Abstract
Statistical modeling of the corrosion inhibition process by twenty-one pyridazine derivatives for mild steel in acidic medium was investigated by the quantitative structure property relationship (QSPR) approach. This modeling was established by the correlation between the corrosion inhibition efficiency (IE %) and a number of the electronic and structural properties of these inhibitors such as: the EHOMO (highest occupied molecular orbital energy), the ELUMO (lowest unoccupied molecular orbital energy), the energy gap (EL-H), the dipole moment (μ), the hardness (η), the softness (σ), the absolute electronegativity (χ), the ionization potential (IP), the electron affinity (EA), the fraction of electrons transferred (ΔN), the electrophilicity index ω the molecular volume (Vm), the logarithm of the partition coefficient (Log P), and the molecular mass (M), in addition to the inhibitor concentration (Ci). The structure electronic properties was calculated by the use of the density functional theory method (DFT), at B3LYP/6-31G (d, p) level of theory and the analysis of dimensionality and redundancy as well as the test of collinearity between descriptors are carried out using principal component analysis (PCA). Whereas, the correlation between EI % and molecular structure is performed through the development of tree mathematical models, based-QSPR approaches: the partial least squares regression (PLS), the principal component regression (PCR) and the artificial neural networks (ANN). Indeed, the statistical quantitative results revealed that PCR and ANN were the most relevant and predictive models in comparison with the PLS model. This pertinence was demonstrated by using leave one-out cross-validation as an efficient method for testing the internal stability and predictive capability of said models with a high cross-validated determination coefficient R2cv= 0.92 and predicted determination coefficient R2pred= 0.92 and R2pred= 0.90 for PCR and ANN respectively; in addition to an extrapolation test set as an external validation with a significant external coefficient of determination: R2test= 0.94 and R2test= 0.92, for the two correspondingly models.
Keywords: Materials chemistry, Theoretical chemistry, Corrosion inhibition, DFT calculations, Principal components analysis, Partial least squares regression, Principal component regression, Artificial neural networks, Cross validation
Materials chemistry, Theoretical chemistry, Corrosion inhibition; DFT calculations; Principal components analysis; Partial least squares regression; Principal component regression; Artificial neural networks; Cross validation
1. Introduction
The corrosion of the metal compounds and alloys as well as the deterioration of its characteristics is a major industrial problem. It is considered a worrying source of pollution for the environment; especially, in carbon steel industry, where acids are being used to pickling, descaling and cleaning. Therefore, scientists are encouraged to look for solutions to protect these materials and reduce their environmental impact [1, 2, 3, 4, 5, 6, 7]. For this reason, the use of corrosion inhibitors to control the metal dissolution and to limit the corrosion rate is one of the common techniques [8, 9, 10, 11, 12, 13, 14]. Indeed, the use of corrosion inhibitors differs according to the nature of the corrosive environment. For example, in acidic mediums, organic molecules remain the most common class of inhibitors to remedy this problem. The action and effectiveness of these compounds depends, to a large extent, on their structural and electronic properties. They generally have chemical structures containing multiple, double and triple bonds, as well as heteroatoms such as nitrogen, oxygen, sulphur or phosphorus embedded in aromatic rings, which give them better adsorption on the surface of the material, and therefore high reactivity [15, 16, 17, 18]. In this optic, previous studies suggest that pyridazine derivatives are considered as a potential class of corrosion inhibitors in electrolytic media, and especially, in acidic solutions [19, 20, 21, 22, 23]. In fact, the choice of this class of molecules for inhibiting corrosion applications is reported by several authors, such as, Chetouani et al. [24], Bouklah et al. [25], Bentiss et al. [26], Zerga et al. [27], Khadiri et al. [28], Mashuga et al. [29] and El-Hajjaji et al. [30]. Experimentally, these organic derivatives can provide electrons to the metal surface to form coordinate covalent bonds and can also accept free electrons from the metal surface by using their lower unoccupied orbital, which facilitates their adsorption onto the metallic surface. This adsorption allows the blocking of active sites by reducing the steel dissolution rate along with the discharge of proton, which increases the coverage ratio, reflected by an elevation of inhibition effect of these compounds [27,29,31, 32, 33, 34, 35, 36, 37].
Parallel to the experimental study, the quantum chemical approaches have proven to be very useful in determining the molecular structure as well as elucidating the electronic structure and the characteristics of the reactive sites [38,39] in order to explain the mechanism of reactivity for corrosion inhibition process, and to understand the relationship between the corrosion inhibition efficiency and a number of molecular indices of these inhibitors [40,41]. On the other hand, quantitative structure-property relationship (QSPR) [42,43] has been widely used recently, to provide quantitative analysis of corrosion inhibition process, as attempts to find consistent relationship between the variations in the values of molecular properties and the inhibitor activity for a series of compounds. Thereby, the application of this approach in corrosion inhibition research, especially, in acidic media, was reported by several authors: F.B. Growcock et al. [45,54], P.G. Abdul-Ahad et al. [46], P. Dupin et al. [47] and I. Lukovits et al. [48,49].
The use of this mathematical technique can supply useful qualitative and quantitative information, for a better understanding of this corrosion inhibition phenomenon. The objective of the present work is to attempt an established a quantitative structure-activity/property relationship between the corrosion inhibition efficiency (IE %) and a number of molecular indices such as EHOMO, ELUMO, EL-H, μ, η, σ, χ, IP, EA, ΔN, ω, Vm, Log P, and (M), in addition to the inhibitor concentration (Ci), computed, in aqueous phases by the means of the DFT at B3LYP methods with 6-31G (d, p) basis set, for twenty one (n = 21) pyridazine derivatives denoted hereafter, P1 to P21, illustrated in Figure 1, already studied experimentally as mild steel corrosion inhibitors in 1.0 M HCl for a various range of inhibitor concentration with distinguishable efficiencies [27,29,31, 32, 33, 34, 35, 36, 37]. This statistical investigation is evaluated by a comparative study between three mathematical regression models: the partial least squares regression (PLS), the principal component regression (PCR) and the artificial neural networks (ANN); in order to deeply understand the corrosion inhibition mechanism and to determine the relevant molecular indices influencing the variation of the inhibition efficiency for the studied compounds. As well as, the exploitation of the established equations makes it furthermore possible to estimate the inhibition properties of other similar compounds, in the absence of experimental data, and consequently to orient the organic partner to their synthesis due to the predicted promising character.
Figure 1.
2D un-optimized molecular structures of the studied pyridazine derivatives.
2. Materials and methods
2.1. Experimental data
In the present study, twenty-one pyridazine derivatives were selected in order to establish a quantitative structure property relationship (QSPR) between their inhibitive potential and their molecular structure. This work is considered as a complementary statistical study and a second part of several searches already published for the proposed pyridazine compounds differently substituted and studied as mild steel corrosion inhibitors in 1.0 M HCl at different concentration with distinguishable efficiencies [27,29,31, 32, 33, 34, 35, 36, 37]. The studied compounds names and their corresponding experimental corrosion inhibition efficiencies are summarized in Table 1.
Table 1.
Names and experimental corrosion inhibition efficiency values (IEexp%) of the studied compounds at different concentrations.
