Abstract
OVATE family proteins (OFPs) are the plant-specific transcription factors, and have significant functions in regulating plant growth, development and resistance. The OFP genes have been investigated in several plants, but they still lack a systematic analysis of OFP genes in Chinese pear and some other five Rosaceae genomes. Here, 28 PbrOFPs were identified within Chinese pear and compared them with those of other five Rosaceae genomes. Evolutionary tree revealed that all OFP genes from six Rosaceae genomes were divided into eight groups. Seventeen conserved microsynteny regions were detected in Chinese pear genome, suggested that these PbrOFP genes might be considered to have originated from the large-scale duplication events., indicating these PbrOFP genes might contain specialized regulatory mechanisms in these tissues, such as flower, ovary and fruit. Remarkably, two PbrOFP genes (Pbr010426.1 and Pbr010427.1) were up-regulated under Venturia nashicola treatment, and five PbrOFP genes were up-regulated under PEG treatment, suggesting that these genes might play crucial roles in defence to environmental stresses. Our data presented a systematic analysis and might aid in the selection of appropriate PbrOFPs for further functional studies in Chinese pear, especially in response to the mechanism of biotic and abiotic stresses.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00866-3) contains supplementary material, which is available to authorized users.
Keywords: OFPs, Chinese pear, Expression, Microsynteny
Introduction
OVATE family protein (OFP), a novel plant-specific transcription factor, belongs to plant growth inhibitory protein and plays important roles in regulating plant growth and development due to a conserved OVATE domain (also known as DUF623) in its C-terminal (Liu et al. 2002; Wang et al. 2007; Wang et al. 2011). OVATE was identified for the first time as a quantitative trait locus, which mainly controls the shape of tomato fruit, and it can also negatively regulate the growth and development of tomato leaves flowers (Liu et al. 2002; Wang et al. 2007; Wang et al. 2011). Recently, many OFPs have been cloned and identified in Arabidopsis, rice and tomato, etc. A total of 18, 31 and 31 of the OFP family members exists in the genomes of Arabidopsis, rice and tomato, respectively, which exert vital effects on plant growth and development (Huang et al. 2013; Yu et al. 2015). Among them, most AtOFPs play a transcriptional inhibitory role in Arabidopsis transient protoplast expression system (Wang et al. 2011). Besides, 31 OsOFPs have different spatial expression profiles, and mainly are expressed at the seed development stage, suggesting that OsOFPs may participate in multiple processes of rice growth and development, especially the seed development (Yu et al. 2015). Three amino acid loop extension (TALE) protein family is a typical ring structure composed of three amino acids, which can specifically interact with other protein molecules. The Yeast Two-Hybrid System showed that nine AtOFPs could interact with TALE homologous domain proteins (Hackbusch et al. 2005). AtOFP1 and AtOFP5 could jointly regulate the subcellular localization of BEL1 like homedomain (BLH1), which is the TALE homologous domain protein. When AtOFP1 and AtOFP5 were co-expressed in the tobacco leaves, BLH1 was relocated from the nucleus to the cytoplasm (Hackbusch et al. 2005). The accumulation of gibberellin can restore the character of shorter shoot tissue length to a certain extent. However, overexpression of AtOFP1 can inhibit the expression of AtGA20ox1 (a gibberellin synthesis gene), leading to the irrecoverable trait (Hackbusch et al. 2005; Wang et al. 2007). AtOFP1 also interacts with AtKu, a DNA double-strand break repair-related gene, to jointly participate in the regulation of DNA double-strand break repair process (Wang et al. 2010). AtOFP5, as a negative regulator of BLH1-KNAT3, plays a role in the early stage of embryo sac development, and they coordinate to regulate embryo sac development (Pagnussat et al. 2007). AtOFP4 is involved in the plant secondary cell wall formation via the interaction with KNAT7 (Li et al. 2011). AtOFP1 and AtOFP4 can mutually regulate the BLH6-KNAT7 multi-protein complex, which is required for the formation of the secondary cell wall (Liu and Douglas 2015). In contrast to OVATEs in tomato, the deletion of AtOFP1, AtOFP4, AtOFP8, AtOFP10, AtOFP15 and AtOFP16 alone showed no growth defects in the mutant Arabidopsis, suggesting that the above genes have functional redundancy (Wang et al. 2011). These studies suggest that OFP may regulate plant growth and development by directly or indirectly affecting the transcriptional regulation of target genes.
