor INU-101 enhanced insulin sensitivity, lowered fasting blood glucose level, reduced body weight, ameliorated the lipid profile, and is therefore considered a novel drug candidate for T2D treatment [110]. Compared with current antidiabetic agents, 11-β-HSD1 inhibitors may have a major advantage of weight loss or a lack of weight gain, which is beneficial for components of metabolic syndromes such as dyslipidemia, obesity, and hypertension [108, 111].
G-Protein-Coupled Receptor 119 Agonists
G-protein-coupled receptor 119 (GPR119), a new therapeutic target for diabetes, is highly expressed in both pancreatic β-cells and intestinal endocrine cells. GPR119 activation has been found to stimulate insulin secretion through a glucose-dependent pathway in pancreatic β-cells, and to promote GLP-1 release in the gastrointestinal tract, and in turn improves blood glucose control [112, 113]. GPR119 agonists might have an advantage in preserving or enhancing pancreatic β-cell function in patients with T2D [112, 114].
Many GPR119 agonists, such as PSN821, GSK1292263, MBX-2982, and LEZ763, have been developed and entered clinic trials, but later discontinued because of some limitations and adverse events [113]. Phase II clinical trials of the GPR119 agonist DS-8500a have been completed, and DS-8500a is expected to be a promising treatment for diabetes. Clinical studies have shown a greater reduction in FPG, postprandial glucose, and 2-h postprandial glucose compared with placebo, and the aforementioned effects do not diminish. Moreover, total cholesterol, LDL-c, and triglycerides are significantly decreased in the DS-8500a group. DS-8500a enhances insulin secretory capacity of pancreatic β-cells and is well tolerated without major adverse events, hypoglycemia, or discontinuation due to adverse events among Japanese patients with T2D, which provides support for conducting further phase III clinical studies [114–117]. GPR119 agonists provide favorable blood glucose control through a dual action mechanism and have the potential to preserve pancreatic β-cell function [112].
Glucagon Receptor Antagonists
Glucagon is secreted by pancreatic α-cells; it mainly promotes gluconeogenesis and glycogenolysis and inhibits glycogen synthesis in the liver in order to increase blood glucose levels. Glucagon and insulin are antagonistic, and their interaction maintains the stability of blood glucose. Glucagon suppression reduces hyperglycemia. After glucagon receptor knockout or inhibition, mice with insulin deficiency or resistance have shown blood glucose levels similar to those of normal mice [118, 119]. Glucagon antagonists exert significant glucose-lowering effects, which cannot be achieved by many current hypoglycemic drugs.
Furthermore, the antagonistic effect of glucagon reduces the demand for insulin, the rate of long-term complications, and the risk of metabolic disorders of diabetes [120, 121]. Phase II clinical trials of PF-06291874 (Pfizer) and LGD-6972 (Ligand Pharmaceuticals), which are small-molecule glucagon receptor (GCGr) antagonists, are completed. Moreover, monoclonal antibodies against GCGr, including LY2786890 (Eli Lilly), PF-06293620 (RN909, Pfizer), and REMD-477 (REMD Biotherapeutics), and antisense oligonucleotides including ISIS-GCGRRx (ISIS 449884, Ionis Pharmaceuticals) and ISIS 325568 have also successfully entered clinic studies [122]. Though these clinical trials have shown some degree of inhibition of GCGr, risks and safety concerns have been reported. Nevertheless, because no severe hypoglycemia event has been reported, glucagon receptor antagonists may be an effective treatment for T2D and are expected to be capable of targeting multiple and complementary pathways [121–123].
In general, phase II clinical trials of a number of drugs have been initiated or completed. These drugs have novel targets and unique pathways of action and advantages in treating diabetes (Table 1). They represent advances in diabetes treatment and will be beneficial for the treatment of diabetes.
Table 1.
Potential drugs with new targets for diabetes
| Class | Typical drug(s) | Hypoglycemic mechanism | Completed phase of clinical trial | Advantage(s) |
|---|---|---|---|---|
| GKAs |
Dorzagliatin (HMS5552) TTP399 |
Sense glucose level Mediates secretion of insulin and glucagon ↑ Glucose uptake and hepatic glycogen synthesis |
Phase II | As a novel and unique physiological pathway |
| 11-β-HSD1 inhibitors | BI 135585 | ↓ Active cortisol and glucocorticoid | Phase I | Weight loss or a lack of weight gain |
| GPR119 agonists | DS-8500a |
↑ Insulin secretion ↑ GLP-1 release |
Phase II |
Dual action mechanism Favorable blood glucose control Preserve pancreatic β-cell |
| Glucagon receptor antagonists |
PF-06291874 LGD-6972 |
↓ Gluconeogenesis and glycogenolysis ↑Glycogen synthesis |
Phase II |
Significant glucose lowering effects No severe hypoglycemia Reduce risk of metabolic disorders and demand for insulin |
Conclusion and Future Research Directions
Pharmacogenomics studies have been conducted on almost all current antidiabetic agents, with a focus on commonly used drugs such as metformin and sulfonylureas. Pharmacogenomic studies have demonstrated the important role of these drugs in the treatment of diabetes, and have provided references for precision medicine for diabetes in the future. Many drugs with new targets for diabetes have also been developed; some of them have entered clinical trials and have potential advantages, which will make up for shortcomings of current therapeutic drugs and provide more treatment choices to patients with diabetes. There is still a long way to go to improve diabetes treatment to achieve therapeutic goals better and faster. Investigating pharmacogenomics of all antidiabetic agents in greater depth, accumulating definite clinical evidence, and developing more drugs according to new targets for diabetes are necessary in the future.