| Inhibitor | IUPAC name | Ci/M | IEexp% | Reference |
|---|---|---|---|---|
| P1 | 6-methyl-4,5-dihydro-2H-pyridazine-3-one | 0.5. 10−3 | 17.70 | [31] |
| P2 | 6-phenyl-2H-pyridazine-3-one | 0.5. 10−3 | 25.60 | [31] |
| P3 | 6-phenyl-2H-pyridazine-3-thione | 0.5. 10−3 | 98.50 | [31] |
| P4 | 3-Chloro-6-(1H-pyrazol-1-yl) pyridazine | 2.8. 10−3 | 69.05 | [32] |
| P5 | 1-(6-Chloro pyridazin-3-yl) piperidin-4-ol | 2.3. 10−3 | 82.42 | [32] |
| P6 | 3-Chloro-6-(3,5-dimethyl-1H-pyrazol-1-yl) pyridazine | 2.2. 10−3 | 83.04 | [32] |
| P7 | Ethyl (6-methyl-3-oxopyridazin-2-yl) acetate | 1.10–3 | 81.10 | [33] |
| P8 | Ethyl[4-(2-chlorobenzyl)-3-methyl-6-thioxopyridazin-1(6H)-yl] acetate | 1.10–3 | 97.00 | [27,34] |
| P9 | Ethyl[4-(2-chlorobenzyl)-3-methyl-6-oxopyridazin-1(6H)-yl] acetate | 1.10–3 | 91.00 | [27,34] |
| P10 | 5-(2-chlorobenzyl)-2-(2-hydroxyethyle)-6-methylpyridazin-3(2H)-one | 1.10–3 | 87.00 | [27,34] |
| P11 | 5-(2-chlorobenzyl)-2-(2-hydroxyethyle)-6-methylpyridazin-3(2H)-thione | 1.10–3 | 93.00 | [27,34] |
| P12 | 5-benzyl-6-methyl pyridazine-3-thione | 1.10–4 | 90.00 | [35] |
| P13 | 5-benzyl-6-methyl pyridazine-3-one | 1.10–4 | 65.00 | [35] |
| P14 | stet 5-benzyl-6-methylpyridazin-3-yl thioethanoic | 1.10–4 | 87.00 | [36] |
| P15 | (5-benzyl-6-methyl–3-Oxopyridazin-3-yl)Ethonoate d'ethyle | 1.10–4 | 69.00 | [36] |
| P16 | (5-benzyl-6-methylpyridazin-3-yl)Thioethonoate d'ethyle | 1.10–4 | 83.00 | [36] |
| P17 | 2-(6-chloropyridazin-3-yl)-2-phenylacetonitrile | 2.2. 10−3 | 95.84 | [29] |
| P18 | 3-(6-chloro-3-pyridazinyl)-1H-indole | 2.2. 10−3 | 95.67 | [29] |
| P19 | 4-(6-chloropyridazin-3-yl) benzoic acid | 2.1. 10−3 | 72.64 | [29] |
| P20 | 3-(6-chloropyridazin-3-yl) benzoic acid | 2.1. 10−3 | 85.20 | [29] |
| P21 | 1,4-bis(2-pyridyl)-5H-pyridazino[4,5-b] indole | 1.10–4 | 90.20 | [37] |
2.2. Methodology
2.2.1. Computational methods
In terms of the structure and the molecular properties, the geometry of the corrosion inhibitors affects directly the characteristics and the nature of the interface inhibitor/metal and plays a major role in the mechanism of adsorption of the molecule on the metal surface. In this context, the 3D molecular structures were generated by the Gauss View 03 software, and the quantum chemical calculations were performed with complete geometry optimizations using the density functional theory (DFT) with the Beck's three parameter exchange functional along with the Lee-Yang-Parr non local correlation functional (B3LYP) at 6-31G (d, p) basis set [50, 51, 52], by the exploitation of the options of the Gaussian-03 software [53]; in attempt to determine the pertinent theoretical parameters, and to identify the inhibition properties of the undertaken derivatives. On the other hand, ChemSketch program, version 12.0 [54] was also employed to calculate the others molecular descriptors, which are selected to correlate the inhibition activity to their chemical structures of the under investigated compounds.
2.2.2. Statistical analysis
The statistical study was carried out on twenty-one pyridazine derivatives in order to establish a quantitative relationship structure-property between the corrosion inhibition efficiency (IE %) and the intrinsic electronic and structural properties of these compounds [45,55,56].
The proposed approach for this study is to perform a principal components analysis (PCA), which allows us to check redundancy and collinearity between the descriptors studied and to conduct a comparative statistical study between three proposed mathematical models: the partial least squares regression (PLS), the principal component regression (PCR) and the artificial neural networks (ANN); in order to correlate the anticorrosion activity to the molecular structure.
After their calculations, and by using XLSTAT software, version 2014 [57], the selected descriptors were used to establish statistical models that relate the inhibitive activity of compounds to their chemical structures such as electronic and structural molecular indices; by separating the data set into a training set, which are used to establish the three QSPR models and a test set which are exploited to assess the performance of these obtained models.
2.2.3. Validation
To evaluate the quality of explicability and the degree of prediction of each QSAR/QSPR model, validation tests must be carried out. In this context, internal and external validations are necessary. Internal validation is based, on the one hand, on several statistical parameters, such as: the determination coefficient R2, the adjusted determination coefficient R2adj, the predicted determination coefficient R2pred, the predicted residual sum of squares PRESS, the standard deviation SD and the significance level p-value; on the other hand, it is justified by the means of the leave one out (LOO) cross validation method, using the cross-validated coefficient of determination R2cv as an indicator to evaluate the pertinence of this procedure. The R2cv is expressed as [58, 59, 60]:
| (1) |
where and are the experimental and calculated values for the dependent variables, respectively, and is the average experimental value.
On the other hand, the external validation was verified by earmarking 20 % of the sample as a test set, using external coefficient of determination R2ext, which is determined as follows [58, 59, 60]:
| (2) |
where and are the calculated and experimental values for the test set, respectively; and is the average experimental value for the training set of the dependent variables.
3. Results and discussion
3.1. Quantum calculation
3.1.1. Molecular geometry
The molecular geometries of the investigated compounds are determined by optimizing all structural parameters in aqueous phases, using the Density Functional Theory (DFT) at B3LYP/6-31G (d, p) level of theory [50, 51, 52]. The obtained geometries were qualified by the minimum energy of the molecular structures, which is proved by the absence of imaginary frequencies and by the comparison of the bond lengths and the pertinent dihedral angel values with the referential values. The final optimized geometries are listed in Figure 2.
Figure 2.
Optimized geometries obtained by B3LYP/6-31G (d, p) of the studied molecules.
3.1.2. Calculations of molecular descriptors
Because of the dependence of this corrosion inhibition phenomenon on various factors, the experimental study alone cannot explain the metal/solution interface behavior; which implies a coupling of this study with a quantum and/or statistical study such as a quantitative structure property relationship (QSPR). In this context, the QSPR approach focuses essentially on the correlation of the intrinsic properties of each molecule with its inhibitory potential. To this end, many models have been developed by several researchers in order to be able to link the inhibition efficiency of molecules to some of their properties. So, the inhibition efficiency can be predicted from molecular descriptors and elucidate the mechanism of inhibition. Thus, according to the literature, the most relevant descriptors that can influence the adsorption of the molecular inhibitors onto the metallic surface and their inhibition efficiencies are mainly the electronic, structural and lipophilic index. The pertinent descriptors considered in this work were: the highest occupied molecular orbital energy (EHOMO), the lowest unoccupied molecular orbital energy (ELUMO), the energy gap (EL-H), the dipole moment (μ), the hardness (η), the softness (σ), the absolute electronegativity (χ), the ionization potential (IP), the electron affinity (EA), the fraction of electrons transferred (ΔN), the electrophilicity index (ω), the molecular volume (Vm), the logarithm of the partition coefficient (Log P), and the molecular mass (M), in addition to the inhibitor concentration (Ci). The obtained values of the used descriptors are illustrated in Table2.
Table 2.
The values of Molecular descriptor calculated at B3LYP/6-31G∗∗ in aqueous phase with the IEexp% for different concentrations.
| Compound | Ci/M |
IEexp % |
EHOMO(eV) | ELUMO(eV) | EL-H(eV) | μ (D) | IP (eV) | EA (eV) | χ (eV) | Ƞ (eV) | σ (eV−1) | ΔN | ω (eV) | Log P | M g.mol−1 | Vmcm3.mol−1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 0.5 10−3 | 17.70 | -6.727 | -0.949 | 5.77 | 4.090 | 6.727 | 0.949 | 3.840 | 2.89 | 0.345 | 0.547 | 2.55 | 0.770 | 110.113 | 89.90 |
| P2 | 0.5 10−3 | 25.60 | -6.511 | -2.274 | 4.24 | 4.130 | 6.511 | 2.274 | 4.390 | 2.19 | 0.456 | 0.615 | 4.400 | 0.980 | 172.183 | 143.10 |