The Rosaceae genomes, including Chinese pear (Pyrus bretschneideri), European pear (Pyrus communis), peach (Prunus persica), strawberry (Fragaria vesca), Chinese plum (Prunus mume), and apple (Malus domestica) have been published and released to date (Chagné et al. 2014; Daccord et al. 2017; Shulaev et al. 2011; Verde et al. 2013; Wu et al. 2013). These data lay a foundation for performing a systematic analysis of the OFP genes in these Rosaceae genomes. Here, we identified the OPF family members in Chinese pear and other five Rosaceae genomes, and then analyzed their phylogeny relationships, microsynteny, gene duplication events, and expression patterns. Finally, we found several PbrOFP genes might be associated with Venturia nashicola and drought resistance. These data provide valuable information and insights into the evolution of OFPs in Rosaceae genomes and will contribute to the future functional studies of OFPs in Chinese pear.
Materials and methods
Database search for Chinese pear and other five Rosaceae genomes
The Chinese pear genome with protein, CDS and GFF files were obtained from the GigaDB database (http://gigadb.org/) (Wu et al. 2013). The European pear, apple, strawberry and peach with proteins, CDS and GFF files were downloaded from GDR database (https://www.rosaceae.org/) (Chagné et al. 2014; Daccord et al. 2017; Shulaev et al. 2011; Verde et al. 2013). The Chinese plum with protein, CDS and GFF files obtained from the GitHub (https://github.com/lileiting/prunusmumegenome) (Zhang et al. 2018a, b). The A. thaliana AtOFP genes were obtained and downloaded from TAIR (https://www.arabidopsis.org/). The HMM profile of OFP domain (PF04844) was downloaded from the Pfam database and then used the HMMER 3.1 software to determine OFP genes in Chinese pear and other five Rosaceae genomes (e-value ≤ 1e−3) (Finn et al. 2011). Finally, these OFPs encoded proteins were discarded manually if they lacked a complete or core OFP domain (PF04844) which confirmed by Pfam (Punta et al. 2011), SMART (Letunic et al. 2011), and InterPro (Jones et al. 2014).
Phylogenetic analyses
The multiple alignment of OFP proteins from Chinese pear and other five Rosaceae genomes were executed by MAFFT software (Katoh and Standley 2013). The neighbour-joining (NJ) tree was constructed by MEGA version 5.1 software with the following parameters: Poisson correction, pairwise deletion and a bootstrap test for 1000 replicate (Tamura et al. 2011).
Microsynteny analysis
The MicroSyn software was used to detect microsynteny of OFP genes in Chinese pear and other five Rosaceae genome with a threshold e-value of < 1e−5 (Cai et al. 2011). In this study, we determined a syntenic block which this region including three or more conserved homolog genes were distributed within 15 genes upstream and downstream of OFP genes, as described by Cao et al. (2019b, 2020).
RNA-seq expression analysis
The RNA-seq reads, including ovary, petal, flower, leaves, sepal, bud, stem, and seven pear fruit development stages, were obtained from NCBI (PRJNA49877 and PRJNA309745) (Cao et al. 2019a). The HISAT2 was used to map the paired reads to the Chinese pear genome with defaults parameters (Kim et al. 2015). The StringTie was used to calculate the FPKM (fragments per kilobase of exon model per million reads mapped) values of differently expressed genes (Pertea et al. 2016).
Results and discussion
Identification of OFP genes in Chinese pear and other five Rosaceae genomes
To determine the best candidates of OFP genes in six Rosaceae genomes, we downloaded all AtOFP sequences from TAIR database and then identified the OFP genes in local databases, as described by Cao et al. (2020). Finally, we identified 133 OFP genes in six Rosaceae genomes, including 28 PbrOFPs in Chinese pear, 30 PcOFPs in European pear, 34 MdOFPs in apple, 17 PmOFPs in Chinese plum, 15 PpOFPs in peach, and 9 FvOFPs in strawberry, respectively (Table 1 and Table S1). The numbers of OFPs in Chinese pear, European pear, and apple was more than that in Chinese plum, peach, and strawberry, thus revealing a greater expansion in the OFP genes of Chinese pear, European pear and apple (Table 1). As previously reported, the recent whole-genome duplication events (WGD) (~ 40 MYA) was shared by Chinese pear, European pear and apple, but not appeared in Chinese plum, peach, and strawberry (Chagné et al. 2014; Daccord et al. 2017; Shulaev et al. 2011; Verde et al. 2013; Wu et al. 2013), indicating that the OFP gene family members might have undergone an expansion corresponding to the members of the WGD events.
Table 1.
