Abstract
Objectives:
To investigate potential diagnostic model for predicting benign or malignant status of subcentimeter pulmonary ground-glass nodules (SPGGNs) (≤1 cm) based on CT texture analysis.
Methods:
A total of 89 SPGGNs from 89 patients were included; 51 patients were diagnosed with adenocarcinoma, and 38 were diagnosed with inflamed or infected benign SPGGNs. Analysis Kit software was used to manually delineate the volume of interest of lesions and extract a total of 396 quantitative texture parameters. The statistical analysis was performed using R software. The SPGGNs were randomly divided into a training set (n = 59) and a validation set (n = 30). All pre-normalized (Z-score) feature values were subjected to dimension reduction using the LASSO algorithm,and the most useful features in the training set were selected. The selected imaging features were then combined into a Rad-score, which was further assessed by ROC curve analysis in the training and validation sets.
Results:
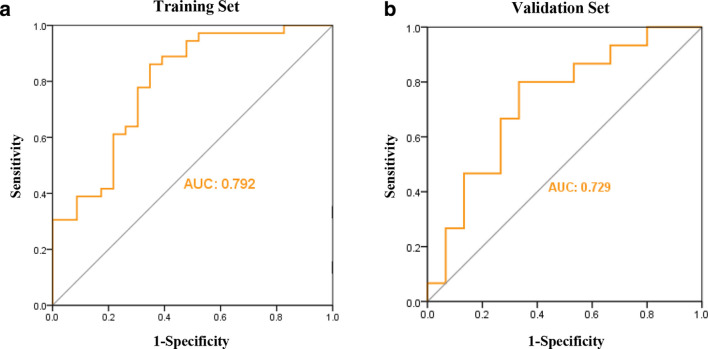
Four characteristic parameters (ClusterShade_AllDirection_offset4_SD, ShortRunEmphasis_angle45_offset1, Maximum3DDiameter, SurfaceVolumeRatio) were further selected by LASSO (p < 0.05). As a cluster of imaging biomarkers, the above four parameters were used to form the Rad-score. The AUC for differentiating between benign and malignant SPGGNs in the training set was 0.792 (95% CI: 0.671, 0.913), and the sensitivity and specificity were 86.10 and 65.20%, respectively. The AUC in the validation set was 72.9% (95% CI: 0.545, 0.913), and the sensitivity and specificity were 86.70 and 60%, respectively.
Conclusion:
The present diagnostic model based on the cluster of imaging biomarkers can preferably distinguish benign and malignant SPGGNs (≤1 cm).
Advances in knowledge:
Texture analysis based on CT images provide a new and credible technique for accurate identification of subcentimeter pulmonary ground-glass nodules.
Introduction
Pulmonary nodules (PNs) are commonly encountered in clinical practice and are well-circumscribed, radiographic opacities measuring <3 cm in diameter; they are of vital importance in light of the high mortality rate associated with lung cancer.1 With the ever-improving resolution and popularization of CT, an increasing number of small PNs are being identified in the clinic.2 Although most PNs are confirmed to be benign and only a small fraction of PNs progress to lung cancer,3 it is a challenge for radiologists to determine the probability of a PN being malignant, especially subcentimeter pulmonary nodules (SPNs) with ground-glass appearance; in addition, PNs are only classified into three different types according to the appearance of the nodule, i.e. a pure ground-glass appearance, a pure solid appearance or a mixed ground-glass and solid appearance.4 This classification system has certain value for the diagnosis of PNs, but it still cannot accurately determine the degree of malignancy of a PN, especially the subcentimeter pulmonary ground-glass nodules (SPGGNs), which are also the focus of present study.
For the accurate diagnosis of SPGGNs, transthoracic needle biopsy and observation by serial radiographs are often applied, and both approaches have advantages and disadvantages.5 The former is the gold-standard for defining a SPGGN as specifically benign or malignant but is invasive and potentially risky; furthermore, SPGGNs are clinically difficult to locate and puncture.6 Serial CT, MRI, and PET/CT observations are safe and noninvasive compared to the pathological examination of biopsies, and among them, serial CT scans are recommended by clinical practice guidelines.7 In addition, the observed growth rate, size and location of PNs are highly correlated with the degree of malignancy,8–10 but there are delays in making an accurate diagnosis and administering treatment when malignancy is truly present, and a substantial portion of results are false negative or false positive,11 especially for SPGGNs.