| P3 | 0.5 10−3 | 98.50 | -6.013 | -2.496 | 3.51 | 5.310 | 6.013 | 2.496 | 4.250 | 1.76 | 0.568 | 0.780 | 5.131 | 1.590 | 188.248 | 154.50 |
| P4 | 2.8 10−3 | 69.05 | -7.088 | -2.271 | 4.817 | 6.317 | 7.088 | 2.271 | 4.679 | 2.409 | 0.415 | 0.481 | 4.545 | 0.860 | 180.594 | 122.30 |
| P5 | 2.3 10−3 | 82.42 | -6.253 | -1.808 | 4.445 | 6.203 | 6.253 | 1.808 | 4.031 | 2.222 | 0.450 | 0.668 | 3.656 | 0.420 | 213.664 | 157.00 |
| P6 | 2.2 10−3 | 83.04 | -5.691 | -1.967 | 3.724 | 5.733 | 5.691 | 1.967 | 3.829 | 1.862 | 0.537 | 0.851 | 3.936 | 0.570 | 222.674 | 169.00 |
| P7 | 1.0 10−3 | 81.10 | -6.228 | -1.577 | 4.711 | 2.433 | 6.228 | 1.577 | 3.902 | 2.325 | 0.430 | 0.666 | 3.274 | 0.380 | 196.207 | 163.70 |
| P8 | 1.0 10−3 | 97.00 | -6.068 | -2.095 | 3.973 | 8.399 | 6.068 | 2.095 | 4.081 | 1.986 | 0.503 | 0.736 | 4.192 | 3.140 | 336.836 | 268.70 |
| P9 | 1.0 10−3 | 91.00 | -6.476 | -1.714 | 4.572 | 5.787 | 6.476 | 1.714 | 4.095 | 2.381 | 0.419 | 0.610 | 3.521 | 2.530 | 320.770 | 257.20 |
| P10 | 1.0 10−3 | 87.00 | -6.558 | -1.742 | 4.816 | 6.438 | 6.558 | 1.742 | 4.150 | 2.408 | 0.415 | 0.592 | 3.576 | 1.280 | 278.734 | 219.50 |
| P11 | 1.0 10−3 | 93.00 | -6.068 | -2.095 | 3.973 | 6.550 | 6.558 | 2.095 | 4.326 | 2.231 | 0.448 | 0.599 | 4.194 | 1.880 | 294.799 | 230.90 |
| P12 | 1.0 10−4 | 90.00 | -6.038 | -1.985 | 4.053 | 8.900 | 6.038 | 1.985 | 4.011 | 2.026 | 0.493 | 0.737 | 3.970 | 2.150 | 216.302 | 185.90 |
| P13 | 1.0 10−4 | 65.00 | -6.392 | -1.523 | 4.869 | 5.438 | 6.392 | 1.523 | 3.957 | 2.869 | 0.348 | 0.530 | 2.728 | 1.540 | 200.236 | 174.50 |
| P14 | 1.0 10−4 | 87.00 | -6.392 | -1.360 | 5.032 | 2.976 | 6.392 | 1.360 | 3.876 | 2.516 | 0.397 | 0.620 | 2.985 | 2.280 | 274.338 | 209.30 |
| P15 | 1.0 10−4 | 69.00 | -6.446 | -1.632 | 4.814 | 4.519 | 6.446 | 1.632 | 4.039 | 2.407 | 0.415 | 0.615 | 3.388 | 1.940 | 286.325 | 247.90 |
| P16 | 1.0 10−4 | 83.00 | -6.392 | -1.360 | 5.032 | 2.525 | 6.392 | 1.360 | 3.876 | 2.516 | 0.397 | 0.620 | 2.985 | 2.810 | 302.391 | 250.50 |
| P17 | 2.2 10−3 | 95.84 | -7.380 | -2.460 | 4.920 | 7.050 | 7.380 | 2.460 | 4.920 | 2.46 | 0.390 | 0.422 | 4.920 | 1.180 | 229.665 | 177.80 |
| P18 | 2.2 10−3 | 95.67 | -5.920 | -1.820 | 4.100 | 6.880 | 5.920 | 1.820 | 3.870 | 2.05 | 0.487 | 0.763 | 3.652 | 1.910 | 229.665 | 165.50 |
| P19 | 2.1 10−3 | 72.64 | -7.190 | -2.550 | 5.360 | 2.870 | 7.190 | 2.550 | 4.870 | 2.68 | 0.373 | 0.397 | 4.424 | 1.760 | 234.638 | 165.60 |
| P20 | 2.1 10−3 | 85.20 | -7.090 | -2.220 | 4.870 | 6.230 | 7.090 | 2.220 | 4.660 | 2.44 | 0.409 | 0.479 | 4.449 | 1.840 | 234.638 | 165.60 |
| P21 | 1.0 10−4 | 90.20 | -7.631 | -1.910 | 5.721 | 6.356 | 7.631 | 1.910 | 4.770 | 2.860 | 0.349 | 0.389 | 3.977 | 3.267 | 323.350 | 239.40 |
3.2. QSPR study
3.2.1. Principal component analysis
Principal component analysis (PCA) [61] is a statistical qualitative analysis procedure. This descriptive method can be used to reduce the dimensionality of large data set, by transforming a large set of variables into a smaller one that are uncorrelated. These new variables are called “main components”, or main axes. It allows the practitioner to reduce the number of variables and make the information less redundant [62].
Accordingly, in this work, the principal component analysis (PCA) was performed to the fourteen descriptors with the molecular concentration of the twenty-one molecules. The fifteen principal obtained components are presented in Figure 3.
Figure 3.
The principal components and their variances.
The contributions of the descriptors to the principal components F1, F2 and F3 were summarized in Table 3. According to these results, the descriptors EHOMO, EL-H, IP, ƞ, σ and ΔN have the most significant contributions to F1, while the descriptors, ELUMO, EA, χ, and ω have the most significant contributions to F2, whereas the descriptors Log P, M, Vm and Ci have the most significant contributions to F3. On the contrary, the descriptor μ represents a low contribution to the three principal components; with a slight contribution value to F2.
Table 3.
Descriptor contributions to the first three principal components F1, F2 and F3.
| Descriptor | F1 |
F2 |
F3 |
|||
|---|---|---|---|---|---|---|
| Correlation | Contribution |
Correlation | Contribution |
Correlation | Contribution |
|
| % | % | % | ||||
| Ci/M | 0.0998 | 0.1711 | -0.4971 | 5.5329 | -0.4409 | 6.0253 |
| EHOMO/eV | 0.8526 | 12.4849 | 0.4827 | 5.2167 | 0.0058 | 0.0010 |
| ELUMO/eV | -0.2350 | 0.9487 | 0.9364 | 19.6318 | 0.1284 | 0.5112 |
| EL-H/eV | -0.9591 | 15.7973 | 0.1617 | 0.5854 | 0.0269 | 0.0224 |
| μ/D | 0.3296 | 1.8659 | -0.4534 | 4.6026 | 0.2460 | 1.8754 |
| IP/eV | -0.8431 | 12.2079 | -0.5141 | 5.9179 | 0.0294 | 0.0268 |
| EA/eV | 0.2350 | 0.9487 | -0.9364 | 19.6318 | -0.1284 | 0.5112 |
| Χ/eV | -0.4476 | 3.4407 | -0.8859 | 17.5709 | -0.0530 | 0.0870 |
| ƞ/eV | -0.9578 | 15.7567 | 0.1968 | 0.8670 | 0.0602 | 0.1123 |
| σ/eV−1 | 0.9662 | 16.0331 | -0.1679 | 0.6313 | -0.0736 | 0.1681 |
| ΔN | 0.9149 | 14.3753 | 0.3799 | 3.2306 | -0.0170 | 0.0090 |
| ω/eV | 0.4053 | 2.8212 | -0.7382 | 12.2001 | -0.1616 | 0.8093 |
| Log P | -0.0928 | 0.1478 | -0.1628 | 0.5934 | 0.8744 | 23.7032 |
| M/g.mol−1 | 0.0269 | 0.0124 | -0.1627 | 0.5928 | 0.9449 | 27.6791 |
| Vm/cm3.mol−1 | 0.0825 | 0.1168 | -0.0147 | 0.0048 | 0.9589 | 28.5038 |
According to the projection of the variables in the plane of the three first principal components F1, F2 and F3 and based on their percentage contribution in the two correlation circles illustrate in Figure 4, these axes account for as much of the variability in the data as possible. They represent respectively, (36.39 %; 27.92 % and 20.16 %) of the total variance and the total information is estimated to a percentage of 84.47 %, were enough to describe the information represented by the data set.
Figure 4.
Correlation circles between the principle compounds F1–F2 (a) and F1–F3 (b).
The principal component analysis (PCA) was also performed to detect and identify the correlation between the different variables. This correlation was presented in Table 4.
Table 4.
The correlation matrix (Pearson (n)) between different descriptors.
| Variable | Ci | EHOMO | ELUMO | EL-H | μ | IP | EA | Χ | ƞ | σ | ΔN | ω | Log P | M | Vm | IEexp % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ci | 1.000 | |||||||||||||||
| EHOMO | -0.147 | 1.000 | ||||||||||||||
| ELUMO | -0.481 | 0.231 | 1.000 | |||||||||||||
| EL-H | -0.143 | -0.766 | 0.401 | 1.000 | ||||||||||||
| μ | 0.221 | 0.046 | -0.389 | -0.402 | 1.000 | |||||||||||
| IP | 0.144 | -0.978 | -0.260 | 0.726 | -0.017 | 1.000 | ||||||||||
| EA | 0.481 | -0.231 | -1.000 | -0.401 | 0.389 | 0.260 | 1.000 | |||||||||
| Χ | 0.374 | -0.745 | -0.746 | 0.273 | 0.210 | 0.837 | 0.746 | 1.000 | ||||||||
| ƞ | -0.224 | -0.700 | 0.415 | 0.939 | -0.350 | 0.693 | -0.415 | 0.242 | 1.000 | |||||||
| σ | 0.164 | 0.720 | -0.392 | -0.936 | 0.343 | -0.719 | 0.392 | -0.273 | -0.984 | 1.000 | ||||||
| ΔN | -0.085 | 0.943 | 0.146 | -0.794 | 0.122 | -0.963 | -0.146 | -0.747 | -0.815 | 0.841 | 1.000 | |||||
| ω | 0.215 | -0.036 | -0.742 | -0.498 | 0.276 | 0.049 | 0.802 | 0.487 | -0.539 | 0.572 | 0.103 | 1.000 | ||||
| Log P | -0.429 | -0.158 | -0.032 | 0.093 | 0.285 | 0.175 | 0.032 | 0.138 | 0.116 | -0.093 | -0.143 | 0.020 | 1.000 | |||
| M | -0.202 | -0.042 | -0.044 | -0.026 | 0.234 | 0.089 | 0.044 | 0.086 | -0.019 | -0.019 | -0.053 | -0.086 | 0.779 | 1.000 | ||
| Vm | -0.386 | 0.078 | 0.065 | -0.074 | 0.182 | -0.035 | -0.065 | -0.061 | -0.041 | 0.006 | 0.045 | -0.136 | 0.756 | 0.963 | 1.000 | |
| IEexp % | 0.183 | 0.163 | -0.313 | -0.379 | 0.380 | -0.135 | 0.313 | 0.085 | -0.408 | 0.374 | 0.213 | 0.320 | 0.391 | 0.619 | 0.550 | 1.000 |
The correlation coefficients in the obtained matrix provide the information about the high or low interrelationship between the used descriptors. Generally, to decrease the redundancy existing in the used data matrix, the descriptors that are highly correlated (R ≥ 0.75), were excluded [63, 64, 65]. Accordingly, a perfectly negative correlation was observed between ELUMO and EA (R = -1) and a strongly negative correlations was noticed between EHOMO and IP (R = - 0.98); EL-H and σ (R = - 0.94); IP and ΔN (R = - 0.96); σ and ƞ (R = - 0.98); EL-H and EHOMO (R = - 0.76); ΔN and EL-H (R = - 0.79) and ΔN and ƞ (R = - 0.82). On the other hand, a positive collinearity was also observed between M and Log P (R = 0.78); Vm and M (R = 0.96); Vm and Log P (R = 0.76); ΔN and EHOMO (R = 0.94); ƞ and EL-H (R = 0.93); X and IP (R = 0.84); ΔN and σ (R = 0.84) and ω and EA (R = 0.80) which implies that these variables are redundant.
