Abstract
Remdesivir (RDV) exerts anti-severe acute respiratory coronavirus 2 activity following metabolic activation in the target tissues. However, the pharmacokinetics and tissue distributions of the parent drug and its active metabolites have been poorly characterized to date. Blood and tissue levels were evaluated in the current study. After intravenous administration of 20 mg/kg RDV in mice, the concentrations of the parent drug, nucleotide monophosphate (RMP) and triphosphate (RTP), as well as nucleoside (RN), in the blood, heart, liver, lung, kidney, testis, and small intestine were quantified. In blood, RDV was rapidly and completely metabolized and was barely detected at 0.5 h, similar to RTP, while its metabolites RMP and RN exhibited higher blood levels with increased residence times. The area under the concentration versus time curve up to the last measured point in time (AUC0-t) values of RMP and RN were 4558 and 136,572 h∙nM, respectively. The maximum plasma concentration (Cmax) values of RMP and RN were 2896 nM and 35,819 nM, respectively. Moreover, RDV presented an extensive distribution, and the lung, liver and kidney showed high levels of the parent drug and metabolites. The metabolic stabilities of RDV and RMP were also evaluated using lung, liver, and kidney microsomes. RDV showed higher clearances in the liver and kidney than in the lung, with intrinsic clearance (CLint) values of 1740, 1253, and 127 mL/(min∙g microsomal protein), respectively.
Keywords: remdesivir, pharmacokinetics, tissue distribution, nucleotide monophosphate, nucleotide triphosphate, nucleoside
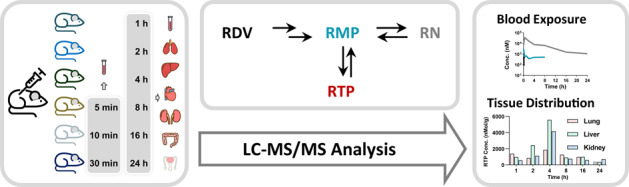
Introduction
The current novel coronavirus disease, named coronavirus disease 2019 (COVID-19), has triggered an outbreak and spread rapidly across the world since December 2019 [1–3]. It has been declared a global pandemic by the World Health Organization (WHO). To date, hundreds of millions of people have suffered from this disease [4]. Most patients (~80%) have common respiratory and gastrointestinal signs and symptoms, such as fever, dry cough, nausea, vomiting and diarrhea, similar to other viral infections, and can self-recover without hospital treatment [1, 3]. The remaining patients progress to severe acute respiratory disorders or multiple organ failure, and some of them suffer serious outcomes [5]. Unfortunately, there is still no effective therapy aimed at the etiology of COVID-19 [5]. Because severe acute respiratory coronavirus 2 (SARS-CoV-2) is proven to be a pathogen, the antivirus strategy is naturally considered in drug development and therapy [6].
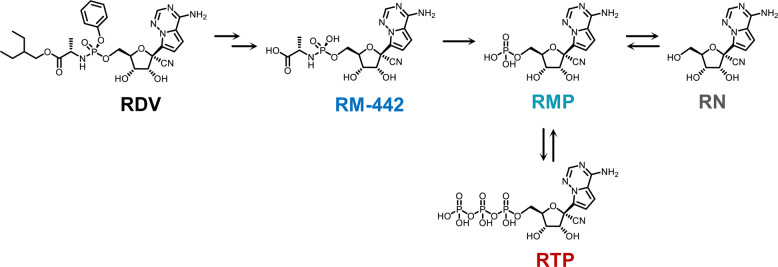
Remdesivir (RDV, Fig. 1) is a 2-ethylbutyl L-alaninate phosphoramidate prodrug of a 1′-cyano-substituted adenine C-nucleoside ribose analog (remdesivir nucleoside, RN). RDV undergoes multistep and extensive metabolism (Fig. 1) [7]. It exhibits antiviral activity against a number of RNA viruses, including Ebola virus, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV), by interfering with the activity of viral RNA-dependent RNA-polymerases (RdRp) through its triphosphate metabolite (RTP) [6, 8]. Recently, RDV was shown to have strong inhibitory activities against SARS-CoV-2 in in vitro and animal studies, such as studies of monkeys and mice [6]. Several clinical trials and case reports on patients also support that RDV is a promising candidate for the treatment of COVID-19 [9, 10]. However, there are only a few preliminary studies on the in vivo processes of RDV. After intravenous (iv) administration of 10 mg/kg RDV in rhesus monkeys (n = 3), RDV exhibited fast systemic elimination with a short plasma half-life (t1/2 = 0.39 h), while its nucleoside metabolite RN showed more persistent plasma levels (t1/2 > 14 h) [7]. Another study observed that after subcutaneous administration of RDV in mice and marmosets (n = 3 for each species), RTP was the predominant nucleotide metabolite in the lungs [11]. To the best of our knowledge, detailed tissue distribution information about RDV and its metabolites is lacking, which is an obstacle to eliciting a therapeutic effect and enhancing the drug development of RDV for COVID-19 treatment.
