Abstract
Introduction
COVID-19 pneumonia is characterized by ground-glass opacities (GGOs) and consolidations on Chest CT, although these CT features cannot be considered specific, at least on a qualitative analysis. The aim is to evaluate if Quantitative Chest CT could provide reliable information in discriminating COVID-19 from non-COVID-19 patients.
Materials and methods
From March 31, 2020 until April 18, 2020, patients with Chest CT suggestive for interstitial pneumonia were retrospectively enrolled and divided into two groups based on positive/negative COVID-19 RT-PCR results. Patients with pulmonary resection and/or CT motion artifacts were excluded. Quantitative Chest CT analysis was performed with a dedicated software that provides total lung volume, healthy parenchyma, GGOs, consolidations and fibrotic alterations, expressed both in liters and percentage. Two radiologists in consensus revised software analysis and adjusted areas of lung impairment in case of non-adequate segmentation. Data obtained were compared between COVID-19 and non-COVID-19 patients and p < 0.05 were considered statistically significant. Performance of statistically significant parameters was tested by ROC curve analysis.
Results
Final population enrolled included 190 patients: 136 COVID-19 patients (87 male, 49 female, mean age 66 ± 16) and 54 non-COVID-19 patients (25 male, 29 female, mean age 63 ± 15). Lung quantification in liters showed significant differences between COVID-19 and non-COVID-19 patients for GGOs (0.55 ± 0.26L vs 0.43 ± 0.23L, p = 0.0005) and fibrotic alterations (0.05 ± 0.03 L vs 0.04 ± 0.03 L, p < 0.0001). ROC analysis of GGOs and fibrotic alterations showed an area under the curve of 0.661 (cutoff 0.39 L, 68% sensitivity and 59% specificity, p < 0.001) and 0.698 (cutoff 0.02 L, 86% sensitivity and 44% specificity, p < 0.001), respectively.
Conclusions
Quantification of GGOs and fibrotic alterations on Chest CT could be able to identify patients with COVID-19.
Keywords: COVID-19, Chest CT, Quantitative Chest CT analysis, Interstitial pneumonia
Introduction
From the last months of 2019 the novel coronavirus, severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) [1], has led a pandemic diffusion all over the World [2] and the disease associated was named Coronavirus Disease 2019 (COVID-19) [3]. Guidelines recommend an upper respiratory specimen that should be tested with next-generation sequencing or real-time reverse transcription polymerase chain reaction (RT-PCR) methods for diagnosis of COVID-19 [4], but the early diagnosis is affected by low sensitivity (60–71%) [5, 6] of the test, due to errors in laboratory work or sample collection [7]; moreover, RT-PCR is not universally available, especially in countries with limited resources.
CT was demonstrated to have a pivotal role in the COVID-19 pandemic thanks to a sensitivity of 97% [8], and might be helpful in the management [9] and follow-up of COVID-19 patients [10]; however, due to the low specificity, false positive patients were encountered in clinical practice leaving CT unreliable for the diagnosis [11, 12]. A recent consensus statement from the Fleischner Society pointed out as imaging is indicated for medical triage of suspected COVID-19 patients presenting moderate-severe clinical features and a high pre-test probability of disease [13].
Recently, severity lung scores [14, 15] were proposed from a visual and qualitative evaluation of CT features, but several limitations were reported (i.e., subjectivity, lack of standardization and reproducibility) based on radiologist’s experience [16, 17].
Quantitative Chest CT analysis has the ability to provide quantitative and objective assessment of the healthy lungs, consolidations and ground-glass opacities (GGOs) necessary to quantify the disease burden [17, 18]. In addition, a quantitative analysis could provide relevant information on the progression of the disease and the response to therapy in the follow-up examinations.
During the pandemic, GGOs and consolidations were frequently observed; nevertheless, these CT features were not specific for COVID-19. Thus, the aim of our study was to evaluate if Quantitative Chest CT could provide reliable information in discriminating RT-PCR COVID-19 from non-COVID-19 patients.
Materials and methods
Patient population
This retrospective study was approved by our local institutional review board (IRB) and written informed consent was obtained from all patients or in case of inability from their relatives or the admitting physicians. Patients admitted at the Emergency Department of Sant'Andrea Hospital in Rome from March 31, 2020 until April 18, 2020 were retrospectively included in the study.
