Abstract
Purpose
During the 2019 coronavirus disease (COVID-19) pandemic, oncologists face new challenges, and they need to adjust their cancer management strategies as soon as possible to reduce the risk of SARS-CoV-2 infection and tumor recurrence. However, data on cancer patients with SARS-CoV-2 infection remains scarce.
Methods
We conducted a retrospective study on 223 cancer patients with SARS-CoV-2 from 26 hospitals in Hubei, China. An individualized nomogram was constructed based on multivariate Cox analysis. Considering the convenience of the nomogram application, an online tool was also created. The predictive performance and clinical application of nomogram were verified by C-index, calibration curve and decision curve analysis (DCA).
Results
Among cancer patients with SARS-CoV-2, there were significant differences in clinical characteristics between survivors and non-survivors, and compared with patients with solid tumors including lung cancer, patients with hematological malignancies had a worse prognosis. Male, dyspnea, elevated PCT, increased heart rate, elevated D-dimers, and decreased platelets were risk factors for these patients. Furthermore, a good prediction performance of the online tool (dynamic nomogram: https://covid-19-prediction-tool.shinyapps.io/DynNomapp/) was also fully demonstrated with the C-indexes of 0.841 (95% CI 0.782–0.900) in the development cohort and 0.780 (95% CI 0.678–0.882) in the validation cohort.
Conclusion
Overall, cancer patients with SARS-CoV-2 had unique clinical features, and the established online tool could guide clinicians to predict the prognosis of patients during the COVID-19 epidemic and to develop more rational treatment strategies for cancer patients.
Electronic supplementary material
The online version of this article (10.1007/s00432-020-03420-6) contains supplementary material, which is available to authorized users.
Keywords: Cancer, COVID-19, SARS-CoV-2, Nomogram, Prognosis
Introduction
Since the outbreak of coronavirus disease (COVID-19) in December 2019, the number of infected cases has been increasing, which has seriously affected the normal life of human beings. Recently, increased studies have suggested that cancer patients were more susceptible to SARS-CoV-2 infection (Dai et al. 2020; Liang et al. 2020). Thus, oncologists face the new challenge during the COVID-19 pandemic. They need to adjust their cancer management strategies as soon as possible to reduce the risk of SARS-CoV-2 infection and tumor recurrence without affecting the therapeutic effect. However, to date, data on SARS-CoV-2 infected cancer patients are still limited, and their prognosis is poorly understood. Given this, we performed a retrospective analysis based on the data on 223 cancer patients with SARS-CoV-2 infection from 26 clinical centers in Hubei province, China. In addition, we constructed an online nomogram to predict the prognosis of these patients, which might help clinicians develop more rational treatment strategies for cancer patients during the COVID-19 epidemic.
Materials and methods
Study subject and ethical statements
All cancer patients with SARS-CoV-2 infection were hospitalized in 26 hospitals in Hubei province, China from December 27, 2019 to March 19, 2020. A total of 296 patients were enrolled, of whom 62 patients lacking a SARS-CoV-2 PCR test were excluded from this study. In addition, a one-year-old child was also excluded from this study. Eventually, 223 patients were selected in this study, who met the diagnostic criteria of the WHO interim guidance (https://www.who.int/publications-detail/clinical-managementof-severeacute-respiratory-infection-when-novelcoronavirus-(ncov)-infection-issuspected) and were confirmed by the nucleic acid test. This study was approved by the ethics committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Approval No. 20/061-2257).
Data collection and extraction
The clinicopathological characters of 223 patients included in this study were collected from their medical records. General data included age, sex, BMI, smoking history, comorbidities (such as diabetes mellitus, arterial hypertension, etc.), vital signs, tumor types, dyspnea, fever and other symptoms, survival time and the interval to the negative conversion of SARS-Cov-2 nucleic acid test. Laboratory findings included procalcitonin (PCT), white blood cell count, neutrophil count, lymphocyte count, monocyte count, eosinophil count, basophil count, platelet count, aspartate aminotransferase (AST), alamine aminotransferase (ALT), C reactive protein (CRP), hypersensitive C reactive protein (hs-CRP), creatinine, serum amyloid A (SAA), creatine kinase isozyme (CK-MB), direct bilirubin, total bilirubin, prothrombin time (PT), activation of partial thrombin time (APTT) and D-dimer. Besides, treatment included antitumor therapy, antiviral therapy, antibacterial therapy, hormone therapy, and traditional Chinese medicine treatment.
Statistical analysis
Categorical variables were represented by a number (percentage). The continuous variables conforming to the normal distribution were represented by mean (standard deviation, SD) otherwise by median (interquartile range, IQR). The unpaired independent sample t test was used for the continuous variables with homogeneous variances and normal distribution. Non-parametric tests (Mann–Whitney U and/or Kruskal–Wallis H) were used for those with uneven variances or non-normal distribution. The Chi-square test or Fisher’s exact test was used to compare the categorical variables between groups, as appropriate. The nucleic acid test time of diagnosis was taken as the starting point of survival time, and the Kaplan–Meier method was used for survival analysis. In this study, all patients were randomly divided into a development cohort and a validation cohort at a ratio of 7:3. Based on the results of the univariate and multivariate Cox analysis in the development cohort, a nomogram was constructed with the “rms” package in R. To evaluate the prediction performance of the nomogram, we calculated the C-indexes of the development cohort and validation cohort with bootstraps method. At the same time, calibration curves were also used to judge the consistency between the model prediction value and the actual observed value. In this study, all statistical analysis was carried out in R software (version 3.6.0). P value < 0.05 was considered statistically significant.