Fig. 1.
Proposed metabolic pathways of RDV.
Therefore, in this study, we aimed to investigate blood and major target tissue levels of RDV and its metabolites in mice and provide valuable information for drug development and clinical practices related to COVID-19 treatment.
Materials and methods
Materials
RDV (≥98%) was synthesized according to the literature [12]. The nucleotide monophosphate RMP (purity of 93%) and nucleotide triphosphate RTP (purity of 98%) were synthesized as previously reported [13]. Nucleoside RN (purity of 98%) was purchased from Target Molecule Corp (Boston, MA, USA). Dimethyl sulfoxide (DMSO), 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB, ≥ 98%), Trizma® base (Tris, ≥99.9%) and magnesium chloride hexahydrate (MgCl2·6H2O, 99%–102%) were purchased from Sigma-Aldrich (St. Louis, MO, USA). PhosSTOP EASYpack (PhosSTOP) was purchased from Roche (Basel, Switzerland). Alamethicin was purchased from J&K Chemical, Ltd. (Beijing, China). Potassium chloride (KCl, ≥99.5%), ethylenediaminetetraacetic acid disodium salt dihydrate (Na2-EDTA·2H2O, ≥99.0%) and sucrose were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All other reagents were of the highest quality commercially available.
Animals
Cesarean Derived-1 (CD-1) mice (7–8 weeks old, 18–22 g) were purchased from Shanghai Super-B&K Laboratory Animal Co., Ltd. (Shanghai, China). All mice were allowed free access to food and water prior to the experiment. All animal experiment procedures were conducted in accordance with the guidelines of the National Research Council. The experimental protocol was approved by the Animal Care and Use Committee of the Shanghai Institute of Materia Medica (No.: 2020-02-YY-10).
Pharmacokinetics and tissue distribution study
Thirty male CD-1 mice were randomly divided into six groups (n = 5). All mice received a single iv administration of 20 mg/kg RDV, which was dissolved in DMSO-ethanol-PEG300-saline (5/5/40/50, v/v/v/v). For mice in group 4 to group 6, a 50 μL blood sample was collected via orbital venous plexus into a heparinized tube at 0.083, 0.167, and 0.5 h post-dosing (one sampling per mouse). Then, mice in group 1 to group 6 were anesthetized at 1, 2, 4, 8, 16, and 24 h post-dosing, and blood samples were collected by cardiac puncture. Next, various tissues, including the heart, liver, lung, kidney, testis, and small intestine, were perfused and harvested. All blood and tissue samples were immediately prepared for further analyses.
Sample preparation
Heparinized blood samples were divided into two parts: (i) For quantification of RDV and RN, 20 μL of blood was immediately added to a 1.5 mL polyethylene tube containing 200 μL of methanol-acetonitrile (MeOH-ACN, 1:1, v/v) and then vortexed thoroughly and centrifuged at 4 °C and 11,363 × g for 5 min. The supernatant was used for analysis. (ii) For quantification of RMP and RTP, 30 μL of blood was immediately added to a 1.5 mL polyethylene tube containing 3 μL of PhosSTOP, 10 μL of DTNB and 90 μL of 2% formic acid aqueous solution, vortexed thoroughly and centrifuged at 4 °C and 11,363 × g for 3 min to remove the cell debris. Then, 110 μL of the supernatant was mixed with an equivalent volume of water to carry out the solid-phase extraction with a pre-equilibrated weak anion exchange plate (Waters Oasis WAX μElution plate). After washing with 200 μL of 5 mM ammonium acetate (NH4Ac) aqueous solution (pH 4.5) and 200 μL of MeOH, the analytes were eluted with 200 μL of 10% NH3·H2O in MeOH-ACN (4:6, v/v). Finally, the eluate was gently evaporated to dryness under a gentle stream of air, and the residue was reconstituted with 100 μL of ACN-H2O (3:7, v/v) for analysis.
Tissue samples were individually homogenized with 2 or 4 times equivalent volumes of water containing PhosSTOP and DTNB. The preparation procedure of the produced homogenates was similar to that of the blood sample.