From the descriptive analysis of the PCA, it appeared that there are strong collinearities between the descriptors, i.e., some explanatory variables are linear combinations of the others. In this frame, and in view of this perfect collinearity, the matrix (X'X)−1 is not invertible. Therefore, the least squares estimator is not determinable. Accordingly, the research of a linear (MLR) or polynomial (MPR) regression model is not possible for the prediction of anticorrosion activity IE % from the structure of the molecules studied, due to the loss of a large part of the useful information for the construction of the desired model. which prompted us to evaluate other statistical models, such as: the partial least squares regression (PLS), the principal component regression (PCR) and the artificial neural networks (ANN).
3.2.2. Partial least squares regression (PLS)
Partial least squares regression (PLS) was proposed to predict quantitatively the inhibitive activity of the studied compounds.
The PLS model is expressed in the following Eq. (3):
| (3) |
where a0 is a constant of regression; a1, a2, a3, a4, a5, a6, a7, a8, a9, a10, a11, a12, a13, a14 and a15 represent the regression coefficients and EHOMO, ELUMO, EL-H, μ, IP, EA, χ, , , , , Log P, M and represent the variables; Ci denotes the inhibitor concentration.
The expression of the established PLS model, accompanied with the values of the statistical parameters, is represented by the following Eq. (4):
| (4) |
| N = 17; R2= 0.89; R2adj= 0.88; SD = 4.75; PRESS = 475.32; R2pred= 0.84 |
The values of the analysis of variance table (ANOVA) for the obtained PLS equation are provided in Table 5.
Table 5.
ANOVA for PLS model.
| Source | DF | SS | MS | p-value |
|---|---|---|---|---|
| Regression | 1 | 2627.552 | 2627.552 | <0.001 |
| Residual error | 15 | 338.576 | 22.571 | |
| Total | 16 | 2966.128 |
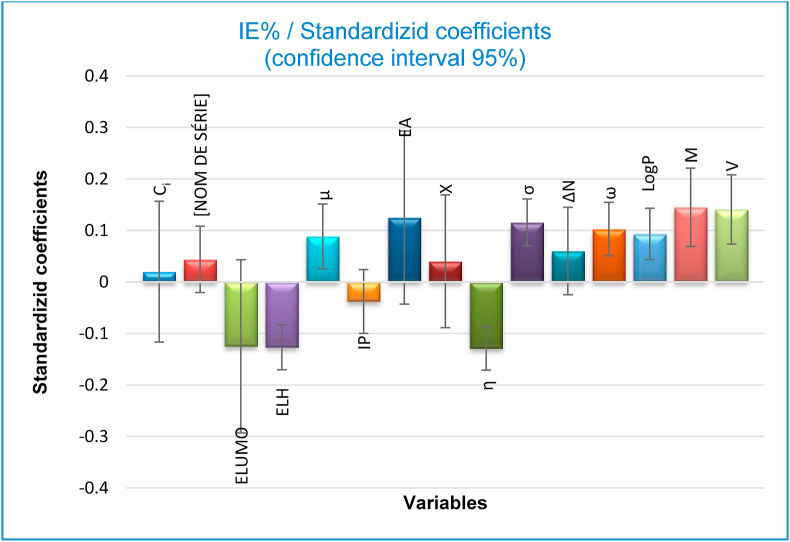
according to the descriptors mentioned in the partial least squares regression Eq. (4), the significance of each individual descriptor vs. the standardized regression coefficients is presented in Figure 5.
Figure 5.
Standardized coefficients versus variables in the proposed PLS model.
It can be seen from Figure 5, that the significance of anticorrosion activity based on molecular structure differs from one descriptor to another. However, because the descriptors in final PLS model have not the same units, these standardized coefficients are estimates with no real scale, which implies that these normalized coefficients cannot be employed to determine the real relative importance and significance of each descriptor in the regression analysis. Therefore, their usefulness is limited to the determination of the positive or negative effect of the molecular indices on the anti-corrosion property under investigation.
Returning to the statistical parameters, and from the values of the determination coefficient (R2 = 0.89), the adjusted determination coefficient (R2adj = 0.88) and the standard deviation (SD = 4.75), it appears that PLS does not have a great robustness in predicting the values of the inhibition efficiency. This insufficiency appeared very clearly in Figure 6, when representing the predictive values versus the experimental values of IE % where the residual error is very important.
Figure 6.
Correlation of experimental vs. calculated IE % efficiencies obtained by the PLS model.
The unsatisfactory statistical results of the PLS model prompted us to evaluate other statistical models, such as: the Principal components regression PCR and the artificial neural networks ANNs models.
3.2.3. Principal components regression (PCR)
To increase the quality of the prediction and degree of the correlation between the corrosion inhibition efficiency and the molecular structure, the selected descriptors were exploited to evaluate a principal components regression PCR, where the general expression of its equation is the identical to that of the PLS regression.
The obtained PCR equation accompanied with the statistical parameters calculated by the means of the following ANOVA table is expressed in Eq. (5) as:
| (5) |
| N = 17; R2= 0.97; R2adj= 0.97; SD = 3.33; PRESS = 475.32; R2pred= 0.92 |
The values of the analysis of variance table are summarized in ANOVA Table 6:
Table 6.
ANOVA for PCR model.
| Source | DF | SS | MS | p-value |
|---|---|---|---|---|
| Regression | 1 | 5587.965 | 5587.965 | <0.001 |
| Residual error | 15 | 166.053 | 11.070 | |
| Total | 16 | 5754.019 |
By the inspection of statistical results, it appeared that the principal components regression (PCR) has a good quality and best prediction of the inhibition efficiency compared with the partial least squares regression (PLS), with it is characterized by a fairly considerable values of determination coefficient (R2 = 0.97) and adjusted determination coefficient (R2adj = 0.97) and a low value of the standard deviation: SD = 3.33.
The obtained values of the inhibition efficiency predicted by the PCR are fitted with the experimental IE % in the Figure 7.
Figure 7.
Correlation of experimental vs. Calculated IE % obtained by the PCR model.
Despite the good quality of significance and prediction of the PCR model, we have proposed artificial neural networks (ANN) as a multiple non-linear model and a suitable concept for better modeling the inhibition efficiency with the molecular structure for studied inhibitors.
3.2.4. Artificial neural networks (ANN)
In order to increase the degree of prediction the inhibition activity of the studied compounds based on the molecular structures, an artificial neural networks (ANN) was tested [66, 67, 68]. Is one of the commonly used methods in the machine learning field. It is a non-linear mathematical approach, which can be investigated to model in a complicated way the inhibitor structures with corrosion inhibition efficiency. Generally, this process involves using three layers: one input layer, which are represents by the number of the selected descriptors. One hidden layer, where the number of nodes in which is an important factor determining network performance. Previous studies have proposed a parameter ρ, leading to the determination of the number of hidden neurons, which has a major role in determining the best network architecture. ρ is defined as a ratio between the number of data in the training set and the sum of the number of connections. It should have a value between 1 and 3 [69,70]. Accordingly, to determine the relevant descriptors that will be used for a non-linear ANN regression, considering the correlations and mathematical relationships between these different indices, several attempts have been made. Indeed, the best non-linear combination obtained is of the type: (7-2-1), where the number of weights is 17 and ρ = 1, which is considered an acceptable value to build the appropriate ANN. In addition, one output layer [71], represents the prediction results. Thereby, the ANN configuration is illustrated in Figure 8.
Figure 8.
The architecture of the selected artificial neural networks (ANN).