Results
The clinicopathological features of SARS-Cov-2 infected cancer patients
Of the 223 patients finally included in this study, 186 patients survived, and 37 patients were not (Table 1). 52.1% of patients were male, 59.7% of patients presented dyspnea, and 75.7% of patients had a fever. Compared with survivors, non-survivors were older (67 years vs. 62.5 years), more male (70.3% vs. 48.4%), and more likely to have dyspnea (75.7% vs. 66.7%). Moreover, non-survivors had a faster heart rate than survivors (100 bpm vs. 88 bpm). In the vast majority of laboratory findings, non-survivors had higher detection values, such as white blood cell count, neutrophil count, PCT, CRP, hs-CRP, direct bilirubin, PT, APTT, creatinine, CK-MB, and D-dimers. While platelet count and lymphocyte count were lower in the non-survivors, the differences were statistically significant (all p < 0.05). Of these 223 patients, there were 82 patients with arterial hypertension and 32 patients with diabetes mellitus. The proportion of non-survivors with underlying diseases was higher than that of survivors, as shown in Figure S1. Among these 223 patients, 171 patients turned nucleic acid to be negative before the follow-up deadline, with a median time of 13 days. However, nucleic acid switching negative was not found in 37 non-survivors. Patients without nucleic acid negative conversion had more dyspnea than those with nucleic acid switching negative (p < 0.001). Similarly, patients without nucleic acid switching negative had higher PCT, HR, neutrophils, lymphocytes, direct bilirubin and indirect bilirubin (Table S1). In addition, more patients received antibacterial treatment (94.6% vs. 80.1%) and glucocorticoid therapy (75.7% vs. 36.0%) in the non-survivors than in the survivors, with p values of 0.034 and < 0.001, respectively (Table 1).
Table 1.
Demographic characteristics of SARS-CoV-2—infected cancer patients in the entire cohort
| Variables | All patients (n = 223) | Survivors (n = 186) | Non-survivors (n = 37) | p value |
|---|---|---|---|---|
| Age (years) | 63 (56, 71) | 62.5 (56, 70) | 67 (54.5, 75) | 0.240 |
| Sex | 0.015 | |||
| Female | 107 (47.9) | 96 (51.6) | 11 (29.7) | |
| Male | 116 (52.1) | 90 (48.4) | 26 (70.3) | |
| BMI (kg/m2) | 0.183 | |||
| < 18.5 | 13 (5.8) | 12 (6.5) | 1 (2.7) | |
| 18.5–24.9 | 73 (32.8) | 65 (34.9) | 8 (21.6) | |
| ≥ 25 | 30 (13.5) | 26 (14.0) | 4 (10.8) | |
| Unknown | 107 (47.9) | 83 (44.6) | 24 (64.9) | |
| Smoke | 0.351 | |||
| Never | 170 (72.6) | 144 (77.4) | 26 (70.3) | |
| Previous/present | 53 (27.4) | 42 (22.6) | 11 (29.7) | |
| Tumor types | 0.001 | |||
| Hematological malignancies | 15 (6.7) | 7 (3.7) | 8 (21.6) | |
| Lung cancer | 39 (17.5) | 31 (16.7) | 8 (21.6) | |
| Other solid tumors | 169 (75.8) | 148 (79.6) | 21 (56.8) | |
| Anti-tumor therapy | 0.049 | |||
| Continous | 126 (56.5) | 111 (59.7) | 15 (40.5) | |
| Discontinous | 30 (13.4) | 21 (11.3) | 9 (24.4) | |
| Unknown | 67 (30.1) | 54 (29.0) | 13 (35.1) | |
| Basic diseases * | 0.608 | |||
| Without | 105 (47.1) | 89 (47.9) | 16 (43.2) | |
| With | 118 (52.9) | 97 (52.1) | 21 (56.8) | |
| Fever | 0.687 | |||
| Without | 54 (24.3) | 46 (24.7) | 8 (21.6) | |
| With | 169 (75.7) | 140 (75.3) | 29 (78.4) | |
| Dyspnea | < 0.001 | |||
| Without | 90 (40.3) | 62 (33.3) | 9 (24.3) | |
| With | 133 (59.7) | 124 (66.7) | 28 (75.7) | |
| Other symptoms * | 0.643 | |||
| Without | 28 (12.5) | 22 (11.8) | 6 (16.2) | |
| With | 195 (87.5) | 164 (88.2) | 31 (83.8) | |
| PCT (ng/ml) | < 0.001 | |||
