Abstract
RNAs are a promising class of therapeutics given their ability to regulate protein concentrations at the cellular level. Developing safe and effective strategies to deliver RNAs remains important for realizing their full clinical potential. Here, we develop lipid nanoparticle formulations that can deliver short interfering RNAs (for gene silencing) or messenger RNAs (for gene upregulation). Specifically, we study how the tail length, tail geometry, and linker spacing in diketopiperazine lipid materials influences LNP potency with siRNAs and mRNAs. Eight lipid materials are synthesized, and 16 total formulations are screened for activity in vitro; the lead material is evaluated with mRNA for in vivo use and demonstrates luciferase protein expression in the spleen. In undertaking this approach, not only do we develop synthetic routes to delivery materials, but we also reveal structural criteria that could be useful for developing next-generation delivery materials for RNA therapeutics.
Graphical Abstract

Short interfering RNAs (siRNAs) and messenger RNAs (mRNAs) can regulate intracellular protein concentrations by either silencing (siRNA) or upregulating (mRNA) genes at the post-transcriptional level.[1] Their use as therapeutic entities, however, is limited due to challenges with their intracellular delivery.[2] When RNAs are administered intravenously, for example, circulating nucleases in the blood stream can cause degradation, pattern recognition receptors can trigger an immune response, and non-specific uptake into off-target tissues can limit their clinical translatability.[3] Additionally, cellular membranes are largely impermeable to naked RNAs given their size and charge, limiting passive diffusion of therapeutic RNAs into the cytoplasm of target cells.[2a] Towards this end, there is interest in developing delivery materials to improve the therapeutic potential of RNAs, with possible applications ranging from diabetes and cancer management to protein replacement therapies and immune tolerization, amongst others.[4]
RNAs can be formulated with ionizable lipids, a class of amphiphilic amine-based small molecules that can reversibly bind RNAs via electrostatic interactions.[5] This complexation can include other accessory excipients to form lipid nanoparticles (LNPs), a class of non-viral nucleic acid delivery vehicle with demonstrated efficacy in animals and humans.[6] Questions still remain as to how the chemical structure of the ionizable lipid affects the overall delivery properties of the LNP which can aid in the design and creation of next-generation RNA delivery materials.[3, 5a, 6b, 6h, 7] Specifically, the influence of tail length, tail geometry, and linker spacing on LNP potency have not yet been explored for degradable diketopiperazine lipids with both siRNA and mRNA cargoes.[6d]
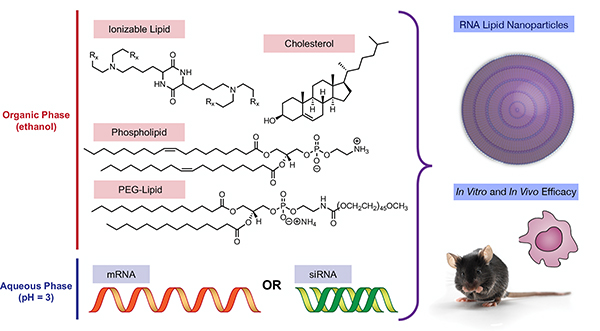
Towards this end, we hypothesized that we could systematically vary ionizable lipid structure when formulating siRNA and mRNA formulations to better understand the delivery properties of LNPs (Figure 1). Our protocol would involve three steps: first, we would synthesize a series of ionizable lipids of precisely varied molecular structure around a previously reported lead material; second, we would formulate each of these ionizable lipid materials into both siRNA- and mRNA-containing LNPs; and third, we would evaluate the potency of each resultant siRNA and mRNA LNP, correlating the protein-knockdown or expression of each LNP system with the structure of the synthesized ionizable lipids.
Figure 1.
