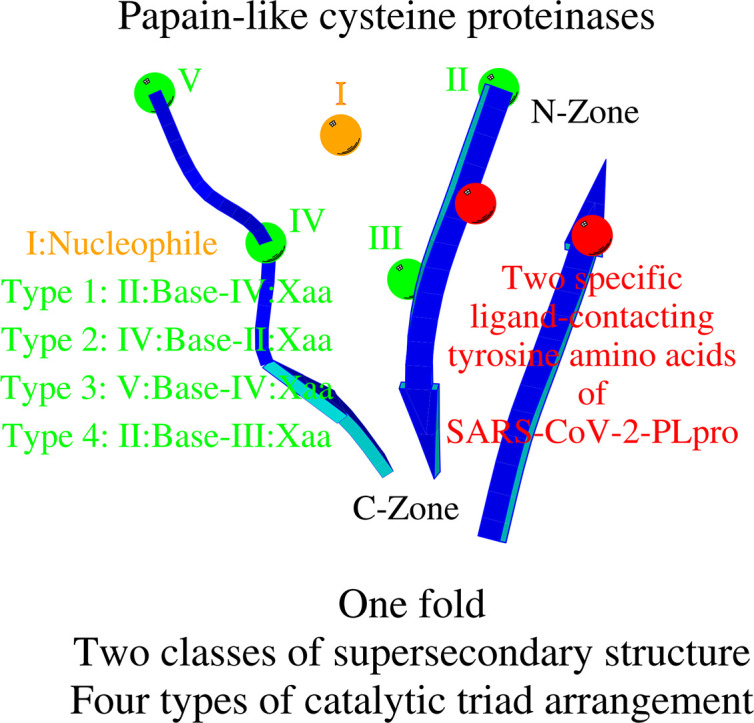
Abstract
There are several families of cysteine proteinases with different folds – for example the (chymo)trypsin fold family and papain-like fold family – but in both families the hydrolase activity of cysteine proteinases requires a cysteine residue as the catalytic nucleophile. In this work, we have analyzed the topology of the active site regions in 146 three-dimensional structures of proteins belonging to the Papain-like Cysteine Proteinase (PCP) superfamily, which includes papain as a typical representative of this protein superfamily. All analyzed enzymes contain a unique structurally closed conformation – a “PCP-Zone” – which can be divided into two groups, Class A and Class B. Eight structurally conserved amino acids of the PCP-Zone form a common Structural Core. The Structural Core, catalytic nucleophile, catalytic base and residue Xaa – which stabilizes the side-chain conformation of the catalytic base – make up a PCP Structural Catalytic Core (PCP-SCC). The PCP-SCC of Class A and Class B are divided into 5 and 2 types, respectively. Seven variants of the mutual arrangement of the amino-acid side chains of the catalytic triad – nucleophile, base and residue Xaa – within the same fold clearly demonstrate how enzymes with the papain-like fold adapt to the need to perform diverse functions in spite of their limited structural diversity. The roles of both the PCP-Zone of SARS-CoV-2-PLpro described in this study and the NBCZone of SARS-CoV-2-3CLpro presented in our earlier article (Denesyuk AI, Johnson MS, Salo-Ahen OMH, Uversky VN, Denessiouk K. Int J Biol Macromol. 2020;153:399-411) that are in contacts with inhibitors are discussed.
Keywords: Papain, Cysteine proteinases, Catalytic triad, Fold, Zone, Structural catalytic core, COVID-19, SARC-CoV-2
Graphical abstract
1. Introduction
The catalytic triad (Acid-Base-Nucleophile) is the widely known structural motif found in the active site of many enzymes [1,2]. Serine and cysteine are the two most common nucleophilic residues in these enzymes. In our previous work, we analyzed the structural environment of the catalytic triad (Structural Catalytic Core (SCC)) in the α/β hydrolase [3] and (chymo)trypsin-like [4] fold enzymes. In proteins from these two superfamilies, serine is the most frequently occurring amino acid used as a nucleophile in the active sites.
The superfamily of papain-like cysteine proteinases (Fold d.3 in Structural Classification of Proteins — extended (SCOPe) https://scop.berkeley.edu/ [5]) is a classic example of enzymes that use a cysteine residue as a nucleophile. Papain-like cysteine proteases are found in most organisms, including animals, plants, protozoa, yeast, bacteria, and virus [6]. Papain-like family enzymes are involved in numerous physiological and pathological processes, such as antigen presentation, extracellular matrix remodeling, immune invasion, hormone processing, parasite invasion, processing surface proteins, cardiovascular diseases and cancer [[7], [8], [9], [10]]. This year, as the pandemic of COVID-19 began, it became particularly important to understand the molecular mechanisms behind the function of the cysteine proteases because the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has only two functionally important proteinases: papain-like (PLpro) and (chymo)trypsin-like (3CLpro) (https://swissmodel.expasy.org/repository/species/2697049), both belonging to the family of cysteine proteinases [11,12].
The comparison of Nucleophile-Base Catalytic Zone (NBCZone) among 169 eukaryotic, prokaryotic, and viral (chymo)trypsin-like proteases showed that: 1) the majority of eukaryotic and prokaryotic proteins have a Cys42T-Cys58T disulfide bond near the catalytic triad, while viral proteases do not have it, and 2) the vast majority of eukaryotic proteases have glycine residues at position 43T. In almost all prokaryotic and viral proteases, the amino acid at position 43T is not glycine. Only one group of viral cysteine proteases (mainly coronavirus) has the amino acid asparagine in position 43T [4].
The importance of the results obtained on the fundamental structural differences in the active sites of viral (chymo)trypsin-like proteases from eukaryotic and bacterial ones prompted us to carry out a similar study on papain-like cysteine proteases. In addition, in the annotation to papain-like cysteine proteases superfamily, SCOPe notes that the families included in the superfamily are distinguished “by insertion into and circular permutation of the common catalytic core made of one α-helix and 3-strands of β-sheet” [5]. Nothing of the kind is observed in (chymo)trypsin-like proteases. The structural analysis of the active site of the proteins of the papain-like cysteine protease superfamily carried out below is a next important step in the systematic study of enzymes with various folds using the catalytic triad in the enzymatic mechanism.
2. Results and discussion
2.1. PCP-Zone, Class A
2.1.1. Zone and SCC of papain
We begin the presentation of our results with an analysis of the structural environment of the catalytic triad in papain (PDB ID 1PPN) [13]. The tertiary structure of papain is well known and this fundamental structure can be found in enzymes that are classified as Fold d.3: Cysteine proteinases, Superfamily d.3.1: Cysteine proteinases, Family d.3.1.1: Papain-like [5]. The catalytic triad of papain consists of three amino acids: catalytic nucleophile Cys25, catalytic base His159 and residue Xaa (Asn175) (Table 1 ). Structure visualization and structural analysis of interactions between amino acids in proteins was carried out using the Discovery Studio Modeling Environment (Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016) and the Ligand-Protein Contacts (LPC) software [14].
Table 1.
