Abstract
Background
Exosomal long non-coding RNAs (lncRNAs) have been recognised as promising stable biomarkers in cancers. The aim of this study was to identify an exosomal lncRNA panel for diagnosis and prognosis of esophageal squamous cell carcinoma (ESCC).
Materials and Methods
Exosomes were isolated from serum by ExoQuick Solution. To validate the exosomes, exosomal markers and characterization of nanoparticle were performed. Quantitative real-time PCR was used to measure the levels of lncRNAs in exosomes from ESCC patients and healthy subjects. In the training set, exosomal lncRNA profiles from 404 samples were conducted and established new models by multivariate logistic regression. In the validation set, the diagnostic performance of the panel was further validated in 222 additional individuals with a receiver operating characteristic curve (ROC). Kaplan–Meier and multivariate Cox proportional hazards analysis were applied to assess the correlation between lncRNAs and survival rate of ESCC patients.
Results
A 4-lncRNA panel (UCA1, POU3F3, ESCCAL-1 and PEG10) in exosomes for ESCC diagnosis was developed by logistic regression model. The diagnostic accuracy of panel was evaluated with AUC value of 0.844 and 0.853 for training and validation stage, respectively. The corresponding AUCs for patients with TNM stage I–II and III were 0.820 and 0.935, significantly higher than squamous cell carcinoma antigen (P<0.001), which were 0.652 and 0.642, respectively. Kaplan–Meier analysis indicated that patients with higher level of UCA1 and POU3F3 had lower survival rate (P<0.001). Additionally, POU3F3 might be as an independent prognostic factor for ESCC patients (P=0.004).
Conclusion
These findings suggested that serum exosomal 4-lncRNA panel has considerable value for ESCC diagnosis, and POU3F3 may serve as a novel and independent prognostic predictor in clinical applications.
Keywords: biomarker, diagnosis, exosome, esophageal squamous cell carcinoma; ESCC, long non-coding RNA; lncRNA
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive malignancies worldwide, and the majority of cases mainly occur in China.1 Early detection and timely intervention of ESCC could reduce the mortality and improve the patient outcome. In clinical practice, barium esophagography is a widely used tool in early detection of ESCC. However, this test is not prone to detect minimal lesion due to low sensitivity.2 Endoscopy with histopathological biopsy is the gold-standard technique for diagnosis of ESCC, but they are invasive and uncomfortable for patients, which rendered them impractical for mass cancer screening.3 Currently, squamous cell carcinoma antigen (SCCA) and CYFRA 21-1 are commonly used as tumor markers in management of ESCC patients.4 Owing to lack of sufficiently high sensitivity and specificity, they have limited utility for detection of ESCC. Therefore, novel and reliable molecular biomarkers to complement and improve on current ESCC screening strategies are urgently needed.
Long non-coding RNAs (lncRNAs) of greater than 200 nucleotides in length play a crucial role in regulating gene expression.5,6 It has been observed that lncRNA expression signatures may serve as potential tumor biomarkers for diagnosis and prognosis of ESCC.7 Recently, accumulating studies have revealed that exosomal lncRNAs are enriched and more stable in circulatory system and protected from RNase degradation.8,9 The identification of exosomal lncRNAs in bodily fluids suggested their predictive application in diagnosis or prognosis for different types of cancer.10,11 Intriguingly, our previous work demonstrated that circulating exosomal lncRNAs were used as promising diagnostic indicators in colorectal cancer10 and bladder cancer.12 Nonetheless, until now only little was known about exosomal lncRNA profiles in serum for ESCC diagnosis and prognosis.
Based on previous studies, we performed a careful literature review to search for candidate lncRNAs, which have been reported to be significantly dysregulated in ESCC tissues.13–25 Herein, exosomes from serum of ESCC patients and healthy controls samples were extracted and verified. We conducted a three-stage study to identify serum exosomal lncRNAs for detecting ESCC by evaluating their expression based on quantitative real-time PCR. A 4-lncRNA panel using logistic regression model was established as promising biomarker of ESCC diagnosis. In addition, the correlation between the four lncRNAs and ESCC prognosis was further assessed.
Materials and Methods
Clinical Samples
Thirty-four pairs of ESCC tissue samples and matched normal tissues were derived from surgery patients and used to verify the levels of identified lncRNAs. All these tissues were immediately frozen in liquid nitrogen until total RNA extraction. Additionally, 313 ESCC serum specimens and 313 control individuals without ESCC history were obtained from Qilu Hospital of Shandong University between January, 2012 and April, 2014. In brief, 5 mL of venous blood from each participant was collected by vena puncture before any treatment. Serum was separated via centrifugation at 1,600×g for 10 min at 4°C within 2 hours after collection, followed by a second centrifugation at 16,000×g for 10 min at 4°C to remove residual cells debris. Each serum supernatant was transferred into RNase free tubes and stored at −80°C until use. Written informed consent was obtained from each participant prior to blood and tumor samples collection. The study protocol was approved by the Clinical Research Ethics Committee of Qilu Hospital of Shandong University.
Study Design
Overall, a multiphase, case-control study was designed to investigate the potential of serum exosomal lncRNAs as biomarkers for ESCC patients. Based on previous studies, 24 dysregulated lncRNAs were selected as candidates for diagnostic markers. In screening phase, paired tissues from 34 ESCC and 34 normal controls were chosen to identify candidate lncRNAs by qRT-PCR. The ESCC patients and healthy controls were divided into two stages: the training and validation stage. In the following two sets, the total exosomal RNA was extracted to monitor lncRNAs expression in ESCC and healthy subjects. In the training set, paired sera from 202 ESCC and 202 healthy subjects were assigned to identify lncRNAs from the screening phase. In the validation set, the lncRNA-signature was further confirmed with an additional cohort including 111 ESCC and 111 normal controls. Detailed clinical data are summarized in Supplementary Table S1.