The genome information for six Rosaceae genomes
| Species | Genome genes | Chromosomes | OFP genes | Gene name |
|---|---|---|---|---|
| Chinese pear (Pyrus bretschneideri) | 42,341 | 34 | 28 | PbrOFPs |
| European pear (Pyrus communis) | 43,419 | 34 | 30 | PcOFPs |
| Apple (Malus domestica) | 63,541 | 34 | 34 | MdOFPs |
| Chinese plum (Prunus mume) | 31,390 | 16 | 17 | PmOFPs |
| Peach (Prunus persica) | 27,864 | 16 | 15 | PpOFPs |
| Strawberry (Fragaria vesca) | 32,831 | 14 | 9 | FvOFPs |
Phylogenetic analysis of OFP genes in Chinese pear and other five Rosaceae genomes
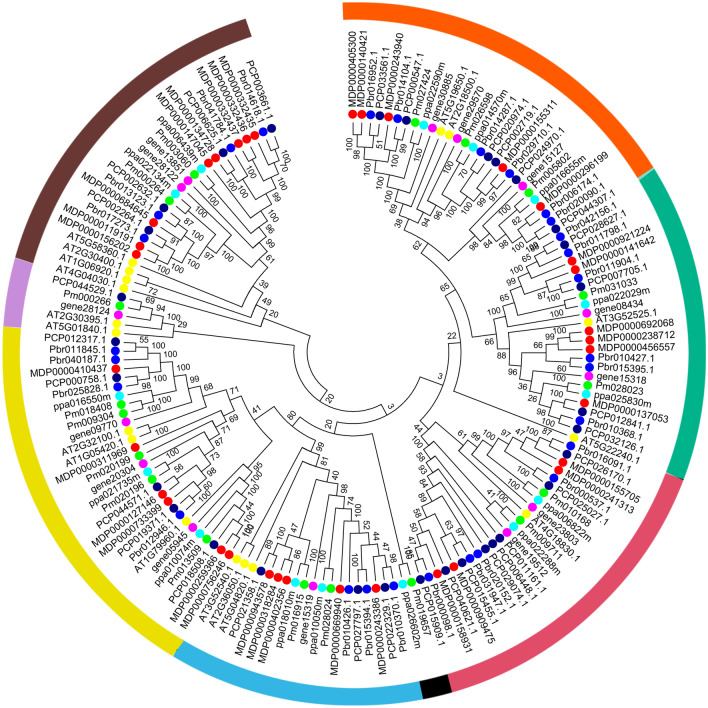
To determine the phylogenetic relationship among the OFP genes in Chinese pear and other five Rosaceae genomes, we constructed the neighbour-joining phylogenetic tree using the total length amino acid sequences of these proteins with 1000 bootstrap values. As shown in Fig. 1, all OFP genes were divided into eight groups, including group-I, group-II, group-III, group-IV, group-V, group-VI, group-VII, and group-VIII. Group-IV had the fewest OFPs (two), while group-VI contained the most OFPs (twenty-nine), followed by group-VIII (twenty-six). Each of these six Rosaceae species contributed at least one OFP to group-I, group-II, group-III, group-V, group-VI and group-VIII, but the members of the group-IV and group-VII included two or four Rosaceae species, indicating that there may be divergent evolutionary mechanisms in different Rosaceae species. Subsequently, we found that the members of OFP gene family from Chinese pear and European pear showed higher similarity according to the genetic distance, which was consistent with the previous reports showing the closer evolutionary relationships of Chinese pear and European pear (Cao et al. 2016). Also, thirty-nine OFP orthologous gene pairs were found among these six Rosaceae genomes, such as Pbr016952.1 and PCP000547.1, and Pm027424 and ppa022590m.
Fig. 1.
The phylogenetic relationships of OFP genes in Chinese pear and other five Rosaceae genomes. The Neighbor-Joining method was used to construct the phylogenetic tree using all OFP genes with 1000 bootstrap replicates. The numbers beside the branches suggest the values of bootstrap that support the adjacent node
Conserved microsynteny of OFP genes in Chinese pear and other five Rosaceae genomes
The genomes of Chinese pear, European pear, and apple had undergone two WGD, including a specific recent WGD (about 30–45 MYA) and an ancient WGD (about 140 MYA) (Chagné et al. 2014; Daccord et al. 2017; Shulaev et al. 2011; Verde et al. 2013; Wu et al. 2013). However, the other Rosaceae genomes of Chinese plum, peach, and strawberry only experienced an ancient WGD (about 140 MYA) (Chagné et al. 2014; Daccord et al. 2017; Shulaev et al. 2011; Verde et al. 2013; Wu et al. 2013). To further understand the expansion and evolution of OFP genes, a performed microsynteny analysis of Chinese pear, European pear, apple, Chinese plum, peach, and strawberry was carried out by MicroSyn software (Cai et al. 2011). In the present study, we defined a WGD or large-scale duplication as these two regions which three or more of the 15 upstream, as well as downstream neighboring genes are homologous pairs.