| ≤ 0.5 | 163 (73.0) | 144 (77.4) | 19 (51.4) | |
| > 0.5 | 30 (13.5) | 17 (9.1) | 13 (35.1) | |
| Not application | 30 (13.5) | 25 (13.5) | 5 (13.5) | |
| Heart rate (bpm) | 88 (78, 100) | 86 (77, 97.25) | 100 (88.5, 112.5) | < 0.001 |
| SBP (mmHg) | 130 (120, 140) | 130 (120, 140.25) | 128 (109, 140.5) | 0.314 |
| DBP (mmHg) | 78.33 (11.128) | 77.8602 (11.132) | 80.703 (10.952) | 0.156 |
| Respiratory rate (braths/min) | 20 (19, 22) | 20 (19, 22) | 20 (20, 23) | 0.152 |
| Temperature (℃) | 36.7 (36.5, 37.5) | 36.7 (36.5, 37.5) | 36.9 (36.4, 38.05) | 0.703 |
| WBC count (10E9/L) | 5.28 (4.05, 7.18) | 5.025 (4.005, 6.423) | 7.23 (5.035, 11.785) | < 0.001 |
| Neutrophil count (10E9/L) | 3.64 (2.59, 5.45) | 3.42 (2.563, 4.793) | 6.62 (3.25, 10.345) | < 0.001 |
| Lymphocyte count (10E9/L) | 0.90 (0.61, 1.42) | 0.95 (0.658, 1.433) | 0.69 (0.495, 1.125) | 0.024 |
| Platelet count (10E9/L) | 188 (117, 249) | 196.5 (130.75, 260.25) | 142 (51, 203.5) | < 0.001 |
| Monocyte count (10E9/L) | 0.43 (0.30, 0.62) | 0.42 (0.3, 0.59) | 0.64 (0.28, 1.005) | 0.041 |
| Eosinophils count (10E9/L) | 0.03 (0, 0.09) | 0.05 (0.01, 0.09) | 0 (0, 0.02) | < 0.001 |
| Basophil count (10E9/L) | 0.01 (0.01, 0.02) | 0.01 (0.01, 0.02) | 0.01 (0, 0.03) | 0.739 |
| ALT (U/L) | 22 (13, 36) | 23 (14, 37) | 18 (10.5, 26.5) | 0.068 |
| AST (U/L) | 28 (18, 39) | 26.5 (18, 38) | 35 (20, 51) | 0.061 |
| Creatinine (umol/L) | 65 (52.7, 85) | 63.5 (52.9, 81.025) | 94.1 (51.85, 120.5) | 0.018 |
| CRP (mg/L) | 28.85 (7.7, 77.925) (n = 176) | 25.4 (5.225, 55.5) (n = 144) | 94.8 (39.45, 139.525) (n = 32) | < 0.001 |
| SAA (mg/L) | 148.39 (26.8, 300) (n = 35) | 128.21 (16.25, 300) (n = 25) | 183.195 (79.483, 300) (n = 10) | 0.811 |
| hs-CRP (mg/L) | 27.4 (5, 71.9) (n = 159) | 21.965 (5, 59.3) (n = 130) | 84.1 (55.22, 141.15) (n = 29) | < 0.001 |
| Total bilirubin (umol/L) | 11.1 (8,14.6) | 11 (7.8, 14.6) | 12.4 (9.35, 15.15) | 0.185 |
| Direct bilirubin (umol/L) | 3.9 (2.8, 5.5) | 3.7 (2.6, 5.3) | 5.1 (3.775, 7.8) | < 0.001 |
| PT (s) | 12.9 (11.9, 14.0) (n = 219) | 12.8 (11.9, 13.6) (n = 182) | 14.4 (12.3, 15.75) | < 0.001 |
| APTT (s) | 34.6 (29.1, 39.7) (n = 216) | 33.9 (28.8, 38.65) (n = 181) | 37.8 (30.7, 46.2) (n = 35) | 0.010 |
| Creatine kinase-MB (U/L) | 1.12 (0.525, 4.525) (n = 153) | 1.03 (0.5, 3.745) (n = 133) | 3.645 (1.025, 14.738) (n = 20) | 0.020 |
| D-dimer (mg/L) | 0.837 (0.37, 2.14) | 0.69 (0.329, 1.9) | 1.63 (0.465, 5.865) | 0.001 |
| Nucleic acid negative time | 13 (9, 22) | 13 (9, 22) | ||
| Antiviral therapy | 0.385 | |||
| No | 19 (8.5) | 14 (7.5) | 5 (13.5) | |
| Yes | 204 (91.5) | 172 (92.5) | 32 (86.5) | |
| Antibacterial therapy | 0.034 | |||
| No | 39 (17.5) | 37 (19.9) | 2 (5.4) | |
| Yes | 184 (82.5) | 149 (80.1) | 35 (94.6) | |
| Hormone therapy | < 0.001 | |||
| No | 128 (57.4) | 119 (64.0) | 9 (24.3) | |
| Yes | 95 (42.6) | 67 (36.0) | 28 (75.7) | |
| Immunoglobulin application | 0.052 | |||
| No | 151 (67.7) | 131 (70.4) | 20 (54.1) | |
| Yes | 72 (32.3) | 55 (29.6) | 17 (45.9) | |
| Traditional Chinese medicine treatment | 0.129 | |||
| No | 27 (16.6) | 34 (18.3) | 3 (8.1) | |
| Yes | 186 (83.4) | 152 (81.7) | 34 (91.9) | |
All variables with missing values are marked with a specific number of samples
Other symptoms include cough, expectoration, fatigue, headache, myalgia, sore throat, diarrhea, nausea, sneezing, nasal congestion, anorexia, night sweats, etc. Basic diseases include hypertension, diabetes, cardiovascular and cerebrovascular diseases, chronic obstructive pulmonary disease, chronic bronchitis, emphysema, chronic liver disease, chronic kidney disease, and neuropsychiatric diseases