Acknowledgements
Funding
This study and the Rapid Service Fee were funded by the Scientific Research Funds of Huaqiao University (Z16Y0017, T.S.), Natural Science Foundation of Fujian Province of China (2018J01585, S.H.), and National Natural Science Foundation of China (31771141).
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Zhiwei Zeng, Shi-Ying Huang and Tao Sun have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Footnotes
Digital Features
To view digital features for this article go to 10.6084/m9.figshare.12854816.
References
- 1.WHO 2019. Classification of diabetes mellitus. Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO.
- 2.Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14:88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 3.IDF 2019. International Diabetes Federation. IDF Diabetes Atlas, 9th edn. Brussels, Belgium: 2019. https://www.diabetesatlas.org. Accessed 22 Nov 2019.
- 4.American Diabetes A. 9. Pharmacologic approaches to glycemic treatment: standards of medical care in diabetes—2020. Diabetes Care. 2020;43:S98–S110. doi: 10.2337/dc20-S009. [DOI] [PubMed] [Google Scholar]
- 5.Edelman SV, Polonsky WH. Type 2 diabetes in the real world: the elusive nature of glycemic control. Diabetes Care. 2017;40:1425–1432. doi: 10.2337/dc16-1974. [DOI] [PubMed] [Google Scholar]
- 6.Kazemian P, Shebl FM, McCann N, Walensky RP, Wexler DJ. Evaluation of the cascade of diabetes care in the United States, 2005–2016. JAMA Intern Med. 2019;179:1376–1385. doi: 10.1001/jamainternmed.2019.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khunti K, Ceriello A, Cos X, de Block C. Achievement of guideline targets for blood pressure, lipid, and glycaemic control in type 2 diabetes: a meta-analysis. Diabetes Res Clin Pract. 2018;137:137–148. doi: 10.1016/j.diabres.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Chung WK, Erion K, Florez JC, et al. Precision medicine in diabetes: a consensus report from the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) Diabetes Care. 2020;43:1617–1635. doi: 10.2337/dci20-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inzucchi SE, Bergenstal RM, Buse JB, et al. Management of hyperglycemia in type 2 diabetes: a patient-centered approach: position statement of the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) Diabetes Care. 2012;35:1364–1379. doi: 10.2337/dc12-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gloyn AL, Drucker DJ. Precision medicine in the management of type 2 diabetes. Lancet Diabetes Endocrinol. 2018;6:891–900. doi: 10.1016/S2213-8587(18)30052-4. [DOI] [PubMed] [Google Scholar]
- 11.Xie F, Chan JC, Ma RC. Precision medicine in diabetes prevention, classification and management. J Diabetes Investig. 2018;9:998–1015. doi: 10.1111/jdi.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor A, Cozma A, Suharoschi R, Sitar-Taut A, Roman G. Clinical and genetic predictors of diabetes drug’s response. Drug Metab Rev. 2019;51:408–427. doi: 10.1080/03602532.2019.1656226. [DOI] [PubMed] [Google Scholar]
- 13.Kahn SE, Haffner SM, Heise MA, et al. Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N Engl J Med. 2006;355:2427–2443. doi: 10.1056/NEJMoa066224. [DOI] [PubMed] [Google Scholar]
- 14.Cascorbi I. Significance of pharmacogenomics in precision medicine. Clin Pharmacol Ther. 2018;103:732–735. doi: 10.1002/cpt.1052. [DOI] [PubMed] [Google Scholar]
- 15.Weinshilboum RM, Wang L. Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc. 2017;92:1711–1722. doi: 10.1016/j.mayocp.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heo CU, Choi CI. Current progress in pharmacogenetics of second-line antidiabetic medications: towards precision medicine for type 2 diabetes. J Clin Med. 2019;8:393. doi: 10.3390/jcm8030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rena G, Hardie DG, Pearson ER. The mechanisms of action of metformin. Diabetologia. 2017;60:1577–1585. doi: 10.1007/s00125-017-4342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sansome DJ, Xie C, Veedfald S, Horowitz M, Rayner CK, Wu T. Mechanism of glucose-lowering by metformin in type 2 diabetes: role of bile acids. Diabetes Obes Metab. 2020;22:141–148. doi: 10.1111/dom.13869. [DOI] [PubMed] [Google Scholar]
- 19.Wu T, Horowitz M, Rayner CK. New insights into the anti-diabetic actions of metformin: from the liver to the gut. Expert Rev Gastroenterol Hepatol. 2017;11:157–166. doi: 10.1080/17474124.2017.1273769. [DOI] [PubMed] [Google Scholar]
- 20.Liang X, Giacomini KM. Transporters involved in metformin pharmacokinetics and treatment response. J Pharm Sci. 2017;106:2245–2250. doi: 10.1016/j.xphs.2017.04.078. [DOI] [PubMed] [Google Scholar]