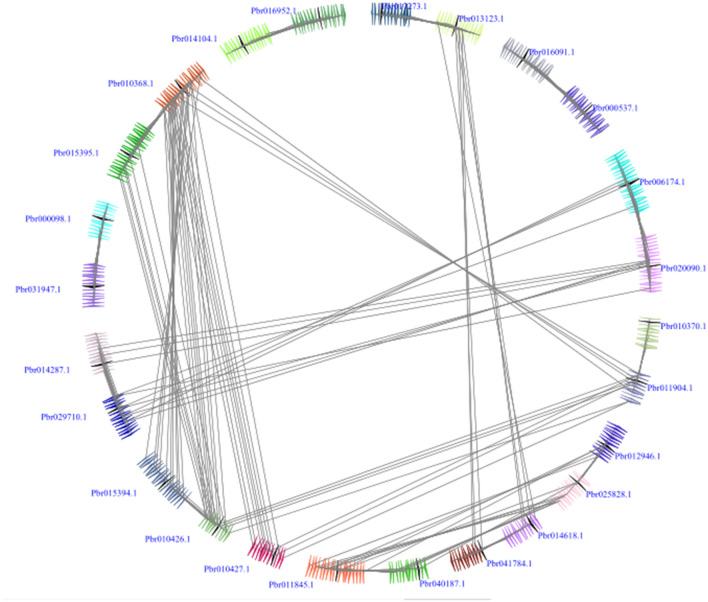
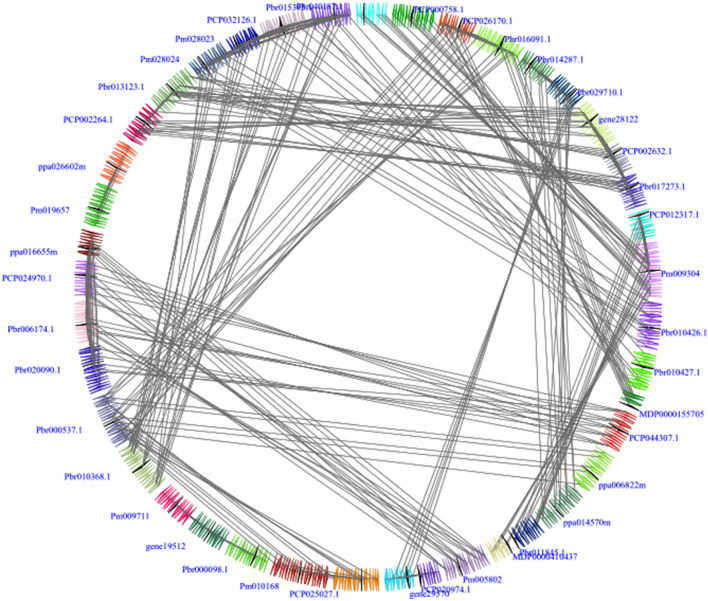
In our study, we detected 17 collinear gene pairs in Chinese pear (Fig. 2). Among them, Pbr010368.1 was collinear with four genes, including Pbr015394.1, Pbr010426.1, Pbr010427.1 and Pbr011904.1. Pbr010426.1 was collinear with three genes, including Pbr015395.1, Pbr010368.1and Pbr011904.1, followed by Pbr029710.1 was collinear with Pbr006174.1 and Pbr020090.1, Pbr011845.1 was collinear with Pbr025828.1 and Pbr0012946.1, Pbr013123.1 was collinear with Pbr041784.1 and Pbr014618.1, Pbr020090.1 was collinear with Pbr014287.1 and Pbr029710.1, and Pbr011904.1 was collinear with Pbr010426.1 and Pbr010368.1 (Table S2). These series of several-for-one collinear relationships indicated that they play key roles in the expansion of the OFP gene family. Interestingly, all these collinear gene pairs contained more than three pairs of conserved flanking genes, indicating that these gene pairs evolved from the large-scale duplication events. Subsequently, we analyzed the interspecies microsynteny between the Chinese pear and European pear, apple, Chinese plum, peach, as well as strawberry (Fig. 3). Among these OFP genes, we scanned 31 conserved syntenic regions, including 13 orthologous gene pairs in the Chinese pear and European pear, 9 gene pairs in Chinese pear and Chinese plum, 4 pairs in Chinese pear and peach, 4 gene pairs in Chinese pear and strawberry, and only one gene pair in Chinese pear and apple (Table S2), which was consistent with the previous papers that showed closer evolutionary relationships of Chinese pear and European pear, versus the Chinese pear and apple/Chinese plum/peach/strawberry (Cao et al. 2016; Wu et al. 2013). The Ka/Ks ratios of OFP genes were calculated between Chinese pear and other five Rosaceae genomes, and these data suggested that these OFP genes were evolving under purifying selection during evolution (Table 2). These data might provide a novel material for studying the OFP genes evolution in different genomes.