The geometric characteristics of the contacts of the amino acids that make up two classes of the Papain-like Cysteine Proteinase Zone (PCP-Zone) and seven types of the PCP Structural Catalytic Core (PCP-SCC) in 7 representative enzymes with the cysteine proteinase fold.
| N | Protein | PDB_ID Res. (Å) | PCP-Zone |
Xaa-Base Nucleophile-(Base+1) Xaa/Asx-turn |
Nucleophile Xaa |
Ref. | ||
|---|---|---|---|---|---|---|---|---|
| N-Zone | N-Zone | C-Zone | ||||||
| SCOPe 2.07, Fold d.3: Cysteine proteinases, Superfamily d.3.1: Cysteine proteinases | ||||||||
| Papain-like Cysteine Proteinases Zone (PCP-Zone): Class A | ||||||||
| PCP Structural Catalytic Core (PCP-SCC): I. Type C(H)X | ||||||||
| 1 | Papain | 1PPN_A 1.60 | N/H159-O/L134 3.1 O/H159-N/L134 3.0 |
N/V161-O/V132 2.9 O/V161-N/V132 3.1 |
N/A162-O/K174 3.3 O/A162-N/K174 2.9 |
OD1/N175-NE2/H159 2.6 N/A160-SG/C25 3.5 |
Cys25 Asn175 |
[34] |
| II. Type (CH)X | ||||||||
| 2 | Ubiquitin carboxyl-terminal esterase L3 | 1XD3_A 1.45 | N/H169-O/F56 2.9 O/H169-N/F56 2.9 |
N/I171-O/L54 3.0 O/I171-N/L54 2.9 |
N/A172-O/L183 3.1 O/A172-N/L183 2.8 |
OD1/D184-NE2/H169 2.8 N/F170-SG/C95 3.7 OD1/D184-N/R186 2.8 |
Cys95 Asp184 |
[35] |
| III. Type (H)XC | ||||||||
| 3 | Adenain | 4WX4_A 1.03 |
N/H54-OG1/T45 3.5 O/H54-N/T45 2.9 |
N/M56-O/V43 2.9 O/M56-N/V43 2.8 |
N/A57-O/F70 2.9 O/A57-N/F70 2.8 |
OD1/D71-NE2/H54 2.8 N/W55-SG/C122 3.5 OD1/D71-N/F73 2.9 |
Cys122 Asp71 |
[36] |
| IV. Type C(X)H | ||||||||
| 4 | Microbial transglutaminase | 5M6Q_A 1.98 |
N/D175-O/R131 3.0 O/D175-N/R131 2.9 |
N/G177-O/S129 2.8 O/G177-N/S129 2.8 |
N/W178-O/W187 2.8 O/W178-N/W187 2.9 |
ND1/H188-OD2/D175 3.1 N/Y176-SG/C46 3.6 |
Cys46 His188 |
[37] |
| V. Type CXH | ||||||||
| 5 | Cysteine protease ATG4B | 2CY7_A 1.90 |
N/S262-O/K259 2.8 O/S262-HOH425 2.6 HOH425-N/K259 2.9 |
O/H264-N/G257 3.2 N/H264-O/G257 2.9 |
N/Y265-O/L277 3.2 O/Y265-N/L277 2.9 |
OD1/D278-ND1/H280 2.6 N/A263-SG/C74 3.6 OD1/D278-N/H280 2.6 |
Cys74 Asp278 |
[38] |
| PCP-Zone: Class B | ||||||||
| PCP-SCC: VI. Type XC(H) | ||||||||
| 6 | Papain-like protease 2 |
4IUM_A 1.45 |
N/H332-O/D329 3.0 O/H332-N/D329 2.9 |
N/R334-O/I327 2.9 O/R334-N/I327 2.8 |
N/V335-O/Y262 2.8 O/V335-N/Y262 2.9 |
OD1/N263-NE2/H332 2.9 N/W333-SG/C270 3.7 |
Cys270 Asn263 |
[39] |
| VII. Type C(HX) | ||||||||
| 7 | Ubiquitin thioesterase OTU1 | 3BY4_A 1.55 |
N/H222-OD1/N219 2.9 O/H222-N/N219 3.1 |
N/D224-O/L217 3.0 O/D224-N/L217 2.8 |
N/S225-O/H112 3.1 O/S225-N/H112 2.7 |
CA/P113-OD1/D224 3.4 (2.4) 152° N/V114-OD1/D224 3.2 OD1/D224-NE2/H222 2.7 N/Y223-SG/C120 3.4 |
Cys120 Asp224 |
[40] |
| Not classified Fold | ||||||||
| VCP-Zone: Class C | ||||||||
| VCP-SCC: Type C(H)X | ||||||||
| 8 | Papain-like cysteine protease | 3MTV_A 2.80 |
N/H159-O/Q148 2.7 O/H159-N/Q148 2.6 |
N/K161-O/V146 3.2 O/K161-N/V146 3.0 |
N/R158-O/W201 2.9 O/R158-N/G203 3.2 |
OXT/G203-CD2/H159 2.6 (1.6) 163° OXT/G203-SG/C90 2.9 N/L160-SG/C90 3.6 |
Cys90 Gly203 |
[27] |
| VCP-Zone: Class D | ||||||||
| VCP-SCC: Type C(HX) | ||||||||
| 9 | Venezuelan equine encephalitis virus protease | 5EZS_A 2.16 |
N/H546-O/R543 3.0 O/H546-N/R543 3.0 |
N/D548-O/S541 2.8 O/D548-N/S541 2.9 |
N/W547-SG/C477 3.2 | OD2/D548-ND1/H546 3.9 | Cys477 Asp548 |
[41] |
For CH…O contacts, in addition to the distance between the C and O atoms, the distance between the H and O atoms (in brackets), and the CHO angle are also presented. The standard criteria for contact CH…O to be considered a weak hydrogen bond are as follows: C-O ≤ 4.1 Å, H-O ≤ 3.1 Å and ∠CHO ≥ 130° [42].