In the validation set, ESCC patients have been followed up at intervals of 3 months during the first 2 years and 6 months up to the fifth year. The date of latest record retrieved was May 31, 2019. The median follow-up time was 42 months (range, 4–79 months).
Serum Exosomes Isolation and RNA Extraction
Exosomes were extracted from serum samples using ExoQuick™ Solution (EXOQ5A-1; SBI System Biosciences, USA) according to manufacturer’s instructions. Briefly, serum was thawed on ice and centrifuged at 3,000×g for 15 min to remove cells and cell debris. Next, 250μL supernatant was mixed with 63μL of ExoQuick Solution and incubated at 4°C for 30 min after up and down mix, followed by centrifugation at 1,500×g for 30min. Then, the supernatant was removed by careful aspiration, followed by another 5 min of centrifugation to remove residual liquid. The exosome-containing pellet was subsequently re-suspended in 250μL phosphate buffered saline (PBS). The final pellets, containing exosomes, were collected for characterization and RNA isolations. Extraction of RNA from exosomes was performed using the commercial miRNeasy Mini Kit (QIAGEN, #217004), and RNA extraction from tissue samples was performed using Trizol (Invitrogen, Carlsbad, CA). All RNA elution steps were carried out at 12,000×g for 15s, and the RNA was finally eluted in 15μL RNase-free ultra pure water.
RT-PCR and Quantitative Real-Time PCR
RT and qPCR kits were used to evaluate the expression of candidate lncRNAs in serum exosomes. The 20μL RT reactions were performed using a PrimeScript® RT reagent kit (Takara, Dalian, China) and incubated for 30 min at 37°C, 5s at 85°C, and then maintained at 4°C. For quantitative real-time PCR, 2µL of diluted RT product was mixed with 12.5μL of 2×SYBR®Premix Ex Taq™, 0.5µL of 50×ROX reference Dye II (Takara, Dalian, China), 2μL forward and reverse primers (10μM), and 8μL nuclease-free water to a final volume of 25μL. All reactions were carried out under the following conditions: 95°C for 30s, followed by 45 cycles of 95°C for 5s and 60°C for 30s. The qRT-PCR was run on CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, USA). At the end of the PCR cycles, melting curve analysis were performed to confirm the specificity of PCR products. All the reactions were carried out in triplicate. The levels of lncRNAs were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the comparative 2 −ΔΔCt method. The primers used in this study are shown in Supplementary Table S2.
Transmission Electron Microscopy (TEM)
The exosomes were resuspended in 250μL PBS and a drop of the suspension was placed on a sheet of parafilm. The grids and samples were incubated for 10 min at room temperature. Then, the grids were removed and excess liquid was drained by touching the grids' edges against a piece of clean filter paper. Samples were fixed with 3% glutaraldehyde for 5 min and washed with double-distilled water 10 times every 2 min, contrasted for 10 min with 4% uranyl acetate. The grids were allowed to dry for several minutes and then examined using a JEM-1200 EX microscope (JEOL, Akishima, Japan) at 80 kilo electron volts.
Nanoparticle Tracking Analysis (NTA)
The size distribution and concentration of exosomes was analyzed by NTA and its corresponding software. Exosomes were diluted (1:1000) in physiological saline until individual nanoparticles could be tracked. We then collected dynamic images and analyzed the concentration and distribution of the exosomes.
Western Blot Analysis
Exosomes were washed once in PBS and the total proteins were isolated from exosome pellets using radioimmunoprecipitation assay (RIPA) lysis buffer. Protein concentration was then measured by BCA Protein Assay kit (Thermo Scientific Pierce). Equal amount of protein (30μg) was loaded on a 10% SDS-PAGE gel and transferred onto nitrocellulose membranes (Life Technologies, Carlsbad, CA, USA) by electroblotting. Subsequently, the membranes were blocked with blocking buffer for 1 hour at room temperature (LI-COR Biosciences, Lincoln, NE, USA), and incubated with rabbit antihuman heat shock protein 70 antibody and CD9 antibody (1:1000; CST, USA) overnight at 4°C.
Analysis of Exosomes by Flow Cytometry
For the immunoprecipitation of exosomes and their subsequent flow cytometry analysis, 4μm-diameter aldehyde or sulphate latex beads (Interfacial Dynamics) were incubated with purified anti-human CD9 antibody (BD Biosciences, #555370) at 22°C overnight, as previously described.26
SCCA Assay
Serum SCCA were measured by chemiluminescent assay with Roche Cobas e601 Analyzer (Roche AG), and the limit of normality were defined as 1.5 ng/mL according to manufacturer’s instruction.
Statistical Analysis
The Kolmogorov–Smirnov test was applied for data analysis with the distribution of each group. Data were presented as median (interquartile range). Comparisons between two groups were performed by nonparametric Mann–Whitney U-tests. Receiver operating characteristic (ROC) curves were established to discriminate ESCC from controls. MedCalc 9.3.9.0 (MedCalc, Mariakerke, Belgium) was used for ROC analysis, Matlab software (Matlab, R2013a) was employed for logistic regression analysis and others were calculated using SPSS version 19.0 software (SPSS, Chicago, IL). Univariate analysis was used to explore clinicopathological factors (i.e., age, gender, tumor stage) related with overall survival (OS). Then, significant clinicopathological factors, which were measured in univariate analysis, were further selected for multivariate analysis to reduce related factors. Cox proportional hazards analysis was used to calculate the hazard ratio (HR) and 95% confidence interval (CI). In addition, Kaplan–Meier survival analysis was applied to test the correlations between expression level and patients’ survival. A two-tailed P<0.05 was considered statistically significant.