By the use of Matlab software, version 9.0 [72], the obtained predicted inhibition efficiency IEcal % accompanied with the residual error (RE.-ANN) values are listed in Table 8. In addition, the correlation between IEexp % and IEcal % accompanied with the statistical parameters is illustrated in Figure 9.
| N = 17; R2 = 0.95; SD = 4.19; PRESS = 475.32; R2pred= 0.90 |
Table 8.
Ecal% vs. Eexp% obtained by PLS, PCR, ANN and CV models, proposed for DFT-B3LYP/6-31G∗∗ along with the residual error (RE).
| Inhibitor | Ci/M | IEexp% |
IEcal% Training set |
IEcal% |
Residual Error (RE) |
||||
|---|---|---|---|---|---|---|---|---|---|
| PLS | PCR | ANN | CV | RE.-PLS | RE.-PCR | RE.-ANN | |||
| P1 | 0.5. 10−3 | 17.70 | 40.49 | 19.87 | 23.56 | 21.48 | -22.79 | -2.17 | -5.86 |
| P3 | 0.5. 10−3 | 98.50 | 91.20 | 98.11 | 87.86 | 86.82 | 7.30 | 0.39 | 10.64 |
| P4 | 2.8. 10−3 | 69.05 | 76.70 | 71.13 | 73.43 | 66.00 | -7.65 | -2.08 | -4.38 |
| P5 | 2.3. 10−3 | 82.42 | 78.79 | 81.03 | 78.23 | 75.19 | 3.63 | 1.39 | 4.19 |
| P6 | 2.2. 10−3 | 83.04 | 84.82 | 87.36 | 86.59 | 89.59 | -1.78 | -4.32 | -3.55 |
| P7 | 1.10–3 | 81.10 | 68.33 | 73.89 | 77.21 | 76.78 | 12.77 | 7.21 | 3.89 |
| P8 | 1.10–3 | 97.00 | 90.19 | 97.40 | 93.97 | 94.83 | 6.81 | -0.40 | 3.03 |
| P9 | 1.10–3 | 91.00 | 88.23 | 92.59 | 87.74 | 87.10 | 2.77 | -1.59 | 3.26 |
| P11 | 1.10–3 | 93.00 | 95.41 | 93.00 | 90.41 | 88.51 | -2.41 | 0.00 | 2.59 |
| P12 | 1.10–4 | 90.00 | 93.76 | 90.73 | 95.40 | 97.52 | -3.76 | -0.73 | -5.40 |
| P13 | 1.10–4 | 65.00 | 63.79 | 64.15 | 62.98 | 63.49 | 1.21 | 0.85 | 2.02 |
| P14 | 1.10–4 | 87.00 | 84.26 | 92.83 | 93.57 | 95.00 | 2.74 | -5.83 | -6.57 |
| P17 | 2.2. 10−3 | 95.84 | 89.79 | 98.49 | 96.84 | 96.04 | 6.05 | -2.65 | -1.00 |
| P18 | 2.2. 10−3 | 95.67 | 88.99 | 90.75 | 91.00 | 92.64 | 6.68 | 4.92 | 4.67 |
| P19 | 2.1. 10−3 | 72.64 | 77.09 | 73.68 | 73.49 | 75.69 | -4.45 | -1.04 | -0.85 |
| P20 | 2.1. 10−3 | 85.20 | 82.01 | 82.85 | 83.04 | 83.00 | 3.19 | 2.35 | 2.16 |
| P21 | 1.10–4 | 90.20 | 88.30 | 86.53 | 87.95 | 86.31 | 1.90 | 3.67 | 2.25 |
RE.-PLS: Residual error between Eexp % and Ecal % obtained by using PLS.
RE.-PCR: Residual error between Eexp % and Ecal % obtained by using PCR.
RE.-ANN: Residual error between Eexp % and Ecal % obtained by using ANN.
Figure 9.
Correlation of experimental vs. calculated IE % efficiencies obtained by the ANN model.
The values of the analysis of variance table are summarized in ANOVA Table 7:
Table 7.
ANOVA for ANN model.
| Source | DF | SS | MS | p-value |
|---|---|---|---|---|
| Regression | 1 | 4643.236 | 4643.236 | <0.001 |
| Residual error | 15 | 263.145 | 17.543 | |
| Total | 16 | 4906.381 |
The obtained values of the inhibition efficiency predicted by the ANN are fitted with the experimental IE % in the Figure 9.
The analysis of the fitting parameters shows that the ANN models bring globally a significant amount of information, which means that the inhibition efficiency is directly related to the selected molecular descriptors, and which implies that the good quality and best prediction of this model, demonstrated by the high determination coefficient R2 = 0.95 and the very low value of the standard deviation SD = 4.19.
The comparison between the experimental and calculated efficiency values is exemplified in Figure 10.
Figure 10.
Comparison diagram of IE % values obtained with PLS, PCR and ANN models.
A comparative analysis between the histograms illustrated in Figure 10 and the values of the residual errors (RE) arranged in Table 8 reveals that the PCR and ANN models explain the inhibition efficiency with a high level of significance when compared to PLS model. This relevance is demonstrated by the best concordance and the lowest values of (RE-PCR) and (RE-ANN) visualized between the IEexp % and IEcal %. The explanatory power of these two models is also highlighted by the statistical parameters such as R2, R2adj, R2pred and SD.
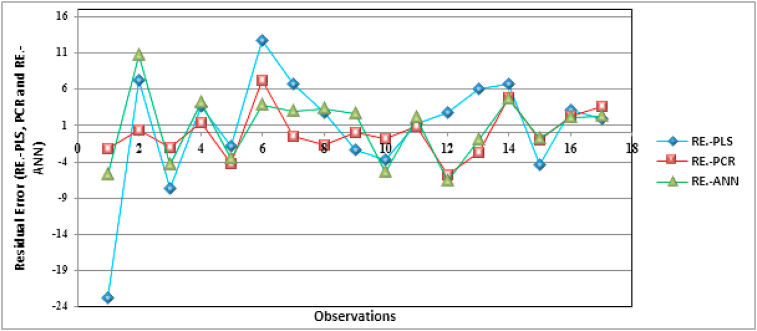
Moreover, it is very important to check for the presence of systematic errors that contravene the basic assumptions of the regression model. If there is a high systematic error in the model, then this model should be discarded [73] and performing any internal or external validation tests is of no use on this model. In this order, the residual error (RE) versus each observation of the training set was listed in Figure 11.
Figure 11.
The residual error (RE) between the experimental and the predicted IE % calculated with PLS, PCR and ANN models.
According to the Figure 11, the random distribution of the residuals shows that they are independent of each other, which implies that the residual errors follow a normal law with a homogeneity of their variance and a nullity of their expectations, and justifies the validity of such statistical models.
3.2.5. Internal and external validation models
3.2.5.1. Cross Validation (CV)
The most common technique used to determine the stability of the predictive model is to test the influence of each sample on the final model. Indeed, a Leave One Out (LOO) cross validation technique is used [74]. For a simple of N observations, this process consists to exclude each time an individual from the training test and to start the calculation. The excluded individual is considered as a test set regarding the rest [75,76]. In fact, this operation is repeated N times, and exploited to predict the IE % for all items in the studied sample.
Accordingly, in this work and by the means of Matlab software version 9.0 [72], this process is repeated 17 consecutive executions in order to predict the values of all the training set compounds. Indeed, the CV results are reported in Table 8 and the correlation between the experimental and the cross-validation predicted values of IE % coupled with the statistical characteristics are displayed in Figure 12.
| N = 17; R2cv = 0.92; SD = 5.26; p-value < 0.0001. |
Figure 12.
Correlation of experimental vs. calculated IE % efficiencies obtained by the LOO cross validation.
As can be seen from the lowest value of the standard deviation SD = 5.26; that the mean square error between the observed and the predicted efficiency values is minimum. This are reflected by the highly significant cross-validated coefficient of determination (R2cv = 0.92); which indicates a significant accordance between the results of the PLS, PCR and ANN as a QSPR models and the results of CV as an internal validation model and justifies that these proposed models, especially, possess a significantly statistical quality.
3.2.5.2. External validation
To assess the quality of the prediction and the degree of performance of the final extrapolation models, the external test of validation was carried out. For this purpose, and according to the above analyses and discussions, the dataset was divided into two samples: N = 17 compounds as a training set which is investigated to developing the QSPR models and k = 4 compounds as a test set which is exploited for an external validation. Following this sequential order, the external validation results are collected in Table 9.
Table 9.
Predicted vs. observed inhibition efficiency along with the residual error (RE) obtained by the external validation (test set).
| Inhibitor | Ci/M | IEexp% |
IEcal% Test set |
Residual Error (RE) |
||||
|---|---|---|---|---|---|---|---|---|
| PLS | PCR | ANN | RE.-PLS (test) | RE.-PCR (test) | RE.-ANN(test) | |||
| P2 | 0.5. 10−3 | 25.60 | 65.56 | 34.04 | 29.87 | -39.96 | -8.44 | -4.27 |
| P10 | 1.10–3 | 87.00 | 79.78 | 81.61 | 75.35 | 7.22 | 5.39 | 11.65 |
| P15 | 1.10–4 | 69.00 | 77.83 | 77.08 | 74.48 | -8.83 | -8.08 | -5.48 |
| P16 | 1.10–4 | 83.00 | 72.72 | 75.46 | 72.74 | 10.28 | 7.54 | 10.26 |
The exploitation of the Table 9 allows to extract the statistical parameters related to the external validation which are listed in Table 10.