Medicinal chemistry approaches for evaluating the effect of ionizable lipid structure on LNP function in siRNA and mRNA LNPs
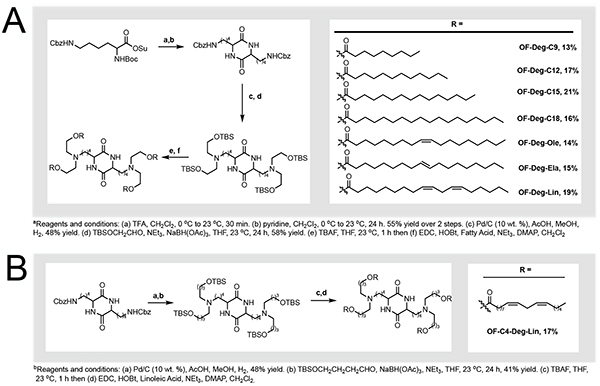
We synthesized a series of degradable diketopiperazine ionizable lipid materials (Figure 2). For our small molecule design criteria, we sought to incorporate three structural motifs: i. bis-lysine diketopiperazine based amine cores, as they have yielded potent synthetic intermediates for RNA delivery in the past including OF-Deg-Lin;[6b–d] ii. ester-based linkages, a degradable chemical functional group which can improve tolerability;[6d, 6g] and iii. a common core approach, where a single synthetic intermediate could be chemically derivatized into multiple ionizable lipids using identical methodologies and starting materials. The resultant series of eight ionizable lipids spans several areas of chemical space including variations in tail lengths, tail geometry, degree of tail unsaturation, and carbon linker lengths (C2 vs. C4).
Figure 2.
a) Synthesis of OF-Deg-CX derivatives, a series of degradable diketopiperazine lipids with varied tail lengths, tail geometries, and total number of unsaturations. b) Synthesis of OF-C4-Deg-Lin, an ionizable lipid with varied linker lengths.
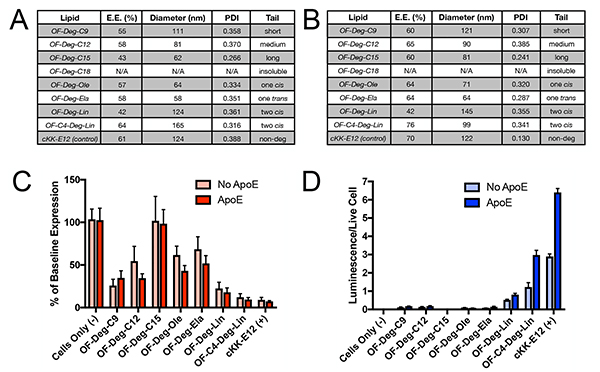
Although RNAs can be directly complexed with ionizable lipids, their potency is superior when they are formulated into LNPs containing cholesterol, lipid anchored polyethylene glycol, and a phospholipid.[2a, 6a, 8–10] Accordingly, each ionizable lipid was then formulated in turn using microfluidic mixing approaches with either siRNA against firefly luciferase or mRNA coding for firefly luciferase while keeping the molar ratio of all LNP components constant and only varying the identity of the ionizable lipid in each formulation (Figure 1).[6a, 6d, 6g] In using this approach, we were able to successfully formulate each ionizable lipid into LNPs, with both siRNA and mRNA cargoes, with the exception of the ionizable lipid OF-Deg-C18. We attribute this finding to a limited solubility profile of OF-Deg-C18 in ethanol, the organic solvent for our LNP formulations. cKK-E12, which shares the same diketopiperazine core as our new lipids and has previously been established as a potent nucleic acid delivery vehicle, was selected as a positive control and also formulated with siRNA and mRNA.[6b, 6c] The size, polydispersity index, and entrapment efficiency for each of the siRNA and mRNA LNPs were then determined and tabulated for siRNA and mRNA LNPs composed of each ionizable lipid (Figure 3A for siRNA; Figure 3B for mRNA). In analyzing these data, there are two apparent trends. First, the nanoparticle diameter for both siRNA and mRNA LNPs decreased as the length of alkyl tails in the ionizable lipid increases (see OF-Deg-C9, OF-Deg-C12, and OF-Deg-C15). Second, the cis/trans geometry of the tails had minimal impact on the LNP properties for singly unsaturated ionizable lipid tails independent of the nucleic acid cargo (see OF-Deg-Ela vs. OF-Deg-Ole for both siRNA and mRNA).