- 21.Umamaheswaran G, Praveen RG, Damodaran SE, Das AK, Adithan C. Influence of SLC22A1 rs622342 genetic polymorphism on metformin response in South Indian type 2 diabetes mellitus patients. Clin Exp Med. 2015;15:511–517. doi: 10.1007/s10238-014-0322-5. [DOI] [PubMed] [Google Scholar]
- 22.Xiao D, Guo Y, Li X, et al. The impacts of SLC22A1 rs594709 and SLC47A1 rs2289669 polymorphisms on metformin therapeutic efficacy in chinese type 2 diabetes patients. Int J Endocrinol. 2016;2016:4350712. doi: 10.1155/2016/4350712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Takizawa M, Chen E, et al. Genetic polymorphisms in organic cation transporter 1 (OCT1) in Chinese and Japanese populations exhibit altered function. J Pharmacol Exp Ther. 2010;335:42–50. doi: 10.1124/jpet.110.170159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Ye W, Wang Y, et al. Genetic variants of OCT1 influence glycemic response to metformin in Han Chinese patients with type-2 diabetes mellitus in Shanghai. Int J Clin Exp Pathol. 2015;8:9533–9542. [PMC free article] [PubMed] [Google Scholar]
- 25.Shokri F, Ghaedi H, Ghafouri Fard S, et al. Impact of ATM and SLC22A1 polymorphisms on therapeutic response to metformin in Iranian diabetic patients. Int J Mol Cell Med. 2016;5:1–7. [PMC free article] [PubMed] [Google Scholar]
- 26.Dawed AY, Zhou K, van Leeuwen N, et al. Variation in the plasma membrane monoamine transporter (PMAT) (encoded by SLC29A4) and organic cation transporter 1 (OCT1) (encoded by SLC22A1) and gastrointestinal intolerance to metformin in type 2 diabetes: an IMI DIRECT Study. Diabetes Care. 2019;42:1027–1033. doi: 10.2337/dc18-2182. [DOI] [PubMed] [Google Scholar]
- 27.Dujic T, Causevic A, Bego T, et al. Organic cation transporter 1 variants and gastrointestinal side effects of metformin in patients with type 2 diabetes. Diabet Med. 2016;33:511–514. doi: 10.1111/dme.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Eitan LN, Almomani BA, Nassar AM, Elsaqa BZ, Saadeh NA. Metformin pharmacogenetics: effects of SLC22A1, SLC22A2, and SLC22A3 polymorphisms on glycemic control and HbA1c levels. J Pers Med. 2019;9:17. doi: 10.3390/jpm9010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resendiz-Abarca CA, Flores-Alfaro E, Suarez-Sanchez F, et al. Altered glycemic control associated with polymorphisms in the SLC22A1 (OCT1) gene in a Mexican population with type 2 diabetes mellitus treated with metformin: a cohort study. J Clin Pharmacol. 2019;59:1384–1390. doi: 10.1002/jcph.1425. [DOI] [PubMed] [Google Scholar]
- 30.Sundelin E, Gormsen LC, Jensen JB, et al. Genetic polymorphisms in organic cation transporter 1 attenuates hepatic metformin exposure in humans. Clin Pharmacol Ther. 2017;102:841–848. doi: 10.1002/cpt.701. [DOI] [PubMed] [Google Scholar]
- 31.Hou W, Zhang D, Lu W, et al. Polymorphism of organic cation transporter 2 improves glucose-lowering effect of metformin via influencing its pharmacokinetics in Chinese type 2 diabetic patients. Mol Diagn Ther. 2015;19:25–33. doi: 10.1007/s40291-014-0126-z. [DOI] [PubMed] [Google Scholar]
- 32.Tzvetkov MV, Vormfelde SV, Balen D, et al. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther. 2009;86:299–306. doi: 10.1038/clpt.2009.92. [DOI] [PubMed] [Google Scholar]
- 33.Moeez S, Khalid Z, Jalil F, et al. Effects of SLC22A2 (rs201919874) and SLC47A2 (rs138244461) genetic variants on metformin pharmacokinetics in Pakistani T2DM patients. J Pak Med Assoc. 2019;69:155–163. [PubMed] [Google Scholar]
- 34.Phani NM, Vohra M, Kakar A, et al. Implication of critical pharmacokinetic gene variants on therapeutic response to metformin in type 2 diabetes. Pharmacogenomics. 2018;19:905–911. doi: 10.2217/pgs-2018-0041. [DOI] [PubMed] [Google Scholar]
- 35.Tkáč I, Klimčáková L, Javorský M, et al. Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes Metab. 2013;15:189–191. doi: 10.1111/j.1463-1326.2012.01691.x. [DOI] [PubMed] [Google Scholar]
- 36.Moeez S, Riaz S, Masood N, et al. Evaluation of the rs3088442 G>A SLC22A3 gene polymorphism and the role of microRNA 147 in groups of adult Pakistani populations with type 2 diabetes in response to metformin. Can J Diabetes. 2019;43:128–135.e3. doi: 10.1016/j.jcjd.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes. 2009;58:745–749. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He R, Zhang D, Lu W, et al. SLC47A1 gene rs2289669 G>A variants enhance the glucose-lowering effect of metformin via delaying its excretion in Chinese type 2 diabetes patients. Diabetes Res Clin Pract. 2015;109:57–63. doi: 10.1016/j.diabres.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Liang H, Xu W, Zhou L, Yang W, Weng J. Differential increments of basal glucagon-like-1 peptide concentration among SLC47A1 rs2289669 genotypes were associated with inter-individual variability in glycaemic response to metformin in Chinese people with newly diagnosed type 2 diabetes. Diabet Med. 2017;34:987–992. doi: 10.1111/dme.13351. [DOI] [PubMed] [Google Scholar]
