Abstract
Background
Secretory carrier membrane proteins 3 (SCAMP3) is an endocytosis-associated protein involved in regulating endosomal pathways and the trafficking of vital signaling receptors. This study aimed to comprehensively assess the role of SCAMP3 in hepatocellular carcinoma (HCC) by integrated bioinformatics analysis.
Methods
In this study, bioinformatics databases were used to explore the differential expression status and prognostic value of SCAMP3 gene in HCC, and bioinformatics analyses of survival data and interactors of SCAMP3 were conducted to predict the prognostic value of SCAMP3 in HCC.
Results
Using the TCGA data, our data shows that SCAMP3 mRNA expression is most significantly different between liver and hepatocellular carcinoma tissues and higher expression of SCAMP3 has unfavorable prognostic significance in HCC. Tumor grade, stage, and gender also showed a significant relevance with SCAMP3 expression. High SCAMP3 expression of males revealed significantly poorer survival and progression compared with low SCAMP3 expression of males. BioGRID statistics explores 79 unique interactions with SCAMP3 and multiple post translational modifications. Further analysis finds that SOCS2 may negatively correlate with SCAMP3, while GBA, MX1, and DDOST positively correlate with SCAMP3. Moreover, ncRNA analysis shows that SCAMP3 gene expression is positively associated with lncRNA SBF2-AS1 and negatively related with Has-miR-145. The expressions of SBF2-AS1 and Has-miR-145 are also significantly related with survival in HCC.
Discussion
SCAMP3 expression can be affected by multiple genes or ncRNAs expression that are associated with survival, thus suggesting that SCAMP3 can be used as a clinical diagnosis and prognostic biomarker in HCC.
Keywords: SCAMP3, hepatocellular carcinoma, prognosis, interaction, ncRNA, Kaplan–Meier plotter
Introduction
Due to its higher invasive and metastatic potential, hepatocellular carcinoma (|HCC) is one of the most leading causes of cancer-related death globally and leads to poorer prognosis. Although considerable progress has been undertaken to elucidate the mechanism of hepatocarcinogenesis and to develop treatment techniques for HCC, the prognosis of HCC remains unclear. Therefore, to improve the survival rate of HCC patients, it is eagerly needed to detect novel biomarkers for early diagnosis and advanced treatment for HCC patients, as well as therapeutic strategies.
A previous study has shown that SCAMP3 is overexpressed in HCC tissues and associated with HCC progression.1 Expression profiles of genes by array-based comparative genomic hybridization show that the expression of SCAMP3 gene located on 1q22 is significantly up-regulated in HCC.2 SCAMP3 is a regulator of endosomal structure and composition, and plays a critical role in the biogenesis of multivesicular endosomes through an interaction with endosomal sorting complex required for transport (ESCRT) proteins,3 and SCAMP3 knockdown can accelerate the degradation of epidermal growth factor receptor (EGFR) to promote receptor recycling.4 In addition, a report found that SCAMP3 was remarkably upregulated in gliomas compared with normal tissues and could act as an independent risk factor of glioma prognosis.5 SCAMP3 also has an important role in the invasion of Inflammatory Breast Cancer (IBC),6 suggesting the tumor-related role of SCAMP3.
Hence, in this study, we aimed to acknowledge the role of SCAMP3 in HCC based on the TCGA database. Integrated bioinformatics analysis was used to comprehensively evaluate the value of SCAMP3 as a clinical diagnosis and prognostic biomarker in HCC. Furthermore, the regulatory network of SCAMP3 and ncRNA-SCAMP3 interaction in HCC was also investigated.
Materials and Methods
Genes Expression Analysis
The mRNA levels of SCAMP3 in diverse cancer types were analyzed by UALCAN, GEPIA2, and STARBASE databases, which are publicly accessible online databases to facilitate novel discovery from genome-wide expression analyses.
UALCAN Database
UALCAN (http://ualcan.path.uab.edu) is a comprehensive and interactive online database that provides transcriptome data, methylation predication, clinical data of a series of cancers from TCGA database, and external links of other databases.7 In this study, UALCAN was used to analyze the mRNA expressions of related genes in hepatocellular carcinoma and their association with clinicopathologic parameters.
GEPIA2 (Gene Expression Profiling Interactive Analysis)
GEPIA2 (http://gepia2.cancer-pku.cn/) is an updated version of GEPIA that includes TCGA and GTEx’ RNA sequencing expression data to provide differential expression analysis, patient survival analysis, and correlation analysis by using a standard processing pipeline.8
Starbase Analysis
mRNA-ncRNA interactions were analyzed by using Starbase database (http://starbase.sysu.edu.cn), which is an open-source platform for studying the miRNA-ncRNA, miRNA-mRNA, ncRNA-RNA, and RNA-RNA interactions from CLIP-seq, degradome-seq, and RNA-RNA interactome data.9,10 This database was employed to analyze the expression association between mRNA or ncRNA and SCAMP3. Similarly, mRNA- SCAMP3 or ncRNA-SCAMP3 interactions with R<−0.1 and P-value<0.05 were regarded as significant.