Figure 3.
a) LNP size and entrapment efficiencies for Luciferase siRNA cargoes measured by dynamic light scattering and Quant-IT RiboGreen RNA Assay Kits. b) LNP size and entrapment efficiencies for Luciferase mRNA cargoes measured by dynamic light scattering and Quant-IT RiboGreen RNA Assay Kits c) Knockdown evaluation of a target luciferase gene in HeLa cells with 50 ng doses of OF-Deg-CX siRNA LNPs per well (n=5). d) Luciferase protein expression in HeLa cells with 50 ng doses of OF-Deg-CX mRNA LNPs per well (n=5)
Having established that our series of ionizable lipid materials could indeed be formulated into siRNA and mRNA LNPs, our efforts were then focused on evaluating their in vitro potency. Given that Apolipoprotein E (ApoE) has been shown to impact the efficacy of some LNPs,[6c] we sought to compare cellular transfection of both our siRNA and mRNA LNPs in the presence of additional ApoE than is endogenously present in media. Towards this end, HeLa cells that stably expressed two reporter luciferase proteins (Luciferase – the target gene, and Renilla – the control gene) were treated with a 50 ng dose of LNPs containing the anti-Firefly luciferase siRNA sequence per well. After 24 hours, the normalized firefly luciferase activities were calculated and plotted as the percent knockdown of the luciferase gene relative to baseline expression levels (Figure 3C).[6g] In a similar fashion, mRNA potency was evaluated by treating non-reporter HeLa cells with a 50 ng dose of LNPs containing mRNA coding for luciferase protein per well. After 24 hours, the luminescence was quantified (Figure 3D).[6d] It is important to note that, at the doses studied in this experiment, limited toxicity was observed as there was minimal reduction in Renilla luciferase levels (siRNA, Figure S1) and a Promega cell viability kit (mRNA, Figure S2) respectively.
In analyzing the siRNA and mRNA in vitro data, several trends emerge. From a general perspective, it was observed that the chemical structure of the ionizable lipid was more critical for biological effect for mRNA delivery than for siRNA delivery within this series of LNPs at this dose. Indeed, only doubly unsaturated tail derivatives OF-Deg-Lin and OF-C4-Deg-Lin resulted in meaningful levels of protein production relative to the signal observed in untreated cells. By contrast, only one ionizable lipid (OF-Deg-C15) did not result in significant levels of silencing using anti-luciferase siRNA relative to untreated cells. As a second point, the presence of ApoE affected potency more for mRNA LNPs than for siRNA LNPs. For example, the presence of ApoE for OF-Deg-Lin and OF-C4-Deg-Lin increased the total amount of protein expression by approximately 140%, whereas for siRNA delivery it had minimal impact. This result suggests that these LNPs may undergo different cellular uptake mechanisms. Moreover, increasing the linker length from a two carbon to a four carbon aliphatic chain increased the potency of both siRNA and mRNA LNPs (see OF-Deg-Lin and OF-C4-Deg-Lin). Finally, increasing the tail length of fully aliphatic systems decreased the potency of the siRNA-LNPs (OF-Deg-C9, OF-Deg-C12, and OF-Deg-C15). While it is difficult to discern why these specific trends are observed, this empirical data may be useful in generating next generation materials for RNA delivery given that chemists can use empirical structural criteria to rationally design new substrates.[11]
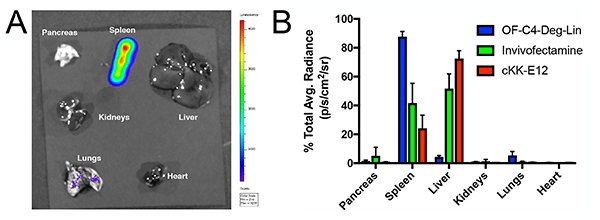
Having established that our systems are functional in vitro, we sought to test a representative LNP formulation with a preliminary study in mice. The in vivo delivery of RNA therapeutics has application in cancer therapy, protein replacement therapies, immune tolerization, and vaccine development, amongst others.[2b, 5b, 6e, 12] Towards this end, we selected OF-C4-Deg-Lin, the most potent ionizable lipid for both siRNA and mRNA delivery in vitro, to serve as our initial platform for in vivo studies. OF-C4-Deg-Lin was first formulated with cyanine 5 (Cy5) labeled mRNA to assess particle distribution. Following intravenous injection, euthanasia, organ resection and fluorescence quantification, it was revealed that these particles predominantly distribute to the liver (Figure S3). By contrast, OF-C4-Deg-Lin LNPs entrapping mRNA coding for luciferase induce the majority of protein expression in the spleen, with minimal translation in the liver, and negligible translation in other organs (Figure 4A). Indeed, quantification of this data suggests that more than 85% of luminescence occurred within the spleen (Figure 4B). This expression profile is unique as compared to those of other mRNA delivery systems including cKK-E12 and Invivofectamine [Figure 4B (quantification), Figure S4 (resected organs)]. These formulations were selected as positive controls given their literature precedent[6b, 6d] and commercial availability respectively, and unlike the OF-C4-Deg-Lin system, both of these positive controls induce the majority of protein expression in the liver.[6b] Nevertheless, the expression profile for OF-C4-Deg-Lin mRNA LNPs is consistent with some other published formulations that also target the spleen, but not others that instead target the liver and lungs.[6a, 6b, 6d, 6f, 13] Future work will aim to study the mechanism of why this biodistribution profile is observed and will also aim to explore molecular cargoes beyond firefly luciferase.