Lung, liver, and kidney microsome metabolism
Liver, kidney, and lung microsomes of mice were prepared as described in the literature [14]. In brief, fresh liver, kidney, and lung were collected from four CD-1 mice (7–8 weeks old, 18–22 g) and rinsed with ice-cold PBS. The tissues were homogenized in ice-cold 50 mM Tris-HCl buffer containing 150 mM KC1 and 2 mM Na2-EDTA (pH 7.4) to produce ~30% homogenates. Next, the homogenates were centrifuged at 12,000 × g for 20 min at 4 °C. Then, the supernatants were centrifuged at 105,000 × g for 60 min at 4 °C with a CP70ME ultracentrifuge (Hitachi, Tokyo, Japan). The supernatants were discarded, and the microsomal pellets were suspended in 250 mM sucrose and stored at −70 °C. The protein concentrations of the microsomes were determined by a bicinchoninic acid (BCA) protein assay kit (Pierce, Thermo Fisher, Rockford, IL, USA).
The in vitro metabolism study for RDV or RMP was performed using these prepared tissue microsomes. Each tissue microsome (lung, liver, and kidney, final concentration 0.5 mg/mL), alamethicin at 50 μg/mL, and MgCl2 at 5 mM in 0.1 M Tris buffer (pH 7.4) were incubated at 37 °C for 2 min. For the RDV metabolism study, the reaction was started by the addition of RDV (final concentration 10 μM, 0.1% DMSO). Aliquots were sampled at 0, 2, 5, 15, 30, and 60 min; then, the same equivalent volume of ice-cold MeOH was added to terminate the reaction. For the RMP metabolism study, the reaction was started by the addition of RMP (final concentration 1 μM, 0.1% DMSO). Aliquots were sampled at 0, 5, 15, 30, and 60 min; then, the same equivalent volume of ice-cold MeOH was added to terminate the reaction. The in vitro samples were then prepared and analyzed similarly to blood samples.
UPLC-MS/MS analysis of RDV and its metabolites
Two different ultraperformance liquid chromatography coupled triple-quadrupole mass spectrometry (UPLC-MS/MS) methods were developed and validated for the quantification of RDV and its metabolites RN, RMP, and RTP. The UPLC-MS/MS system comprised a Waters AQUITY UPLC system with a thermostatted autosampler (kept at 15 °C) and an ultrahigh performance binary pump (I-class, Waters, Milford, MA, USA) coupled to a triple quadrupole mass spectrometer with an electrospray ionization (ESI) source (Xevo TQ-S, Waters, Milford, MA, USA). Data were captured and analyzed using MassLynx software (version 4.1, Waters, Milford, MA, USA).
For quantification of RDV and RN, chromatographic separation was performed on an ACQUITY HSS T3 column (2.1 mm × 50 mm, 1.8 μm; Waters) at a temperature of 45 °C. Mobile phase A consisted of water with 0.1% formic acid, and mobile phase B consisted of ACN-MeOH (9:1, v/v) with 0.1% formic acid. A gradient elution was set as 5%–10% B at 0–0.8 min, 10%–90% B at 0.8–2 min, 90%–95% B at 2–2.5 min, and 95%–95% B at 2.5–3 min with a flow rate of 0.5 mL/min. The injection volume was 2 μL. The mass spectrometer was operated in the positive ion detection with multiple reaction monitoring (MRM) mode. The following transitions were monitored: m/z 603.2 → m/z 200.1 for RDV and m/z 292.1 → m/z 163.0 for RN. Mass spectrometry settings were as follows: capillary voltage 3.0 kV, source offset 50 V, desolvation gas flow 1000 L/h, cone gas flow 150 L/h, nebulizer pressure 7.0 bar, collision gas (argon) flow 0.14 mL/min, desolvation temperature 500 °C.
For quantification of RMP and RTP, chromatographic separation was performed on a BioBasic AX column (2.1 mm × 50 mm, 4.6 μm; Thermo Fisher) at a temperature of 45 °C. Mobile phase A consisted of ACN-H2O (3:7, v/v) with 10 mM NH4Ac (pH 6.0), and mobile phase B consisted of ACN-H2O (3:7, v/v) with 1 mM NH4Ac (pH 10.5). An isocratic elution was set as 30% B for 2 min with a flow rate of 0.50 mL/min. The injection volume was 5 μL. The following transitions were monitored: m/z 372.0 → m/z 202.1 for RMP and m/z 532.1 → m/z 202.1 for RTP. Except for the MRM transitions, the mass spectrometer was operated identically as described above.