CT image-based computerized texture analysis is an emerging and promising tool for the characterization and differentiation of PNs. Texture analysisis a quantitative imaging-based tool that enables a more detailed and reproducible quantitative assessment of lesion characteristics than visual analysis by human observers.12 Many attempts to classify PNs based on two-dimensional (2D) and three-dimensional (3D) shape, 3D texture and combinations of 2D shape and texture CT features have been made.13 However, CT-based texture analysis for the identification of the degree of malignancy of SPGGNs is still insufficient, which directly affects clinical decisions regarding the therapeutic strategy, such as sublobar surgery or follow-up without surgery.
In this context, the aim of our study was to determine potential diagnostic model for the differentiation of benign and malignant SPGGNs based on CT texture analysis.
Methods and materials
Patients
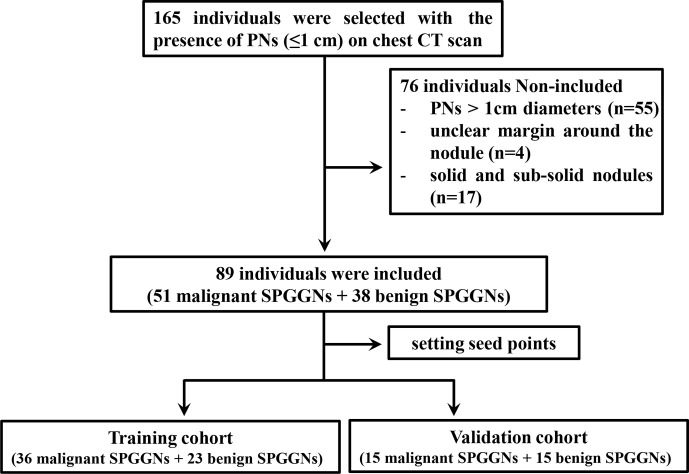
The study protocols were approved for this retrospective study by the Institutional Review Board of our Lishui hospital of Zhejiang University, and the requirement for informed consent was waived. From March 2014 to Mach 2016, 165 patients were selected based on the presence of PNs on chest CT scans obtained when the patients visited the PN clinic (Figure 1), and the final clinical diagnoses of all subjects were confirmed by the histopathological examination of specimens obtained by CT-guided transthoracic needle biopsy, transbronchial biopsy, video-assisted thoracoscopic surgery, or surgical resection. The pure ground-glass nodules in CT performance were selected as SPGGNs for further in-depth analysis. 76 patients were excluded due to the following factors: (1) PN >1 cm in diameter (n = 55); (2) unclear margin owing to interstitial pneumonia around the nodule (n = 4); and (3) solid and subsolid nodules (n = 17). Finally, 89 patients with 89 SPGGNs were included. Of the 89 subjects, according to the pathological results, 51 had malignant SPGGNs and were diagnosed with adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA) or invasive adenocarcinoma (IA), and 38 had benign SPGGNs and were diagnosed with inflammation (n = 18), nonspecific inflammatory changes (n = 7), or lung infections (n = 13). Representative CT images and photomicrographs of the histopathology images for benign and malignant SPGGNs were shown in Figure 2.
Figure 1.
Flowchart of study enrollment. PNs, pulmonary nodules; SPGGNs, subcentimeter pulmonary ground-glass nodules.
Figure 2.
Representative CT and histopathology images for benign (A & B) and malignant (C & D) SPGGNs. A. a 52-year-old female with a SPGGNs in the right lobe showed a 0.9 cm lesion. Pathological diagnosis confirmed atypical adenomatous hyperplasia, which was benign SPGGNs. B. A 60-year-old female with a SPGGNs in the left lobe showed a 0.9 cm lesion, and the pathological diagnosis also confirmed it was atypical adenomatous hyperplasia. C. A 54-year-old female with a SPGGNs in the right lobe showed a 0.8 cm lesion with an irregular shape. Pathological diagnosis confirmed microinvasive adenocarcinoma, which was a malignant SPGGNs. D. A 62-year-old male with a SPGGNs in the right lobe, and the pathological diagnosis also confirmed it was microinvasive adenocarcinoma. SPGGNs, subcentimeter pulmonary ground-glass nodules.