Fig. 2.
Extensive microsynteny of OFP genes in Chinese pear. The gene’s orientations and genomic fragments were represented by a series of triangles. The homologous genes on two fragments were connected by grey lines
Fig. 3.
Microsynteny related to OFP families in Chinese pear and other five Rosaceae genomes. The gene’s orientations and genomic fragments were represented by a series of triangles. The homologous genes on two fragments were connected by grey lines
Table 2.
Ka/Ks ratios of OFP genes between Chinese pear and other five Rosaceae genomes
| Gene_1 | Gene_2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Pbr017273.1 | PCP002632.1 | 0.062923687 | 0.154329561 | 0.407722844 |
| Pbr017273.1 | gene28122 | 0.235799225 | 0.702508661 | 0.335653121 |
| Pbr029710.1 | Pbr014287.1 | 0.036366602 | 0.207456604 | 0.175297395 |
| Pbr016091.1 | PCP026170.1 | 0.006452809 | 0.011897143 | 0.542383082 |
| Pbr040187.1 | Pm009304 | 0.142841862 | 0.600583384 | 0.237838518 |
| Pbr015395.1 | PCP032126.1 | 0.107262272 | 0.148685487 | 0.721403779 |
| Pbr015395.1 | Pm028023 | 0.089555799 | 0.823959217 | 0.108689601 |
| Pbr015395.1 | Pm028024 | 0.717059236 | 3.032288451 | 0.236474613 |
| Pbr013123.1 | gene28122 | 0.242829789 | 0.780391728 | 0.31116397 |
| Pbr013123.1 | PCP002264.1 | 0.064801076 | 0.165327892 | 0.391954892 |
| Pbr013123.1 | Pbr017273.1 | 0.066710615 | 0.159215945 | 0.418994558 |
| Pbr006174.1 | Ppa016655m | 0.151979947 | 0.73500298 | 0.2067746 |
| Pbr020090.1 | Ppa016655m | 0.141309608 | 0.634304716 | 0.222778744 |
| Pbr000537.1 | Pbr016091.1 | 0.104971588 | 0.27995433 | 0.374959688 |
| Pbr000537.1 | PCP026170.1 | 0.102081361 | 0.28466833 | 0.358597535 |
| Pbr006174.1 | PCP024970.1 | 0.021926044 | 0.020925507 | 1.047814207 |
| Pbr015395.1 | Pbr010368.1 | 0.026103777 | 0.069682579 | 0.374609799 |
| Pbr010368.1 | PCP032126.1 | 0.040311278 | 0.055865742 | 0.721574205 |
| Pbr000098.1 | gene19512 | 0.219017723 | 0.746997251 | 0.293197495 |
| Pbr000537.1 | Pm010168 | 0.152051512 | 0.520109183 | 0.292345371 |
| Pbr013123.1 | PCP002632.1 | 0.010050402 | 0.020479088 | 0.490764122 |
| Pbr013123.1 | PCP002632.1 | 0.010050402 | 0.020479088 | 0.490764122 |
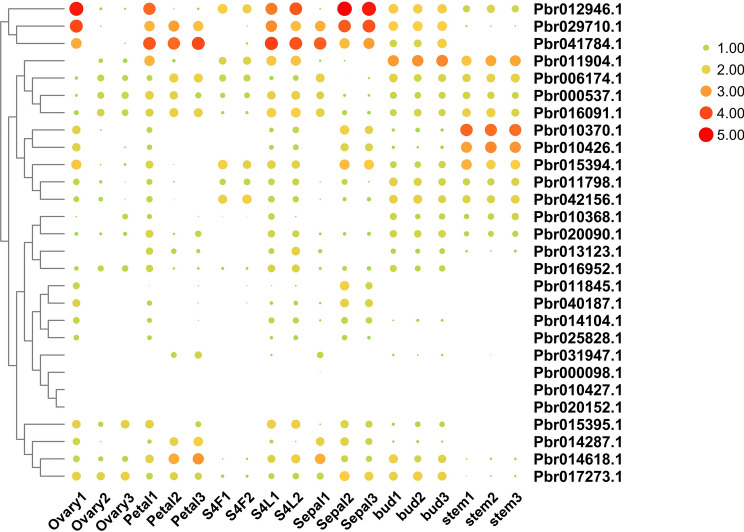
The expression pattern of PbrOFP genes in various tissues
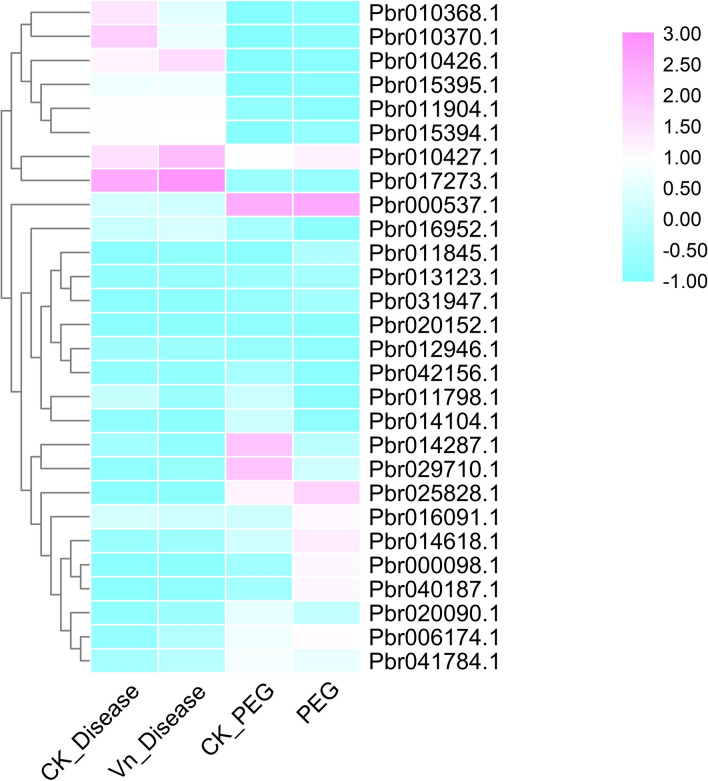