Inclusion criteria were (a) diagnosis of interstitial pneumonia on Chest CT and (b) availability of RT-PCR results for COVID-19. Patients with history of lung malignancy who required pulmonary resection and oncohaematologic patients were excluded from the analysis. Image datasets affected by severe respiratory artifacts in whom the software was unable to process the data were also excluded.
Based on RT-PCR results, our population was divided in COVID-19 and non-COVID-19.
Clinical data
According to the hospital internal protocol, at the time of admission suspected COVID-19 patients presenting moderate-severe clinical features and a high pre-test probability of disease (fever defined as > 37.5 °C and respiratory symptoms or direct contact with a confirmed COVID-19 patient) underwent nasopharyngeal and oropharyngeal swabs for SARS-CoV-2. Every patient was tested with two nasopharyngeal and oropharyngeal swabs, the first swab at the entrance and the second after 24 h. The positivity to SARS-CoV-2 was obtained with RT-PCR (Charitè, Berlin, Germany) [19], whereas patients were considered SARS-CoV-2 negative after two consecutive negative RT-PCR results. Demographic data and laboratory results were also collected.
CT acquisition technique
After the first swab, all suspected COVID-19 patients underwent Chest CT to evaluate the presence of interstitial pneumonia. Chest CT was acquired without contrast medium and in supine position during end-inspiration. Each patient was studied using a COVID-19 dedicated 128-slice CT (GE Revolution EVO 64 Slice CT Scanner, GE Medical Systems, Milwaukee, WI, USA). CT scan technical parameters: tube voltage: 120 kV; tube current modulation: 100-250 mAs; spiral pitch factor: 0.98; collimation width: 0.625. Reconstruction images were performed with convolution kernel BONEPLUS at a slice thickness of 1.25 mm. After completion of the CT examination the decontamination of the room was made for every patient, it was performed throughout passive air exchange and disinfecting the surfaces with 0.1% sodium hypochlorite or 62-71% ethanol.
Quantitative Chest CT analysis
Two radiologists in consensus (MZ and FP with 5 and 4 years of experience), blinded to clinical data and RT-PCR results, performed Quantitative Chest CT analysis by using a dedicated software (Thoracic VCAR v13.1, GE). Before segmentation, attenuation value < − 1000 HU was used to exclude trachea air from the analysis. Quantification was performed on naive acquisition using a lung window with a width of 1500 HU and a level of − 600 HU. The software automatically calculated the following parameters: healthy parenchyma, GGOs, consolidation, fibrotic alterations (including fibrotic stripes and subpleural lines) using an adaptive mean based on grayscale, expressed both in liters and percentages, whereas the total lung volume was reported in liters. Vessel was automatically selected and delated. In case of non-adequate automatic segmentation, readers were free to adjust the area of lung impairment segmented by the software.
Statistical analysis
Statistical analysis was performed using MedCalc Statistical Software version 17.9.7 (MedCalc Software bvba, Ostend, Belgium), and p values < 0.05 were considered statistically significant. All data are expressed as mean ± standard deviation (SD). To compare the two group of COVID-19 and non-COVID-19 patients, Kolmogorov–Smirnov test was used to assess data distribution. In case of Gaussian distribution, data were tested with Student’s t test, while Wilcoxon test was applied for non-Gaussian distributed data. Receiver operating characteristic (ROC) curves and Area under the curve (AUC) were calculated for predicting the performance of the quantitative analysis. Results are provided both in liters and percentages.
Results
Patient population and clinical data
Two-hundred and sixteen patients met the inclusion criteria, whereas 26 patients were excluded because of image artefacts. Thus, our final population comprised 190 patients: 136 COVID-19 patients (87 males and 49 females, mean age 66 ± 16 years) and 54 non-COVID-19 patients (25 males and 29 females, mean age 63 ± 15 years) as shown in Fig. 1.
Fig. 1.