BMI body mass index, PCT procalcitonin, SBP systolic blood pressure, DBP diastolic blood pressure, WBC white blood cell, AST aspartate aminotransferase, ALT alanine transaminase, CRP C-reactive protein, hs-CRP hypersensitive c-reactive protein, SAA Serum amyloid A, PT prothrombin time, APTT activation of partial thrombin time
The difference in the short-time survival among different cancer patients with SARS-CoV-2 infection
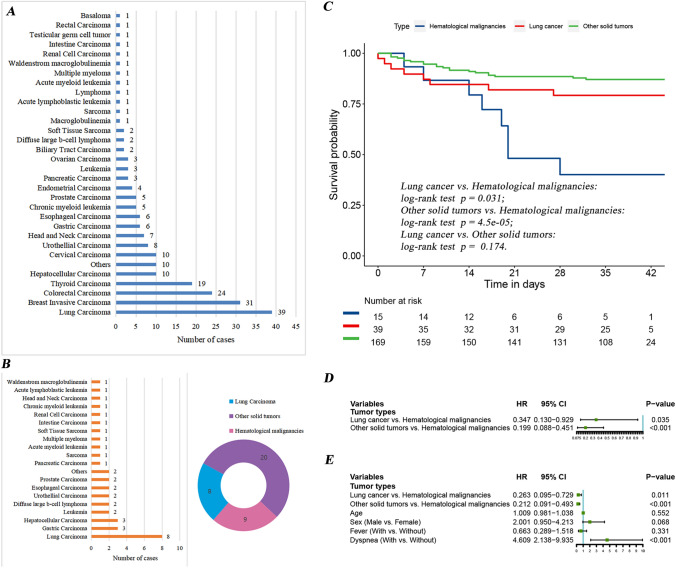
Nearly 18% of the patients were lung cancer patients (n = 39), ranking first among all the 223 cancer patients with SARS-CoV-2 infection (Fig. 1a). Similarly, lung cancer patients accounted for 21.6% (n = 8), which was the highest proportion in 37 non-survivors (Fig. 1b). To further explore the differences in short-term survival among patients with different tumor types, we conducted a Kaplan–Meier survival analysis (Fig. 1c). We found that lung cancer patients and other solid tumors patients had a better prognosis than hematological malignancies patients (the p values were 0.031 and < 0.001, respectively). Further a univariate Cox analysis showed the same results (Fig. 1d). This finding was also confirmed in multivariate Cox analysis after adjusting for age, gender, fever and dyspnea (Fig. 1e).
Fig. 1.
a The number of cases with different tumor types in the 223 cancer patients with SARS-CoV-2 infection. b Distribution of tumor types in 37 non-survivors. There were 8 cases of lung cancer, 9 cases of hematological malignancies and 20 cases of other solid tumors. c Kaplan–Meier survival curves of hematological malignancies patients, lung cancer patients and other solid tumors patients infected with SARS-CoV-2. Among cancer patients infected with SARS-CoV-2, compared with patients with solid tumors such as lung cancer, patients with hematological malignancies had a worse prognosis. d Univariate Cox analysis of different tumor types. e Multivariate Cox analysis after adjusting for age, gender, fever and dyspnea
The univariate and multivariate Cox analysis
To explore the risk factors associated with SARS-CoV-2 infection in cancer patients, univariate and multivariate survival analysis were performed based on the Cox proportional hazard model. We randomly divided the 223 patients into a development cohort and a validation cohort according to a ratio of 7:3. Table 2 shows the distribution of the clinical features of the two groups. There was no statistical difference in most clinical characteristics between the two groups, indicating that the baseline levels of the two groups were consistent. With the development cohort data, we performed a univariate Cox analysis of 24 clinical variables and found that gender, dyspnea, PCT, heart rate, white blood cell count, platelet count, neutrophil count, AST, direct bilirubin, total bilirubin, and D-dimer affected patient outcomes (Table 3). Using the mean or median of the continuous variables as the cut-off point, the Kaplan–Meier survival analysis was also performed on the 24 variables. Consistent with the results of univariate Cox analysis, gender, dyspnea, PCT, heart rate, platelet count, and D-dimer were also considered as prognostic factors (Figure S2). Moreover, the multivariate Cox analysis included the variables with p < 0.05 in the univariate Cox analysis, where platelet count (HR = 0.991, p = 0.007) and neutrophil count (HR = 1.047, p = 0008) were considered independent prognostic factors in these patients.
Table 2.