- 40.Mousavi S, Kohan L, Yavarian M, Habib A. Pharmacogenetic variation of SLC47A1 gene and metformin response in type 2 diabetes patients. Mol Biol Res Commun. 2017;6:91–94. [PMC free article] [PubMed] [Google Scholar]
- 41.Raj GM, Mathaiyan J, Wyawahare M, Priyadarshini R. Lack of effect of the SLC47A1 and SLC47A2 gene polymorphisms on the glycemic response to metformin in type 2 diabetes mellitus patients. Drug Metab Pers Ther. 2018;33:175–185. doi: 10.1515/dmpt-2018-0030. [DOI] [PubMed] [Google Scholar]
- 42.Choi JH, Yee SW, Ramirez AH, et al. A common 5′-UTR variant in MATE2-K is associated with poor response to metformin. Clin Pharmacol Ther. 2011;90:674–684. doi: 10.1038/clpt.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon SJ, Oh J, Lee SH, Choi Y, Yu KS, Chung JY. Effect of plasma membrane monoamine transporter genetic variants on pharmacokinetics of metformin in humans. Transl Clin Pharmacol. 2018;26:79–85. doi: 10.12793/tcp.2018.26.2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rathmann W, Strassburger K, Bongaerts B, et al. A variant of the glucose transporter gene SLC2A2 modifies the glycaemic response to metformin therapy in recently diagnosed type 2 diabetes. Diabetologia. 2019;62:286–291. doi: 10.1007/s00125-018-4759-z. [DOI] [PubMed] [Google Scholar]
- 45.Zhou K, Yee SW, Seiser EL, et al. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet. 2016;48:1055–1059. doi: 10.1038/ng.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez-Ibarra HE, Reyes-Cortes LM, Jiang XL, et al. Genotypic and phenotypic factors influencing drug response in Mexican patients with type 2 diabetes mellitus. Front Pharmacol. 2018;9:320. doi: 10.3389/fphar.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, Mao G, Ren X, et al. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese type 2 diabetic patients. Diabetes Care. 2008;31:1939–1944. doi: 10.2337/dc07-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolac N, Simundic AM, Katalinic D, Topic E, Cipak A, Zjacic Rotkvic V. Metabolic control in type 2 diabetes is associated with sulfonylurea receptor-1 (SUR-1) but not with KCNJ11 polymorphisms. Arch Med Res. 2009;40:387–392. doi: 10.1016/j.arcmed.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Klen J, Dolžan V, Janež A. CYP2C9, KCNJ11 and ABCC8 polymorphisms and the response to sulphonylurea treatment in type 2 diabetes patients. Eur J Clin Pharmacol. 2014;70:421–428. doi: 10.1007/s00228-014-1641-x. [DOI] [PubMed] [Google Scholar]
- 50.Castelán-Martínez OD, Hoyo-Vadillo C, Bazán-Soto TB, Cruz M, Tesoro-Cruz E, Valladares-Salgado A. CYP2C9*3 gene variant contributes independently to glycaemic control in patients with type 2 diabetes treated with glibenclamide. J Clin Pharm Ther. 2018;43:768–774. doi: 10.1111/jcpt.12710. [DOI] [PubMed] [Google Scholar]
- 51.Javorsky M, Klimcakova L, Schroner Z, et al. KCNJ11 gene E23K variant and therapeutic response to sulfonylureas. Eur J Intern Med. 2012;23:245–249. doi: 10.1016/j.ejim.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 52.Li Q, Chen M, Zhang R, et al. KCNJ11 E23K variant is associated with the therapeutic effect of sulphonylureas in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol. 2014;41:748–754. doi: 10.1111/1440-1681.12280. [DOI] [PubMed] [Google Scholar]
- 53.Ragia G, Tavridou A, Petridis I, Manolopoulos VG. Association of KCNJ11 E23K gene polymorphism with hypoglycemia in sulfonylurea-treated type 2 diabetic patients. Diabetes Res Clin Pract. 2012;98:119–124. doi: 10.1016/j.diabres.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 54.Holstein A, Hahn M, Stumvoll M, Kovacs P. The E23K variant of KCNJ11 and the risk for severe sulfonylurea-induced hypoglycemia in patients with type 2 diabetes. Horm Metab Res. 2009;41:387–390. doi: 10.1055/s-0029-1192019. [DOI] [PubMed] [Google Scholar]
- 55.Phani NM, Vohra M, Adhikari P, et al. Genetic variants identified from GWAS for predisposition to type 2 diabetes predict sulfonylurea drug response. Curr Mol Med. 2017;17:580–586. doi: 10.2174/1566524018666180222122653. [DOI] [PubMed] [Google Scholar]