Previously published work suggested that the OFP genes play significant roles in plant growth, development and stress response activities (Huang et al. 2013; Liu et al. 2015; Ma et al. 2017; Schmitz et al. 2015; Wang et al. 2016). Thus, differential expression analysis of PbrOFPs in Chinese pear is helpful for us to find out specialized functions of these genes in plant growth and development and stress response activities from the practical application point of view (Wang et al. 2016). In the present study, we only considered a PbrOFP was expressed if the value of Fragments Per Kilobase per Million (FPKM) was ≥ 1 in an expression atlas according to the previously published work. The results revealed that three PbrOFP genes were not expressed in any tissue, indicating that these PbrOFPs are pseudogenes or these PbrOFPs may be inducible genes (Fig. 4). In general, most of the PbrOFP genes were expressed in ovary, petal, flower, and sepal, which suggested that these genes contained a direct involvement with these tissues in Chinese pear. Interestingly, Pbr010370.1 and Pbr010426.1 were specifically expressed in the stem, and remaining PbrOFPs were diversely expressed in several tissues, suggesting that these PbrOFPs might contribute to these tested tissues of Chinese pear.
Fig. 4.
Expression patterns of PbrOFP genes family in various tissues. The colour bar in each panel represents log2 expression values. The colour scale for each value is present on the upper right corner
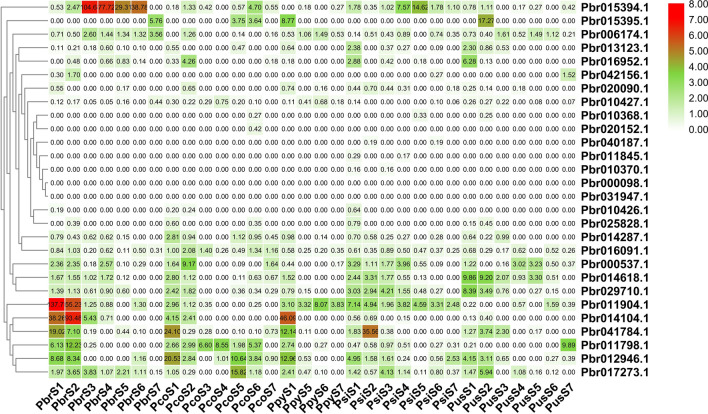
Subsequently, we further investigated the expression pattern of PbrOFPs in five different pear cultivars (i.e. ‘Yali’, ‘Starkrimson’, ‘Hosui’,‘Kuerlexiangli’, ‘Nanguoli’) during fruit development. Among them, some PbrOFP genes were expressed in different developmental stages of these five pear cultivars. The number of PbrOFP genes expressed in ‘Yali’ pear was the largest (15), followed by ‘Starkrimson’ pear (14) and ‘Nanguoli’ pear (14) (Fig. 5). These data suggested that the PbrOFP gene family members contained diverse functions in different pear cultivars. Noteworthy, we found that most of PbrOFP genes were highly expressed in the early stages of fruit development (DAF15 and DAF30), such as Pbr01904.1 and Pbr01401.1, indicating that these PbrOFP genes play important roles in the early stages of fruit development. Additionally, some genes, such as Pbr0161798.1, Pbr016952.1, Pbr015395.1, Pbr014614.1 and Pbr029710.1, were also found to be highly expressed in ‘ Nanguoli ‘ with small fruit shape index, suggesting that these genes might affect the size of fruit during pear fruit development.