Clinical characteristics of SARS-CoV-2 infected cancer patients in the development and validation cohorts
| Variables | Development cohort (n = 159) | Validation cohort (n = 64) | p value |
|---|---|---|---|
| Age (years) | 62 (55, 70) | 64 (57, 73) | 0.263 |
| Sex | 0.272 | ||
| Female | 80 (50.3) | 27 (42.2) | |
| Male | 79 (49.7) | 37 (57.8) | |
| BMI (kg/m2) | 0.323 | ||
| < 18.5 | 8 (5.0) | 5 (7.8) | |
| 18.5–24.9 | 57 (35.8) | 16 (25.0) | |
| ≥ 25 | 19 (11.9) | 11 (17.2) | |
| Unknown | 75 (47.3) | 32 (50.0) | |
| Smoke | 0.143 | ||
| Never | 117 (73.5) | 53 (82.8) | |
| Previous/present | 42 (26.5) | 11 (17.2) | |
| Anti-tumor therapy | 0.476 | ||
| Continous | 93 (58.5) | 33 (51.6) | |
| Discontinous | 22 (13.8) | 8 (12.5) | |
| Unknown | 44 (27.7) | 23 (35.9) | |
| Basic diseases | 0.069 | ||
| Without | 81 (50.9) | 24 (37.5) | |
| With | 78 (49.1) | 40 (62.5) | |
| Fever | 0.863 | ||
| Without | 39 (24.5) | 15 (23.4) | |
| With | 120 (75.5) | 49 (76.6) | |
| Dyspnea | 0.724 | ||
| Without | 96 (60.3) | 37 (57.8) | |
| With | 63 (39.7) | 27 (42.2) | |
| PCT (ng/ml) | 0.114 | ||
| ≤ 0.5 | 111 (69.8) | 52 (81.2) | |
| > 0.5 | 22 (13.8) | 8 (12.5) | |
| Not application | 26 (16.4) | 4 (6.3) | |
| Heart rate (bpm) | 88 (77, 100) | 87.5 (78, 100) | 0.829 |
| SBP (mmHg) | 129.7 (18.58) | 131.1 (19.09) | 0.622 |
| DBP (mmHg) | 78 (70, 85) | 80 (71, 85) | 0.895 |
| Respiratory rate (braths/min) | 20 (19, 22) | 20 (20, 22.75) | 0.267 |
| Temperature (°C) | 36.7 (36.5, 37.5) | 36.8 (36.5, 38) | 0.798 |
| WBC count (10E9/L) | 5.1 (3.8, 7.08) | 5.65 (4.385, 7.215) | 0.048 |
| Neutrophil count (10E9/L) | 3.35 (2.48, 5.32) | 4.27 (3.01, 5.858) | 0.010 |
| Lymphocyte count (10E9/L) | 0.88 (0.62, 1.44) | 0.96 (0.553, 1.275) | 0.892 |
| Platelet count (10E9/L) | 186.8 (96.71) | 200 (89.69) | 0.340 |
| ALT (U/L) | 21 (12, 35) | 25 (15.25, 38.75) | 0.120 |
| AST (U/L) | 26 (18, 38) | 32.5 (22, 44) | 0.013 |
| Total bilirubin (umol/L) | 11.2 (7.9, 14.8) | 10.85 (8.475, 14.15) | 0.731 |
| Direct bilirubin (umol/L) | 3.9 (2.7, 5.6) | 3.95 (2.8, 5.3) | 0.914 |
| Creatinine (umol/L) | 63 (51, 82) | 71.5 (56, 92.75) | 0.022 |
| D-dimer (mg/L) | 0.86 (0.4, 2.14) | 0.74 (0.33, 1.848) | 0.544 |
BMI body mass index, PCT procalcitonin, SBP systolic blood pressure, DBP diastolic blood pressure, WBC white blood cell, AST aspartate aminotransferase, ALT alanine transaminase
Table 3.
Univariate and multivariate COX analysis of prognosis in the development cohort
| Variables | Univariate Cox analysis | Multivariate Cox analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age (years) | 1.017 (0.983–1.053) | 0.332 | ||
| Sex (male vs. female) | 2.513 (1.042–6.059) | 0.042 | 2.996 (0.861–10.43) | 0.085 |
| BMI | ||||
| 18.5–24.9 vs. < 18.5 | 0.965 (0.119–7.846) | 0.973 | ||
| ≥ 25 vs. < 18.5 | 0.777 (0.070–8.578) | 0.837 | ||
| Unknown vs. < 18.5 | 1.491 (0.671–11.34) | 0.700 | ||
| Smoke (previous/present vs. never) | 1.405 (0.601–3.283) | 0.432 | ||
| Anti-tumor therapy | ||||
| Discontinous vs. continous | 2.313 (0.432–6.769) | 0.126 | ||
| Unknown vs. continous | 2.020 (0.495–4.972) | 0.126 | ||
| Basic_diseases | ||||
| With vs. without | 1.217 (0.821–2.718) | 0.631 | ||
| Fever (with vs. without) | 0.933 (1.072–2.350) | 0.883 | ||
| Dyspnea (with vs. without) | 5.044 (0.198–12.71) | 0.001 | 2.942 (0.934–9.266) | 0.065 |
| PCT | ||||
| > 0.5 vs. ≤ 0.5 | 6.316 (0.158–15.26) | 0.000 | 1.789 (0.598–5.348) | 0.298 |
| Not application vs. ≤ 0.5 | 1.824 (0.548–5.816) | 0.310 | 1.619 (0.445–5.887) | 0.465 |
| Heart rate | 1.051 (1.024–1.079) | 0.000 | 1.018 (0.987–1.050) | 0.264 |
| SBP | 0.993 (0.971–1.016) | 0.545 | ||
| DBP | 0.972 (0.936–1.008) | 0.129 | ||
| Respiratory rate | 1.034 (0.939–1.140) | 0.496 | ||
| Temperature | 0.949 (0.577–1.554) | 0.828 | ||
| WBC count | 1.177 (1.063–1.305) | 0.002 | 1.115 (0.947–1.312) | 0.192 |
| Neutrophil count | 1.035 (1.012–1.059) | 0.003 | 1.047 (1.012–1.084) | 0.008 |
| Lymphocyte count | 1.571 (0.248–1.313) | 0.187 | ||
| Platelet count | 0.993 (0.988–0.998) | 0.006 | 0.991 (0.984–0.997) | 0.007 |
| ALT | 0.993 (0.973–1.013) | 0.500 | ||
| AST | 1.008 (1.001–1.014) | 0.021 | 1.010 (0.998–1.020) | 0.094 |
| Total bilirubin | 1.031 (1.014–1.048) | 0.000 | 0.961 (0.872–1.058) | 0.413 |
| Direct bilirubin | 1.043 (1.022 –1.065) | 0.000 | 1.071 (0.938–1.223) | 0.314 |
| Creatinine | 1.001 (0.999–1.003) | 0.146 | ||
| D-dimer | 1.037 (1.007–1.067) | 0.014 | 1.026 (0.985–1.068) | 0.225 |
Construction and evaluation of the prognostic nomogram
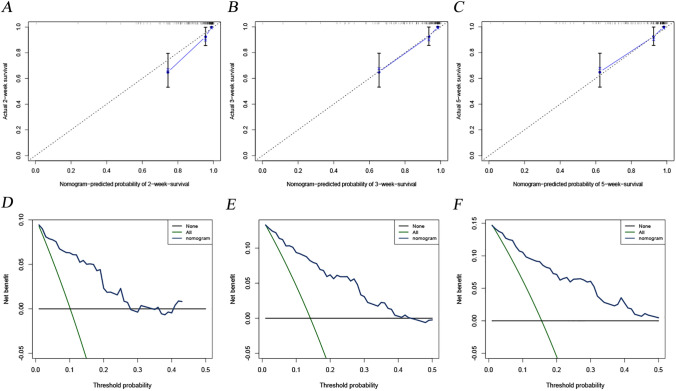