The linear ranges of RDV, RMP, RTP, and RN in blood and different tissues are listed in Supplementary Table S1. The intrabatch accuracy and precision of RDV and its metabolites were acceptable as guidance defined by the Food and Drug Administration [15] and are listed in Supplementary Tables S2 and S3.
Data analysis
The analysis of the pharmacokinetic and tissue distribution data was performed using WinNolin 6.4 (Pharsight, Princeton, NJ, USA). The main pharmacokinetic parameters were calculated using a noncompartmental model. The maximum blood or tissue concentration (Cmax) and time to Cmax (tmax) were determined from the observed data. The area under the concentration versus time curve up to the last measured point in time (AUC0-t) was calculated using the trapezoidal rule. The area under the concentration versus time curve from time zero to infinity (AUC0-∞) was generated by extrapolating AUC0-t to infinity using the trapezoidal rule-extrapolation method. The terminal elimination half-life (t1/2) was calculated by dividing ln2 by the terminal elimination rate constant λz, which was determined by linear regression of at least three data points from the terminal portion of the concentration versus time plots.
The analysis of the in vitro metabolic stability data was performed using Excel 2019 (Microsoft, Redmond, WA, USA). The incubation time was plotted by the natural logarithm of the residual rate of the substrate in the incubation system, and then, the slope k was obtained by linear regression. The intrinsic clearance (CLint) was calculated by dividing k by the concentration of microsomes.
Results
Pharmacokinetics of RDV and its metabolites
After an iv bolus of 20 mg/kg of RDV to mice, the concentrations of the parent drug and its three metabolites RMP, RTP, and RN were quantified by two validated LC-MS/MS methods. In blood samples, both RDV and RTP were only detected up to 0.5 h post-dosing, while RMP and RN were detected up to 8 h and 24 h post-dosing, respectively. The mean plasma concentration versus time profiles of RDV and its three metabolites RMP, RTP, and RN are shown in Fig. 2. The main pharmacokinetic parameters of RMP and RN are listed in Table 1. The Cmax and AUC0-t values of RMP and RN were 2896 nM and 4558 h∙nM and 35,819 nM and 136,572 h∙nM, respectively.
Fig. 2.
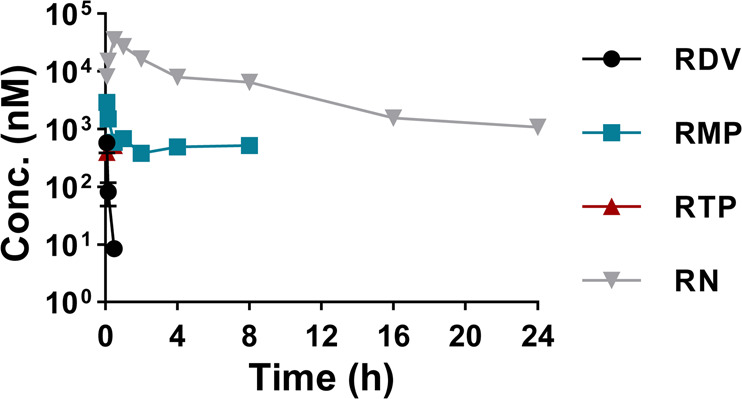
Blood concentration curves of RDV and its metabolites RMP, RTP, and RN after iv administration of RDV at 20 mg/kg in CD-1 mice (n = 5, mean ± SD).
Table 1.
Main pharmacokinetic parameters of RDV’s metabolites RMP and RN in mouse blood after iv administration of RDV 20 mg/kg (n = 5).