Fig. 5.
Gene expression pattern for five different pear cultivars (i.e. ‘Yali’, ‘Starkrimson’, ‘Hosui’,‘Kuerlexiangli’, ‘Nanguoli’). Pbr, Pco, Ppy, Psi, Pus indicates ‘Yali’, ‘Starkrimson’, ‘Hosui’,‘Kuerlexiangli’, ‘Nanguoli’, respectively. The colour bar in each panel represents log2 expression values. The color scale for each value is present on the upper right corner
The expression pattern of PbrOFP genes in response to biotic and abiotic stresses
Environmental stresses, including biotic and abiotic stresses, can affect the regulation of important genes, and finally, influence the health and growth of plants (Ghorbani et al. 2020; Li et al. 2019; Shafiei-Koij et al. 2020). Many stress-related genes can be induced under adverse conditions to help plants cope with stresses (Fan et al. 2018; Kavas et al. 2019). For example, the OFP29 from Oryza sativa reached a peak of expression after 24 h in the ABA treatment (Kavas et al. 2019). The OFP11, OFP14 and OFP20 from apple were upregulated under the NaCl treatment, and OFP6 and OFP14 were upregulated under the mannitol treatment (Li et al. 2019). Drought and V. nashicola is an important factor affecting pear fruit quality and yield. Firstly, we investigated the expressed characteristics of PbrOFPs in pear showing resistance to V. nashicola (Fig. 6). Subsequently, we found that the only two PbrOFP genes (Pbr010426.1 and Pbr010427.1) were up-regulated, and the remaining PbrOFP genes were almost unchanged under V. nashicola treatment. At the same time, we also surveyed the expression pattern of PbrOFPs in pear under drought treatment. Here, the expression levels of five PbrOFPs were higher compared with the control, while two PbrOFPs were down-regulated than that of the control under PEG treatment. Our results suggested that these PbrOFP genes might play key roles in response to biotic and abiotic stresses, and also could increase the understanding of PbrOFP genes resistance to stresses and molecular breeding in Chinese pear.
Fig. 6.
Expression of PbrOFP genes family in response to disease and drought stresses. The colour bar in each panel represents log2 expression values. The colour scale for each value is present on the upper right corner
Conclusion
Our study provided the first comprehensive analysis of OFP in Chinese white pear and compared them with those of other five Rosaceae species, including European pear, apple, Chinese plum, peach and strawberry. We observed the large-scale duplication events in Chinese pear and other five Rosaceae species OFP genes. Transcriptome data analysis of PbrOFP genes exhibited different expression pattern and revealed that these genes might play key roles during pear fruit development. Our data provided the first integrative analysis about these OFP genes and opened up new perspectives for studying the function of these genes in different processes, including different tissues and fruit development in the economical fruit crops (Chinese pear).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
The funding was provided by the Shanxi Province Science Foundation for Youths (Grant No. 201801D221290).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethical approval
This article did not include any experiments with human participants or animals performed by all the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Cai B, Yang X, Tuskan GA, Cheng Z-M. MicroSyn: a user-friendly tool for detection of microsynteny in a gene family. BMC Bioinform. 2011;12:79. doi: 10.1186/1471-2105-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Han Y, Li D, Lin Y, Cai Y, Fips J. MYB transcription factors in Chinese pear (Pyrus bretschneideri Rehd.): genome-wide identification, classification, and expression profiling during fruit development. Front Plant Sci. 2016;7:577. doi: 10.3389/fpls.2016.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li X, Jiang L. Integrative analysis of the core fruit lignification toolbox in pear reveals targets for fruit quality bioengineering. Biomolecules. 2019;9:504. doi: 10.3390/biom9090504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, et al. Integrative analysis reveals evolutionary patterns and potential functions of SWEET transporters in Euphorbiaceae. Int J Biol Macromol. 2019;139:1–11. doi: 10.1016/j.ijbiomac.2019.07.102. [DOI] [PubMed] [Google Scholar]
- Cao Y, Xu X, Jiang L. Integrative analysis of the RNA interference toolbox in two Salicaceae willow species, and their roles in stress response in poplar (Populus trichocarpa Torr. & Gray) Int J Biol Macromol. 2020;162:1127–1139. doi: 10.1016/j.ijbiomac.2020.06.235. [DOI] [PubMed] [Google Scholar]