A nomogram was constructed based on the final multivariate Cox analysis. Five factors closely related to prognosis were included in this model: sex, dyspnea, platelet count, neutrophil count and AST. According to the multivariate Cox analysis, each factor (in the nomogram) was assigned a score. We obtained the total score of nomogram from the sum of individual scores of all predictors. Based on the total score, patients’ survival at 2, 3, and 5 weeks could be calculated by projection downward (Fig. 2). In this study, the C-index was 0.841 (95% CI 0.782–0.900) in the development cohort and was 0.780 (95% CI 0.678–0.882) in the validation cohort, indicating that the constructed model had reliable prediction performance. From the calibration curves of the development cohort (Fig. 3a–c) and the verification cohort (Figure S3a–c), it can be seen that the predicted value of the model is in good agreement with the observed value. Moreover, the clinical decision curves of the development (Fig. 3d–f) and validation (Figure S3d–f) cohorts for 2, 3, and 5 weeks also suggested that this nomogram had good clinical application significance.
Fig. 2.
A prognostic nomogram including significant clinical parameters for 2-week, 3-week, and 5-week OS in cancer patients with SARS-CoV-2 infection. By adding the scores obtained by projecting the corresponding ‘Points’ of each variable to the ‘Total Point’ axis, the total score can correspond to the corresponding prediction results
Fig. 3.
Construction and evaluation of the prognostic nomogram. Calibration curves for 2-week (a), 3-week (b) and 5-week (c) OS in the development cohort. It could be seen that all calibration curves are close to the ideal 45° dotted line. This indicates that the predicted value of the model had good consistency with the actual observed value. DCA curves for 2-week (d), 3-week (e) and 5-week (f) OS in the development cohort. In a large range of threshold probability, the net benefit of patients is higher than that of other two extreme cases (all and none), which shows that the nomogram model has good clinical applicability
Development of an online tool to facilitate the clinical application of our constructed model
To make it easier for clinicians to use this model, we created a dynamic web version of nomogram (https://covid-19-prediction-tool.shinyapps.io/DynNomapp/). The interfaces of this web version are shown in Figure S4. On the right side of the interfaces, by inputting the corresponding data of patients and clicking the button at the top of the page, the results would appear. Next, we could obtain the corresponding survival curves, survival probabilities of 2, 3 and 5 weeks and 95% confidence intervals.
Discussion
Although some studies have confirmed that cancer patients were more susceptible to SARS-CoV-2 than the general population (Dai et al. 2020; Liang et al. 2020), the clinical features and short-term prognosis of cancer patients infected with SARS-CoV-2 were still unclear. This study showed that compared to survivors, non-survivors were mostly male and the elder, most of whom suffered from basic diseases and dyspnea. Moreover, the heart rate of the non-survivors was faster than that of the survivors, and the detection values of non-survivors were higher in most laboratory findings, such as white blood cell count, neutrophil count, PCT, CRP, hs-CRP, direct bilirubin, PT, APTT, creatinine, CK-MB, and D-dimer, except for platelet counts and lymphocyte counts. These findings were consistent with previous researches on patients with SARS-CoV-2 infection (Chen et al. 2020a, b; Yang et al. 2020). Although cancer patients infected with SARA-CoV-2 shared common epidemiological characteristics with the general population, they might also have unique clinical features. Thus, this study performed further exploration of cancer patients infected with SARS-CoV-2 to evaluate the potential impact of COVID-19 on cancer patients.
The study of Dai et al. has demonstrated that patients with lung cancer were at the highest risk among patients with solid tumors infected with SARS-CoV-2 (Dai et al. 2020). They suspected that reduced pulmonary reserve and severe infection were responsible for poor outcomes in these patients. In this study, lung cancer patients accounted for the highest proportion of all cancer patients with SARS-CoV-2 infection. Similarly, lung cancer patients remained the most in 37 non-survivors, which might be associated with the high prevalence of lung cancer. The lung is the leading site of SARS-CoV-2 infection-induced lesions, and it remains unclear whether lung cancer patients have a worse prognosis than other cancer patients during the COVID-19 epidemic. In view of this, we divided these patients into hematological malignancies group, lung cancer group and other solid tumors group. Kaplan–Meier survival analysis, univariate and multivariate Cox analysis showed that the prognosis of patients with hematological malignancies was worse than that of patients with lung cancer and other solid tumors. Hematological malignancies in our study included leukemia, diffuse large B-cell lymphoma, acute lymphoblastic leukemia, acute myeloid leukemia, multiple myeloma, waldenström macroglobulinemia, and chronic myeloid leukemia. Because the immune function of malignant or dysfunctional plasma cells, lymphocytes or leukocytes in hematological malignancies was reduced (Lainey et al. 2013; Raab et al. 2009), all hematological malignancies patients were prone to severe infection complications, which might be the main reason for the high mortality rate of hematological cancer patients.