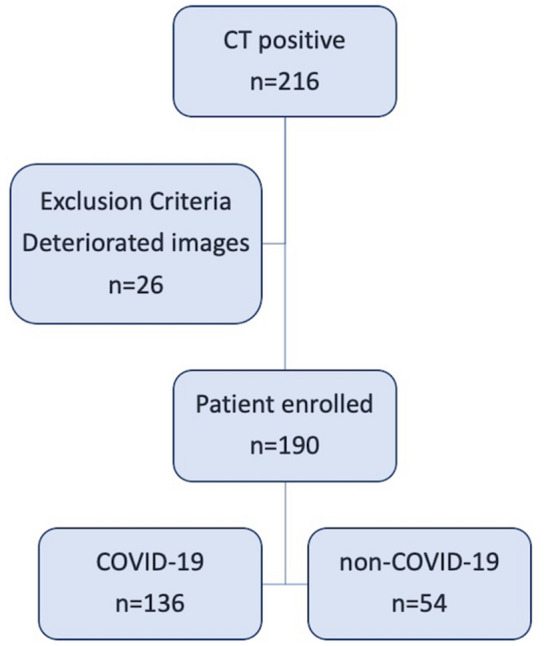
Flowchart of the study. From the initial population of 216 Chest CT positive for interstitial pneumonia, we enrolled 136 patients COVID-19 and 54 patients non-COVID-19
Considering COVID-19 patients, 80/136 patients (59%) presented fever (> 37.5 °C), 49/136 (36%) cough, and 47/136 (34%) dyspnea. Laboratory tests showed lymphocytopenia in 106/136 patients (78%); high values (> 0.50 mg/dL) of C-reactive protein (CRP), lactate dehydrogenase (LDH) (> 220 U/L) and D-dimer (> 243 ng/ml) were found in 128/136 patients (94%), 122/136 patients (90%) and 87/136 patients, respectively.
Regarding non-COVID-19 patients, 26/54 patients (48%) had fever (> 37.5 °C), 20/54 (37%) cough and 13/54 (24%) dyspnea. Lymphocytopenia was reported in 32/54 patients (59%); high values (> 0.50 mg/dL) of CRP, LDH (> 220 U/L) and D-dimer (> 243 ng/ml) were found in 41/54 patients (76%), 40/54 patients (74%) and 33/54 patients (61%), respectively. Full results are listed in Table 1.
Table 1.
Clinical data
| COVID-19 | Non-COVID-19 | |||
|---|---|---|---|---|
| N. patients | % | N. patients | % | |
| Patients demographics | ||||
| Mean age | 66 ± 16 year | 63 ± 15 year | ||
| Years (range) | 18-97 | 18-96 | ||
| Number patients | 136 | 100 | 54 | 100 |
| Male | 87/136 | 64 | 25/54 | 46 |
| Female | 49/136 | 36 | 29/54 | 54 |
| Blood test | ||||
| C-reactive protein (mg/L; normal range 0.00–0.50) | ||||
| Increased | 128/136 | 94 | 41/54 | 76 |
| Normal | 8/136 | 7 | 13/54 | 24 |
| Lactic Acid Dehydrogenase (U/L; range 125–220) | ||||
| Increased | 122/136 | 90 | 40/54 | 74 |
| Normal | 14/136 | 10 | 14/54 | 26 |
| Lymphocytes (× 103/mm3, normal range 1.5–3.0) | ||||
| Increased | 1/136 | 0.7 | 4/54 | 7 |
| Decreased | 106/136 | 78 | 32/54 | 59 |
| Normal | 29/136 | 21.3 | 18/54 | 34 |
| D-dimer (ng/ml, normal < 243) | ||||
| Increased | 87/136 | 64 | 33/54 | 61 |
| Normal | 49/136 | 36 | 21/54 | 39 |
| Symptoms | ||||
| Fever (> 38°) | 80/136 | 59 | 26/54 | 48 |
| Cough | 49/136 | 36 | 20/54 | 37 |
| Dyspnea | 47/136 | 34 | 13/54 | 24 |
Quantitative Chest CT analysis
Considering results expressed in liters, significant differences were reported for GGOs (0.55 ± 0.26 L vs 0.43 ± 0.23 L, p = 0.0005) and fibrotic alterations (0.05 ± 0.03 L vs 0.04 ± 0.03 L, p < 0.0001) (Fig. 2). No significant differences were found for total lung volume (4.09 ± 1.49 L vs 4.20 ± 1.28 L, p = 0.37), healthy parenchyma (3.36 ± 1.58 L vs 3.64 ± 1.33 L, p = 0.11) and consolidations (0.06 ± 0.04 L vs 0.06 ± 0.03 L, p = 0.31). No significant differences were found in the evaluations of lung parenchyma expressed in percentage. Full results are shown in Table 2.