- 56.Cho HJ, Lee SY, Kim YG, et al. Effect of genetic polymorphisms on the pharmacokinetics and efficacy of glimepiride in a Korean population. Clin Chim Acta. 2011;412:1831–1834. doi: 10.1016/j.cca.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 57.Zhou K, Donnelly L, Burch L, et al. Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes: a Go-DARTS study. Clin Pharmacol Ther. 2010;87:52–56. doi: 10.1038/clpt.2009.176. [DOI] [PubMed] [Google Scholar]
- 58.Schroner Z, Dobrikova M, Klimcakova L, et al. Variation in KCNQ1 is associated with therapeutic response to sulphonylureas. Med Sci Monit. 2011;17:Cr392-6. doi: 10.12659/MSM.881850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duan F, Guo Y, Zhang L, et al. Association of KCNQ1 polymorphisms with gliclazide efficacy in Chinese type 2 diabetic patients. Pharmacogenet Genom. 2016;26:178–183. doi: 10.1097/FPC.0000000000000204. [DOI] [PubMed] [Google Scholar]
- 60.Li Q, Tang TT, Jiang F, et al. Polymorphisms of the KCNQ1 gene are associated with the therapeutic responses of sulfonylureas in Chinese patients with type 2 diabetes. Acta Pharmacol Sin. 2017;38:80–89. doi: 10.1038/aps.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prudente S, Morini E, Lucchesi D, et al. IRS1 G972R missense polymorphism is associated with failure to oral antidiabetes drugs in white patients with type 2 diabetes from Italy. Diabetes. 2014;63:3135–3140. doi: 10.2337/db13-1966. [DOI] [PubMed] [Google Scholar]
- 62.Seeringer A, Parmar S, Fischer A, et al. Genetic variants of the insulin receptor substrate-1 are influencing the therapeutic efficacy of oral antidiabetics. Diabetes Obes Metab. 2010;12:1106–1112. doi: 10.1111/j.1463-1326.2010.01301.x. [DOI] [PubMed] [Google Scholar]
- 63.Schroner Z, Javorsky M, Tkacova R, et al. Effect of sulphonylurea treatment on glycaemic control is related to TCF7L2 genotype in patients with type 2 diabetes. Diabetes Obes Metab. 2011;13:89–91. doi: 10.1111/j.1463-1326.2010.01324.x. [DOI] [PubMed] [Google Scholar]
- 64.Zhou X, Chen C, Yin D, et al. A variation in the ABCC8 gene is associated with type 2 diabetes mellitus and repaglinide efficacy in Chinese type 2 diabetes mellitus patients. Intern Med. 2019;58:2341–2347. doi: 10.2169/internalmedicine.2133-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He YY, Zhang R, Shao XY, et al. Association of KCNJ11 and ABCC8 genetic polymorphisms with response to repaglinide in Chinese diabetic patients. Acta Pharmacol Sin. 2008;29:983–989. doi: 10.1111/j.1745-7254.2008.00840.x. [DOI] [PubMed] [Google Scholar]
- 66.Yu M, Xu XJ, Yin JY, et al. KCNJ11 Lys23Glu and TCF7L2 rs290487(C/T) polymorphisms affect therapeutic efficacy of repaglinide in Chinese patients with type 2 diabetes. Clin Pharmacol Ther. 2010;87:330–335. doi: 10.1038/clpt.2009.242. [DOI] [PubMed] [Google Scholar]
- 67.Niemi M, Backman JT, Kajosaari LI, et al. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther. 2005;77:468–478. doi: 10.1016/j.clpt.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 68.Pei Q, Liu JY, Yin JY, et al. Repaglinide-irbesartan drug interaction: effects of SLCO1B1 polymorphism on repaglinide pharmacokinetics and pharmacodynamics in Chinese population. Eur J Clin Pharmacol. 2018;74:1021–1028. doi: 10.1007/s00228-018-2477-6. [DOI] [PubMed] [Google Scholar]
- 69.Kalliokoski A, Neuvonen M, Neuvonen PJ, Niemi M. The effect of SLCO1B1 polymorphism on repaglinide pharmacokinetics persists over a wide dose range. Br J Clin Pharmacol. 2008;66:818–825. doi: 10.1111/j.1365-2125.2008.03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He J, Qiu Z, Li N, et al. Effects of SLCO1B1 polymorphisms on the pharmacokinetics and pharmacodynamics of repaglinide in healthy Chinese volunteers. Eur J Clin Pharmacol. 2011;67:701–707. doi: 10.1007/s00228-011-0994-7. [DOI] [PubMed] [Google Scholar]
- 71.Kalliokoski A, Backman JT, Neuvonen PJ, Niemi M. Effects of the SLCO1B1*1B haplotype on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. Pharmacogenet Genom. 2008;18:937–942. doi: 10.1097/FPC.0b013e32830d733e. [DOI] [PubMed] [Google Scholar]
- 72.Dai XP, Huang Q, Yin JY, et al. KCNQ1 gene polymorphisms are associated with the therapeutic efficacy of repaglinide in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol. 2012;39:462–468. doi: 10.1111/j.1440-1681.2012.05701.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhou X, Zhu J, Bao Z, et al. A variation in KCNQ1 gene is associated with repaglinide efficacy on insulin resistance in Chinese type 2 diabetes mellitus patients. Sci Rep. 2016;6:37293. doi: 10.1038/srep37293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu W, Hu C, Zhang R, et al. Effects of KCNQ1 polymorphisms on the therapeutic efficacy of oral antidiabetic drugs in Chinese patients with type 2 diabetes. Clin Pharmacol Ther. 2011;89:437–442. doi: 10.1038/clpt.2010.351. [DOI] [PubMed] [Google Scholar]