Oncomine
The ONCOMINE database (www.oncomine.org) is an interactive web resource based on cancer microarray, which aims to facilitate discovery from the gene-wide expression analyses.11 In this study, five different HCC data from the ONCOMINE database were used to analyze the transcriptional expression of SCAMP3 gene between different cancer tissues and the adjacent normal control samples. Difference of transcriptional expression was compared by Students’ t-test.
The Kaplan–Meier Plotter Survival Analysis
Prognostic values of related genes and ncRNA were analyzed by using Kaplan–Meier plotter (http://kmplot.com/analysis/), an online tool that includes gene expression data and clinical data on diverse cancer types from GEO and TCGA.12 In this report, we mainly utilize overall survival patient information to draw the survival curves of patients on the basis of high and low expression levels. Hazard ratios with 95% confidence intervals and log-rank P-value were calculated to assess the statistical difference.
Protein–Protein Interaction (PPI) Network Analysis Construction
Protein–protein interaction (PPI) was analyzed using BioGRID and STRING pathway analysis tools. Biological General Repository for Interaction Datasets database (BioGRID, https://thebiogrid.org)13 was used to identify the known protein interaction network of SCAMP3. Further, the PPI network of SCAMP3 was further identified by the STRING network (Search Tool for the Retrieval of Interacting Genes/Proteins).14
LncRNA2Target
LncRNA2Target (http://123.59.132.21/lncrna2target/index.jsp) is developed to provide a comprehensive online resource of lncRNA-target relationships confirmed by immunoprecipitation assays, RNA pull-down assays and luciferase reporter assays.15
SCAMP3-miRNA Interaction Prediction
SCAMP3-miRNA interaction was predicted using four databases: miRWalk (http://mirwalk.umm.uni-heidelberg.de/),16 microRNA.org (http://www.microrna.org/)17, miRDB (http://mirdb.org),18 and TargetScan (http://www.targetscan.org).19
DIANA-LncBase V2
DIANA-LncBase v2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=site%2Ftools) provides a database of miRNA-lncRNA interactions through experimentally supported and in silico predicted miRNA Recognition Elements (MREs) on lncRNAs.20 In this study, we analyze the miRNAs of SCAMP3 and its related lncRNAs and construct the network of miRNA-lncRNA-SCAMP3. The Venny 2.1 Online Tool (http://bioinfogp.cnb.csic.es) was used to find overlapping miRNAs targeted by SCAMP3 and SCAMP3-interacted lncRNAs. The miRNA-lncRNA-SCAMP3 regulatory network was depicted and visualized using Cytoscape software.
Statistical Analysis
All of the statistical analysis has been done by the bioinformatic tools themselves mentioned above. Difference of transcriptional expression was compared by Students’ t-test. A value of P<0.05 was considered statistically significant.
Results
SCAMP3 Expression Associated with Patients Survival in Hepatocellular Carcinoma
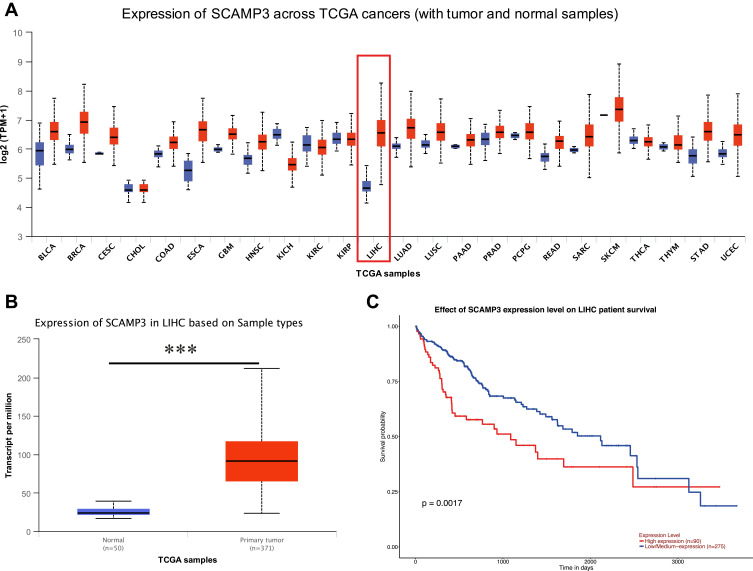
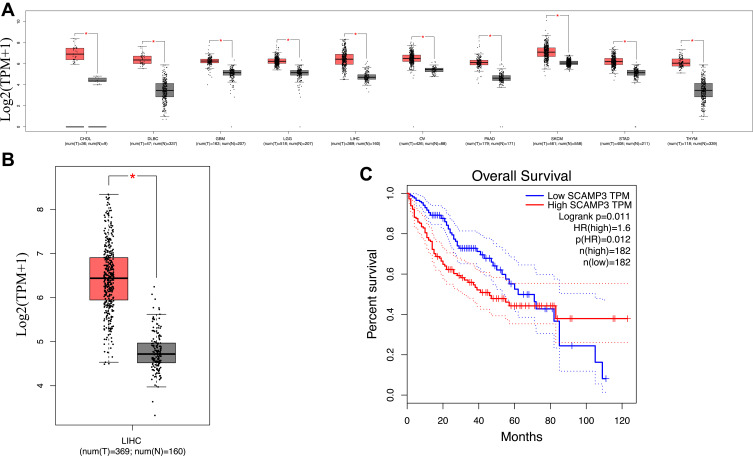