Results
Identification of Exosomes in Serum
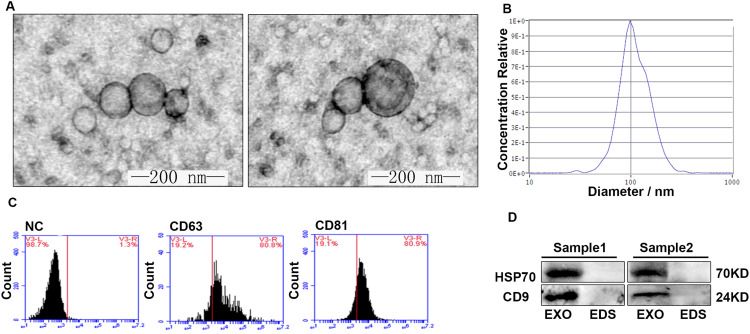
Exosomes were isolated from serum with the ExoQuick kit according to the protocol. To confirm whether exosomes were extracted successfully by the method, exosomes were characterized by TEM. The representative micrograph showed vesicles with round or oval membrane, and a diameter of 30–150 nm under TEM (Figure 1A). The size and concentration of exosomes was measured using NTA. As shown in Figure 1B, the size of most particles was observed at 103.0 nm. The original concentration of 6.6 ×1010 Particles/mL has been isolated. The exosomal markers were analyzed by Flow Cytometry. The results showed that serum-derived exosomes contained enriched proteins such as CD63 and CD81 (Figure 1C). Moreover, we further confirmed the presence of the other exosomal markers (HSP70 and CD9) by Western blot analysis, which showed specific bands in exosomes pellets, but not in exosome-depleted supernatant (EDS) (Figure 1D). Taken together, these observations suggested that the particles characteristics of exosomes and confirmed the efficacy of our protocol for extracting exosomes from serum samples.
Figure 1.
Characterization of exosomes isolated from serum samples. TEM images of exosomes. HV = 100.0kv, Direct Mag: 100,000× (scale bars 200 nm). Exosomes were winkled oval or spherical in shape under TEM (A). Size distribution and concentration of exosomes were analyzed by NTA (B). Flow cytometry detection of surface molecules on exosomes. Exosomes were captured onto anti-CD9 beads and immunostained by monoclonal antibodies against CD63 and CD81 (C). Exosomal protein markers (HSP70 and CD9) detection by Western blot. Purified EXO showed enriched HSP70 and CD9 compared with EDS (D).
Abbreviations: EXO, exosomes; EDS, exosome-depleted supernatant; HV, high voltage; NTA, nanoparticle tracking analysis; TEM, transmission electron microscopy.
Expression Profiles of Serum Exosomal lncRNAs in ESCC Patients
Based on previous studies, 24 dysregulated lncRNAs were selected as candidates for diagnostic markers. In screening phase, we measured the expression levels of these lncRNAs by qRT-PCR in 34 paired ESCC and adjacent normal tissues, and found 20 of them were significantly changed. The other lncRNAs did not show significantly differential expression between ESCC tissues and normal samples (Table 1).
Table 1.
The Selected 24 lncRNA Concentrations in ESCC Tissues Compared with Matched Adjacent Normal Tissues [Median (Interquartile Range)]
| lncRNA | Normal | ESCC | P |
|---|---|---|---|
| SPRY4-IT1 | 0.93 (0.72–1.50) | 1.46 (1.07–1.98) | 0.0018 |
| TUG1 | 1.06 (0.87–1.48) | 1.49 (0.82–2.33) | 0.0244 |
| XLOC013014 | 1.30 (0.60–1.60) | 2.12 (1.39–2.58) | < 0.0001 |
| HOTAIR | 1.08 (0.72–1.51) | 1.48 (1.22–2.18) | 0.0071 |
| MALAT-1 | 1.02 (0.40–1.79) | 1.40 (0.93–2.85) | 0.0268 |
| LET | 1.58 (1.13–2.10) | 1.18 (0.64–1.46) | 0.0064 |
| UCA1 | 0.96 (0.67–1.49) | 3.19 (2.12–4.41) | < 0.0001 |
| PEG10 | 1.10 (0.71–1.42) | 2.23 (1.40–3.28) | < 0.0001 |
| ESCCAL-1 | 1.12 (0.66–1.61) | 4.50 (3.85–5.75) | < 0.0001 |
| POU3F3 | 1.15 (0.55–1.89) | 1.63 (1.06–3.63) | 0.0188 |
| PCAT-1 | 0.95 (0.74–1.53) | 1.43 (0.94–2.13) | 0.0121 |
| CCAT2 | 0.92 (0.61–1.31) | 1.31 (0.94–1.95) | 0.0026 |
| ANRIL | 1.06 (0.64–1.45) | 1.45 (0.95–2.42) | 0.0012 |
| FOXCUT | 1.28 (0.63–1.63) | 1.74 (1.13–2.60) | 0.0021 |
| H19 | 1.00 (0.79–1.52) | 1.50 (1.07–2.20) | 0.0066 |
| ZEB1-AS1 | 1.11 (0.70–1.45) | 0.21 (0.11–0.42) | < 0.0001 |
| TINCR | 1.12 (0.61–1.70) | 1.96 (0.77–2.77) | 0.0017 |
| SOX2OT | 1.12 (0.76–1.57) | 1.42 (0.93–2.35) | 0.0252 |
| PlncRNA-1 | 1.20 (0.66–1.58) | 1.79 (1.17–2.35) | 0.0004 |
| AFAP1-AS1 | 1.18 (0.57–1.89) | 1.59 (1.17–2.94) | 0.0072 |
| NONHSAT112918 | 1.08 (0.71–1.51) | 1.25 (1.08–1.47) | 0.0894 |
| TP73-AS1 | 1.05 (0.67–1.53) | 1.32 (0.79–2.00) | 0.1237 |
| Epist | 1.22 (0.73–1.59) | 1.36 (0.79–1.71) | 0.3235 |
| HNF1A-AS1 | 0.99 (0.68–1.76) | 1.18 (0.62–1.83) | 0.8396 |
Abbreviation: ESCC, esophageal squamous cell carcinoma.