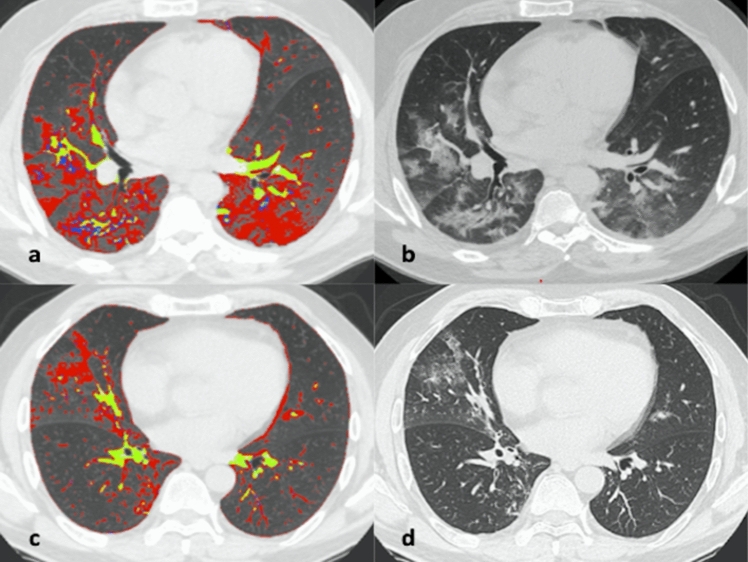
Fig. 2.
57-year-old man with COVID-19 (a, b) and 58-year-old man non-COVID-19 (c, d). Axial unenhanced quantified Chest CT scans that show diffuse bilateral ground-glass opacities (GGOs) and some fibrotic alterations in COVID-19 patient (a, b) and rare ground-glass opacities in non-COVID-19 patient (c, d). Chest CT semi-automatic quantification shows GGOs in red, vessels in yellow and fibrotic alterations in blue, these findings are more represented in COVID-19 patient (a) then in non-COVID-19 patient (c)
Table 2.
Quantitative Chest CT analysis
| COVID-19 | Non-COVID-19 | P values | |
|---|---|---|---|
| Mean | Mean | ||
| Quantitative analysis in liters | |||
| Total lung volume | 4.09 ± 1.49 | 4.20 ± 1.28 | 0.37 |
| Healthy parenchyma | 3.36 ± 1.58 | 3.64 ± 1.33 | 0.11 |
| Ground-glass opacities | 0.55 ± 0.26 | 0.43 ± 0.23 | 0.0005 |
| Fibrotic alterations | 0.05 ± 0.03 | 0.04 ± 0.03 | < 0.0001 |
| Consolidations | 0.06 ± 0.04 | 0.06 ± 0.03 | 0.31 |
| Quantitative analysis in percentage | |||
| Healthy parenchyma | 79.35 ± 13.54 | 85.09 ± 9.35 | 0.36 |
| Ground-glass opacities | 15.59 ± 9.65 | 11.54 ± 7.47 | 0.47 |
| Fibrotic alterations | 1.40 ± 1.07 | 0.93 ± 0.77 | 0.06 |
| Consolidations | 1.75 ± 1.47 | 1.53 ± 1.03 | 0.91 |
Significant differences are shown in bold
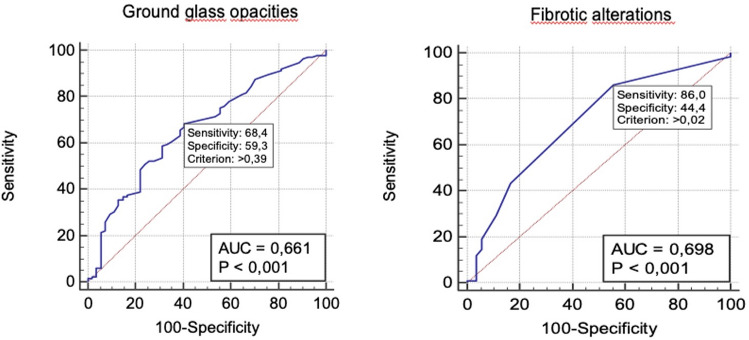
ROC analysis performed for GGOs and fibrotic alterations expressed in liters returned an area under the curve (AUC) of 0.661 for GGOs and 0.698 for fibrotic alterations. When the cutoff of GGOs was set at 0.39L (p < 0.001), sensitivity and specificity were 68% and 59%, respectively, while sensitivity and specificity of fibrotic alterations were 86% and 44%, respectively with a cutoff of 0.02L (p < 0.001) (Fig. 3).