- 75.Nanjan MJ, Mohammed M, Prashantha Kumar BR, Chandrasekar MJN. Thiazolidinediones as antidiabetic agents: a critical review. Bioorg Chem. 2018;77:548–567. doi: 10.1016/j.bioorg.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 76.Aquilante CL, Bushman LR, Knutsen SD, Burt LE, Rome LC, Kosmiski LA. Influence of SLCO1B1 and CYP2C8 gene polymorphisms on rosiglitazone pharmacokinetics in healthy volunteers. Hum Genomics. 2008;3:7–16. doi: 10.1186/1479-7364-3-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dawed AY, Donnelly L, Tavendale R, et al. CYP2C8 and SLCO1B1 variants and therapeutic response to thiazolidinediones in patients with type 2 diabetes. Diabetes Care. 2016;39:1902–1908. doi: 10.2337/dc15-2464. [DOI] [PubMed] [Google Scholar]
- 78.Kalliokoski A, Neuvonen M, Neuvonen PJ, Niemi M. No significant effect of SLCO1B1 polymorphism on the pharmacokinetics of rosiglitazone and pioglitazone. Br J Clin Pharmacol. 2008;65:78–86. doi: 10.1111/j.1365-2125.2007.02986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsieh MC, Lin KD, Tien KJ, et al. Common polymorphisms of the peroxisome proliferator-activated receptor-gamma (Pro12Ala) and peroxisome proliferator-activated receptor-gamma coactivator-1 (Gly482Ser) and the response to pioglitazone in Chinese patients with type 2 diabetes mellitus. Metabolism. 2010;59:1139–1144. doi: 10.1016/j.metabol.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 80.Namvaran F, Azarpira N, Rahimi-Moghaddam P, Dabbaghmanesh MH. Polymorphism of peroxisome proliferator-activated receptor γ (PPARγ) Pro12Ala in the Iranian population: relation with insulin resistance and response to treatment with pioglitazone in type 2 diabetes. Eur J Pharmacol. 2011;671:1–6. doi: 10.1016/j.ejphar.2011.09.158. [DOI] [PubMed] [Google Scholar]
- 81.Priya SS, Sankaran R, Ramalingam S, Sairam T, Somasundaram LS. Genotype phenotype correlation of genetic polymorphism of PPAR gamma gene and therapeutic response to pioglitazone in type 2 diabetes mellitus—a pilot study. J Clin Diagn Res. 2016;10:Fc11-4. doi: 10.7860/JCDR/2016/16494.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramírez-Salazar M, Pérez-Luque E, Fajardo-Araujo M, Garza SM, Malacara JM. Effect of the Pro12Ala polymorphism of the PPAR gamma 2 gene on response to pioglitazone treatment in menopausal women. Menopause. 2008;15:1151–1156. doi: 10.1097/gme.0b013e31816d5b2d. [DOI] [PubMed] [Google Scholar]
- 83.Florez JC, Jablonski KA, Sun MW, et al. Effects of the type 2 diabetes-associated PPARG P12A polymorphism on progression to diabetes and response to troglitazone. J Clin Endocrinol Metab. 2007;92:1502–1509. doi: 10.1210/jc.2006-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stage TB, Christensen MM, Feddersen S, Beck-Nielsen H, Brøsen K. The role of genetic variants in CYP2C8, LPIN1, PPARGC1A and PPARγ on the trough steady-state plasma concentrations of rosiglitazone and on glycosylated haemoglobin A1c in type 2 diabetes. Pharmacogenet Genom. 2013;23:219–227. doi: 10.1097/FPC.0b013e32835f91fc. [DOI] [PubMed] [Google Scholar]
- 85.van de Laar FA, Lucassen PL, Akkermans RP, van de Lisdonk EH, Rutten GE, van Weel C. Alpha-glucosidase inhibitors for type 2 diabetes mellitus. Cochrane Database Syst Rev. 2005;2:003639. doi: 10.1002/14651858.CD003639.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andrulionyte L, Kuulasmaa T, Chiasson JL, Laakso M. Single nucleotide polymorphisms of the peroxisome proliferator-activated receptor-alpha gene (PPARA) influence the conversion from impaired glucose tolerance to type 2 diabetes: the STOP-NIDDM trial. Diabetes. 2007;56:1181–1186. doi: 10.2337/db06-1110. [DOI] [PubMed] [Google Scholar]
- 87.Ahrén B. GLP-1 for type 2 diabetes. Exp Cell Res. 2011;317:1239–1245. doi: 10.1016/j.yexcr.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 88.Chedid V, Vijayvargiya P, Carlson P, et al. Allelic variant in the glucagon-like peptide 1 receptor gene associated with greater effect of liraglutide and exenatide on gastric emptying: a pilot pharmacogenetics study. Neurogastroenterol Motil. 2018;30:e13313. doi: 10.1111/nmo.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Luis DA, Diaz Soto G, Izaola O, Romero E. Evaluation of weight loss and metabolic changes in diabetic patients treated with liraglutide, effect of RS 6923761 gene variant of glucagon-like peptide 1 receptor. J Diabetes Complicat. 2015;29:595–598. doi: 10.1016/j.jdiacomp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 90.Yu M, Wang K, Liu H, Cao R. GLP1R variant is associated with response to exenatide in overweight Chinese type 2 diabetes patients. Pharmacogenomics. 2019;20:273–277. doi: 10.2217/pgs-2018-0159. [DOI] [PubMed] [Google Scholar]