CT protocol
All images were acquired using a 256-slice Brilliance iCTsystem (Philips Healthcare, Best, The Netherlands).CT data were acquired in full inspiration and without contrast enhancement. The data acquisition parameters were as follows: detector collimation, auto; detector configuration, 128 × 0.625 mm; beam pitch, 0.758; rotation time, 0.75 s; FOV, 350 mm; slicethickness, 1.0 mm; slice increment, 1.0 mm, matrix 512 × 512; tube voltage, 120 kVp; tube current exposure time product, 110 mAs. All images were reviewed on a commercially available workstation (IntelliSpace Portal 5.0, Philips Healthcare, Cleveland, OH).
Histological evaluation
Histological evaluations were performed by examining formalin-fixed paraffin-embedded tissue sections stained with hematoxylin and eosin. The histological diagnosesof AIS, MIA, IA and other benign lesions were based on the WHO Classification fourth Edition and made by an experienced pulmonary pathologist.
Texture feature extraction
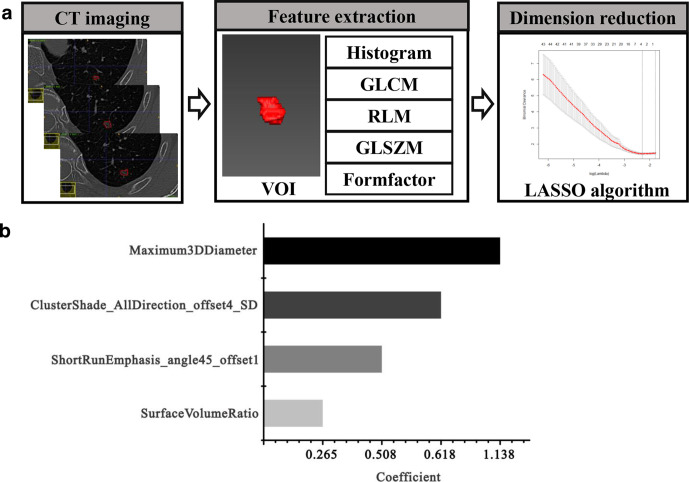
All features were extracted using Analysis Kit (A.K., GE Healthcare, China) software based on the radiomic method. This method analyzes the target region to obtain a series of imaging features according to the following basic steps (Figure 3A): (1) the CT images (DICOM format) were imported into the A.K. software to access all aspects of the PNs; (2) two experienced radiologists (both with more than 10 years of experience) then semi-automatically delineated the volume of interest (VOI) and fused the lesions at each level; (3) the quantitative imaging features of the VOI were calculated. A total of 396 texture features, including tumor size, tumor shape, first-order statistics of descriptor values (e.g. histogram features) and high-order texture features (e.g. gray level cooccurrence matrix (GLCM) and gray level run length matrix (GLRLM)) were extracted from the CT images.
Figure 3.
Workflow for the radiomic process. (A) First, the volume of interest was segmented on CT images; second, different texture features, including histogram, GLCM, GLRLM, GLSZM and Formfactor, were extracted; third, radiomic features were selected using the LASSO regression model. (B) 10-fold cross-validation was applied to select the most useful prognostic features. GLCM = gray level cooccurrence matrix, GLRLM = gray level run length, GLSZM = gray level size zone matrix, LASSO = least absolute shrinkage and selection operator.
Selection of radiomic features and building of the radiomic signature
The extracted texture features were standardized before feature dimension reduction, which can remove the unit limits of the data of each feature. The abnormal values were replaced by the median of the parameter in all cases, and all feature values were normalized (Z-score). Then, the patients were randomly divided into a training set (n = 59) and a validation set (n = 30) by setting seed points. The least absolute shrinkage and selection operator (LASSO) algorithm (Figure 3A) and 10-fold cross-validation (Figure 3B) were applied to reduce the dimensions and select the most useful prognostic features in the training data set. Finally, the best dimension reduction method was used to build the radiomic signature and calculate the radiomic score (Radscore) for each patient. The Rad score was computed in each case by a linear combination of selected features weighted by their respective coefficients. The predictive accuracy of the radiomic signature was quantified by the AUC in both the training and validation sets.
Intra- and interobserver agreement
The intra- and interobserver agreement of the extracted features was evaluated using the intraclass correlation coefficient (ICC). We initially chose 50 random CT images for region of interest (ROI) segmentation and feature extraction. ROI segmentation was performed by two experienced radiologists independently. The intraobserver ICC was computed by comparing two extractions performed by Reader A (with 10 years of experience). The interobserver ICC was computed by comparing the extraction performed by a second reader (Reader B, with 15 years of experience) with the first extraction performed by Reader A. An ICC greater than 0.75 was considered to indicate good agreement; the remaining image segmentation was performed by Reader A.