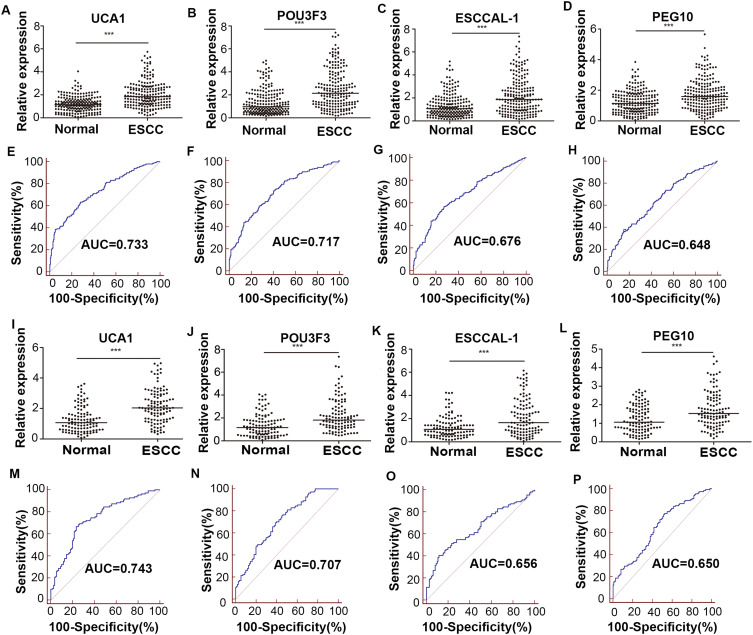
In training phase, the expression levels of 20 candidate lncRNAs were detected in exosomes (202 ESCC patients and 202 controls) by qRT-PCR analysis. Only the lncRNAs with cycle threshold (Ct) value < 35 and detection rate of >75% in the panel were eligible for further analysis. After filtering and selecting these lncRNAs, there were only 4 lncRNAs (POU3F3, UCA1, PEG10 and ESCCAL-1) consistently up-regulated in exosomes of ESCC patients compared with normal controls (P<0.001, Figure 2A–D and Table 2). To estimate the probability of being diagnosed with ESCC, we conduct the separate ROC curves for four lncRNAs (Figure 2E–H). The AUCs for UCA1, POU3F3, ESCCAL-1 and PEG10 were 0.733 (95% confidence interval [CI] =0.687–0.776), 0.717 (95% CI= 0.670–0.760), 0.676 (95% CI=0.628–0.720), 0.648 (95% CI=0.599–0.694), respectively. Therefore, these four lncRNAs were selected for further validation.
Figure 2.
The concentration and diagnostic values of candidate lncRNA biomarkers in exosomes for detecting ESCC. Scatter dot plots of UCA1 (A), POU3F3 (B), ESCCAL-1 (C) and PEG10 (D) lncRNA concentration analyzed by qRT-PCR and ROC curve analysis for detection of ESCC using UCA1 (E), POU3F3 (F), ESCCAL-1 (G), PEG10 (H) in healthy (n=202) and ESCC (n=202) patients’ serum exosomes in the training set. At the same methods, Scatter dot plots of UCA1 (I), POU3F3 (J), ESCCAL-1 (K) and PEG10 (L) lncRNA concentration, ROC curve of UCA1 (M), POU3F3 (N), ESCCAL-1 (O), PEG10 (P) in patients with ESCC (n=111) and control individuals (n=111) in the validation set.***p<0.001.
Abbreviations: ESCC, esophageal squamous cell carcinoma; ESCCAL-1, ESCC associated lncRNA-1; lncRNA, long non-coding RNA; POU3F3, POU class 3 homeobox 3; PEG10, paternally expressed 10; ROC, receiver operating characteristic; UCA1, urothelial carcinoma antigen 1.
Table 2.
Expression of Four lncRNAs in Exosomes of ESCC Patients and Normal Controls in the Training and Validation Set [Median (Interquartile Range)]
| LncRNA | Training Set | Validation Set | ||||
|---|---|---|---|---|---|---|
| Normal (n = 202) | ESCC (n = 202) | P | Normal (n = 111) | ESCC (n = 111) | P | |
| UCA1 | 1.14 (0.75–1.62) | 1.84 (1.20–2.68) | < 0.001 | 1.07 (0.61–1.64) | 2.03 (1.28–2.93) | < 0.001 |
| POU3F3 | 0.97 (0.54–2.01) | 2.14 (1.17–3.57) | < 0.001 | 1.15 (0.54–1.81) | 1.81 (1.22–2.88) | < 0.001 |
| ESCCAL-1 | 1.06 (0.58–1.81) | 1.86 (0.94–3.02) | < 0.001 | 1.07 (0.60–1.64) | 1.65 (0.90–3.11) | < 0.001 |
| PEG10 | 1.14 (0.63–1.73) | 1.59 (0.96–2.24) | < 0.001 | 1.06 (0.66–1.77) | 1.53 (1.04–2.36) | < 0.001 |
Abbreviation: ESCC, esophageal squamous cell carcinoma.
The Stability and Enrichment of Four lncRNAs in Exosomes
Since better stability is a critical prerequisite for tumor markers, we next tested the stability of four lncRNAs in exosomes. A total of 18 ESCC exosomes were divided into five groups. The exosomes were exposed to different conditions including incubation at room temperature or 4°C for 0, 3, 6, 12 and 24h, repeated freeze-thaw (0, 3, 5, 10) cycles, low (pH=1) or high (pH=13) pH solution and RNase A digestion for 3 h at room temperature. The levels of most lncRNAs were not significantly changed in any of experimental conditions (Figure S1A–E) indicating that these lncRNAs were stable in exosomes. However, the Ct values of POU3F3 could detect more than 35 and even not detect when treated for 3h in low PH solution. This data suggests exosomal POU3F3 is not resistant to strong acid for 3h using qRT-PCR analysis.