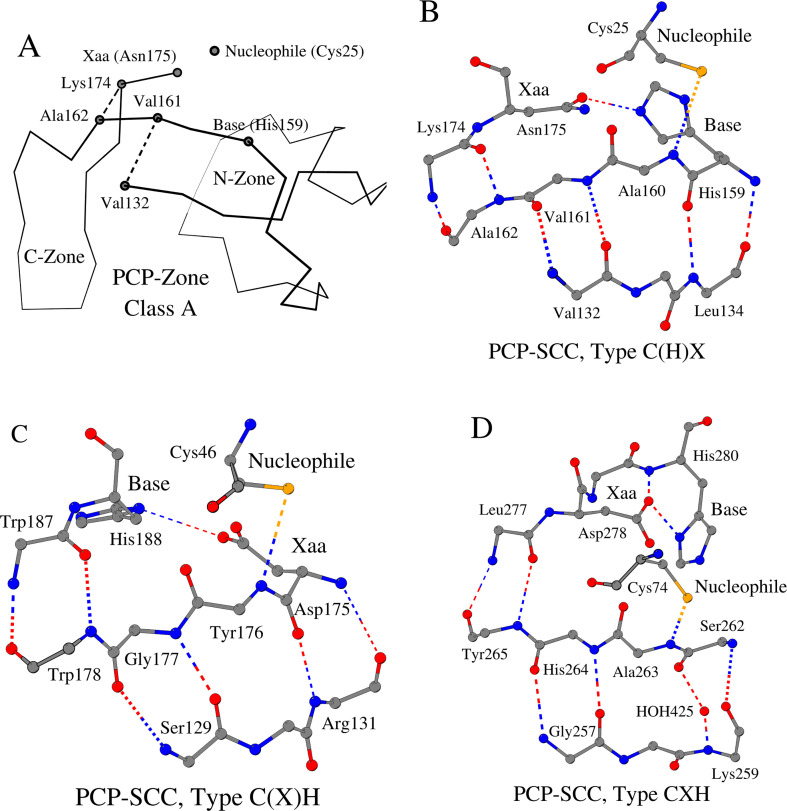
As a result of the visual analysis of the tertiary structure of papain in the area of its active site, we found two connected zones, a 30 amino-acid “N-Zone” Val132-Val161 and a 13 amino-acid “C-Zone” Ala162-Lys174 (Fig. 1A, Table 1). We refer to the structural union of these two Zones as the “Papain-Zone”. The Papain-Zone is formed by a 43-residue-long continuous fragment (Class A). A characteristic structural feature of the N-Zone is the presence of four main-chain hydrogen bonds between the tripeptide Val132-Val133-Leu134 at the N-terminal end of the zone and the tripeptide His159-Ala160-Val161 at the C-terminal end of the zone (Fig. 1B). A characteristic structural feature of the C-Zone is the presence of two hydrogen bonds between Ala162 at the N-terminal end of the zone and Lys174 at the C-terminal end of the zone. The three-dimensional structure of papain in the region of the Papain-Zone represents a unique mutual arrangement (papain structural core) of eight amino acids: six residues, including catalytic base (histidine), from the N-Zone and two residues from the C-Zone.
Fig. 1.
PCP-Zone Class A as a structural union of the N-Zone and C-Zone. A) A schematic representation of a PCP-Zone Class A using papain three-dimensional structure as an example. Two dashed lines connect the amino acids that define the boundaries of the N- and C-Zones. Positions of the catalytic triad (Nucleophile, Base and residue Xaa) are also shown. B), C), D) Contact schemes of ten amino acids of PCP-SCC of three types: C(H)X, C(X)H and CXH, respectively.
The nucleophile Cys25 and residue Asn175Xaa of the catalytic triad are located outside of the papain structural core, nevertheless, these two amino acids and the structural core interact with each other. The amino acids Ala160 and Asn175Xaa support the spatial arrangement of the catalytic cysteine and catalytic histidine, respectively (Fig. 1B, Tables 1 and S1). Ala160 directly follows base His159 in the amino acid sequence (position Base+1) and, like His159, is involved in formation of the papain structural core; and Asn175Xaa immediately follows the Val132-Lys174 fragment of the Papain-Zone in the amino-acid sequence. Ten amino acids, including eight residues of the papain structural core, catalytic cysteine and residue Xaa, form the Papain Structural Catalytic Core (Papain-SCC). According to the sequential numbering of the residues of the catalytic triad of papain, we will refer to the Cys25…His159…Asn175Xaa triad type of SCC as “C(H)X”, where H in brackets signifies that only the catalytic histidine is located within the N-Zone.
In addition to the ten residues of the SCC, papain contains Trp177, which is located at position Xaa + 2 of the amino-acid sequence. Trp177 is in contact with both His159 and Asn175Xaa [6,15]. Trp177 takes part in substrate binding [16] as does the N-zone, the importance of which is shown in several three-dimensional structures of papain-inhibitor complexes [[17], [18], [19], [20]].
2.1.2. Cysteine proteinase fold enzymes: PCP-Zone, Class A and PCP-SCC, Type C(H)X
The superfamily of cysteine proteinases contains 24 families (SCOPe) [5]. Eighteen families (88 proteins) have a structural organization similar to the arrangement of a Class A Zone and Type C(H)X SCC as found in papain (Table S1), referred to as the Papain-like Cysteine Proteinase Zone (PCP-Zone) and PCP-SCC. In addition to the 88 three-dimensional structures, Table S1 also represents the data for 22 X-ray structures extracted from RSCB PDB [21,22]. Variation is found among these structures: the length of the fragment of the amino-acid sequence that forms the N-Zone range from 8 to 73 residues and residue Xaa of the triad can not only be asparagine, but also aspartic acid, glutamine, glutamic acid, histidine, serine, cysteine and even tryptophan. In two proteins, the catalytic nucleophile is serine (Table S1, enzymes numbered 77 and 105).
In many cases, the amino acid Xaa and the residue at position Xaa + 2 of the amino-acid sequence form a widely observed Asn-turn motif [23] (Tables 1 and S1). The equivalent structural-functional principle of using the Asx-turn motif to coordinate the catalytic histidine and substrate binding was described earlier for the catalytic acid in the unrelated α/β hydrolase fold enzymes [3].
Often, instead of the amino acid at position Xaa + 2 of the amino acid sequence, the residue at position Xaa + 1 takes part in coordinating the conformation of the side chain of the catalytic histidine. An exception to the rule of using the residue at position Xaa + 1 or Xaa + 2 of the amino acid sequence to coordinate catalytic histidine are cysteine proteinases belonging to the Family d.3.1.23: Papain-like viral protease catalytic domain, including proteases from SARS coronavirus (Table S1, enzymes numbered 79 and 80).
Not only is the N-Zone important for the functioning of cysteine proteinases, but the C-Zone also can play some other significant roles. For example, for the major allergens Der p 1 and f 1, amino acids from the C-Zone have been proposed to take part in recognition by the human IgE antibody [24,25].
2.1.3. Type (CH)X of PCP-SCC
Enzymes with the cysteine proteinase fold demonstrate that the N-Zone can contain not only the catalytic histidine, but also the catalytic cysteine. This (CH)X type of organization of the PCP-SCC is observed in the Family d.3.1.6: Ubiquitin carboxyl-terminal hydrolase UCH-L (Tables 1 and S1). Strictly speaking, Types C(H)X and (CH)X of the PCP-SCC are not structurally different from each other. However, because type (CH)X enzymes contain a large insertion, up to 118 residues, the cysteine of the catalytic triad is positioned within the N-zone.
2.1.4. Type (H)XC of PCP-SCC
As noted above, the location of the catalytic cysteine preceding the N-Zone or within the N-Zone may not affect the principles of the PCP-SCC organization. From our structural analysis of enzymes with the Papain-like cysteine proteinase fold, we identified a third variation, in which the catalytic nucleophile is located after the N-Zone. This arrangement was found in 14 proteins: Family d.3.1.7: Adenain-like, Family d.3.1.21: YiiX-like and 6 cysteine peptidases from the RSCB PDB (Tables 1 and S1). For these 14 proteins, we found the following regularity: if the N-Zone contains only the catalytic histidine of the triad, then the length of the N-Zone is approximately the same as that in Type C(H)X of PCP-SCC. In one of the cysteine proteinases (Table S1, enzyme numbered 128) the residue Xaa is an aromatic amino acid. In this case, the structural coordination of the side chain of the catalytic histidine is achieved via the aromatic/hydrophobic interactions.