Statistical analysis
All statistical analyses were performed using R software (v. 3.3). The distribution of each variable was examined using the Kolmogorov–Smirnov test, and Student's t-test wasused to determine whether characteristic features were significantly different between the two groups (benign and malignant) for normally distributed features; otherwise, the Mann–Whitney test was used. A significance threshold of p < 0.05 was set. The LASSO logistic regression model was used with penalty parameter tuning that was conducted by 10-fold cross-validation based on minimum criteria. Finally, logistic linear regression was applied to link the Radscores of the selected features. ROC curves were plotted to evaluate the diagnostic efficiency of the model.
Results
Clinicopathological characteristics
In this study, all patients received a pathological diagnosis based on a pre-operative biopsy or surgically resected specimen, and the selected SPGGNs were all smaller than 1 cm. The baseline characteristics in the training and validation cohorts are listed in Table 1. In the training cohort, 36 patients had malignant SPGGNs; 30.6% of these patients were male, and 69.4% were female. In the training cohort, 23 patients had benign SPGGNs; 30.4% of these patients were male, 69.6% were female. There were no significant differences in sex, age, or nodule size between the patients with malignant and benign SPGGNs in the training cohort (p > 0.05). In the validation cohort, there were 15 patients with malignant and benign nodules, with no significant differences in sex, age, or nodule size between the patients with malignant and benign SPGGNs (p > 0.05).
Table 1.
Baseline characteristics in the training and validation cohorts
| Characteristics | Training cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|
| Malignant PNs (n = 36) | Benign PNs (n = 23) | p value | Malignant PNs (n = 15) | Benign PNs (n = 15) | p value | |
| Age | 53.72 ± 8.44 | 50.57 ± 9.10 | 0.18 | 50.33 ± 14.55 | 50.53 ± 12.08 | 0.296 |
| Sex | 0.992 | 0.136 | ||||
| M | 11 (30.6%) | 7 (30.4%) | 4 (26.7%) | 8 (53.3%) | ||
| F | 25 (69.4%) | 16 (69.6%) | 11 (73.3%) | 7 (46.7%) | ||
| Nodule size (cm) | ||||||
| Medium (SD) | 0.88 (0.15) | 0.82 (0.17) | 0.216 | 0.82 (0.17) | 0.93 (0.14) | 0.056 |
| Range | 0.4–1 | 0.3–1 | 0.4–1 | 0.5–1 | ||
| Histological type | ||||||
| AIS | 7 | 7 | ||||
| MIA | 13 | 3 | ||||
| IA | 16 | 5 | ||||
There were 7 patients with AIS, 13 patients with MIA, and 16 patients with IA in the training cohort, and thesenumbers were 7, 3 and 5, respectively, in the validation cohort. 12 of the SPGGNs exhibited inflammation, 5 of the SPGGNs exhibited nonspecific inflammatory changes, and 6 of the SPGGNs exhibited lung infection in the training cohort, and the corresponding numbers in validation cohort were 6, 2 and 7, respectively.
Inter- and intraobserver reproducibility of radiomic feature extraction
The intraobserver ICC calculated based on two measurements obtained by Reader A ranged from 0.819 to 0.964. The interobserver agreement between the two readers ranged from 0.783 to 0.918. These results indicate favorable intra- and interobserver feature extraction reproducibility.
Feature selection for the radiomic signature
A total of 396 radiomic features were extracted using the A.K.software, and the LASSO model with 10-fold cross-validation was applied to reduce the radiomic features (Figure 3). Finally, four potential predictors were selected in the training cohort (p < 0.05): ClusterShade_AllDirection_offset4_SD, ShortRunEmphasis_angle45_offset, Maximum3DDiameter and SurfaceVolumeRatio (Figure 4A), and the formulas for calculating the four texture features are shown in Table 2.The logistic regression model was trained with the four potential predictors, and the AUCs were 0.632, 0.607, 0.691 and 0.656, respectively (Figure 4B) (Table 3).
Figure 4.
Verification of selected radiomic features. (A) Significance of the imaging biomarkers. (B) ROC curves of the four potential predictors. ROC = receiver operating characteristic.
Table 2.