| Parameters | RMP | RN |
|---|---|---|
| t1/2 | ||
| (h) | 5.3 | 5.7 |
| tmax | ||
| (h) | 0.083 | 0.5 |
| Cmax | ||
| (ng/mL) | 1075 | 10,433 |
| (nM) | 2896 | 35,819 |
| AUC0-t | ||
| (h∙ng/mL) | 1692 | 39,779 |
| (h∙nM) | 4558 | 136,572 |
| AUC0-∞ | ||
| (h∙ng/mL) | 3149 | 42,426 |
| (h∙nM) | 8482 | 145,660 |
Tissue distribution of RDV and its metabolites
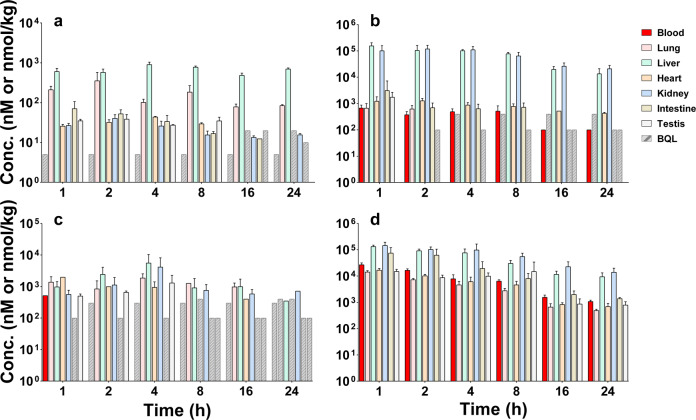
The concentrations of RDV and its metabolites (RMP, RTP, and RN) at different time points (1, 2, 4, 8, 16, and 24 h) in various tissues are shown in Fig. 3. Their Cmax, tmax, and AUC0-t values in various tissues are summarized in Table 2. After iv dosing, the parent drug RDV could be detected in all investigated tissues, among which the liver was the most predominant organ, followed by the lung. The values of RDV in the liver and lung were 15,732 and 3116 h∙nmol/kg, respectively. RMP was mainly distributed in the liver and kidney with AUC0-t values of 1,358,265 and 1,316,427 h∙nmol/kg, respectively. RTP, the active metabolite, could be detected in most tissues, except for the small intestine. The liver, kidney, and lung showed the highest levels of RTP, with AUC0-t values of 33,576, 25,238, and 20,732 h∙nmol/kg, respectively. RN had the highest level in kidney with an AUC0-t value of 1,232,816 h∙nmol/kg, which was 8.0-fold higher than that of blood.
Fig. 3. Blood and tissue concentrations of RDV and its metabolites RMP, RTP, and RN after iv administration of RDV 20 mg∙kg−1 in CD-1 mice (n = 5, Mean ± SD).
a RDV. b RMP. c RTP. d RN. BQL below lower limit of quantitation.
Table 2.
Main pharmacokinetic parameters of RDV and its metabolites RMP, RTP, and RN in mouse tissues after iv administration of RDV 20 mg/kg (n = 5).
| Tissue | RDV | RMP | RTP | RN | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cmax | tmax | AUC0-t | Cmax | tmax | AUC0-t | Cmax | tmax | AUC0-t | Cmax | tmax | AUC0-t | |
| Lung | 355 | 2 | 3116 | 673 | 1 | 1347 | 1871 | 4 | 20,732 | 14,237 | 1 | 73,218 |
| Liver | 905 | 4 | 15,732 | 155,705 | 1 | 1,358,265 | 5588 | 4 | 33,576 | 132,351 | 1 | 877,811 |
| Heart | 43.6 | 4 | 275 | 1,283 | 2 | 17,003 | 1969 | 1 | 6191 | 16,893 | 1 | 96,353 |
| Kidney | 40.3 | 2 | 438 | 119,679 | 2 | 1,316,427 | 4166 | 4 | 25,238 | 145,954 | 1 | 1,232,816 |
| Intestine | 71.0 | 1 | 441 | 3132 | 1 | 12,981 | N.D. | N.D. | N.D. | 74,548 | 1 | 323,631 |
| Testis | 38.5 | 2 | 261 | 1747 | 1 | 1747 | 1298 | 4 | 3035 | 14,985 | 1 | 145,768 |
If the density of each tissue was presumed as 1 kg/L, the units of Cmax and tmax could be considered as nM and h∙nM.
Units: Cmax: nmol/kg; tmax: h; AUC: h∙nmol/kg.
N.D. not detected.
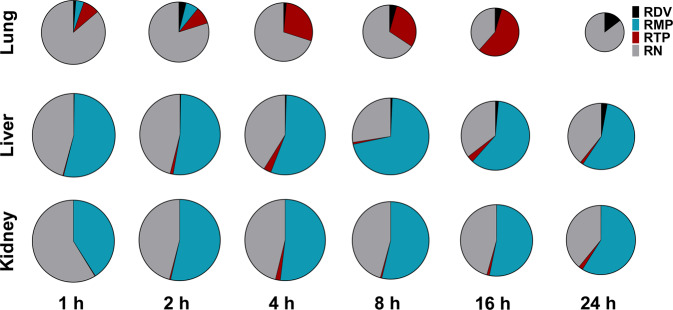
The amounts and proportions of RDV and its metabolites in the lung, liver, and kidney at each time point (from 1–24 h post-dosing) are shown in Fig. 4. In lung exposure (AUC0-t), RN was the most abundant compound, followed by RTP. However, both RDV and RMP had low levels in the lung. Notably, at 4 h, 8 h, and 16 h post-dosing, RTP accounted for a significant proportion. In contrast, in liver and kidney exposures (AUC0-t), RMP was the most abundant compound, followed by RN. The RTP levels in both the liver and kidney were low.