- 91.Lin CH, Lee YS, Huang YY, Hsieh SH, Chen ZS, Tsai CN. Polymorphisms of GLP-1 receptor gene and response to GLP-1 analogue in patients with poorly controlled type 2 diabetes. J Diabetes Res. 2015;2015:176949. doi: 10.1155/2015/176949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferreira MC, da Silva MER, Fukui RT, Do Carmo Arruda-Marques M, Azhar S, Dos Santos RF. Effect of TCF7L2 polymorphism on pancreatic hormones after exenatide in type 2 diabetes. Diabetol Metab Syndr. 2019;11:10. doi: 10.1186/s13098-019-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Javorský M, Gotthardová I, Klimčáková L, et al. A missense variant in GLP1R gene is associated with the glycaemic response to treatment with gliptins. Diabetes Obes Metab. 2016;18:941–944. doi: 10.1111/dom.12682. [DOI] [PubMed] [Google Scholar]
- 94.Űrgeová A, Javorský M, Klimčáková L, et al. Genetic variants associated with glycemic response to treatment with dipeptidylpeptidase 4 inhibitors. Pharmacogenomics. 2020;21:317–323. doi: 10.2217/pgs-2019-0147. [DOI] [PubMed] [Google Scholar]
- 95.Han E, Park HS, Kwon O, et al. A genetic variant in GLP1R is associated with response to DPP-4 inhibitors in patients with type 2 diabetes. Medicine (Baltimore) 2016;95:e5155. doi: 10.1097/MD.0000000000005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gotthardová I, Javorský M, Klimčáková L, et al. KCNQ1 gene polymorphism is associated with glycaemic response to treatment with DPP-4 inhibitors. Diabetes Res Clin Pract. 2017;130:142–147. doi: 10.1016/j.diabres.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 97.Jamaluddin JL, Huri HZ, Vethakkan SR. Clinical and genetic predictors of dipeptidyl peptidase-4 inhibitor treatment response in type 2 diabetes mellitus. Pharmacogenomics. 2016;17:867–881. doi: 10.2217/pgs-2016-0010. [DOI] [PubMed] [Google Scholar]
- 98.Zimdahl H, Ittrich C, Graefe-Mody U, et al. Influence of TCF7L2 gene variants on the therapeutic response to the dipeptidylpeptidase-4 inhibitor linagliptin. Diabetologia. 2014;57:1869–1875. doi: 10.1007/s00125-014-3276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zimdahl H, Haupt A, Brendel M, et al. Influence of common polymorphisms in the SLC5A2 gene on metabolic traits in subjects at increased risk of diabetes and on response to empagliflozin treatment in patients with diabetes. Pharmacogenet Genomics. 2017;27:135–142. doi: 10.1097/FPC.0000000000000268. [DOI] [PubMed] [Google Scholar]
- 100.Francke S, Mamidi RN, Solanki B, et al. In vitro metabolism of canagliflozin in human liver, kidney, intestine microsomes, and recombinant uridine diphosphate glucuronosyltransferases (UGT) and the effect of genetic variability of UGT enzymes on the pharmacokinetics of canagliflozin in humans. J Clin Pharmacol. 2015;55:1061–1072. doi: 10.1002/jcph.506. [DOI] [PubMed] [Google Scholar]
- 101.Lloyd DJ, St Jean DJ, Kurzeja RJ, et al. Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors. Nature. 2013;504:437–440. doi: 10.1038/nature12724. [DOI] [PubMed] [Google Scholar]
- 102.Agius L. Lessons from glucokinase activators: the problem of declining efficacy. Expert Opin Ther Pat. 2014;24:1155–1159. doi: 10.1517/13543776.2014.965680. [DOI] [PubMed] [Google Scholar]
- 103.Toulis KA, Nirantharakumar K, Pourzitaki C, Barnett AH, Tahrani AA. Glucokinase activators for type 2 diabetes: challenges and future developments. Drugs. 2020;80:467–475. doi: 10.1007/s40265-020-01278-z. [DOI] [PubMed] [Google Scholar]
- 104.Egan A, Vella A. TTP399: an investigational liver-selective glucokinase (GK) activator as a potential treatment for type 2 diabetes. Expert Opin Investig Drugs. 2019;28:741–747. doi: 10.1080/13543784.2019.1654993. [DOI] [PubMed] [Google Scholar]
- 105.Zhu D, Gan S, Liu Y, et al. Dorzagliatin monotherapy in Chinese patients with type 2 diabetes: a dose-ranging, randomised, double-blind, placebo-controlled, phase 2 study. Lancet Diabetes Endocrinol. 2018;6:627–636. doi: 10.1016/S2213-8587(18)30105-0. [DOI] [PubMed] [Google Scholar]
- 106.Zhu XX, Zhu DL, Li XY, et al. Dorzagliatin (HMS5552), a novel dual-acting glucokinase activator, improves glycaemic control and pancreatic β-cell function in patients with type 2 diabetes: a 28-day treatment study using biomarker-guided patient selection. Diabetes Obes Metab. 2018;20:2113–2120. doi: 10.1111/dom.13338. [DOI] [PubMed] [Google Scholar]