The classification and calculation formula of four texture features
| Category | Feature | Formula | |
| Formfactor | ClusterShade_AllDirection_offset4_SD |
*g is a GLCM Where i,j are the spatial coordinates of g (i,j). |
|
| Maximum3DDiameter | The largest pairwise Euclidean distance | ||
| RLM | SurfaceVolumeRatio | ||
| GLCM | ShortRunEmphasis_angle45_offset1 |
GLCM, gray level cooccurrence matrix; RLM, run length matrix.
Table 3.
The predictive performance of different imaging biomarkers in the training cohort and the Rad score in the training and validation cohorts.
| Parameters | AUC | Sensitivity | Specificity | 95% CI | Cut-off value |
| ClusterShade_AllDirection_offset4_SD | 0.632 | 73.70% | 44% | [0.511, 0.754] | −0.414 |
| ShortRunEmphasis_angle45_offset1 | 0.607 | 50% | 58.80% | [0.489, 0.725] | −0.026 |
| Maximum3DDiameter | 0.691 | 71% | 64.70% | [0.582, 0.8] | −0.134 |
| SurfaceVolumeRatio | 0.656 | 73.70% | 49.00% | [0.543, 0.770] | −0.146 |
| Rad score in training and validation cohorts | |||||
| Training cohort | 0.792 | 86.10% | 65.20% | [0.671, 0.913] | −0.174 |
| Validation cohort | 0.729 | 86.70% | 60% | [0.545, 0.913] | −0.215 |
AUC, area under the curve; CI, confidence interval.
The above four parameters were used as a cluster of imaging biomarkers to form the radiomic signature score (Radscore) for each SPN. The formula for calculating theRadscore was as follows:
Radscore = 0.201–0.618 × ClusterShade_AllDirection_offset4_SD + 0.508×ShortRunEmphasis_angle45_offset1 – 1.138 × Maximum3DDiameter + 0.265×SurfaceVolumeRatio
The Radscore for each patient is shown in a waterfall plot (Figure 5), which indicates that the Radscore was significantly different between malignant and benign SPGGNs in both the training and verification cohorts.
Figure 5.
Waterfall plot of the Radscore for each patient in the training (A) and validation (B) cohorts.The Radscore was significantly different between malignant and benign SPGGNs in both the training and verification cohorts. SPGGNs, subcentimeter pulmonary ground-glass nodules.
Performance of the radiomic signature
In both training and validation cohorts, the logistic regression model was trained with the Radscore(Figure 6). The AUC between benign and malignant SPGGNs in the training cohort was 0.792 (95% CI: 0.671, 0.913), and the sensitivity and specificity were 86.10 and 65.20%, respectively (Table 3). The AUC in the validation cohort was 72.9% (95% CI: 0.545, 0.913), and the sensitivity and specificity were 86.70 and 60%, respectively (Table 3).
Figure 6.
Logistic regression models in both the training (A) and validation (B) cohorts.The AUC for distinguishing between benign and malignant SPGGNs in the training cohort was 0.792, and the AUC in the validation cohort was 0.729.
AUC = area under the receiver operating characteristic curve, SPGGNs = subcentimeter pulmonary ground-glass nodules.
Discussion
PNs are the most common lung disease, and it is difficult to distinguish benign from malignant PNs, especially SPGGNs, and this difficulty affects clinical decisions.12 The noninvasive diagnosis of PNs has thus been a hot research topic,14,15 and an increasing number of studies have found that radiomics has great potential for achieving this goal.16–18 In the present study, we developed a quantitative prediction model based on the radiomic features of CT images for the differential diagnosis of benign and malignant SPGGNs (≤1 cm).