This study revealed that more patients were treated with antibacterial therapy and glucocorticoid therapy in the non-survivors than in the survivors, suggesting that non-survivors were often associated with bacterial infections and often accompanied by severe symptoms. Higher PCT values and neutrophil count for non-survivors also appeared to indicate this. PCT, the procalcitonin peptide synthesized from thyroid C cells and released from leukocytes, is a highly specific indicator of bacterial infection and is closely related to the prognosis of the disease (de Jong et al. 2016). In this study, PCT values of non-survivors were significantly higher than that of survivors, and PCT significantly affected the prognosis of patients. This was in line with the study of Chen et al. (2020a, b). Notably, although the participants of this study (cancer patients with SARS-CoV-2 infection) was different from that of Chen et al. (all patients with COVID-19) (Chen et al. 2020a, b), PCT was considered to significantly affect patient outcomes in both studies. In addition, as the first defense barrier against suppurative infection, neutrophils play an essential role in the defense and protection function of the human body and are increased significantly in patients with bacterial infections. In this study, the median neutrophil count of non-survivors was 6.62*10E9, significantly higher than that of the survivors (3.42*10E9). These findings further suggested that patients with bacterial infections were at a higher risk of death and required more attention from clinicians.
Of the 223 patients included in this study, 133 patients had dyspnea and up to 75.7% of the 37 non-survivors had dyspnea. If not correctly managed, dyspnea may aggravate hypoxia and lead to acute respiratory failure and other serious complications. In this study, dyspnea was identified to affect the prognosis of patients in univariate Cox analysis and Kaplan–Meier survival analysis. As with dyspnea, sex, heart rate, platelet count, D-dimer, and AST were also demonstrated to be important factors affecting patient outcomes. Moreover, platelet count, as well as neutrophil count, was considered independent prognostic factors in these patients. These patients with low platelets had a higher risk of death, similar to ordinary COVID-19 patients (Bi et al. 2020; Lippi et al. 2020). The factors mentioned above could significantly affect the prognosis of patients, so clinicians should focus on the changes in these indicators when managing the patients with SARS-CoV-2 infection. We believe that sufficient identification of prognostic factors in such patients would help clinicians to determine the prognosis of patients in the early stage and adjust treatment strategies timely.
Nomogram, a graphical prediction tool, can use statistical regression to assess the impact of various clinicopathological parameters on the likelihood of events occurring. Compared with the traditional staging system, nomogram has more accurate risk assessment methods, which is helpful to the proper individual treatment of clinical patients (Balachandran et al. 2015). According to the results of the multivariate Cox analysis, we also constructed a prognostic nomogram that accurately predicted the overall survival of patients at 2, 3, and 5 weeks based on individual characteristics of patients. Importantly, the predictive performance and clinical utility of this model were also well-validated in an independent validation cohort. Furthermore, to facilitate clinicians to use the model established in this study, we created a dynamic nomogram on the web. Consequently, clinicians could access the website directly through mobile phone or computer anytime and anywhere, and inputting the corresponding information of patients to predict the survival of patients. This would undoubtedly simplify the application process and facilitate clinical use.
Indeed, this study has the following limitations. First, the nature of retrospective research inevitably led to selection bias. Second, despite our efforts to collect some essential clinical information after feedback, it was inevitable to omit some data that may inform our analysis. In addition, although the study used the patient’s data from 26 clinical centers in Hubei province, Hubei was only one of the epicenter of the outbreak. It was still less representative than the cohort from the whole country or even several countries. Thus, it is still necessary to strengthen the in-depth cooperation among countries as soon as possible and carry out international large-scale or prospective researches.
In conclusion, this study provided evidence that male, dyspnea, elevated PCT, increased heart rate, elevated D-dimers, and decreased platelets were risk factors for cancer patients with SARS-CoV-2 infection. Abnormalities of a series of laboratory findings at admission were more common in non-survivors than survivors. Among cancer patients infected with SARS-CoV-2, compared with patients with solid tumors such as lung cancer, patients with hematological malignancies had a worse prognosis. In addition, the nomogram proposed in this study had good predictive performance, which would assist clinicians in predicting the prognosis of patients early and performing more reasonable and effective treatment strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1: Distribution of underlying diseases among survivors and non-survivors. a Prevalence of underlying diseases of all patients, survivors and non survivors (case number representation). b The percentage of patients with corresponding underlying diseases (for example, 12 out of 37 patients had diabetes, the ratio was 32.4%). It could be seen that the proportion of non-survivors with underlying diseases was higher than that of survivors (TIF 356 kb)
Figure S2: Overall survival for cancer patients with SARS-CoV-2 infection stratified by sex (a), dyspnea (b), PCT (c), heart rate (d), platelet counts (e) and D-dimer (f). Among them, heart rate, platelet counts, and D-dimers were truncated by a median or mean (TIF 1014 kb)
Figure S3: Construction and evaluation of the prognostic nomogram. Calibration curves for 2-week (a), 3-week (b) and 5-week (c) OS in the validation cohort. It could be seen that all calibration curves are close to the ideal 45° dotted line. This indicates that the predicted value of the model had good consistency with the actual observed value. DCA curves for 2-week (d), 3-week (e) and 5-week (f) OS in the validation cohort. In a large range of threshold probability, the net benefit of patients is higher than that of other two extreme cases (all and none), which shows that the nomogram model has good clinical applicability (TIF 797 kb)
Figure S4: Four interfaces (corresponding four modules) of the dynamic version nomogram. a Corresponding ‘Survival Plot’ module. b Corresponding ‘Predicted Survival’ module. c Corresponding ‘Numerical Summary’ module. d Corresponding ‘Model Summary’ module (TIF 1002 kb)
Author contributions
Conception and design: WH, XL and CS. Collection and assembly of data: Data were collected by WH in collaboration with the authors. Data analysis and interpretation: WH, CS and XL. Manuscript writing: CS, HG and XD prepared the first draft of the report. WH, QS and YC co-supervised the project. Final approval of manuscript: WH had full access to all the data and had final responsibility to submit for publication.