Table 10.
Statistical parameters derived from the external validation of statistical models.
| Parameter | Statistical model |
||
|---|---|---|---|
| PLS | PCR | ANN | |
| n | 4 | 4 | 4 |
| R2ext | 0.71 | 0.94 | 0.92 |
| p-value | < 0.001 | <0.001 | < 0.001 |
As a confirmation with the above analyses and discussions, for the PLS, PCR and ANN models and as they indicate their values of the external coefficient of determination R2ext: 0.71, 0.94 and 0.92 respectively, they are superior of 0.5 as a critical value [58, 59, 60]. Which implies that the proposed principal component regression and artificial neural networks models have a highest predictive power. Their extrapolation can be successfully used for the prediction of the corrosion inhibition efficiency of other compounds that are not included in the establishment of these QSPR models.
According to all the obtained statistical results, the degree of robustness and the quality of prediction of the developed models are compared in Table 11.
Table 11.
Comparison of the statistical parameters of PLS, PCR and ANN models.
| Parameter | Statistical model |
||
|---|---|---|---|
| PLS | PCR | ANN | |
| N | 17 | 17 | 17 |
| R2 | 0.89 | 0.97 | 0.95 |
| R2adj | 0.88 | 0.97 | 0.95 |
| R2pred | 0.84 | 0.92 | 0.90 |
| SD | 4.75 | 3.33 | 4.19 |
| R2ext | 0.71 | 0.94 | 0.92 |
| Fobs | 116.408 | 504.774 | 264.677 |
| p-value | <0.001 | <0.001 | <0.001 |
The overall analysis of the statistical results of PLS, PCR and ANN from the Table 11 shows that PCR and ANN are the most predictive models for estimating the anticorrosion activity of the inhibitors selected in this work, compared to the PLS model. This robustness in prediction is justified by the best statistical parameters associated to these two models, especially, the predicted determination coefficient, where R2pred = 0.92 and R2pred = 0.90 for PCR and ANN respectively. Furthermore, an extensive comparison of the current results with those of similar studies reported in the literature by I. Lukovits et al. [77], E. S. H. El Ashry et al. [78], H. El Sayed et al. [79], F. Bentiss et al. [80,81], El H. El Assiri et al. [1,2], M. R. Fissa et al. [82], A. Aouidate et al. [83], A. Ghaleb et al. [84], P. Toropova et al. [85], shows that that the arsenal of quantum chemistry can be used to establish correlations between molecular structure and activity using the Quantitative Structure Activity/Property Relationship (QSAR/QSPR) approach, by developing mathematical models to predict inhibition efficiency and to select compounds with desired properties. This statistical technique based on the correlation between the inhibition properties of molecules and their molecular structures can also provide useful qualitative and quantitative information for a better understanding of the corrosion inhibition process.
The exploitation of the established equations also makes it possible to estimate the inhibition properties of other similar compounds in the absence of the data and consequently, guide the organic partner to their synthesis through the promising character predicted.
4. Conclusion
From the above analyses and discussions, the following main conclusions can be drawn:
-
-
Twenty-one pyridazine derivatives have been theoretically and statistically investigated as corrosion inhibitors for carbon steel in 1.0 M HCl solution.
-
-
The calculation of the molecular descriptors by the means of DFT quantum method allows to correlate the corrosion inhibition efficiency IE % to the molecular structure using the QSPR approach.
-
-
A qualitative analysis with the principal component analysis (PCA) allowed us to examine redundancy and collinearity between the proposed descriptors.
-
-
The established regressions show that the anticorrosion activity of the molecules studied can be explained in terms of the electronic and structural properties of such inhibitors.
-
-
The inspection of the quantitative analyses results demonstrates that PCR and ANN are the most robust models to predict the inhibition efficiency of the selected compounds in comparison with PLS model. Indeed, the relevance of these models is reflected by the means of the internal validation including the leave one out cross-validation and by the external validation using a test set as an extrapolation of these proposed models.
-
-
PCR and ANN results can be suggested to predict the corrosion inhibition efficiency of new pyridazine compounds, and to encourage the collaboration between experimenters, theorists and industrialists. As well as to assist the organic chemist in the synthesis of promising molecules as corrosion inhibitors.
Declarations
Author contribution statement
El Hassan El Assiri, Majid Driouch, Jamila Lazrak: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Zakariae Bensouda, Ali Elhaloui: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Mouhcine Sfaira, Taoufiq Saffaj, Mustapha Taleb: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors gratefully thank the ‘‘Association Marocaine des Chimistes Théoriciens’’ (AMCT) for the means of the calculations.
References
- 1.El Assiri El H., Driouch M., Bensouda Z., Beniken M., El haloui A., Sfaira M., Saffaj T. Computational study and QSPR approach on the relationship between corrosion inhibition efficiency and molecular electronic properties of some benzodiazepine derivatives on C-steel surface. Anal. Bioanal. Electrochem. 2019;11:373–395. [Google Scholar]
- 2.El Assiri El H., Driouch M., Bensouda Z., Jhilal F., Saffaj T., Sfaira M., Abboud Y. Quantum chemical and QSPR studies of bis-benzimidazole derivatives as corrosion inhibitors by using electronic and lipophilic descriptors. Desalination Water Treat. 2018;111:208–225. [Google Scholar]
- 3.Bensouda Z., Elassiri E., Galai M., Sfaira M., Farah A., Ebn Touhami M. Corrosion inhibition of mild steel in 1 M HCl solution by Artemisia Abrotanum essential oil as an eco-friendly inhibitor. J. Mater. Environ. Sci. 2018;9:1851–1865. [Google Scholar]
- 4.Beniken M., Driouch M., Sfaira M., Hammouti B., Ebn Touhami M., Mohsin M.A. Anticorrosion activity of a polyacrylamide with high molecular weight on C-steel in acidic media: Part 1. J. Bio Tribo-Corrosion. 2018;4:38. [Google Scholar]
- 5.Beniken M., Driouch M., Sfaira M., Hammouti B., Ebn Touhami M., Mohsin M. Kinetic–thermodynamic properties of a polyacrylamide on corrosion inhibition for C-steel in 1.0 M HCl medium: Part 2. J. Bio Tribo-Corrosion. 2018;4:34. [Google Scholar]
- 6.Bensouda Z., Driouch M., Belakhmima R.A., Sfaira M., Ebn Touhami M., Farah A. Thymus sahraouian essential oil as corrosion eco-friendly inhibitor for mild steel in a molar hydrochloric acid solution. Port. Electrochim. Acta. 2018;36:339–364. [Google Scholar]
- 7.Bensouda Z., Driouch M., Sfaira M., Farah A., Ebn Touhami M., Hammouti B., Emran K.M. Effect of Mentha Piperita essential oil on mild steel corrosion in hydrochloric acid. Int. J. Electrochem. Sci. 2018;13:8198–8221. [Google Scholar]
- 8.Schmitt G. Of inhibitors for acid media: report prepared for the European federation of corrosion working party on inhibitors. Br. Corrosion J. 1984;19:165–176. [Google Scholar]
- 9.Bockris J.M., Yang B. The mechanism of corrosion inhibition of iron in acid solution by acetylenic alcohols. J. Electrochem. Soc. 1991;138:2237–2252. [Google Scholar]
- 10.Pillali C., Narayan R. Anodic dissolution of mild steel in HCl solutions containing thio-ureas. Corrosion Sci. 1983;23:151–166. [Google Scholar]
- 11.Growcock F.B., Lopp V.R. The inhibition of steel corrosion in hydrochloric acid with 3-phenyl-2-propyn-1-ol. Corrosion Sci. 1988;28:397–410. [Google Scholar]
- 12.Hackerman Bartos N. A study of inhibition action of propargyl alcohol during anodic dissolution of iron in hydrochloric acid. J. Electrochem. Soc. 1992;139:3429–3433. [Google Scholar]
- 13.Zucchi F., Trabanelli G., Brunoro G. The influence of the chromium content on the inhibitive efficiency of some organic compounds. Corrosion Sci. 1992;33:1135–1139. [Google Scholar]
- 14.Granese S.L. Study of the inhibitory action of nitrogen-containing compounds. Corrosion. 1988;44:322–327. [Google Scholar]
- 15.El Adnani Z., Mcharfi M., Sfaira M., Benzakour M., Benjelloun A.T., Touhami M.E. DFT study of 7-R-3methylquinoxalin-2 (1H)-ones (R= H; CH3; Cl) as corrosion inhibitors in hydrochloric acid. Int. J. Electrochem. Sci. 2012;7:6738–6751. [Google Scholar]
- 16.Eddy N.O., Odoemelam S.A. Ethanol extract of Musa acuminate peel as an eco-friendly inhibitor for the corrosion of mild steel in [H. sub. 2] S [O. sub. 4] Adv. Nat. Appl. Sci. 2008;2:35–43. [Google Scholar]
- 17.Umoren S.A., Obot I.B., Ebenso E.E., Obi-Egbedi N.O. Synergistic inhibition between naturally occurring exudate gum and halide ions on the corrosion of mild steel in acidic medium. Int. J. Electrochem. Sci. 2008;3:1029–1043. [Google Scholar]
- 18.Bensouda Z., El Assiri El H., Sfaira M., Touhami M.E., Farah A., Hammouti B. Extraction, characterization and anticorrosion potential of an essential oil from orange zest as eco-friendly inhibitor for mild steel in acidic solution. J. Bio Tribo-Corrosion. 2019;5:84. [Google Scholar]