To explore whether these lncRNAs were enriched enough to be detected, we examined the expression of them in exosomes and EDS from ESCC patients. The average of Cq value for these lncRNAs used the same amount of total RNA. The concentrations of them were significantly higher in exosomes than EDS (P<0.01, Figure S1F), suggesting that these lncRNAs are enriched in exosomes.
Establishing the Predictive lncRNA Panel in ESCC and Control Group
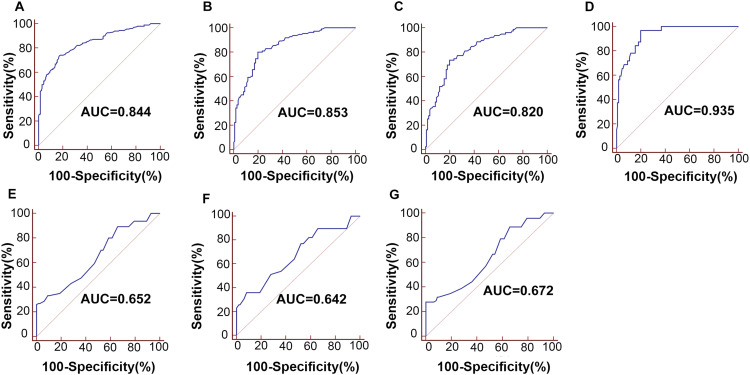
Due to their stability and enriched in exosomes, a logistic regression model was constructed and applied using parameters in the training set. The following equation was used to construct the ROC curve: logit (P=ESCC) = - 1.5035 + 0.3643 × UCA1 + 0.1481 × PEG10 + 0.1778 × ESCCAL-1 + 0.1919 × POU3F3. The AUC for the 4-lncRNA panel in discriminating ESCC patients from healthy controls was 0.844 (95% CI =0.805–0.878; Figure 3A), the sensitivity and specificity for this tumor biomarker was 74.3% and 82.2%, respectively. Our data suggested that the diagnostic value of 4-lncRNA panel was higher than any single lncRNA. Thus, this novel 4-lncRNA panel had high accuracy for predicting ESCC.
Figure 3.
Exploratory comparison of 4-lncRNA panel and serum SCCA for detection of ESCC. ROC curves for detection of ESCC using 4-lncRNA panel in the training set (A) and validation set (B); ROC curves using the 4-lncRNA panel for detection of TNM stage I–II (C) and stage III (D) in validation set; ROC curve analysis using serum SCCA for the detection of ESCC with all stages (E), stage I–II (F) and stage III (G) in the validation set.
Abbreviations: ESCC, esophageal squamous cell carcinoma; lncRNA, long non-coding RNA; ROC, receiver operating characteristic; SCCA, squamous cell carcinoma antigen.
Validation of the Predictive lncRNA Panel
In the validation set, the expression of 4-lncRNA panel was confirmed in an independent cohort including 111 ESCC and 111 normal controls (Figure 2I–L and Table 2). Similarly, the predicted probability was used to construct ROC curves for single lncRNA and the 4-lncRNA panel. The AUCs for UCA1, POU3F3, ESCCAL-1 and PEG10 were 0.743 (95% CI=0.680–0.799), 0.707 (95% CI=0.642–0.760), 0.656 (95% CI=0.590–0.719), 0.650 (95% CI=0.583–0.712), respectively (Figure 2M–P). The AUC of the 4-lncRNA panel was 0.853 (95% CI=0.799–0.897) with sensitivity of 80.20% and specificity of 80.20% (Figure 3B). These results were mostly consistent with the training phase. In addition, the AUCs of panel for the patients diagnosed with ESCC stage I–II and III were 0.820 (95% CI=0.758–0.872, sensitivity=73.40% and specificity=80.20%) and 0.935 (95% CI=0.881–0.969, sensitivity=96.90% and specificity=80.20%), respectively (Figure 3C–D).
The diagnostic performance of panel and SCCA in distinguishing ESCC from controls was compared in the validation set. The AUC of SCCA was 0.652 (95% CI=0.586–0.715, sensitivity=26.10% and specificity=100%) (Figure 3E). The AUCs of the panel for stage I–II and III were significantly higher than those of serum SCCA, which were 0.642 (95% CI=0.568–0.711, sensitivity=27.80% and specificity=100%) and 0.672 (95% CI=0.591–0.747, sensitivity=35.90% and specificity=91.90%), respectively (Figure 3F–G). Therefore, the sensitivity of 4-lncRNA panel was statistically superior to SCCA in discriminating patients with different tumor stages (all at P<0.001).
Expression of Exosomal lncRNAs and Clinicopathological Characteristics
To further explore the potential of exosomal lncRNAs as predictors for ESCC, we evaluated the association between lncRNA levels and clinical characteristics in ESCC patients (Supplementary Table S3). However, there was no significant correlation between the expression of four lncRNAs and the patients’ gender and age (P>0.05). No association was detected between any clinical characteristics and exosomal PEG10 expression (P>0.05). Exosomal UCA1, ESCCAL-1and POU3F3 were significantly correlated with clinical stages. Exosomal UCA1 and POU3F3 expression were higher in ESCC with lymph node metastasis (P<0.01). Among these lncRNAs, POU3F3 and UCA1 were involved in differentiation or T classification of ESCC, respectively (P<0.01). Moreover, the levels of them were elevated in advanced diseases than early stages (P<0.01).