Fig. 4. Proportions of RDV and its metabolites RMP, RTP, and RN in lung, liver, and kidney.
Sizes of the pies were normalized according to the total concentrations of RDV, RMP, RTP, and RN (by taking the natural logarithm).
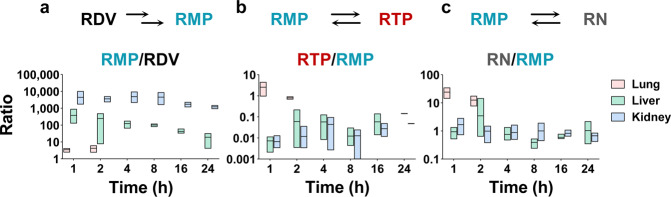
Regarding the relative content of RDV and its metabolites in different tissues (Fig. 5), kidney showed the highest RMP to RDV ratio, which was 10.8-fold and 1265-fold higher than that of the liver and lung at 1 h post-dosing, respectively. The lung showed the highest RTP to RMP ratio and RN to RMP ratio. The RTP to RMP ratio in the lung was more than 350-fold higher than that in the liver and lung at 1 h post-dosing, and the RN to RMP ratio in the lung was more than 15-fold higher than that in the liver. These results indicated higher hydrolysis rates of RDV in the liver and kidney as well as a higher dephosphorylation rate or efficient phosphorylation of RMP in the lung.
Fig. 5. Concentration ratios of RDV and its metabolites in lung, liver, and kidney.
a RMP to RDV. b RTP to RMP. c RN to RMP.
Metabolism of RDV and RMP in the liver, kidney and lung microsomes
To further investigate the metabolic differences of RDV and RMP in the lung, liver, and kidney, we evaluated their microsomal metabolic stabilities. Both RDV and RMP had very poor stabilities in the lung, liver, and kidney microsomes. The t1/2 and CLint values are shown in Table 3. In the three tissue microsomes, RDV could be rapidly and completely metabolized (Fig. 6a). The t1/2 values in the liver and kidney microsomes were only 0.80 min and 1.11 min, respectively, whereas the CLint values were as high as 1740 and 1253 mL/(min∙g microsomal protein), respectively. In lung microsomes, t1/2 for RDV was 10.95 min, and CLint was 127 mL/(min∙g microsomal protein), indicating better stability in the lung microsomes than in the other two microsomes. In addition, the metabolism of RMP was investigated. RMP also underwent rapid and thorough hydrolysis in the lung, liver, and kidney microsomes (Fig. 6b), with t1/2 values of 5.42 min, 4.94 min, and 2.73 min, respectively, and CLint values of 261, 304, and 517 mL/(min∙g microsomal protein), respectively.
Table 3.
Microsomal stability of RDV and RMP in lung, liver, and kidney.
| Tissue | RDV | RMP | ||
|---|---|---|---|---|
| t1/2 | CLint | t1/2 | CLint | |
| Lung | 10.95 ± 0.69 | 127 ± 8 | 5.42 ± 0.95 | 261 ± 44 |
| Liver | 0.80 ± 0.08 | 1740 ± 162 | 4.94 ± 1.80 | 304 ± 33 |
| Kidney | 1.11 ± 0.06 | 1253 ± 71 | 2.73 ± 0.41 | 517 ± 85 |
Units: t1/2: min; CLint: mL/(min∙g microsomal protein).
Fig. 6. Metabolic stability of RDV and RMP in mouse lung, liver, and kidney microsomes.
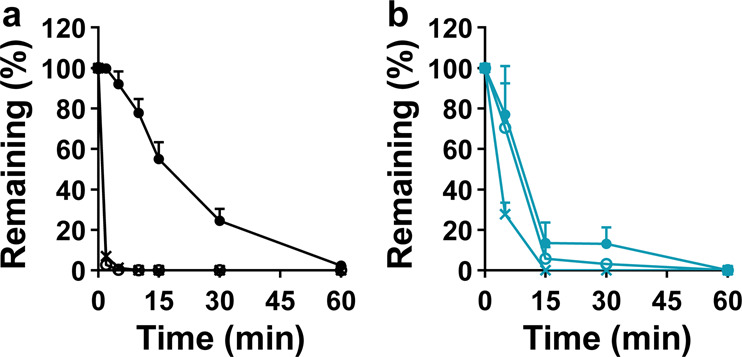
a RDV. b RMP. ● liver. ○ liver. × kidney.