In order to assess SCAMP3 mRNA expression in various tumor types and corresponding normal tissues, three different databases, UALCAN, GEPIA, and Starbase, were used to analyze the difference expression of SCAMP3 between cancers and normal tissues based on TCGA data. First, TCGA data results obtained from UALCAN, GEPIA, and Starbase showed that the expression of SCAMP3 mRNA was only significantly up-regulated in liver hepatocellular carcinoma (LICH), cholangiocarcinoma (CHOL), and stomach adenocarcinoma (STAD) (Figures 1A and 2A and Table 1). Then, further analysis found that SCAMP3 mRNA expression was the lowest in normal liver tissues, while the staggering higher expression in SCAMP3 expression was exhibited in HCC (P<0.001) (Figures 1B and 2B). In addition, five different HCC data from the ONCOMINE database also exhibited a different expression pattern of SCAMP3 between hepatocellular carcinoma and normal liver (Table 2). Further analysis found that HCC patients’ higher survival rate was significantly associated with low SCAMP3 expression (Figures 1C and 2C, Table 3). Taken together, these data indicate that SCAMP3 might be a potential specific prognosis marker for hepatocellular carcinoma.
Figure 1.
SCAMP3 gene expression in different types of cancer diseases and normal tissues in the UALCAN database. (A) Transcriptional expression level of SCAMP3 gene in various cancer was analyzed in the UALCAN database. Blue: normal samples; Red: tumor samples. Red box: hepatocellular carcinoma samples and liver samples. TPM: Transcript per million. All abbreviations mentioned above about cancer names: see Table 1. (B) Box plot showing the correlation between SCAMP3 and Hepatocellular carcinoma in UALCAN database. Blue: normal samples; Red: tumor samples. ***P<0.001. (C) Kaplan–Meier survival curves revealing SCAMP3 expression and survival probability in hepatocellular carcinoma.
Figure 2.
SCAMP3 gene expression in different types of cancer diseases and normal tissues in the GEPIA database. (A) Transcriptional expression level of SCAMP3 gene in various cancers was analyzed in the GEPIA database. Grey: normal samples; Red: tumor samples. (B) Box plot showing the correlation between SCAMP3 and Hepatocellular carcinoma in the GEPIA database. Grey: normal samples; Red: tumor samples. *P<0.0001. (C) Kaplan–Meier survival curves revealing SCAMP3 expression and survival probability in hepatocellular carcinoma.
Table 1.
SCAMP3 mRNA Expression in Various Cancers and Normal Tissues in Starbase Database
| Cancer | Full Name | Cancer Number | Normal Number | Cancer Express | Normal Express | Fold Change | P -value | FDR |
|---|---|---|---|---|---|---|---|---|
| BRCA | Breast Invasive Carcinoma | 1104 | 113 | 46.12 | 22.39 | 2.06 | 4.00E-61 | 1.70E-59 |
| LIHC | Liver Hepatocellular Carcinoma | 374 | 50 | 44.85 | 11.8 | 3.8 | 1.30E-50 | 1.10E-47 |
| LUSC | Lung Squamous Cell Carcinoma | 501 | 49 | 37.91 | 25.05 | 1.51 | 4.70E-17 | 3.70E-16 |
| KICH | Kidney Chromophobe | 65 | 24 | 18.55 | 31.5 | 0.59 | 1.60E-15 | 3.10E-14 |
| LUAD | Lung Adenocarcinoma | 526 | 59 | 40.47 | 24.05 | 1.68 | 2.40E-15 | 2.80E-14 |
| STAD | Stomach Adenocarcinoma | 375 | 32 | 35.69 | 21.99 | 1.62 | 3.80E-15 | 1.30E-13 |
| UCEC | Uterine Corpus Endometrial Carcinoma | 548 | 35 | 40.93 | 21.41 | 1.91 | 3.90E-15 | 6.70E-14 |
| CHOL | Cholangiocarcinoma | 36 | 9 | 60.41 | 10.06 | 6 | 8.30E-14 | 3.00E-12 |
| PRAD | Prostate Adenocarcinoma | 499 | 52 | 37.69 | 29.24 | 1.29 | 9.30E-09 | 9.30E-08 |
| ESCA | Esophageal Carcinoma | 162 | 11 | 32.48 | 18.65 | 1.74 | 4.50E-07 | 1.60E-05 |
| COAD | Colon Adenocarcinoma | 471 | 41 | 30.14 | 21.77 | 1.38 | 4.60E-06 | 2.40E-05 |
Abbreviation: FDR, false discovery rate.