Prognostic Significance of Exosomal lncRNA Expression
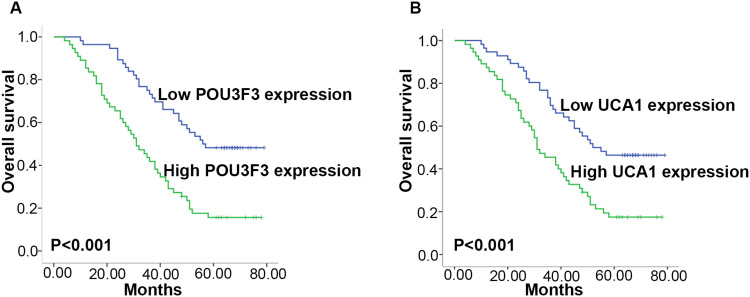
The Kaplan–Meier survival analysis was used to investigate the association between four lncRNAs expression and prognosis of ESCC patients in validation phase. Patients with high UCA1 and POU3F3 expression had a significantly poorer prognosis (P<0.001, Figure 4). Univariate Cox proportional hazards regression model analysis showed that the relative levels of UCA1 (P<0.001) and POU3F3 (P<0.001) expression, as well as tumor stage (P=0.004), lymph node metastasis (P<0.001), and clinical stage (P<0.001) were correlated with the OS rate (Table 3). Parameters significantly associated with OS in the univariate analysis were put into multivariate analysis to identify independent factors for prognosis. Moreover, multivariate analysis revealed that POU3F3 (HR, 2.210; 95% CI=1.293–3.776; P=0.004) and clinical stage (HR, 2.374; 95% CI=1.155–4.881; P= 0.019) were independent prognosis factors that affected OS of ESCC (Table 3).
Figure 4.
Prognostic significance of exosomal lncRNA expression. Kaplan–Meier curves for OS according to the levels of exosomal POU3F3 (A) and UCA1 (B) in the validation set.
Abbreviations: lncRNA, long non-coding RNA; OS, overall survival; POU3F3, POU class 3 homeobox 3; PEG10, paternally expressed 10; UCA1, urothelial carcinoma antigen 1.
Table 3.
Univariate and Multivariate Cox Proportional Hazards Regression Model Analysis of OS in ESCC Patients in the Validation Set
| Parameters | Categories | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age | ≤61 vs. >61 | 0.902 (0.573–1.419) | 0.655 | ||
| Gender | Male vs. Female | 0.764 (0.450–1.299) | 0.320 | ||
| Differentiation | Well vs. Moderate vs. Poor | 1.350 (0.978–1.864) | 0.068 | ||
| Tumor stage | T1-2 vs. T3-4 | 2.257 (1.296–3.930) | 0.004 | 1.525 (0.782–2.974) | 0.216 |
| Lymph node metastasis | N0 vs. N1-3 | 2.303 (1.453–3.652) | <0.001 | 0.928 (0.465–1.852) | 0.832 |
| Clinical Stage | I–II vs. III | 3.280 (2.056–5.234) | <0.001 | 2.374 (1.155–4.881) | 0.019 |
| ESCCAL-1expression | Low vs. High | 1.197 (0.761–1.882) | 0.437 | ||
| PEG10 expression | Low vs. High | 1.053 (0.669–1.656) | 0.823 | ||
| POU3F3 expression | Low vs. High | 2.691 (1.6819–4.307) | <0.001 | 2.210 (1.293–3.776) | 0.004 |
| UCA1 expression | Low vs. High | 2.287 (1.436–3.643) | <0.001 | 1.198 (0.676–2.126) | 0.536 |
Abbreviations: CI, confidence interval; ESCC, esophageal squamous cell carcinoma; HR, hazard ratio; OS, overall survival.
Discussion
In this study, we aimed to investigate circulating exosomal lncRNAs for diagnostic and prognostic significance of ESCC. The aberrantly expressed lncRNAs was screened and identified in ESCC tissues, and explored their expression in a training set of exosomes from ESCC and normal controls. Consequently, a novel 4-lncRNA panel (UCA1, POU3F3, PEG10 and ESCCAL-1) was constructed from logistic regression model to estimate potential diagnostic value in ESCC. These findings demonstrated that circulating exosomal 4-lncRNA signature has significant clinical value for diagnosis of ESCC. Additionally, POU3F3 could be as an independent prognostic factor for ESCC patients.
Several lncRNAs have served as potential diagnostic or prognostic biomarkers in cell-free serum or plasma.27,28 However, these potential biomarkers are often blocked by complexity of bodily fluids. More and more researchers have drawn attention to circulating exosomal noncoding RNA for tumor biomarker. Circulating exosomes containing specific lncRNAs can reflect the characteristics of tumor cells and monitor the tumor progression.29 Emerging evidence8,10,30 has shown that serum-derived exosomal lncRNA profile serves as stable and noninvasive biomarker in various diseases.
To test whether the candidate lncRNAs, UCA1, PEG10, ESCCAL-1 and POU3F3, are mainly existed in exosomes, we compared these lncRNAs expression in exosomes with those in the supernatants. Not surprisingly, we found that their levels were significantly higher in exosomes compared with the supernatants, suggesting that the membrane structures of exosomes could indeed protect lncRNAs from degradation by proteases. Furthermore, we confirmed that these lncRNAs were stable in exosomes. This is consistent with the previous studies which have shown that exosomes in bodily fluids are stable resource of biomarkers. However, the Ct values of POU3F3 could not detect when treated for 3h in low PH solution. The precise mechanism used to explain why circulating exosomal POU3F3 altered after 3 h of exposure to strong acid remains largely unknown. One explanation is that the expression of POU3F3 was actually affected by strong acid. Another possible explanation is that POU3F3 is low abundance, which could not facilitate their detection in serum exosome after treating with low PH. Collectively, we found that most lncRNAs were enriched and stable in exosomes, indicating that exosomal lncRNAs may be suitable for developing diagnostic or prognostic biomarkers of ESCC.