Discussion
The novel coronavirus disease COVID-19 has spread rapidly worldwide. This disease has seriously affected the life and work of human beings. RDV is a promising antiviral agent for COVID treatment [9]. However, the detailed in vivo process of RDV is still unclear, which is an obstacle to eliciting a therapeutic effect and enhancing the drug application of RDV [16]. In this study, we investigated the levels of RDV and its three metabolites, RMP, nucleotide triphosphate (RTP), and nucleoside metabolite (RN), in mouse blood and different tissues after a single iv administration of 20 mg/kg RDV. In blood, RN was the major component, which had the highest concentration among the four compounds at each time point (0–24 h). The low level of the parent drug could be due to the high esterase activities in mouse blood in the current study [17]. However, the level of RDV in human plasma was not low [18], which might be due to the species differences in esterase activities between humans and mice [17]. However, nucleoside metabolites were also the predominant components for other nucleoside prodrugs or in other species [7, 19].
In addition to the lung, which is the major organ invaded by SARS-CoV-2, other organs, including the intestine, kidney, liver, heart, and testis, are susceptible to virus infection [20, 21]. In this study, we investigated RDV and its major metabolite concentrations in all tissues mentioned above. After iv dosing, RDV and its metabolites were widely distributed in various tissues and had relatively high levels in the lung, liver, and kidney. According to the AUC0-t values of RDV, the ranking order of its exposure was liver > lung > kidney. For RMP, RDV hydrolyzed metabolites and precursors of active RTP, the ranking order of its exposure was liver ≈ kidney » lung. Exposure in the liver and kidney was ~1000-fold higher than that in the lung. Regarding the active form RTP, the liver, kidney, and lung had the highest levels with similar exposures. As RTP might be only generated intracellularly, its concentrations in tissues were far higher than those in blood. Intriguingly, the proportions of RTP varied significantly in different tissues, accounting for a considerable proportion in the lung. RN was also considered the pool and precursor of RTP as well as the eliminated form of the drug. Its exposure in the kidney was higher than that in the liver and lung, which was reasonable because the kidney is the major excretory organ of this drug.
Although the metabolic enzymes of RDV have not been fully identified, its biotransformation processes were supposed according to studies on drugs with a similar structure (Fig. 1) [11, 22]. After entering cells of different tissues, RDV is hydrolyzed by carboxylesterase (CES) and/or cathepsin A cleavage to form RM-442. Subsequently, the monophosphate metabolite RMP is released by phosphoramidase. Then, RMP can either be phosphorylated to form the pharmacologically active triphosphate metabolite RTP or be dephosphorylated to form the nucleoside metabolite RN. To obtain preliminary insight into the metabolic capability of RDV or its metabolites in various tissues, we evaluated the metabolic ratios between the product compound and parent compound in mouse tissues (Fig. 5) as well as their microsomal metabolic stabilities. The RMP to RDV ratios were much higher in the kidney and liver than in the lung, which was consistent with the much poorer in vitro metabolic stabilities. CES exists widely in various tissues in mammals. As in humans and rats, CES expression in the liver was higher than that in the lung, and the CES distribution in mouse tissues might be similar [23, 24]. Moreover, according to hydrolase research for the prodrug candesartan cilexetil, mouse kidney microsomes showed higher hydrolase activity than that of the lung [25]. Taken together, the capacity for RDV hydrolysis in the mouse liver and kidney microsomes was probably stronger than that of the lung, which could explain the above results.
The metabolism of RMP was more complicated: first, it is the metabolite of the prodrug RDV; second, it can be dephosphorylated into RN; third, it can be resynthesized from RN in living cells and in vivo; fourth, it is a precursor of the active metabolite RTP. In the current study, RMP showed very poor stability in the lung, liver, and kidney microsomes. However, to explain the high RTP proportion in the lung, researchers should perform more studies.
Conclusion
In this study, the pharmacokinetic behavior and tissue distribution of RDV and its major metabolites RMP, RTP, and RN were evaluated. RDV showed rapid elimination and extensive tissue distribution. Moreover, the proportions and biotransformations of the parent drug and its metabolites in tissues were evaluated.
Supplementary information
Acknowledgements
This study was supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2016263).
Author contributions
JL and BT designed research; WJH, LC, YY, and XW performed research; YCX and JSS contributed chemical synthesis; WJH, LC and JL analyzed data; WJH, BT and JL wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Wen-juan Hu, Lu Chang, Ying Yang
Contributor Information
Bo Tan, Email: tbot@163.com.