Funding
This study was supported by ZhongNan Hospital of Wuhan University Science Technology and Innovation Cultivating Fund (znpy2019088).
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
Editorial support was provided by the ethics committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Approval No.: 20/061-2257).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Congkuan Song, Zhe Dong and Hongyun Gong contributed equally to this study.
Contributor Information
Yuan Chen, Email: chenyuan008@163.com.
Qibin Song, Email: qibinsong@whu.edu.cn.
Weidong Hu, Email: huwd@whu.edu.cn.
References
- Balachandran VP, Gonen M, Smith JJ, DeMatteo RP (2015) Nomograms in oncology: more than meets the eye. Lancet Oncol 16(4):e173–e180. 10.1016/S1470-2045(14)71116-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi XJ, Su ZX, Yan HX, Du JP, Wang JJ, Chen LP et al (2020) Prediction of severe illness due to COVID-19 based on an analysis of initial Fibrinogen to Albumin Ratio and Platelet count. Platelets 31(5):674–679. 10.1080/09537104.2020.1760230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NS, Zhou M, Dong X, Qu JM, Gong FY, Han Y et al (2020a) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395(10223):507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RC, Liang WH, Jiang M, Guan WJ, Zhan C, Wang T et al (2020b) Medical Treatment Expert Group for COVID-19. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest 158(1):97–105. 10.1016/j.chest.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MY, Liu DB, Liu M, Zhou FX, Li GL, Chen Z et al (2020) Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov 10(6):783–791. 10.1158/2159-8290.CD-20-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong E, van Oers JA, Beishuizen A, Vos P, Vermeijden WJ, Haas LE et al (2016) Efficacy and safety of procalcitonin guidance in reducing the duration of antibiotic treatment in critically ill patients: a randomised, controlled, open-label trial. Lancet Infect Dis 16(7):819–827. 10.1016/S1473-3099(16)00053-0 [DOI] [PubMed] [Google Scholar]
- Lainey E, Wolfromm A, Sukkurwala AQ, Micol JB, Fenaux P, Galluzzi L et al (2013) EGFR inhibitors exacerbate differentiation and cell cycle arrest induced by retinoic acid and vitamin D3 in acute myeloid leukemia cells. Cell Cycle 12(18):2978–2991. 10.4161/cc.26016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WH, Guan WJ, Chen RC, Wang W, Li JF, Xu K et al (2020) Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol 21(3):335–337. 10.1016/S1470-2045(20)30096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G, Plebani M, Henry BM (2020) Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta 506:145–148. 10.1016/j.cca.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC (2009) Multiple myeloma. Lancet 374(9686):324–339. 10.1016/S0140-6736(09)60221-X [DOI] [PubMed] [Google Scholar]
- Yang XB, Yu Y, Xu JQ, Shu HQ, Xia JA, Liu H et al (2020) Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8(5):475–481. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Distribution of underlying diseases among survivors and non-survivors. a Prevalence of underlying diseases of all patients, survivors and non survivors (case number representation). b The percentage of patients with corresponding underlying diseases (for example, 12 out of 37 patients had diabetes, the ratio was 32.4%). It could be seen that the proportion of non-survivors with underlying diseases was higher than that of survivors (TIF 356 kb)
Figure S2: Overall survival for cancer patients with SARS-CoV-2 infection stratified by sex (a), dyspnea (b), PCT (c), heart rate (d), platelet counts (e) and D-dimer (f). Among them, heart rate, platelet counts, and D-dimers were truncated by a median or mean (TIF 1014 kb)
Figure S3: Construction and evaluation of the prognostic nomogram. Calibration curves for 2-week (a), 3-week (b) and 5-week (c) OS in the validation cohort. It could be seen that all calibration curves are close to the ideal 45° dotted line. This indicates that the predicted value of the model had good consistency with the actual observed value. DCA curves for 2-week (d), 3-week (e) and 5-week (f) OS in the validation cohort. In a large range of threshold probability, the net benefit of patients is higher than that of other two extreme cases (all and none), which shows that the nomogram model has good clinical applicability (TIF 797 kb)
Figure S4: Four interfaces (corresponding four modules) of the dynamic version nomogram. a Corresponding ‘Survival Plot’ module. b Corresponding ‘Predicted Survival’ module. c Corresponding ‘Numerical Summary’ module. d Corresponding ‘Model Summary’ module (TIF 1002 kb)