- 107.Hollis G, Huber R. 11β-Hydroxysteroid dehydrogenase type 1 inhibition in type 2 diabetes mellitus. Diabetes Obes Metab. 2011;13:1–6. doi: 10.1111/j.1463-1326.2010.01305.x. [DOI] [PubMed] [Google Scholar]
- 108.Gathercole LL, Lavery GG, Morgan SA, et al. 11β-Hydroxysteroid dehydrogenase 1: translational and therapeutic aspects. Endocr Rev. 2013;34:525–555. doi: 10.1210/er.2012-1050. [DOI] [PubMed] [Google Scholar]
- 109.Freude S, Heise T, Woerle HJ, et al. Safety, pharmacokinetics and pharmacodynamics of BI 135585, a selective 11β-hydroxysteroid dehydrogenase-1 (HSD1) inhibitor in humans: liver and adipose tissue 11β-HSD1 inhibition after acute and multiple administrations over 2 weeks. Diabetes Obes Metab. 2016;18:483–490. doi: 10.1111/dom.12635. [DOI] [PubMed] [Google Scholar]
- 110.Hong SP, Han D, Chang KH, Ahn SK. A novel highly potent and selective 11β-hydroxysteroid dehydrogenase type 1 inhibitor, INU-101. Eur J Pharmacol. 2018;835:169–178. doi: 10.1016/j.ejphar.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 111.Anderson A, Walker BR. 11β-HSD1 inhibitors for the treatment of type 2 diabetes and cardiovascular disease. Drugs. 2013;73:1385–1393. doi: 10.1007/s40265-013-0112-5. [DOI] [PubMed] [Google Scholar]
- 112.Ohishi T, Yoshida S. The therapeutic potential of GPR119 agonists for type 2 diabetes. Expert Opin Investig Drugs. 2012;21:321–328. doi: 10.1517/13543784.2012.657797. [DOI] [PubMed] [Google Scholar]
- 113.Ritter K, Buning C, Halland N, Pöverlein C, Schwink L. G protein-coupled receptor 119 (GPR119) agonists for the treatment of diabetes: recent progress and prevailing challenges. J Med Chem. 2016;59:3579–3592. doi: 10.1021/acs.jmedchem.5b01198. [DOI] [PubMed] [Google Scholar]
- 114.Watada H, Shiramoto M, Irie S, et al. G protein-coupled receptor 119 agonist DS-8500a effects on pancreatic β-cells in Japanese type 2 diabetes mellitus patients. J Diabetes Investig. 2019;10:84–93. doi: 10.1111/jdi.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Inagaki N, Chou HS, Tsukiyama S, et al. Glucose-lowering effects and safety of DS-8500a, a G protein-coupled receptor 119 agonist, in Japanese patients with type 2 diabetes: results of a randomized, double-blind, placebo-controlled, parallel-group, multicenter, phase II study. BMJ Open Diabetes Res Care. 2017;5:e000424. doi: 10.1136/bmjdrc-2017-000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Terauchi Y, Yamada Y, Watada H, et al. Efficacy and safety of the G protein-coupled receptor 119 agonist DS-8500a in Japanese type 2 diabetes mellitus patients with inadequate glycemic control on sitagliptin: a phase 2 randomized placebo-controlled study. J Diabetes Investig. 2018;9:1333–1341. doi: 10.1111/jdi.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yamada Y, Terauchi Y, Watada H, et al. Efficacy and safety of GPR119 agonist DS-8500a in Japanese patients with type 2 diabetes: a randomized, double-blind, placebo-controlled, 12-week study. Adv Ther. 2018;35:367–381. doi: 10.1007/s12325-018-0668-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee Y, Wang MY, Du XQ, Charron MJ, Unger RH. Glucagon receptor knockout prevents insulin-deficient type 1 diabetes in mice. Diabetes. 2011;60:391–397. doi: 10.2337/db10-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Okamoto H, Cavino K, Na E, et al. Glucagon receptor inhibition normalizes blood glucose in severe insulin-resistant mice. Proc Natl Acad Sci USA. 2017;114:2753–2758. doi: 10.1073/pnas.1621069114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee YH, Wang MY, Yu XX, Unger RH. Glucagon is the key factor in the development of diabetes. Diabetologia. 2016;59:1372–1375. doi: 10.1007/s00125-016-3965-9. [DOI] [PubMed] [Google Scholar]
- 121.Pearson MJ, Unger RH, Holland WL. Clinical trials, triumphs, and tribulations of glucagon receptor antagonists. Diabetes Care. 2016;39:1075–1077. doi: 10.2337/dci15-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scheen AJ, Paquot N, Lefèbvre PJ. Investigational glucagon receptor antagonists in phase I and II clinical trials for diabetes. Expert Opin Investig Drugs. 2017;26:1373–1389. doi: 10.1080/13543784.2017.1395020. [DOI] [PubMed] [Google Scholar]
- 123.Wu T, Rayner CK, Marathe CS, Jones KL, Horowitz M. Glucagon receptor signalling—backwards and forwards. Expert Opin Investig Drugs. 2018;27:135–138. doi: 10.1080/13543784.2018.1428306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.