Fig. 3.
ROC curves to test the ability of ground-glass opacities and fibrotic alterations in differentiating COVID-19 from non-COVID-19 patients
Discussion
The results of our study showed a good performance of semi-automatic lung quantification in distinguishing symptomatic patients affected by COVID-19 from those with interstitial pneumonia from non-COVID-19 causes. In particular, quantification of GGOs and fibrotic alterations (including fibrotic stripes and subpleural lines), although with moderate sensitivity and specificity, may be helpful, whereas healthy parenchyma and consolidations are not.
Our results disagree with some literature data, where GGOs are described as the most frequent initial findings in COVID-19 pneumonia [8, 20], but they are not considered specific for COVID-19 [8]. In fact, Chen D. and colleagues analyzed both RT-PCR positive and negative patients in a preliminary study involving 21 patients and their results showed presence of GGOs for 20/21 (95%) patients, with no differences between positive and negative RT-PCR results (p = 0.469), while significant lower presence of consolidation (p = 0.04) was observed in negative RT-PCR patients [21]. The discrepancies between our results and literature data might be explained by several factors: firstly, during early disease spread RT-PCR tests were even less sensitive than now, and the possibility of false negative in a population enrolled during January and February 2020 was concrete with consequent bias on Chest CT findings [22]; then reduced sample size analyzed by authors could have influenced the statistical prevalence of results obtained. In addition, the different imaging analysis, visual or qualitative versus quantitative assessment could be explain the differences. Qualitative analysis is also affected by radiologist skill and experience in image interpretation, whereas quantitative evaluation is a reproducible and comparable technique.
Our results, however, differ also from published studies using a quantitative approach. In a recent paper, a combined scoring system (clinical and radiological features) based on visual assessment was proposed to differentiate COVID-19 patients from interstitial pneumonia of other etiology [23]. Authors interestingly concluded that the combined CT features analysis revealed an AUC of 0.854. Our single-parameter showed an AUC of 0.661 for GGOs and of 0.698 for fibrotic alterations. Despite GGOs performance did not reach the AUC of the cited combined model, this single quantified parameter indicated that it could be a good parameter for the identification of COVID-19 patients just with the sole CT imaging technique. This might be useful in an resource-constrained setting when rapid point-of-care COVID-19 testing is not available or negative. As stated by Fleischner Society, in high pre-test probability scenarios, presumptive diagnosis of COVID-19 achieved by imaging features might support medical triage and clinical management [22]. Moreover, despite these indications are not exactly in line with international guidelines which consider RT-PCR as reference standard for diagnosis, direct experience in critical scenarios revealed how Chest CT represents an helpful resource to suspect a COVID-19 case before the RT-PCR tests; in fact, the latter require an unsustainable response time in a scenario where urgent decision-making is a priority with high number of new hospital admissions associated with limited health personnel and infrastructure resources.
Several controversies emerged on interpretation and frequency of fibrotic alterations. Some authors have described fibrotic alterations as atypical findings, observed on Chest CT only in 1.6% of the population studied [24]; on the contrary, Lomoro et al. [25] considered fibrotic alterations as a typical CT findings due to their presence in 50% of COVID-19 patients at hospital admission. In accordance with Lomoro P. and colleagues, our results showed higher percentage of fibrotic alterations in COVID-19 patients than non-COVID-19 (p < 0.0001). Discrepancies among studies might have different explanations: first of all, different CT pulmonary evaluations (visual vs quantitative) could have affect how CT features were assessed; then, a possible bias could be the time range between symptoms onset and hospital admission when CT scans were performed. In fact, the longer the time between the onset of symptoms and the acquisition of CT scan, the higher the probability to detect fibrotic alterations, indicative of a late stage pneumonia.