Recent observations have demonstrated that numerous lncRNAs are present in exosomes of cancers and show great potential as non-invasive tumor markers.31 Meanwhile, a panel of lncRNAs in exosomes may improve the sensitivity and specificity for cancer diagnosis or prognosis.11 However, the contribution of exosomal lncRNAs for ESCC detection remains still unclear. Herein we screened ESCC-related lncRNAs based on differential expression profiles, which has been suggested in previous studies. Through the training and validation stages, four lncRNAs of UCA1, ESCCAL-1, PEG10 and POU3F3 were most markedly upregulated in exosomes of ESCC. The expression levels of them were also significantly higher in patients with advanced disease stage. Using ROC analyses, we evaluated the significance of exosomal lncRNAs as valuable diagnostic biomarkers for ESCC. Several lncRNAs have showed high diagnostic values, for example, the AUCs of UCA1 and POU3F3 were 0.733 and 0.717, respectively. Moreover, we used multivariate logistic regression model to analyze the combined 4-lncRNA panel. The AUCs of panel were 0.844 and 0.853 for the training and validation phase, greatly increased compared to the single predictor. In addition, 4-lncRNA panel also had a higher sensitivity (73.40%) to detect stage I–II tumors, while the sensitivity of SCCA was only 27.80% in the same cohort. Based on these findings, exosomal lncRNA panel provides a much more sensitive diagnosis of ESCC, especially for the early stage disease.
Previously, several studies have shown that dysregulation of lncRNAs in ESCC tissues or plasma was significantly correlated with tumor progression. Li et al21 reported that UCA1 could serve as a prognostic marker for ESCC OS. Meanwhile, Hu et al7 found that POU3F3 in plasma was useful in predicting prognosis of ESCC. In this study, we also described the prognostic relevance of these lncRNAs in exosomes of ESCC patients. Overexpression of UCA1 and POU3F3 were significantly correlated with poor OS. Moreover, further analysis using the Cox regression model confirmed that POU3F3 was an independent factor in predicting OS for ESCC patients.
In present study, the 4-lncRNA panel consisted of several lncRNAs that were known to be involved in carcinogenesis and progression of various cancers. For instance, UCA1 has been known to perform various roles in tumorigenesis and progression of different cancers such as colorectal cancer32 and gastric cancer.33 Interestingly, one study suggested that exosomal transfer of UCA1 from the breast cancer cells might increase the tamoxifen resistance,34 which raised the possibility that exosomal UCA1 to exert a crucial role in cancer pathobiology. Li et al35 found that POU3F3 could promote ESCC development through epigenetically modification with Notch signaling pathway. As for lncRNA PEG10, the levels of PEG10 in diffuse large B cell lymphoma (DLBCL) tissues were significantly higher than that in normal.36 Another candidate molecule, ESCCAL-1 in exosomes was increased of ESCC compared with normal samples in our study. This finding was in concordance with the previous reports that ESCCAL-1 might have a putative oncogenic role by increasing cell proliferation and inhibiting apoptosis.37 Overall, previous studies and our results observed a relationship between lncRNAs expression and ESCC progression, but the mechanisms through which cancer cells secreted exosome-containing specific lncRNAs have unknown in detail. Moreover, levels of exosomal lncRNAs in serum/or plasma were not necessary in the same trend as that in tissues. Further studies are required to clarify the relations between exosomal lncRNAs and the tissue origins of exosomes in ESCC.
There may be different underlying mechanisms controlling the noncoding RNAs packing into exosomes, which could be diagnostic or prognostic biomarkers or therapeutic targets for cancer. Recently, Qu et al38 discovered that the protein heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) was specifically bound to lncARSR through a specific sequence and directed its packaging into exosomes. In addition, exosomal lncARSR could disseminate Sunitinib resistance by intercellular transfer. Previous studies have described that the RNA-binding protein SYNCRIP and hnRNPA2B1 may serve as key players in RNA sorting into exosomes. Moreover, evidence proved that SYNCRIP and hnRNPA2B1 displayed different sequence-specificities in RNA exosomes sorting, and a small mutation in RNA can affect its functions. Santangelo et al39 shed light on the mechanism that allows a cell to load exosomes with a specific repertoire of miRNAs, and they found that SYNCRIP could specifically interact with a miRNA motif (GGCU). The other report suggested that sumoylated hnRNPA2B1 could control the sorting of miRNAs into exosomes by binding to specific motifs (GGAG/CCCU).40 Taken together, further studies are needed to investigate whether these four lncRNAs packaged into exosomes in our study are controlled by hnRNPA2B1 or SYNCRIP, which might act not only as promising biomarkers for ESCC detection but also as therapeutic targets to overcome dug resistance in clinical application.
Conclusions
In summary, our study demonstrates that circulating exosomal 4-lncRNA signature has considerable clinical value for ESCC diagnosis, and POU3F3 might serve as a potential prognostic predictor in clinical applications. This discovery of the lncRNA biomarkers in exosomes could open up new avenues for investigating ESCC progression, recovery and therapy response.
Funding Statement
This work was supported by National Natural Science Foundation of China (No. 81501822 and 81472025), Natural Science Foundation of Shandong (ZR2014HP001), Taishan Scholar Program of Shandong Province.