2.1.5. Type C(X)H of PCP-SCC
In the cases considered above, the location of the catalytic base and residue Xaa in a PCP-SCC did not change relative to each other, but the location of the catalytic nucleophile in the amino-acid sequence can differ. Cysteine proteinases of the Family d.3.1.8: Microbial transglutaminase illustrate one more type of PCP-SCC organization. In fact, despite sharing the common fold, in the microbial transglutaminase from Kutzneria albida, for example, the sequential position of catalytic His188 and Asp175Xaa are interchanged compared to catalytic histidine and amino acid Xaa in the cysteine proteinases analyzed above; i.e., Type C(H)X of PCP-SCC (Fig. 1C, Tables 1 and S1). As a result, residue Xaa is located within the N-Zone, and the catalytic histidine is attached to the C-Zone at its C-terminal end.
2.1.6. Type CXH of PCP-SCC
Family d.3.1.22: the Autophagin-like сysteine proteinase represents Type CXH of PCP-SCC, in which both the catalytic histidine and residue Xaa are located outside of the N-Zone (Fig. 1D, Tables 1 and S1). However, the catalytic histidine in this case is located two residues after Xaa (position Xaa/Asp+2) in the amino acid sequence, forming the Asx-turn motif [23]. Therefore, while in the aforementioned cases the position Xaa/Asp+2 of the amino-acid sequence was auxiliary for the positions of the catalytic triad (see sections “Zone and SCC of papain” and “Cysteine proteinase fold enzymes: PCP-Zone, Class A and PCP-SCC, Type C(H)X”), in this case it is the main one.
2.1.7. Inactive peptidases
In addition to 134 cysteine peptidases containing a catalytic triad, we found 3 inactive peptidases (Table S1). The catalytic nucleophile is absent in two inactive peptidases, and both the catalytic nucleophile and catalytic base are absent in the third inactive peptidase. Despite the lack of catalytic activity, the Zone (Class A) and SCC (Type C(H)X) characteristics of these three proteins are similar to those of active papain-like proteases.
2.2. PCP-Zone, Class B
Of the 137 analyzed structures, 23 out of 24 сysteine proteinase families (SCOPe) [5] corresponded to the types described above. The one remaining family is the Family d.3.1.11: Ubiquitin thiolesterase protein OTUB2 (Otubain-2), and one proteinase: papain-like protease 2 - have the Class B structural organization of PCP-Zone (Fig. 2A, Tables 1 and S1 (enzymes numbered 138–146)). In the PCP-Zone Class B proteinases, the C-Zone is structurally transformed (see below).
Fig. 2.
PCP-Zone Class B. A. A schematic representation of a PCP-Zone Class B using three-dimensional structure of papain-like protease 2 as an example (for the designation of the details of the figure, see the legend to Fig. 1A) B. and C. Contact schemes of ten amino acids of PCP-SCC of two types: XC(H) and C(HX), respectively.
2.2.1. Type XC(H) of PCP-SCC
The papain-like protease 2 is a Class B variant of the structural organization of PCP-Zone (Tables 1 and S1). The C-Zone in this case consists of two dipeptides Gly261-Tyr262 and Val335-Lys336. Val335 that follows the N-Zone is linked by two hydrogen bonds to Tyr262 preceding Asn263Xaa. The structure of this protease represents not only a new class of the Zone, but also a new Type XC(H) of the PCP-SCC (Fig. 2B).
2.2.2. Type C(HX) of PCP-SCC
In the Family d.3.1.11: Ubiquitin thiolesterase protein OTUB2 (Otubain-2) of сysteine proteinases, the C-Zone is also structurally transformed. However, unlike papain-like protease 2, the size of the C-Zone is larger in this case. For example, in ubiquitin thioesterase OTU1, the C-Zone consists of two pentapeptides Val108-…-His112 and Ser225-…-Asn229 (Tables 1 and S1). Furthermore, residue Xaa (Asp, Asn or Ser), which coordinates the conformation of the catalytic histidine, is situated two sequence positions after the catalytic base (position Base+2) within the N-Zone and is classified as Type C(HX) of PCP-SCC (Fig. 2C). Residue Xaa is the last residue of the N-Zone. Pro113, whose position corresponds to the position of the canonical residue Xaa, acts in this case as its structural mediator. In OTU deubiquitinase A20, a water molecule serves as an additional mediator of the structural coordination of the side chain of the catalytic histidine (Table S1). Note that Family d.3.1.11: Ubiquitin thiolesterase protein OTUB2 (Otubain-2) is the only family of the 24 families studied that has the amino acid Asp/Asn (Xaa) at position Base+2 (Table S1).