Despite the interesting results, the present study has some limitations such as the retrospective nature, lack of data regarding symptoms onset from the hospital admission, the small sample of patients with negative RT-PCR included and the lack of a combined clinical and radiological decisional model to discriminate COVID-19 from non-COVID-19 patients.
Conclusions
In conclusion, Chest CT quantification has the potential to improve COVID-19 management for diagnosis; it could increase specificity of CT images and help the identification of COVID-19 patients before the RT-PCR results. By so doing, it could be a very useful tool in difficult scenarios such as resource-constrained setting triage. Future application might also include COVID-19 CT follow-up with a quantitative assessment of lung impairment as expression of worsening, stability or healing of lung parenchyma. To explore these possibilities, larger studies and data interpretation are needed.
CT quantification of ground-glass opacities and fibrotic alterations is able to identify COVID-19 patients with moderate accuracy and may help radiologists to overcome low specificity of Chest CT.
Acknowledgements
We would like also to acknowledge the entire Radiological Medical and Technical Staff of the Radiology Unit of Sant’Andrea Academic Hospital in Rome.
Abbreviations
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- COVID-19
Coronavirus Disease 2019
- RT-PCR
Reverse transcription polymerase chain reaction
- GGOs
Ground-glass opacities
- CRP
C-reactive protein
- LDH
Lactate dehydrogenase
Authors’ contribution
D. Caruso: Project administration, Formal analysis, Writing—review & editing; M. Polici: Data curation, Writing—original draft; M. Zerunian: Methodology, Writing—original draft; F. Pucciarelli: Software; T. Polidori: Visualization; G. Guido: Validation; C. Rucci: Software; B. Bracci: Methodology; E. Muscogiuri: Visualization; C. De Dominicis: Conceptualization; A. Laghi: Supervision, Formal analysis, Writing—review & editing.
Funding
None.
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
Informed consent was obtained from all individual participants included in the study
Consent for publication
The authors affirm that human research participants provided informed consent for publication of the images in Fig. 2.
Code availability
Quantitative Chest CT analysis was performed by using a dedicated software (Thoracic VCAR v13.1, GE).
Ethics approval
This retrospective study was approved by our local institutional review board (IRB) and written informed consent was obtained from all patients or in case of inability from their relatives or the admitting physicians.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Damiano Caruso, Email: damiano.caruso@uniroma1.it.
Michela Polici, Email: michela.polici@uniroma1.it.
Marta Zerunian, Email: marta.zerunian@uniroma1.it.
Francesco Pucciarelli, Email: francesco.pucciarelli@uniroma1.it.
Tiziano Polidori, Email: tiziano.polidori@uniroma1.it.
Gisella Guido, Email: gisella.guido@uniroma1.it.
Carlotta Rucci, Email: carlotta.rucci@uniroma1.it.
Benedetta Bracci, Email: benedetta.bracci@uniroma1.it.
Emanuele Muscogiuri, Email: emanuele.muscogiuri@uniroma1.it.
Chiara De Dominicis, Email: dedominicischiara.1@gmail.com.
Andrea Laghi, Email: andrea.laghi@uniroma1.it.