Abbreviations
AUC, area under the receiver operating characteristic curve; ESCC, esophageal squamous cell carcinoma; ESCCAL-1, ESCC associated lncRNA-1; lncRNA, long non-coding RNA; OS, overall survival; POU3F3, POU class 3 homeobox 3; PEG10, paternally expressed 10; ROC, receiver operating characteristic; SCCA, squamous cell carcinoma antigen; UCA1, urothelial carcinoma antigen 1.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. doi: 10.1016/S0140-6736(12)60643-6 [DOI] [PubMed] [Google Scholar]
- 3.Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol. 2012;9(5):278–287. doi: 10.1038/nrclinonc.2012.35 [DOI] [PubMed] [Google Scholar]
- 4.Sunpaweravong S, Puttawibul P, Sunpaweravong P, Nitiruangjaras A, Boonyaphiphat P, Kemapanmanus M. Correlation between serum SCCA and CYFRA 2 1-1, tissue Ki-67, and clinicopathological factors in patients with esophageal squamous cell carcinoma. J Med Assoc Thai = Chotmaihet Thangphaet. 2016;99(3):331–337. [PubMed] [Google Scholar]
- 5.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24(11):651–663. doi: 10.1016/j.tcb.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17(2–3):135–143. doi: 10.1007/s10142-016-0524-x [DOI] [PubMed] [Google Scholar]
- 7.Hu HB, Jie HY, Zheng XX. Three circulating LncRNA predict early progress of esophageal squamous cell carcinoma. Cell Physiol Biochem. 2016;40(1–2):117–125. doi: 10.1159/000452529 [DOI] [PubMed] [Google Scholar]
- 8.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9(3–4):358–367. doi: 10.1002/prca.201400114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38(9):1076–1079. doi: 10.1002/cbin.10301 [DOI] [PubMed] [Google Scholar]
- 10.Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7(51):85551–85563. doi: 10.18632/oncotarget.13465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L, Lin W, Qi P, et al. Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1158–1166. doi: 10.1158/1055-9965.EPI-16-0006 [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Du L, Wang L, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23(2):1396–1405. doi: 10.1111/jcmm.14042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zang W, Wang T, Wang Y, et al. Knockdown of long non-coding RNA TP73-AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget. 2016;7(15):19960–19974. doi: 10.18632/oncotarget.6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei G, Luo H, Sun Y, et al. Transcriptome profiling of esophageal squamous cell carcinoma reveals a long noncoding RNA acting as a tumor suppressor. Oncotarget. 2015;6(19):17065–17080. doi: 10.18632/oncotarget.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao J, Huang JX, Lin M, et al. Microarray expression profile analysis of aberrant long non-coding RNAs in esophageal squamous cell carcinoma. Int J Oncol. 2016;48(6):2543–2557. doi: 10.3892/ijo.2016.3457 [DOI] [PubMed] [Google Scholar]
- 16.Shafiee M, Aleyasin SA, Vasei M, Semnani SS, Mowla SJ. Down-regulatory effects of miR-211 on long non-coding RNA SOX2OT and SOX2 genes in esophageal squamous cell carcinoma. Cell J. 2016;17(4):593–600. doi: 10.22074/cellj.2016.3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Qiu M, Chen Y, et al. Long noncoding RNA, tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus. 2016;29(8):950–958. doi: 10.1111/dote.12436 [DOI] [PubMed] [Google Scholar]
- 18.Wang YL, Bai Y, Yao WJ, Guo L, Wang ZM. Expression of long non-coding RNA ZEB1-AS1 in esophageal squamous cell carcinoma and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol. 2015;8(9):11871–11876. [PMC free article] [PubMed] [Google Scholar]
- 19.Wang PL, Liu B, Xia Y, Pan CF, Ma T, Chen YJ. Long non-coding RNA-low expression in tumor inhibits the invasion and metastasis of esophageal squamous cell carcinoma by regulating p53 expression. Mol Med Rep. 2016;13(4):3074–3082. doi: 10.3892/mmr.2016.4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zang W, Wang T, Huang J, et al. Long noncoding RNA PEG10 regulates proliferation and invasion of esophageal cancer cells. Cancer Gene Ther. 2015;22(3):138–144. doi: 10.1038/cgt.2014.77 [DOI] [PubMed] [Google Scholar]
- 21.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(11):7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. doi: 10.1136/gutjnl-2013-305806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugihara H, Ishimoto T, Miyake K, et al. Noncoding RNA expression aberration is associated with cancer progression and is a potential biomarker in esophageal squamous cell carcinoma. Int J Mol Sci. 2015;16(11):27824–27834. doi: 10.3390/ijms161126060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan D, Wu Y, Hu L, et al. Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis Esophagus. 2017;30(1):1–9. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Shi X, Yang W, et al. Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA sequencing. Tumour Biol. 2016;37(10):13091–13100. doi: 10.1007/s13277-016-5227-3 [DOI] [PubMed] [Google Scholar]
- 26.Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012;59:e3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, He X, Zheng Z, et al. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol Cancer. 2017;16(1):75. doi: 10.1186/s12943-017-0643-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong YS, Wang XW, Zhou XL, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA. 2017;8(4):e1413. doi: 10.1002/wrna.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang N, Pan J, Fang S, et al. Liquid biopsy: circulating exosomal long noncoding RNAs in cancer. Clin Chim Acta. 2019;495:331–337. doi: 10.1016/j.cca.2019.04.082 [DOI] [PubMed] [Google Scholar]
- 31.Cai C, Zhang H, Zhu Y, et al. Serum exosomal long noncoding RNA pcsk2-2:1 as a potential novel diagnostic biomarker for gastric cancer. Onco Targets Ther. 2019;12:10035–10041. doi: 10.2147/OTT.S229033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y, Yang YN, Yuan HH, et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46(5):396–401. doi: 10.1097/PAT.0000000000000125 [DOI] [PubMed] [Google Scholar]
- 33.Zheng Q, Wu F, Dai WY, et al. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17(8):640–646. doi: 10.1007/s12094-015-1290-2 [DOI] [PubMed] [Google Scholar]
- 34.Xu CG, Yang MF, Ren YQ, Wu CH, Wang LQ. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20(20):4362–4368. [PubMed] [Google Scholar]
- 35.Li W, Zheng J, Deng J, et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146(7):1714–1726 e1715. doi: 10.1053/j.gastro.2014.03.002 [DOI] [PubMed] [Google Scholar]
- 36.Peng W, Fan H, Wu G, Wu J, Feng J. Upregulation of long noncoding RNA PEG10 associates with poor prognosis in diffuse large B cell lymphoma with facilitating tumorigenicity. Clin Exp Med. 2016;16(2):177–182. doi: 10.1007/s10238-015-0350-9 [DOI] [PubMed] [Google Scholar]
- 37.Hao Y, Wu W, Shi F, et al. Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer. 2015;15:168. doi: 10.1186/s12885-015-1179-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi: 10.1016/j.ccell.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 39.Santangelo L, Giurato G, Cicchini C, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17(3):799–808. doi: 10.1016/j.celrep.2016.09.031 [DOI] [PubMed] [Google Scholar]
- 40.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]