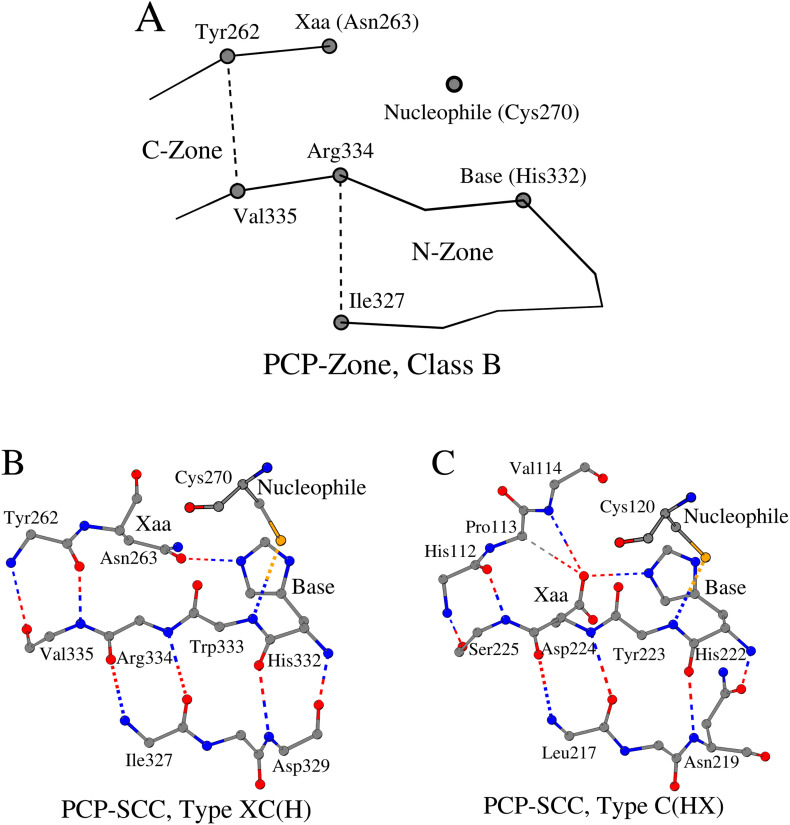
2.3. Viral Cysteine Proteinases (VCPs) of the currently unclassified fold(s)
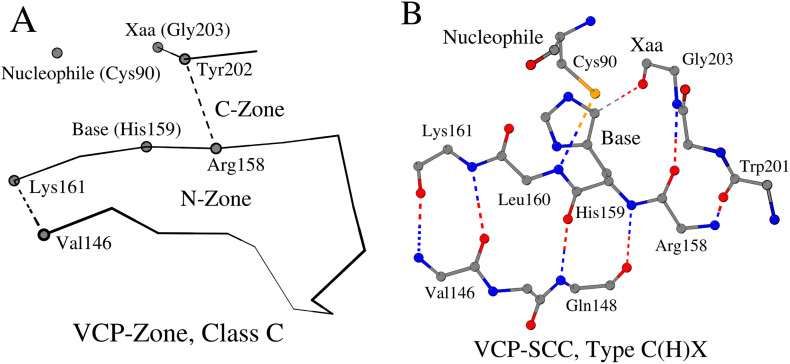
2.3.1. VCP-Zone, Class C and VCP-SCC, Type C(H)X
In the course of our analysis, we identified seven VCPs, the three-dimensional structures of which could not be unambiguously attributed to Fold d.3: Cysteine proteinases (Table S1, enzymes numbered 147–153). The structural analysis divided them into two groups. The first group includes three proteases (Table S1, enzymes numbered 147–149), which demonstrate the third variant (Class C) of the VCP-Zone arrangement (Tables 1 and S1 (enzymes numbered 147–149)). The structural principles of the N-Zone organization have not changed in this group. However, the C-Zone is arranged differently. Firstly, unlike the features seen in Classes A and B, in Class C, the amino acids of the C-Zone directly interact with the amino acids of the N-Zone, and significantly modify position and structure of the ligand binding site (Fig. 3A). Secondly, in connection with the previous point, instead of the antiparallel direction of the amino acid sequence fragments that form the C-Zone, their parallel direction is observed (Fig. 3B). Despite these structural differences, Class C of VCP-SCC and Class A of PCP-SCC share the same C(H)X Type. For example, in papain-like cysteine protease (Fig. 3B), the amino acid Arg158 (as opposed to Leu162, as it would be if the protein belonged to either Class A or B) forms the C-Zone and coordinates the three-dimensional position of amino acid Xaa with the help of two hydrogen bonds. Contact between the N-Zone and the catalytic nucleophile (N/Leu160-SG/Cys90 = 3.6 Å) still takes place. This tertiary organization of the catalytic triad was found to be suitable, for example, for the formation of a zinc-binding site, as observed in non-structural protein Nsp1α (Table S1 (enzyme numbered 148)). The zinc ion is important for proteolytic self-release of Nsp1α [26]. Furthermore, the N-Zone and C-Zone together (VCP-Zone) might be important for the dimerization of some viral cysteine proteinases. An example is given by the nonstructural protein Nsp1β (which cleaves itself from the downstream Nsp2 protein via a C-terminal papain-like cysteine protease (PCP) domain) from the porcine reproductive and respiratory syndrome virus (PRRSV) that forms a functional dimer (PDB ID: 3MTV) [27]. Another example of the N-Zone and C-Zone together playing a role in homooligomerization is an octamer of the autocatalytic cysteine protease domain of potyvirus helper-component proteinase (PDB ID: 3RNV). On the other hand, the homodimer of the PRRSV Nsp1α, which is crucial for the subgenomic mRNA synthesis, serving as a transcription factor, and which utilizes its PCP domain to self-release from the Nsp1β, is organized differently from the aforementioned cases (PDB ID: 3IFU; [26]). In fact, Nsp1α contains the zinc ion in the active site and is characterized by the N-Zone (Tyr141-Ser148), which is noticeably shorter than the N-Zones of the Nsp1β (Val146-Lys161; PDB ID: 3IFU) and the autocatalytic cysteine protease domain of potyvirus helper-component proteinase (Arg406-Val419; PDB ID: 3RNV).
Fig. 3.
VCP-Zone Class C. A. A schematic representation of a VCP-Zone Class C using three-dimensional structure of papain-like cysteine protease as an example (for the designation of the details of the figure, see the legend to Fig. 1A) B. Contact scheme of eleven amino acids of VCP-SCC, Type C(H)X.
2.3.2. VCP-Zone, Class D and VCP-SCC, Type C(HX)
In the second group of proteases, which consists of four enzymes (Tables 1 and S1 (enzymes numbered 150–153)), one of the three strands of the basis β-sheet is absent in the tertiary structures. The loss of the strand results in the absence of the C-Zone in these proteases. However, these proteases have the N-Zone, including catalytic histidine, and also a conservative contact between the N-Zone and the catalytic nucleophile. The loss of the C-Zone also led to the absence of residue Xaa at its most frequently encountered position. Nevertheless, it can be assumed that the residue Asp/Ser that is situated two sequence positions after the catalytic base can play the role of the residue Xaa, as is the case for the proteases of PCP-Zone, Class B, Type C(HX) of PCP-SCC.
2.4. Relevance of structural analysis of cysteine proteinases
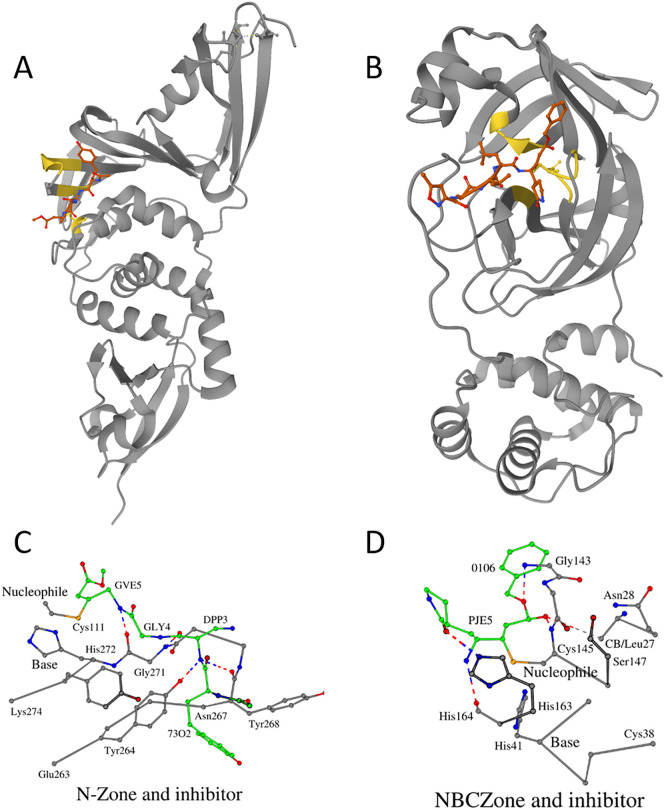
The three-dimensional structures of papain-like proteins have been studied for over 50 years [28]. During that time, structural information for more than 150 proteins was obtained [21,22]. This year, as the pandemic of COVID-19 began, it became particularly important to understand the molecular mechanisms behind the function of various proteins of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [29]. It turned out that the SARS-CoV-2 has only two functionally important proteinases: papain-like (PLpro) and (chymo)trypsin-like (3CLpro) (https://swissmodel.expasy.org/repository/species/2697049), both belonging to the family of cysteine proteinases. Structural organizations of PLpro and 3CLpro complexed with the inhibitors are shown in Fig. 4A and B, respectively. Table S1 (enzyme numbered 80, PDB_ID 6WX4) shows structural data for SARS-CoV-2-PLpro [30]. The three-dimensional structures of papain and SARS-CoV-2-PLpro belong to the same Class A of PCP-Zone and Type C(H)X of PCP-SCC. In section “Zone and SCC of papain”, the importance of the N-Zone for substrates binding by papain was noted. Fig. 4C shows the interaction between N-Zone of SARS-CoV-2-PLpro (fragment of sequence Glu263-Lys274) and an inhibitor. The majority of protein-ligand interactions are mediated through four potential hydrogen bonds: OH/Tyr264-N/DPP3, O/Tyr268-NH/DPP3, N/Gly271-O/DPP3, O/Gly271-NH/GVE5 and van der Waals contacts between residues Tyr264, Tyr268, Tyr273 and 73O2, Gly4. Tetrapeptide Asn267-Cys270 of the N-Zone forms Asx-turn [23]. The C-Zone does not take part in the interaction with the inhibitor.