Figure 4.
a) Representative in vivo expression profile of luciferase mRNA OF-C4-Deg-Lin LNPs in mice following luciferin injection and euthanasia at a 0.75 mg/kg intravenous dose (n=3) b) Quantification of luciferase expression per organ as measured by percent of total average radiance for OF-C4-Deg-Lin, cKK-E12, and Invivofectamine luciferase mRNA formulations at 0.75 mg/kg intravenous doses (n=3).
In summary, we have designed, synthesized, and biologically evaluated a series of ionizable lipid materials for the in vitro and in vivo LNP mediated delivery of model siRNA and mRNA cargoes. These new materials are not only capable of delivering RNAs, but also provide an empirical set of data from which scientists can better design future generations of ionizable lipid materials for RNA delivery.
Supplementary Material
Footnotes
The authors declare no competing financial interests.
ASSOCIATED CONTENT
Supporting information is available free of charge via the Internet at www.onlinelibrary.wiley.com
REFERENCES
- [1].a) Kanasty R, Dorkin JR, Vegas A, Anderson D, Nat Mater 2013, 12, 967–977 [DOI] [PubMed] [Google Scholar]; b) Kauffman KJ, Webber MJ, Anderson DG, J Control Release 2016, 240, 227–234. [DOI] [PubMed] [Google Scholar]
- [2].a) Whitehead KA, Langer R, Anderson DG, Nat Rev Drug Discov 2009, 8, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sahin U, Kariko K, Tureci O, Nat Rev Drug Discov 2014, 13, 759–780. [DOI] [PubMed] [Google Scholar]
- [3].Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Nat Rev Genet 2014, 15, 541–555. [DOI] [PubMed] [Google Scholar]
- [4].Kaczmarek JC, Kowalski PS, Anderson DG, Genome Med 2017, 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Fenton OS, Olafson KN, Pillai PS, Mitchell MJ, Langer R, Advanced Materials 2018, 30, 1705328. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kesharwani P, Gajbhiye V, Jain NK, Biomaterials, 2012, 33, 7138–7150. [DOI] [PubMed] [Google Scholar]
- [6].a) Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, Fenton OS, Anderson DG, Nano Lett 2015, 15, 7300–7306 [DOI] [PubMed] [Google Scholar]; b) Fenton OS, Kauffman KJ, McClellan RL, Appel EA, Dorkin JR, Tibbitt MW, Heartlein MW, DeRosa F, Langer R, Anderson DG, Adv Mater 2016, 28, 2939–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AK, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG, Proc Natl Acad Sci U S A 2014, 111, 3955–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Fenton OS, Kauffman KJ, Kaczmarek JC, McClellan RL, Jhunjhunwala S, Tibbitt MW, Zeng MD, Appel EA, Dorkin JR, Mir FF, Yang JH, Oberli MA, Heartlein MW, DeRosa F, Langer R, Anderson DG, Adv Mater 2017, 29. [DOI] [PubMed] [Google Scholar]; e) Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu sH., Siegwart DJ, Angew Chem Int Ed Engl 2017, 56, 1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Yan Y, Xiong H, Zhang X, Cheng Q, Siegwart DJ, Biomacromolecules 2017, 18, 4307–4315 [DOI] [PubMed] [Google Scholar]; g) Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG, Nat Commun 2014, 5, 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Morrison C, Nat Rev Drug Discov 2018, 17, 156–157. [DOI] [PubMed] [Google Scholar]