- 19.Lukovits I., Shaban A., Kálmán E. Non-linear quantitative structure-efficiency model of corrosion inhibition. Electrochim. Acta. 2005;50:4128–4133. [Google Scholar]
- 20.Lesar A., Milosev I. Density functional study of the corrosion inhibition properties of 1, 2, 4-triazole and its amino deriva-tives. Chem. Phys. Lett. 2009;483:198–203. [Google Scholar]
- 21.Jamalizadeh E., Hosseini S.M.A., Jafari A.H. Quantum chemical studies on corrosion inhibition of some lactones on mild steel in acid media. Corrosion Sci. 2009;51:1428–1435. [Google Scholar]
- 22.Arslan T., Kandemirli F., Ebenso E.E., Love I., Alemu H.M. Quantum chemical studies on the corrosion inhibition of some sulphonamides on mild steel in acidic medium. Corrosion Sci. 2009;51:35–47. [Google Scholar]
- 23.Obot I.B., Obi-Egbedi N.O. Theoretical study of benzimidaz-ole and its derivatives and their potential activity as corrosion inhibitors. Corrosion Sci. 2010;52:657–660. [Google Scholar]
- 24.Chetouani A., Hammouti B., Aouniti A., Benchat N., Benhadda T. New synthesised pyridazine derivatives as effective inhibitors for the corrosion of pure iron in HCl medium. Prog. Org. Coating. 2002;45:373–378. [Google Scholar]
- 25.Boukla M., Benchat N., Aouniti A., Hammouti B., Benkaddour M., Lagrenée M., Vezin H., Bentiss F. Effect of the substitution of an oxygen atom by sulphur in a pyridazinic molecule towards inhibition of corrosion of steel in 0.5 M H2SO 4 medium. Prog. Org. Coating. 2004;51:118–124. [Google Scholar]
- 26.Bentiss F., Gassama F., Barbry D., Gengembre L., Vezin H., Lagrenée M., Traisnel M. Enhanced corrosion resistance of mild steel in molar hydrochloric acid solution by 1, 4-bis (2-pyridyl)-5H-pyridazino [4, 5-b] indole: electrochemical, theoretical and XPS studies. Appl. Surf. Sci. 2006;252:2684–2691. [Google Scholar]
- 27.Zerga B., Saddik R., Hammouti B., Taleb M., Sfaira M., Ebn Touhami M., Al-Deyab S.S., Benchat N. Effect of new synthesised pyridazine derivatives on the electrochemical behaviour of mild steel in 1M HCl solution: Part-1. Int. J. Electrochem. Sci. 2012;7:631–642. [Google Scholar]
- 28.Khadiri A., Saddik R., Bekkouche K., Aouniti A., Hammouti B., Benchat N., Bouachrine M., Solmaz R. Gravimetric, electrochemical and quantum chemical studies of some pyridazine derivatives as corrosion inhibitors for mild steel in 1 M HCl solution. J. Taiwan Inst. Chem. Eng. 2016;58:552–564. [Google Scholar]
- 29.Mashuga M.E., Olasunkanmi L.O., Ebenso E.E. Experimental and theoretical investigation of the inhibitory effect of new pyridazine derivatives for the corrosion of mild steel in 1 M HCl. J. Mol. Struct. 2017;1136:127–139. [Google Scholar]
- 30.El-Hajjaji F., Merimi I., Messali M., Obaid R.J., Salima R., Taleb M., Hammouti B. Experimental and quantum studies of newly synthesized pyridazinium derivatives on mild steel in hydrochloric acid medium. Mater. Today Proc. 2019;13:822–831. [Google Scholar]
- 31.Khadiri A., Saddik R., Bekkouche K., Aouniti A., Hammouti B., Benchat N., Solmaz R. Gravimetric, electrochemical and quantum chemical studies of some pyridazine derivatives as corrosion inhibitors for mild steel in 1 M HCl solution. J. Taiwan Instit. Chem. Eng. 2016;58:552–564. [Google Scholar]
- 32.Olasunkanmi L.O., Mashuga M.E., Ebenso E.E. Surface protection activities of some 6-substituted 3-chloropyridazine derivatives for mild steel in 1 M hydrochloric acid: experimental and theoretical studies. Surf. Interfaces. 2018;12:8–19. [Google Scholar]
- 33.Ghazoui A., Benchat N., El-Hajjaji F., Taleb M., Rais Z., Saddik R., Hammouti B. The study of the effect of ethyl (6-methyl-3-oxopyridazin-2-yl) acetate on mild steel corrosion in 1M HCl. J. Alloys Compd. 2017;693:510–517. [Google Scholar]
- 34.El Adnani Z., Mcharfi M., Sfaira M., Ben-jelloun A.T., Benzakour M., Ebn Touhami M., Hammouti B., Taleb M. Investigation of newly pyridazine derivatives as corrosion inhibitors in molar hydrochloric acid. Part III: computational calculations. Int. J. Electrochem. Sci. 2012;7:3982–3996. [Google Scholar]
- 35.Chetouani A., Hammouti B., Aouniti A., Benchat N., Benhadda T. New synthesised pyridazine derivatives as effective inhibitors for the corrosion of pure iron in HCl medium. Prog. Org. Coating. 2002;45:373–378. [Google Scholar]
- 36.Chetouani A., Aouniti A., Hammouti B., Benchat N., Benhadda T., Kertit S. Corrosion inhibitors for iron in hydrochloride acid solution by newly synthesised pyridazine derivatives. Corrosion Sci. 2003;45:1675–1684. [Google Scholar]
- 37.Bentiss F., Gassama F., Barbry D., Gengembre L., Vezin H., Lagrenée M., Traisnel M. Enhanced corrosion resistance of mild steel in molar hydrochloric acid solution by 1, 4-bis (2-pyridyl)-5H-pyridazino [4, 5-b] indole: electrochemical, theoretical and XPS studies. Appl. Surf. Sci. 2006;252:2684–2691. [Google Scholar]
- 38.Kraka E., Cremer D. A new enediyne warhead. J. Am. Chem. Soc. 2000;122:8245–8264. [Google Scholar]
- 39.Mokhtari O., Hamdani I., Chetouani A., Lahrach A., El Halouani H., Aouniti A., Berrabah M. Inhibition of steel corrosion in 1M HCl by Jatropha curcas oil. J. Mater. Environ. Sci. 2014;5:310–319. [Google Scholar]
- 40.Shivakumar S.S., Mohana K.N. Corrosion behavior and adsorption thermodynamics of some schiff bases on mild steel corrosion in industrial water medium. Int. J. Corrosion. 2013:2013. [Google Scholar]
- 41.El Adnani Z., Mcharfi M., Sfaira M., Benzakour M., Benjelloun A.T., Touhami M.E. DFT theoretical study of 7-R-3methylquinoxalin-2 (1H)-thiones (RH; CH3; Cl) as corrosion inhibitors in hydrochloric acid. Corrosion Sci. 2013;68:223–230. [Google Scholar]
- 42.Hansch C., Muir R.M., Fujita T., Maloney P.P., Geiger F., Streich M. The correlation of biological activity of plant growth regulators and chloromycetin derivatives with Hammett constants and partition coefficients. J. Am. Chem. Soc. 1963;85:2817–2824. [Google Scholar]
- 43.Bodor N. Biochemistry of redox reactions. Curr. Med. Chem. 1988;5:353–380. [Google Scholar]
- 45.Growcock F.B., Frenier W.W., Andreozzi P.A. Inhibition of steel corrosion in HCl by derivatives of cinnamaldehyde: Part II. Structure–activity correlations. Corrosion. 1989;45:1007–1015. [Google Scholar]
- 46.Abdul-Ahad P.G., Al Madfai S.H.F. Elucidation of corrosion inhibition mechanism by means of calculated electronic indexes. Corrosion. 1989;45:978–980. [Google Scholar]
- 47.Dupin P., Vilovia-Vera D.A., de Savignac A., Lattes A., Haicour P. University of Ferrara (Italy); 1980. Proceedings of Fifth European Symposium on Corrosion Inhibitors; p. 301. [Google Scholar]
- 48.Lukovits I., Kalman E., Palinkas G. Nonlinear group-contribution models of corrosion inhibition. Corrosion. 1995;51:201–205. [Google Scholar]
- 49.Lukovits I., Palfi K., Bako I., Kalman E. LKP model of the inhibition mechanism of thiourea compounds. Corrosion. 1997;53:915–919. [Google Scholar]
- 50.Becke A.D. Density functional thermochemistry. I, the effect of the exchange only gradient correction. J. Chem. Phys. 1992;96:2155–2160. [Google Scholar]
- 51.Becke A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 52.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti conelation energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 53.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Montgomery J.A., Jr., Vreven T., Kudin K.N., Burant J.C., Millam J.M., Iyengar S.S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G.A., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J.E., Hratchian H.P., Cross J.B., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Ayala P.Y., Morokuma K., Voth G.A., Salvador P., Dannenberg J.J., Zakrzewski V.G., Dapprich S., Daniels A.D., Strain M.C., Farkas O., Malick D.K., Rabuck A.D., Raghavachari K., Foresman J.B., Ortiz J.V., Cui Q., Baboul A.G., Clifford S., Cioslowski J., Stefanov B.B., Liu G., Liashenko A., Piskorz P., Komaromi I., Martin R.L., Fox D.J., Keith T., Al-Laham M.A., Peng C.Y., Nanayakkara A., Challacombe M., Gill P.M.W., Johnson B., Chen W., Wong M.W., Gonzalez C., Pople J.A. Gaussian, Inc.; Pittsburgh PA: 2003. Gaussian 03, Revision B.01. [Google Scholar]
- 54.ACD/ChemSketch . February 2018. ACD/ChemSketch for Academic and Personal Use: ACD/Labs.Com.http://www.acdlabs.com/resources/freeware/chemsketch/ n.d. [Google Scholar]
- 55.Shahraki M., Dehdab M., Elmi S. Theoretical studies on the corrosion inhibition performance of three amine derivatives on car-bon steel: molecular dynamics simulation and density functional theory approaches. J. Taiwan Inst. Chem. Eng. 2016;62:313–321. [Google Scholar]
- 56.Qiang Y., Zhang S., Guo L., Zheng X., Xiang B., Chen S. Experimental and theoretical studies of four allyl imidazolium-based ionic liquids as green inhibitors for copper corrosion in sulfuric acid. Corros. Sci. 2017;119:68–78. [Google Scholar]
- 57.XLSTAT software version 2014. https://www.xlstat.com/fr/ XLSTAT Company.