Jia Liu, Email: jia.liu@simm.ac.cn.
Supplementary information
The online version of this article (10.1038/s41401-020-00537-9) contains supplementary material, which is available to authorized users.
References
- 1.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–9. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33.. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–36.. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard [updated 2020 Jul 29; cited 2020 Jul 30]. https://covid19.who.int/.
- 5.Bajwah S, Wilcock A, Towers R, Costantini M, Bausewein C, Simon ST, et al. Managing the supportive care needs of those affected by COVID-19. Eur Respir J. 2020;55:2000815. doi: 10.1183/13993003.00815-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–5. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown AJ, Won JJ, Graham RL, Dinnon KH, 3rd, Sims AC, Feng JY, et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–36.. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firstenberg MS, Stahel PF, Hanna J, Kotaru C, Crossno J, Jr, Forrester J. Successful COVID-19 rescue therapy by extra-corporeal membrane oxygenation (ECMO) for respiratory failure: a case report. Patient Saf Surg. 2020;14:20. doi: 10.1186/s13037-020-00245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel D, Hui HC, Doerffler E, Clarke MO, Chun K, Zhang L, et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine c-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J Med Chem. 2017;60:1648–61. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 13.Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–504.. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatini DD. Subcellular fractionation of rough microsomes. Cold Spring Harb Protoc. 2014;2014:932–4. doi: 10.1101/pdb.top074567. [DOI] [PubMed] [Google Scholar]
- 15.Food and Drug Administration (US). Bioanalytical method validation guidance for industry. 2018 May [cited 2020 Jul 30]. https://www.fda.gov/media/70858/download.
- 16.European Medicines Agency. Summary on compassionate use: Remdesivir Gilead. 2020 Apr 3 [cited 2020 Jul 30]. https://www.ema.europa.eu/en/documents/other/summary-compassionate-use-remdesivir-gilead_en.pdf.
- 17.Li B, Sedlacek M, Manoharan I, Boopathy R, Duysen EG, Masson P, et al. Butyrylcholinesterase, paraoxonase, and albumin esterase, but not carboxylesterase, are present in human plasma. Biochem Pharmacol. 2005;70:1673–84.. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen SCJ, Kebriaei R, Dresser LD. Remdesivir: review of pharmacology, pre-clinical data, and emerging clinical experience for COVID-19. Pharmacotherapy. 2020;40:659–71.. doi: 10.1002/phar.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirby BJ, Symonds WT, Kearney BP, Mathias AA. Pharmacokinetic, pharmacodynamic, and drug-interaction profile of the hepatitis C virus NS5B polymerase inhibitor sofosbuvir. Clin Pharmacokinet. 2015;54:677–90.. doi: 10.1007/s40262-015-0261-7. [DOI] [PubMed] [Google Scholar]
- 20.Bourgonje AR, Abdulle AE, Timens W, Hillebrands JL, Navis GJ, Gordijn SJ, et al. Angiotensin-converting enzyme-2 (ACE2), SARS-CoV-2 and pathophysiology of coronavirus disease 2019 (COVID-19) J Pathol. 2020;251:228–48.. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Xu X. scRNA-seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, leydig and sertoli cells. Cells. 2020;9:920. doi: 10.3390/cells9040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babusis D, Curry MP, Kirby B, Park Y, Murakami E, Wang T, et al. Sofosbuvir and ribavirin liver pharmacokinetics in patients infected with hepatitis C virus. Antimicrob Agents Chemother. 2018;62:e02587–17. doi: 10.1128/AAC.02587-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Dean RA, Brzezinski MR, Bosron WF. Gender-specific differences in activity and protein levels of cocaine carboxylesterase in rat tissues. Life Sci. 1996;59:1175–84.. doi: 10.1016/0024-3205(96)00435-3. [DOI] [PubMed] [Google Scholar]
- 24.Gabriele M, Puccini P, Lucchi M, Vizziello A, Gervasi PG, Longo V. Presence and inter-individual variability of carboxylesterases (CES1 and CES2) in human lung. Biochem Pharmacol. 2018;150:64–71. doi: 10.1016/j.bcp.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 25.Ishizuka T, Yoshigae Y, Murayama N, Izumi T. Different hydrolases involved in bioactivation of prodrug-type angiotensin receptor blockers: carboxymethylenebutenolidase and carboxylesterase 1. Drug Metab Dispos. 2013;41:1888–95.. doi: 10.1124/dmd.113.053595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.