Fig. 4.
Structural characterization of the SARS-CoV-2 proteases. A. Crystal structure of SARS-CoV-2-PLpro complexed with the inhibitor (PDB ID: 6WX4; [30]). B. Crystal structure of SARS-CoV-2-3CLpro complexed with the inhibitor (PDB ID: 7BQY [31]). Positions of the residues belonging to the N-Zone and the NBCZone are shown in yellow. Closer look at the complexes of the N-Zone of SARS-CoV-2-PLpro (C) and the NBCZone of SARS-CoV-2-3CLpro (D) with inhibitors. The color of carbon atoms in proteinases is gray, and in inhibitors, green. Contacts (potential hydrogen bonds) between proteinases and inhibitors are shown by dotted lines.
Residues Tyr264 and Tyr273 are located in the immediate vicinity of the beginning and end of the amino acid sequence fragment that defines the N-Zone. Since all papain-like cysteine proteinases have corresponding two amino acid positions, it was decided to check which pair of residues is observed in them. It turned out that in Class A proteinases, only three coronaviral proteinases (Table S1 (enzymes numbered 79–81)) have the same pair of residues. Additionally, three proteinases from the Class B Family d.3.1.11: Ubiquitin thiolesterase protein OTUB2 (Otubain-2) (Table S1 (enzymes numbered 140–142)) also have a tyrosine pair of amino acids in the analogous amino acid positions (data not shown). It is known that the SARS-CoV-2-PLpro is a deubiquitinating enzyme [12]. Amino acids Tyr264 and Tyr273 of PLpro structurally correspond to amino acids Val133 and Ala160 of papain (PDB ID 1PPN). Earlier, in section “Zone and SCC of papain”, it was noted that the amino acid Ala160 supports the three-dimensional arrangement of the catalytic cysteine. Indeed, there are three contacts between these two amino acids: N/Ala160-SG/Cys25 = 3.5 Å, O/Ala160-CB/Cys25 = 3.4 (2.4) 172° and CB/Ala160-O/Cys25 = 3.7 (2.7) 151°. Similar contacts take place between amino acids Tyr273 and Cys111 in PLpro: N/Tyr273-SG/Cys111 = 3.5 Å, O/Tyr273-CB/Cys111 = 3.7 (2.7) 164° and CB/Tyr273-O/Cys111 = 4.0 (3.2) 128°. It can be assumed that this tyrosine pair of amino acids is a functionally specific structural marker of the deubiquitinating enzymes.
At the time of writing about the NBCZone in the superfamily of (chymo)trypsin-like folds proteases [4], the three-dimensional structure of SARS-CoV-2-3CLpro was not known. However, the situation is changed dramatically now and the three-dimensional structures of about 170 variants of this enzyme are currently known (https://swissmodel.expasy.org/repository/species/2697049). We selected one file (PDB_ID 7BQY [31]) from this list to analyze the structural details of the interaction between the NBCZone and the inhibitor (Fig. 4D), as in the case of SARS-CoV-2-PLpro. The NBCZone consists of eleven amino acids: Leu27-Asn28, Cys38-Pro39-Arg40-His41(base)-Val42, Cys145(nucleophile)-Gly146-Ser147 and His163. The Fig. 4D additionally shows the main chain atoms of residue His164, the carbonyl oxygen of which forms a universal weak hydrogen bond [32] with catalytic histidine: O/His164-(CE1...H)/His41, and a dipeptide Gly143-Ser144 containing a first oxyanion nitrogen atom N/Gly143. There are four canonical (N/Gly143-O/0106, N/Cys145(second oxyanion atom)-O7/PJE5, NE2/His163-O8/PJE5 and O/His164-N5H/PJE5) and one weak ((CB…H)/Leu27-O7/PJE5) hydrogen bonds between these fourteen amino acids of the SARS-CoV-2-3CLpro and two residues (PJE5 and 0106) of the inhibitor. The other four residues of the inhibitor (02J1, Ala2, Val3 and Leu4) do not participate in similar bonding interactions. Two intraprotein hydrogen bonds (ND2/Asn28-O/Gly143 and OG/Ser147-O/Ser144) provide the necessary arrangement of the oxyanionic atoms (N/Gly143 and N/Cys145) for such contacts. The results presented hear clearly show that the use of amino acids of both N-Zone and NBCZone for the search for effective inhibitors of SARS-CoV-2-PLpro and SARS-CoV-2-3CLpro will accelerate the production of drugs that stop viral replication.
3. Conclusions
The 146 enzymes with the papain-like cysteine proteinase (PCP) fold have a unique, structurally closed conformation (PCP-Zone) of two Classes: A and B. The PCP-Zone, Class A consists of two Zones (N- and C-Zones) formed by a continuous fragment of the amino acid sequence. In enzymes of Class B, the C-Zone is structurally transformed. Eight structurally conserved amino acids of the PCP-Zone, in both Classes A and B, form the common Structural Core. The Structural Core, catalytic nucleophile, catalytic base and residue Xaa, which coordinates the catalytic base, make up a PCP Structural Catalytic Core (PCP-SCC). Comparison of the PCP-SCC, Class A enzymes with each other showed that they can be divided into 5 types in accordance with the order of arrangement of the residues of the catalytic triad in the amino-acid sequence and their individual location within the N-Zone. Class B enzymes demonstrate two types of the PCP-SCC. In summary, seven variants of the mutual arrangement of the amino acids of the catalytic triad within the same fold clearly demonstrate how the enzymes of the papain-like cysteine proteinases adapt to the need to perform diverse functions in spite of the limited structural diversity.