- 58.Golbraikh A., Tropsha A. Beware of q2. J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 59.Tropsha A. Best practices for QSAR model development validation and exploitation. Mole. Inf. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 60.Gramatica P. Principles of QSAR models validation: internal and external. QSAR Comb. Sci. 2007;26:694–701. [Google Scholar]
- 61.STATITCF Software . 1987. Technical Institute of Cereals and Fodder, Paris, France. [Google Scholar]
- 62.Hmamouchi R., Larif M., Adad A., Bouachrine M., Lakhlifi T. Structure activity and prediction of biological activities of compound (2-methyl-6-phenylethynylpyridine) derivatives relationships rely on electronic and topological descriptors. J. Comput. Methods Mol. Des. 2014;4:61–71. [Google Scholar]
- 63.Henseler J., Ringle C., Sinkovics R. The use of partial least squares path modeling in international marketing. Adv. Int. Market. 2009;20:277–320. [Google Scholar]
- 64.Hair J.F., Ringle C.M., Sarstedt Partial least squares structural equation modeling: rigorous applications, better results and higher acceptance. Long. Range Plan. 2013;46:1–12. [Google Scholar]
- 65.Moore D.S., Notz W.I., Flinger M.A. sixth ed. W. H. Freeman; New York: 2013. The Basic Practice of Statistics. [Google Scholar]
- 66.Kong D.Y., Song S.Z. Analysis of corrosion data for carbon steel and low-alloy steel in seawater by artificial neural network. J. Chin. Soc. Corrosion Protect. 1998;18:289–296. [Google Scholar]
- 67.Liu X.Q., Tang X., Wang J. Correlation between seawater environmental factors and marine corrosion rate using artificial neural network analysis. J. Chin. Soc. Corrosion Protect. 2005;25:11–14. [Google Scholar]
- 68.Liu Y.X., Gao X.C., Zhang G.Y., Guo H.H. BP neural networks used in prediction and analyses of 3C steel corrosion function. J. Mater. Sci. Eng. 2008;26:94–97. [Google Scholar]
- 69.So S.S., Richards W.G. Application of neural networks: quantitative structure-activity relationships of the derivatives of 2,4-diamino-5-(substituted-benzyl) pyrimidines as DHFR inhibitors. J. Med. Chem. 1992;35:3201–3207. doi: 10.1021/jm00095a016. [DOI] [PubMed] [Google Scholar]
- 70.Andrea T.A., Kalayeh H. Applications of neural networks in quantitative structure-activity relationships of dihydrofolate reductase inhibitors. J. Med. Chem. 1991;34:2824–2836. doi: 10.1021/jm00113a022. [DOI] [PubMed] [Google Scholar]
- 71.Zupan J., Gasteiger J. second ed. 1999. Neural Networks for Chemistry and Drug Design: an Introduction. Weinheim, Germany. [Google Scholar]
- 72.MATLAB 9.0 (R2009b) Statistics Toolbox Release. The Math Works, Inc. Natick; Massachusetts, United States: 2011. [Google Scholar]
- 73.Jalali-Heravi M., Kyani A. Use of computer-assisted methods for the modeling of the retention time of a variety of volatile organic compounds: a PCA-MLR-ANN approach. J. Chem. Inf. Comput. Sci. 2004;44:1328–1335. doi: 10.1021/ci0342270. [DOI] [PubMed] [Google Scholar]
- 74.Stone M. Cross-validatory choice and assessment of statistical predictions. J. Roy. Stat. Soc. B. 1974;36:111–133. [Google Scholar]
- 75.Efron B. Estimating the error rate of a prediction rule: improvement on cross-validation. J. Am. Stat. Assoc. 1983;78:316–331. [Google Scholar]
- 76.Efroymson M.A. Multiple regression analysis. Math. Methods Digit. Comput. 1960;1:191–203. [Google Scholar]
- 77.a Lukovits I., Shaban A., Kálmán E. Thiosemicarbazides and thiosemicarbazones: non-linear quantitative structure–efficiency model of corrosion inhibition. Electrochim. Acta. 2005;50:4128–4133. [Google Scholar]; b El Ashry E.S.H., El Nemr A., Esawy S.A., Ragab S. Corrosion Inhibitors-Part II: quantum chemical studies on the corrosion inhibitions of steel in acidic medium by some triazole, oxadiazole and thiadiazole derivatives. Electrochim. Acta. 2006;51:3957–3968. [Google Scholar]
- 78.El Sayed H., El Nemr, Essawy S.A., Ragab S. Corrosion inhibitors part V: QSAR of benzimidazole and 2-substituted derivatives as corrosion inhibitors by using the quantum chemical parameters. Prog. Org. Coating. 2008;61:11–20. [Google Scholar]
- 79.Bentiss F., Traisnel M., Chaibi N., Mernari B., Vezin H., Lagrenée M. 2, 5-Bis (n-methoxyphenyl)-1, 3, 4-oxadiazoles used as corrosion inhibitors in acidic media: correlation between inhibition efficiency and chemical structure. Corrosion Sci. 2002;44:2271–2289. [Google Scholar]
- 80.Bentiss F., Traisnel M., Vezin H., Lagrenée M. Linear resistance model of the inhibition mechanism of steel in HCl by triazole and oxadiazole derivatives: structure–activity correlations. Corrosion Sci. 2003;45:371–380. [Google Scholar]
- 81.Fissa M.R., Lahiouel Y., Khaouane L., Hanini S. QSPR estimation models of normal boiling point and relative liquid density of pure hydrocarbons using MLR and MLP-ANN methods. J. Mol. Graph. Model. 2019;87:109–120. doi: 10.1016/j.jmgm.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 82.Aouidate A., Ghaleb A., Ghamali M., Chtita S., Ousaa A., Sbai A., Bouachrine M., Lakhlifi T. Investigation of indirubin derivatives: a combination of 3D-QSAR, molecular docking and ADMET towards the design of new DRAK2 inhibitors. Struct. Chem. 2018;29:1609–1622. [Google Scholar]
- 83.Ghaleb A., Aouidate A., Ghamali M., Sbai A., Bouachrine M., Lakhlifi T. 3D-QSAR modeling and molecular docking studies on a series of 2, 5 disubstituted 1, 3, 4-oxadiazoles. J. Mol. Struct. 2017;1145:278–284. [Google Scholar]
- 84.Toropova A.P., Toropov A.A. QSPR and nano-QSPR: what is the difference. J. Mol. Struct. 2019;1182:141–149. [Google Scholar]
- 85.Villaverde J.J., Sevilla-Morán B., López-Goti C., Alonso-Prados J.L., Sandín-España P. Considerations of nano-QSAR/QSPR models for nanopesticide risk assessment within the European legislative framework. Sci. Total Environ. 2018;634:1530–1539. doi: 10.1016/j.scitotenv.2018.04.033. [DOI] [PubMed] [Google Scholar]