Our findings demonstrate that a novel radiomic signature based on four radiomic features (ClusterShade_AllDirection_offset4_SD, ShortRunEmphasis_angle45_offset, Maximum3DDiameter and SurfaceVolumeRatio) obtained by CT-based texture analysisis an independent factor for discriminating between benign and malignant SPGGNs (≤1 cm). The inner structure of SPGGNs is heterogeneous and can be reflected quantitatively by texture parameters. The second-order feature ClusterShade clusters similar samples according to their position into the same group, which reflects the heterogeneity of the spatial distribution of voxel intensities. Maximum3DDiameter and SurfaceVolumeRatio represent the three-dimensional size and shape of the tumor region. ShortRunEmphasis refers to the number of pixels with the same grayscale and continuous collinearity. Both of these features reflect the roughness and the similarity of the texture within the tissues, which are significantly correlated with the nodule density and related to invasive tumor biology.12,19
In all, 396 candidate radiomic features were reduced to four potential predictors by reducing the regression coefficients with the LASSO algorithm using 10-fold cross-validation, which is a popular regression method for high-dimensional data and is more effective than conventional methods for choosing predictors based on the intensity of their univariate association with an outcome.20–22 The four selected characteristic features were then combined into a Radscore to further establish the predictive model, and ROC curve analysis was applied to verify the predicted performance. We obtained satisfactory results when the prediction model was applied in both the training and verification cohorts. In addition, the AUC for distinguishing between benign and malignant SPGGNs in the training set was 0.792 (95% CI: 0.671, 0.913), and the sensitivity and specificity were 86.10 and 65.20%, respectively. The AUC in the validation set was 72.9% (95% CI: 0.545, 0.913), and the sensitivity and specificity were 86.70 and 60%, respectively. The diagnostic performance of the logistic regression model using these texture features was good.
Radiomic analysis, which was applied in our study, is an advanced technology that is widely used in the diagnosis of various diseases for status classification23 and has excellent potential for the identification of benign and malignant PNs. Recently, She et al.24 developed a radiomic signature based on five radiomic features, and the signature showed a good ability to discriminate between MIA/AIS and IA in both the primary and validation cohorts, with AUCs of 0.95 and 0.89, respectively. Choi et al.25 developed a support vector machine (SVM)-LASSO model to predict the malignancy of PNs based on two CT radiomic features and demonstrated that the model achieved an accuracy of 84.6%. Chen et al.26 also created a radiomic model based on four different features with 84% accuracy, 92.85% sensitivity and 72.73% specificity in the classification of benign and malignant PNs.As indicated by the above studies, there are many ways to apply radiomics to analyze PNs, but most of these studies have focused on lung nodules larger than 1 cm, with less attention on SPGGNs.SPGGNs are particularly difficult to identify and diagnose clinically, and the present study effectively solves this problem.
Compared with the models in previous studies, the predictive model in our study does not have the highest accuracy for distinguishing between benign and malignant PNs, but this diagnostic efficiency was better than the recent study.27 The reason why the accuracy of the constructed model in the present study wasn’t particularly high may be closely related to the diameter of the PN, with larger lung nodules being easier to identify as malignant; however, this hypothesis requires further study. In addition to CT, other imaging modalities can be used for radiomic analysis to identify benign and malignant PNs. Some of the studies28–30 have reported that PET/CT is highly accurate in distinguishing the characteristics of PNs, but false-positive results remain an issue. We believe that our prediction model has the ability to identify small benign or malignant SPGGNs (<1 cm).
There were several limitations to our study. First, this was a retrospective study and thus has an inherent selection bias. Second, we only used radiomic features to construct the prediction model, which would be more accurate if we used other additional clinical indicators. Third, although our study did not include external validation, our model showed good reliability and reproducibility.
Conclusion
In summary, the diagnostic model based on the cluster of imaging biomarkers identified in our study can be used to preferably distinguish between benign and malignant SPGGNs, which could transform the management of SPGGNs by enabling early diagnosis. At the same time, while the present diagnostic model is not expected to replace tissue diagnosis, it may add to radiologists’ confidence in diagnosing SPGGNs.
Footnotes
Acknowledgment: This study was supported by National Natural Science Foundation of China (No.81803778), and The Key Research and development Project of Zhejiang Province (No. 2018C03024), and the Public Welfare Technology Research Program of Zhejiang Province (LGF19H180009), The Science and Technology Project of Jinhua City (2019A453157).
Competing interests: The authors declare no conflicts of interest.
Patient consent: The study was approved by the institutional review board and human ethics committee of Lishui Hospital of Zhejiang University, and the requirement for informed consent was waived.
The authors Xianghua Hu and Weichuan Ye contributed equally to the work.
Contributors: Jiansong Ji and Minjiang Chen designed the study. Xianghua Hu, Weichuan Ye, Zhongxue Li, Chunmiao Chen, Peipei Pang, and Min Xu conducted all experiments, analyzed the data and wrote the manuscript. Shimiao Cheng, Xiuling Lv, Wei Weng, Jie Li and Qiaoyou Wengassisted with the data analysis. All authors contributed to the manuscript.
Contributor Information
Xianghua Hu, Email: huxianghuare@163.com.
Weichuan Ye, Email: 979747407@qq.com.