Finding and describing local structural similarities, structural motifs, if they exist, in the active sites of proteins from different fold families is one of the fundamental areas of structural biology. Such analysis makes it possible to compare and group proteins without making a superposition of the entire tertiary structures. It serves as the basis for possible classification of proteins based on such local structural similarities. We show here that structures of active sites of different PCPs share a unique common structural organization (Structural Catalytic Core (PCP-SCC)), and can be divided into seven groups. Therefore, the members of the PCP family are characterized by one fold, two classes of supersecondary structure, and seven types of catalytic triad arrangement. The proteins that belong to each group have different tertiary structures, belong to different families and fulfill different functions, and yet their PCP-SCCs are the same. Within each group, the PCP-SCCs incorporates an identical set of conserved interactions and bonds. Furthermore, the PCP-SCCs are not only conserved in papain-like proteases with different tertiary structures and functions, but also play key role in interactions with other proteins. In particular, for PLpro of SARS-CoV-2, it was assumed that tyrosine pair of amino acids near the catalytic triad is a functionally specific structural marker of the deubiquitinating enzymes.
4. Materials and methods
Using the SCOPe classification database [5], the Sequence Similarity (https://www.rcsb.org/pdb/explore/sequenceCluster.do?structureId=1ppn) and Structural Similarities (https://www.rcsb.org/pdb/explore/structureCluster.do?structureId=1ppn) of the Protein Data Bank (PDB, http://www.rcsb.org/ [21,22]), we have retrieved 146 different entries with the papain-like cysteine proteinases fold d.3 which represent 24 families according to SCOPe definition. Additionally, we identified 7 viral cysteine proteinases, the three-dimensional structures of which could not be unambiguously attributed to papain-like cysteine proteinases fold d.3. These 153 entries are representative structures with the highest resolution for each individual proteinase. Nine structures are used to illustrate the structural diversity of the active site observed across the papain-like cysteine proteinases fold families.
Structure visualization and structural analysis of interactions between amino acids in proteins (hydrogen bonds, hydrophobic, other types of weak interactions) was carried out using the Discovery Studio Modeling Environment (Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016) and the Ligand-Protein Contacts (LPC) software [14]. Figures are drawn with MOLSCRIPT [33].
The following is the supplementary data related to this article.
The geometric characteristics of the contacts of the amino acids that make up two classes of the Papain-like Cysteine Proteinase Zone (PCP-Zone) and seven types of the PCP Structural Catalytic Core (PCP-SCC) in 146 enzymes with the cysteine proteinase fold.
Funding
The project was supported by the Sigrid Jusélius Foundation and Joe, Pentti and Tor Borg Memorial Fund (A.I.D. and M.S.J).
CRediT authorship contribution statement
Vladimir N. Uversky: Formal analysis, Methodology, Writing - Original Draft, Writing - Review & Editing; Konstantin Denessiouk: Study design, Formal analysis, Methodology, Visualization, Investigation, Writing - Original Draft, Writing - Review & Editing; Sergei E. Permyakov: Formal analysis, Writing - Review & Editing; Eugene A. Permyakov: Formal analysis, Writing - Review & Editing; Mark S. Johnson: Formal analysis, Methodology, Writing - Original Draft; Alexander I. Denesyuk: Study design, Formal analysis, Methodology, Visualization, Writing - Original Draft, Writing - Review & Editing;
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work is supported by a grant from the Sigrid Jusélius Foundation and Joe, Pentti, and Tor Borg Memorial Fund. We thank the Biocenter Finland Bioinformatics Network (Dr. Jukka Lehtonen) and CSC IT Center for Science for computational support for the project. The Structural Bioinformatics Laboratory is part of the Drug Development and Diagnostics Platform of Åbo Akademi University.
References
- 1.Dodson G., Wlodawer A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998;23(9):347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 2.Buller A.R., Townsend C.A. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proc. Natl. Acad. Sci. U. S. A. 2013;110(8):E653–E661. doi: 10.1073/pnas.1221050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denesyuk A., Dimitriou P.S., Johnson M.S., Nakayama T., Denessiouk K. The acid-base-nucleophile catalytic triad in ABH-fold enzymes is coordinated by a set of structural elements. PLoS One. 2020;15(2) doi: 10.1371/journal.pone.0229376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denesyuk A.I., Johnson M.S., Salo-Ahen O.M.H., Uversky V.N., Denessiouk K. NBCZone: universal three-dimensional construction of eleven amino acids near the catalytic nucleophile and base in the superfamily of (chymo)trypsin-like serine fold proteases. Int. J. Biol. Macromol. 2020;153:399–411. doi: 10.1016/j.ijbiomac.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox N.K., Brenner S.E., Chandonia J.M. SCOPe: structural classification of proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014;42(Database issue):D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novinec M., Lenarcic B. Papain-like peptidases: structure, function, and evolution. Biomol. Concepts. 2013;4(3):287–308. doi: 10.1515/bmc-2012-0054. [DOI] [PubMed] [Google Scholar]
- 7.Baez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurumallesh P., Alagu K., Ramakrishnan B., Muthusamy S. A systematic reconsideration on proteases. Int. J. Biol. Macromol. 2019;128:254–267. doi: 10.1016/j.ijbiomac.2019.01.081. [DOI] [PubMed] [Google Scholar]
- 9.Liu H., Hu M., Wang Q., Cheng L., Zhang Z. Role of papain-like cysteine proteases in plant development. Front. Plant Sci. 2018;9:1717. doi: 10.3389/fpls.2018.01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma S., Dixit R., Pandey K.C. Cysteine proteases: modes of activation and future prospects as pharmacological targets. Front. Pharmacol. 2016;7:107. doi: 10.3389/fphar.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zehra Z., Luthra M., Siddiqui S.M., Shamsi A., Gaur N.A., Islam A. Corona virus versus existence of human on the earth: a computational and biophysical approach. Int. J. Biol. Macromol. 2020;161:271–281. doi: 10.1016/j.ijbiomac.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemente V., D’Arcy P., Bazzaro M. Deubiquitinating enzymes in coronaviruses and possible therapeutic opportunities for COVID-19. Int. J. Mol. Sci. 2020;21(10) doi: 10.3390/ijms21103492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris G.W., Pickersgill R.W., Howlin B., Moss D.S. The segmented anisotropic refinement of monoclinic papain by the application of the rigid-body TLS model and comparison to bovine ribonuclease A. Acta Crystallogr. B. 1992;48(Pt 1):67–75. doi: 10.1107/s0108768191006663. [DOI] [PubMed] [Google Scholar]