References
- 1.Naming the coronavirus disease (COVID-19) and the virus that causes it (2020). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- 2.Coronavirus disease (COVID-2019) situation reports (2020). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 3.Novel Coronavirus – China (2020). https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/
- 4.Interim Guidance: Healthcare Professionals 2019-nCoV|CDC (2020). https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-criteria.html
- 5.Yang Yang MY, Shen Chenguang, Wang Fuxiang, Yuan Jing, Li Jinxiu, Zhang Mingxia, Wang Zhaoqin, Xing Li, Wei Jinli, Peng Ling, Wong Gary, Zheng Haixia, Weibo Wu, Liao Mingfeng, Feng Kai, Li Jianming, Yang Qianting, Zhao Juanjuan, Zhang Zheng, Liu Lei, Liu Yingxia. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020;1:5. doi: 10.1101/2020.02.11.20021493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH. Essentials for radiologists on COVID-19: an update. Radiology. 2020 doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe M, Caruso D, Tuccinardi D, Risi R, Zerunian M, Polici M, Pucciarelli F, Tarallo M, Strigari L, Manfrini S, Mariani S, Basciani S, Lubrano C, Laghi A, Gnessi L. Visceral fat shows the strongest association with the need of intensive care in patients with COVID-19. Metabolism. 2020 doi: 10.1016/j.metabol.2020.154319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol. 2020;1:1–7. doi: 10.2214/ajr.20.22954. [DOI] [PubMed] [Google Scholar]
- 11.Kim H, Hong H, Yoon SH. Diagnostic performance of CT and reverse transcriptase-polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. 2020 doi: 10.1148/radiol.2020201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caruso D, Zerunian M, Polici M, Pucciarelli F, Polidori T, Rucci C, Guido G, Bracci B, de Dominicis C, Laghi A. Chest CT features of COVID-19 in Rome, Italy. Radiology. 2020 doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, Schluger NW, Volpi A, Yim JJ, Martin IBK, Anderson DJ, Kong C, Altes T, Bush A, Desai SR, Goldin J, Goo JM, Humbert M, Inoue Y, Kauczor HU, Luo F, Mazzone PJ, Prokop M, Remy-Jardin M, Richeldi L, Schaefer-Prokop CM, Tomiyama N, Wells AU, Leung AN. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology. 2020 doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, Pan I, Shi LB, Wang DC, Mei J, Jiang XL, Zeng QH, Egglin TK, Hu PF, Agarwal S, Xie F, Li S, Healey T, Atalay MK, Liao WH. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang G, Gong T, Wang G, Wang J, Guo X, Cai E, Li S, Li X, Yu Y, Lin L. Timely diagnosis and treatment shortens the time to resolution of coronavirus disease (COVID-19) pneumonia and lowers the highest and last CT scores from sequential chest CT. AJR Am J Roentgenol. 2020 doi: 10.2214/ajr.20.23078. [DOI] [PubMed] [Google Scholar]
- 16.Shen C, Yu N, Cai S, Zhou J, Sheng J, Liu K, Zhou H, Guo Y, Niu G. Quantitative computed tomography analysis for stratifying the severity of coronavirus disease 2019. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Huang RH, Ai Tao, Pengxin Yu, Kang Han, Tao Qian, Xia Liming. Serial quantitative chest CT assessment of COVID-19: deep-learning approach. Radiology. 2020 doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran Yang XL, Liu Huan, Zhen Yanling, Zhang Xianxiang, Xiong Qiuxia, Luo Yong, Gao Cailiang, Zeng Wenbing. Chest CT severity score: an imaging tool for assessing severe COVID-19. Radiology. 2020 doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.es.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, Cui J, Xu W, Yang Y, Fayad ZA, Jacobi A, Li K, Li S, Shan H. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, Jiang X, Hong Y, Wen Z, Wei S, Peng G, Wei X. Can chest CT features distinguish patients with negative from those with positive initial RT-PCR results for coronavirus disease (COVID-19)? AJR Am J Roentgenol. 2020 doi: 10.2214/ajr.20.23012. [DOI] [PubMed] [Google Scholar]
- 22.Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, Schluger NW, Volpi A, Yim JJ, Martin IBK, Anderson DJ, Kong C, Altes T, Bush A, Desai SR, Goldin J, Goo JM, Humbert M, Inoue Y, Kauczor HU, Luo F, Mazzone PJ, Prokop M, Remy-Jardin M, Richeldi L, Schaefer-Prokop CM, Tomiyama N, Wells AU, Leung AN. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Chest. 2020 doi: 10.1016/j.chest.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo L, Luo Z, Jia Y, Zhou C, He J, Lyu J, Shen X. CT differential diagnosis of COVID-19 and non-COVID-19 in symptomatic suspects: a practical scoring method. BMC Pulm Med. 2020;20(1):129. doi: 10.1186/s12890-020-1170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z, Guo D, Li C, Fang Z, Chen L, Yang R, Li X, Zeng W. Coronavirus disease 2019: initial chest CT findings. Eur Radiol. 2020 doi: 10.1007/s00330-020-06816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomoro P, Verde F, Zerboni F, Simonetti I, Borghi C, Fachinetti C, Natalizi A, Martegani A. COVID-19 pneumonia manifestations at the admission on chest ultrasound, radiographs, and CT: single-center study and comprehensive radiologic literature review. Eur J Radiol Open. 2020;7:100231. doi: 10.1016/j.ejro.2020.100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.