- Chagné D, et al. The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’) PLoS One. 2014;9:e92644. doi: 10.1371/journal.pone.0092644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord N, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49:1099–1106. doi: 10.1038/ng.3886. [DOI] [PubMed] [Google Scholar]
- Fan C, Guo G, Yan H, Qiu Z, Liu Q, Zeng B. Characterization of Brassinazole resistant (BZR) gene family and stress-induced expression in Eucalyptus grandis. Physiol Mol Biol Plants. 2018;24:821–831. doi: 10.1007/s12298-018-0543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucl Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani R, Zakipour Z, Alemzadeh A, Razi H. Genome-wide analysis of AP2/ERF transcription factors family in Brassica napus. Physiol Mol Biol Plants. 2020 doi: 10.1007/s12298-020-00832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Mller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci. 2005;102:4908–4912. doi: 10.1073/pnas.0501181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genom. 2013;288:111–129. doi: 10.1007/s00438-013-0733-0. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavas M, Kızıldoğan AK, Balık HI. Gene expression analysis of bud burst process in European hazelnut (Corylus avellana L.) using RNA-Seq. Physiol Mol Biol Plants. 2019;25:13–29. doi: 10.1007/s12298-018-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucl Acids Res. 2011;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Wang S, Liu Y, Chen JG, Douglas CJ. Ovate Family Protein4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 2011;67:328–341. doi: 10.1111/j.1365-313X.2011.04595.x. [DOI] [PubMed] [Google Scholar]
- Li H, Dong Q, Zhao Q, Ran K. Genome-wide identification, expression profiling, and protein-protein interaction properties of ovate family proteins in apple. Tree Genet Genom. 2019;15:45. doi: 10.1007/s11295-019-1354-5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Douglas CJ. A role for Ovate family protein1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thaliana. Plant Signal Behav. 2015;10:e1033126. doi: 10.1080/15592324.2015.1033126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. Banana Ovate family protein MaOFP1 and MADS-box protein MuMADS1 antagonistically regulated banana fruit ripening. PLoS ONE. 2015;10:e0123870. doi: 10.1371/journal.pone.0123870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yang C, He Y, Tian Z, Li J, Sunkar R. Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. J Exp Bot. 2017;68:4885–4898. doi: 10.1093/jxb/erx309. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Sundaresan V. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell. 2007;19:3578–3592. doi: 10.1105/tpc.107.054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucl Acids Res. 2011;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz AJ, Begcy K, Sarath G, Walia H. Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 2015;241:177–188. doi: 10.1016/j.plantsci.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Shafiei-Koij F, Ravichandran S, Barthet VrJ, Rodrigue N, Mirlohi A, Majidi MM, Cloutier S. Evolution of Carthamus species revealed through sequence analyses of the fad2 gene family. Physiol Mol Biol Plants. 2020;26:419–432. doi: 10.1007/s12298-019-00739-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I, et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 2013;45:487–494. doi: 10.1038/ng.2586. [DOI] [PubMed] [Google Scholar]
- Wang S, Chang Y, Guo J, Chen JG. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007;50:858–872. doi: 10.1111/j.1365-313X.2007.03096.x. [DOI] [PubMed] [Google Scholar]
- Wang Y-K, Chang W-C, Liu P-F, Hsiao M-K, Lin C-T, Lin S-M, Pan R-L. Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair . Plant Mol Biol. 2010;74:453–466. doi: 10.1007/s11103-010-9685-5. [DOI] [PubMed] [Google Scholar]
- Wang S, Chang Y, Guo J, Zeng Q, Ellis BE, Chen J-G. Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS ONE. 2011;6:e23896. doi: 10.1371/journal.pone.0023896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chang Y, Ellis B. Overview of Ovate family proteins, a novel class of plant-specific growth regulators. Front Plant Sci. 2016;7:417. doi: 10.3389/fpls.2016.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. The genome of the pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Jiang W, Liu Q, Zhang H, Piao M, Chen Z, Bian M. Expression pattern and subcellular localization of the ovate protein family in rice. PLoS ONE. 2015;10:e0118966. doi: 10.1371/journal.pone.0118966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, et al. The genetic architecture of floral traits in the woody plant Prunus mume. Nat Commun. 2018;9:1–12. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.