- 14.Sobolev V., Sorokine A., Prilusky J., Abola E.E., Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15(4):327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 15.Hussain S., Khan A., Gul S., Resmini M., Verma C.S., Thomas E.W., Brocklehurst K. Identification of interactions involved in the generation of nucleophilic reactivity and of catalytic competence in the catalytic site Cys/His ion pair of papain. Biochemistry. 2011;50(49):10732–10742. doi: 10.1021/bi201207z. [DOI] [PubMed] [Google Scholar]
- 16.Bromme D., Bonneau P.R., Purisima E., Lachance P., Hajnik S., Thomas D.Y., Storer A.C. Contribution to activity of histidine-aromatic, amide-aromatic, and aromatic-aromatic interactions in the extended catalytic site of cysteine proteinases. Biochemistry. 1996;35(13):3970–3979. doi: 10.1021/bi9523015. [DOI] [PubMed] [Google Scholar]
- 17.Stubbs M.T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990;9(6):1939–1947. doi: 10.1002/j.1460-2075.1990.tb08321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alphey M.S., Hunter W.N. High-resolution complex of papain with remnants of a cysteine protease inhibitor derived from Trypanosoma brucei. Acta Crystallogr. Sec. F Struct. Biol. Cryst. Commun. 2006;62(Pt 6):504–508. doi: 10.1107/S1744309106014849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redzynia I., Ljunggren A., Bujacz A., Abrahamson M., Jaskolski M., Bujacz G. Crystal structure of the parasite inhibitor chagasin in complex with papain allows identification of structural requirements for broad reactivity and specificity determinants for target proteases. FEBS J. 2009;276(3):793–806. doi: 10.1111/j.1742-4658.2008.06824.x. [DOI] [PubMed] [Google Scholar]
- 20.Chu M.H., Liu K.L., Wu H.Y., Yeh K.W., Cheng Y.S. Crystal structure of tarocystatin-papain complex: implications for the inhibition property of group-2 phytocystatins. Planta. 2011;234(2):243–254. doi: 10.1007/s00425-011-1398-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J.D., Zardecki C. The protein data bank. Acta Crystallogr. D Biol. Crystallogr. 2002;58(Pt 6 1):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 23.Wan W.Y., Milner-White E.J. A natural grouping of motifs with an aspartate or asparagine residue forming two hydrogen bonds to residues ahead in sequence: their occurrence at alpha-helical N termini and in other situations. J. Mol. Biol. 1999;286(5):1633–1649. doi: 10.1006/jmbi.1999.2552. [DOI] [PubMed] [Google Scholar]
- 24.Chruszcz M., Pomes A., Glesner J., Vailes L.D., Osinski T., Porebski P.J., Majorek K.A., Heymann P.W., Platts-Mills T.A., Minor W., Chapman M.D. Molecular determinants for antibody binding on group 1 house dust mite allergens. J. Biol. Chem. 2012;287(10):7388–7398. doi: 10.1074/jbc.M111.311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osinski T., Pomes A., Majorek K.A., Glesner J., Offermann L.R., Vailes L.D., Chapman M.D., Minor W., Chruszcz M. Structural analysis of Der p 1-antibody complexes and comparison with complexes of proteins or peptides with monoclonal antibodies. J. Immunol. 2015;195(1):307–316. doi: 10.4049/jimmunol.1402199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y., Xue F., Guo Y., Ma M., Hao N., Zhang X.C., Lou Z., Li X., Rao Z. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha. J. Virol. 2009;83(21):10931–10940. doi: 10.1128/JVI.02579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue F., Sun Y., Yan L., Zhao C., Chen J., Bartlam M., Li X., Lou Z., Rao Z. The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease. J. Virol. 2010;84(13):6461–6471. doi: 10.1128/JVI.00301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drenth J., Jansonius J.N., Koekoek R., Swen H.M., Wolthers B.G. Structure of papain. Nature. 1968;218(5145):929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- 29.Ranga V., Niemela E., Tamirat M.Z., Eriksson J.E., Airenne T.T., Johnson M.S. Immunogenic SARS-CoV-2 epitopes: in silico study towards better understanding of COVID-19 disease-paving the way for vaccine development. Vaccines (Basel) 2020;8(3) doi: 10.3390/vaccines8030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. bioRxiv. 2020. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 32.Derewenda Z.S., Derewenda U., Kobos P.M. (His)C epsilon-H...O=C < hydrogen bond in the active sites of serine hydrolases. J. Mol. Biol. 1994;241(1):83–93. doi: 10.1006/jmbi.1994.1475. [DOI] [PubMed] [Google Scholar]
- 33.Kraulis P.J. Molscript - a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 34.Pickersgill R.W., Harris G.W., Garman E. Structure of monoclinic papain at 1.60 angstroms resolution. Acta Crystallogr. B. 1992;48:59–67. doi: 10.1107/s0108768191006663. [DOI] [PubMed] [Google Scholar]
- 35.Misaghi S., Galardy P.J., Meester W.J., Ovaa H., Ploegh H.L., Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 2005;280(2):1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 36.Grosche P., Sirockin F., Mac Sweeney A., Ramage P., Erbel P., Melkko S., Bernardi A., Hughes N., Ellis D., Combrink K.D., Jarousse N., Altmann E. Structure-based design and optimization of potent inhibitors of the adenoviral protease. Bioorg. Med. Chem. Lett. 2015;25(3):438–443. doi: 10.1016/j.bmcl.2014.12.057. [DOI] [PubMed] [Google Scholar]
- 37.Steffen W., Ko F.C., Patel J., Lyamichev V., Albert T.J., Benz J., Rudolph M.G., Bergmann F., Streidl T., Kratzsch P., Boenitz-Dulat M., Oelschlaegel T., Schraeml M. Discovery of a microbial transglutaminase enabling highly site-specific labeling of proteins. J. Biol. Chem. 2017;292(38):15622–15635. doi: 10.1074/jbc.M117.797811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 2005;280(48):40058–40065. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- 39.van Kasteren P.B., Bailey-Elkin B.A., James T.W., Ninaber D.K., Beugeling C., Khajehpour M., Snijder E.J., Mark B.L., Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110(9):E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messick T.E., Russell N.S., Iwata A.J., Sarachan K.L., Shiekhattar R., Shanks J.R., Reyes-Turcu F.E., Wilkinson K.D., Marmorstein R. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J. Biol. Chem. 2008;283(16):11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu X., Compton J.R., Leary D.H., Olson M.A., Lee M.S., Cheung J., Ye W., Ferrer M., Southall N., Jadhav A., Morazzani E.M., Glass P.J., Marugan J., Legler P.M. Kinetic, mutational, and structural studies of the Venezuelan equine encephalitis virus nonstructural protein 2 cysteine protease. Biochemistry. 2016;55(21):3007–3019. doi: 10.1021/acs.biochem.5b00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derewenda Z.S., Lee L., Derewenda U. The occurrence of C-H...O hydrogen bonds in proteins. J. Mol. Biol. 1995;252(2):248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The geometric characteristics of the contacts of the amino acids that make up two classes of the Papain-like Cysteine Proteinase Zone (PCP-Zone) and seven types of the PCP Structural Catalytic Core (PCP-SCC) in 146 enzymes with the cysteine proteinase fold.