Zhongxue Li, Email: 1279774686@qq.com.
Chunmiao Chen, Email: 439542497@qq.com.
Shimiao Cheng, Email: 261517377@qq.com.
Xiuling Lv, Email: 1085435323@qq.com.
Wei Weng, Email: wengwei318@foxmail.com.
Jie Li, Email: a15990007235@126.com.
Qiaoyou Weng, Email: wengqy99@gmail.com.
Peipei Pang, Email: pangpeipei1221@126.com.
Min Xu, Email: lschrxm@163.com.
Minjiang Chen, Email: minjiangchenv@163.com.
Jiansong Ji, Email: lschrjjs@163.com.
REFERENCES
- 1.Mehta HJ, Ravenel JG, Shaftman SR, Tanner NT, Paoletti L, Taylor KK, et al. The utility of nodule volume in the context of malignancy prediction for small pulmonary nodules. Chest 2014; 145: 464–72. doi: 10.1378/chest.13-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gould MK, Tang T, Liu I-LA, Lee J, Zheng C, Danforth KN, et al. Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med 2015; 192: 1208-14. doi: 10.1164/rccm.201505-0990OC [DOI] [PubMed] [Google Scholar]
- 3.Abraham J. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011; 365: 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ost DE, Gould MK. Decision making in patients with pulmonary nodules. Am J Respir Crit Care Med 2012; 185: 363–72. doi: 10.1164/rccm.201104-0679CI [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen J, Liu Z, Todd NW, Zhang H, Liao J, Yu L, et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer(). 2011; 11: 3742011-08-24. doi: 10.1186/1471-2407-11-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta S, Krishnamurthy S, Broemeling LD, Morello FA, Wallace MJ, Ahrar K, et al. Small (</=2-cm) subpleural pulmonary lesions: short- versus long-needle-path CT-guided Biopsy--comparison of diagnostic yields and complications. Radiology 2005; 234: 631–7. doi: 10.1148/radiol.2342031423 [DOI] [PubMed] [Google Scholar]
- 7.Wang Memoli JS, Nietert PJ, Silvestri GA. Meta-Analysis of guided bronchoscopy for the evaluation of the pulmonary nodule. Chest 2012; 142: 385–93. doi: 10.1378/chest.11-1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko JP, Berman EJ, Kaur M, Babb JS, Bomsztyk E, Greenberg AK, et al. Pulmonary nodules: growth rate assessment in patients by using serial CT and three-dimensional volumetry. Radiology 2012; 262: 662–71. doi: 10.1148/radiol.11100878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saghir Z, Dirksen A, Ashraf H, Bach KS, Brodersen J, Clementsen PF, et al. Ct screening for lung cancer brings forward early disease. The randomised Danish lung cancer screening trial: status after five annual screening rounds with low-dose CT. Thorax 2012; 67: 296–301. doi: 10.1136/thoraxjnl-2011-200736 [DOI] [PubMed] [Google Scholar]
- 10.Cerfolio RJ, Bryant AS, McCarty TP, Minnich DJ. A prospective study to determine the incidence of non-imaged malignant pulmonary nodules in patients who undergo metastasectomy by thoracotomy with lung palpation. Ann Thorac Surg 2011; 91: 1696–701. doi: 10.1016/j.athoracsur.2011.02.075 [DOI] [PubMed] [Google Scholar]
- 11., Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al.National Lung Screening Trial Research Team . Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011; 365: 395–409. doi: 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chae H-D, Park CM, Park SJ, Lee SM, Kim KG, Goo JM. Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology 2014; 273: 285. doi: 10.1148/radiol.14132187 [DOI] [PubMed] [Google Scholar]
- 13.Dhara AK, Mukhopadhyay S, Dutta A, Garg M, Khandelwal N. A combination of shape and texture features for classification of pulmonary nodules in lung CT images. J Digit Imaging 2016; 29: 466–75. doi: 10.1007/s10278-015-9857-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Son JY, Lee HY, Kim J-H, Han J, Jeong JY, Lee KS, et al. Quantitative CT analysis of pulmonary ground-glass opacity nodules for distinguishing invasive adenocarcinoma from non-invasive or minimally invasive adenocarcinoma: the added value of using iodine mapping. Eur Radiol 2016; 26: 43. doi: 10.1007/s00330-015-3816-y [DOI] [PubMed] [Google Scholar]