- [7].Dong Y, Dorkin JR, Wang W, Chang PH, Webber MJ, Tang BC, Yang J, Abutbul-Ionita I, Danino D, DeRosa F, Heartlein M, Langer R, Anderson DG, Nano Lett 2016, 16, 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lu JJ, Langer R, Chen J, Mol Pharm 2009, 6, 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mui BL, Tam YK, Jayaraman M, Ansell SM, Du X, Tam YY, Lin PJ, Chen S, Narayanannair JK, Rajeev KG, Manoharan M, Akinc A, Maier MA, Cullis P, Madden TD, Hope MJ, Mol Ther Nucleic Acids 2013, 2, e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zuhorn IS, Bakowsky U, Polushkin E, Visser WH, Stuart MC, Engberts JB, Hoekstra D, Mol Ther 2005, 11, 801–810. [DOI] [PubMed] [Google Scholar]
- [11].Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ, Nat Biotechnol 2010, 28, 172–176. [DOI] [PubMed] [Google Scholar]
- [12].a) Pack DW, Hoffman AS, Pun S, Stayton PS, Nat Rev Drug Discov 2005, 4, 581–593 [DOI] [PubMed] [Google Scholar]; b) Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP, Proc Natl Acad Sci U S A 1995, 92, 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kozielski KL, Tzeng SY, Green JJ, Chem Commun (Camb) 2013, 49, 5319–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kozielski KL, Tzeng SY, De Mendoza BA, Green JJ, ACS Nano 2014, 8, 3232–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Tzeng SY, Hung BP, Grayson WL, Green JJ, Biomaterials 2012, 33, 8142–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG, J Intern Med 2010, 267, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Merhautova J, Vychytilova-Faltejskova P, Demlova R, Slaby O, Physiol Res 2016, 65, S481–S488 [DOI] [PubMed] [Google Scholar]; h) Pardi N, Secreto AJ, Shan X, Debonera F, Glover J, Yi Y, Muramatsu H, Ni H, Mui BL, Tam YK, Shaheen F, Collman RG, Kariko K, Danet-Desnoyers GA, Madden TD, Hope MJ, Weissman D, Nat Commun 2017, 8, 14630. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Oberli MA, Reichmuth AM, Dorkin JR, Mitchell MJ, Fenton OS, Jaklenec A, Anderson DG, Langer R, Blankschtein D, Nano Lett 2017, 17, 1326–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, Griese M, Bittmann I, Handgretinger R, Hartl D, Rosenecker J, Rudolph C, Nat Biotechnol 2011, 29, 154–157 [DOI] [PubMed] [Google Scholar]; k) Ramaswamy S, Tonnu N, Tachikawa K, Limphong P, Vega JB, Karmali PP, Chivukula P, Verma IM, Proc Natl Acad Sci U S A 2017, 114, E1941–E1950 [DOI] [PMC free article] [PubMed] [Google Scholar]; l) Wang N, Butler JP, Ingber DE, Science 1993, 260, 1124–1127. [DOI] [PubMed] [Google Scholar]
- [13].a) Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC, Meng M, Fritz D, Vascotto F, Hefesha H, Grunwitz C, Vormehr M, Husemann Y, Selmi A, Kuhn AN, Buck J, Derhovanessian E, Rae R, Attig S, Diekmann J, Jabulowsky RA, Heesch S, Hassel J, Langguth P, Grabbe S, Huber C, Tureci O, Sahin U, Nature 2016, 534, 396–401 [DOI] [PubMed] [Google Scholar]; b) Kaczmarek JC, Patel AK, Kauffman KJ, Fenton OS, Webber MJ, Heartlein MW, DeRosa F, Anderson DG, Angew Chem Int Ed Engl 2016, 55, 13808–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) McKinlay CJ; Benner NL; Haabeth OA; Waymouth RM; Wender PA Proc. Natl. Acad. Sci. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) McKinlay CJ; Vargas JR; Blake TR; Hardy JW; Kanada M; Contag CH; Wender PA; Waymouth RM Proc. Natl. Acad. Sci. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.