- 15.Shlomi D, Peled N, Schwarz YA, Soo Hoo GW, Batra RK, Fink G, et al. Non-Invasive early detection of malignant pulmonary nodules by FISH-based sputum test. Cancer Genet 2018; 226-227: 1-10. doi: 10.1016/j.cancergen.2018.04.118 [DOI] [PubMed] [Google Scholar]
- 16.Tu S-J, Wang C-W, Pan K-T, Wu Y-C, Wu C-T, SJ T, YC W, CT W. Localized thin-section CT with radiomics feature extraction and machine learning to classify early-detected pulmonary nodules from lung cancer screening. Phys Med Biol 2018; 63: 065005. doi: 10.1088/1361-6560/aaafab [DOI] [PubMed] [Google Scholar]
- 17.Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RTH, Hermann G, et al. Ct-Based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015; 114: 345–50. doi: 10.1016/j.radonc.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Zhang R, Gan Y, Yang L, Li W. Development and clinical application of radiomics in lung cancer. Radiat Oncol 2017; 12: 154. doi: 10.1186/s13014-017-0885-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang I-P, Park CM, Park SJ, Lee SM, McAdams HP, Jeon YK, et al. Persistent pure Ground-Glass nodules larger than 5 mm: differentiation of invasive pulmonary adenocarcinomas from preinvasive lesions or minimally invasive adenocarcinomas using texture analysis. Invest Radiol 2015; 50: 798–804. doi: 10.1097/RLI.0000000000000186 [DOI] [PubMed] [Google Scholar]
- 20.Chichignoud M, Lederer J. Wainwright M. a practical scheme and fast algorithm to tune the LASSO with optimality guarantees. Statistics 2016; 17: 8162–81. [Google Scholar]
- 21.Schouten TM, Koini M, Vos Fde, Seiler S, Rooij Mde, Lechner A, et al. Individual classification of Alzheimer's disease with diffusion magnetic resonance imaging. Neuroimage 2017; 152: 476. doi: 10.1016/j.neuroimage.2017.03.025 [DOI] [PubMed] [Google Scholar]
- 22.Ramsay IS, Ma S, Fisher M, Loewy RL, Ragland JD, Niendam T, et al. Model selection and prediction of outcomes in recent onset schizophrenia patients who undergo cognitive training. Schizophr Res Cogn 2018; 11: 1–5. doi: 10.1016/j.scog.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, Ehteshami BB, vDP J, Van GB, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017; 318: 2199. doi: 10.1001/jama.2017.14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.She Y, Zhang L, Zhu H, Dai C, Xie D, Xie H, et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur Radiol 2018; 28: 1–8. doi: 10.1007/s00330-018-5509-9 [DOI] [PubMed] [Google Scholar]
- 25.Choi W, JH O, Riyahi S, Liu CJ, Jiang F, Chen W, et al. Radiomics analysis of pulmonary nodules in low‐dose CT for early detection of lung cancer. Medical Physics 2017;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C-H, Chang C-K, Tu C-Y, Liao W-C, Wu B-R, Chou K-T, et al. Radiomic features analysis in computed tomography images of lung nodule classification. PLoS One 2018; 13: e0192002. doi: 10.1371/journal.pone.0192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Li C, Jin L, Gao P, Zhao W, Ma W, et al. Radiomics for lung adenocarcinoma manifesting as pure ground-glass nodules: invasive prediction. European Radiology 2020;: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fletcher JW, Kymes SM, Gould M, Alazraki N, Coleman RE, Lowe VJ, et al. A comparison of the diagnostic accuracy of 18F-FDG PET and CT in the characterization of solitary pulmonary nodules. J Nucl Med 2008; 49: 179–85. doi: 10.2967/jnumed.107.044990 [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Velloso MJ, Bastarrika G, de-Torres JP, Lozano MD, Sanchez-Salcedo P, Sancho L, et al. Assessment of indeterminate pulmonary nodules detected in lung cancer screening: diagnostic accuracy of FDG PET/CT. Lung Cancer 2016; 97: 81–6. doi: 10.1016/j.lungcan.2016.04.025 [DOI] [PubMed] [Google Scholar]
- 30.Hatt M, Majdoub M, Vallières M, Tixier F, RC L, Groheux D, et al. 18F-Fdg PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. Journal of Nuclear Medicine Official Publication Society of Nuclear Medicine 2015; 56: 38. [DOI] [PubMed] [Google Scholar]