Table 2.
Significant Changes in Transcription Level of SCAMP3 between HCC and Normal Liver Analyzed by the ONCOMINE Database
| Types of HCC vs. Liver | Fold Change | P-value | t-test | Ref |
|---|---|---|---|---|
| Hepatocellular Carcinoma vs. Normal Liver | 2.536 | 1.42E-28 | 13.238 | Chen Liver |
| 2.269 | 3.71E-10 | 8.921 | Roessler Liver | |
| 2.116 | 9.79E-71 | 21.680 | Roessler Liver 2 | |
| 1.103 | 4.89E-8 | 7.086 | Guichard Liver 2 | |
| 1.109 | 2.08E-19 | 10.822 | Guichard Liver |
Table 3.
SCAMP3 mRNA Expression was Associated with Survival (Only P-value<0.05 was Exhibited)
| Cancer | Cancer Full Name | Cancer Number | Median | coef | HR | P-value |
|---|---|---|---|---|---|---|
| ACC | Adrenocortical Carcinoma | 79 | 34.11 | 1.42 | 4.12 | 0.00033 |
| LIHC | Liver Hepatocellular Carcinoma | 369 | 39.82 | 0.42 | 1.53 | 0.018 |
Relationship Between Patients’ Clinicopathological Features and SCAMP3 mRNA Expression in HCC
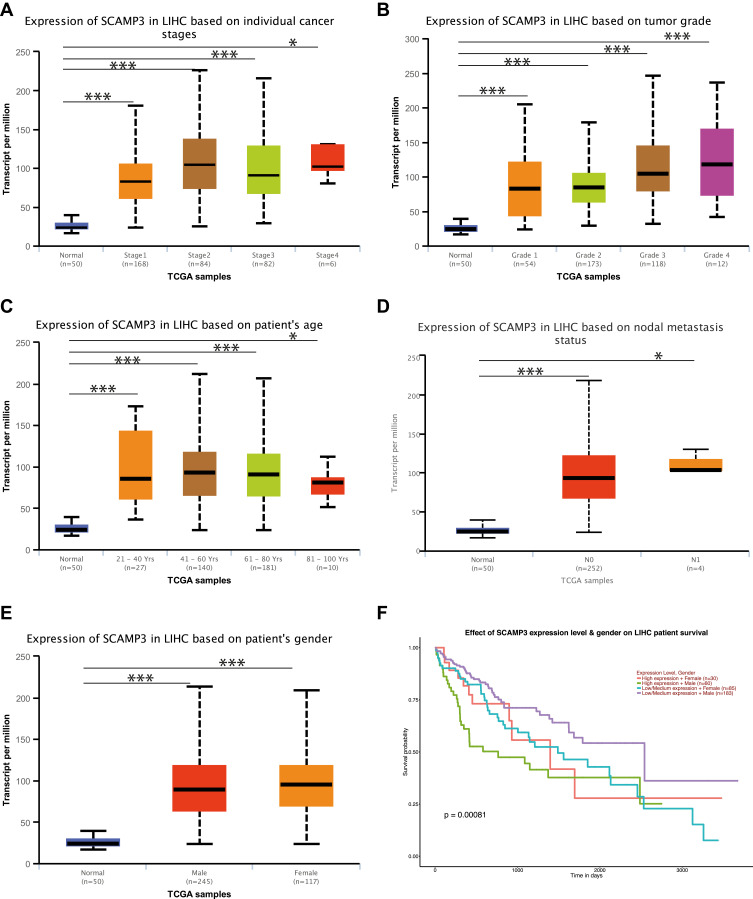
The relationship of SCAMP3 mRNA expression with clinicopathological characteristics of HCC patients was analyzed by UALCAN. As is shown in Figure 3, mRNA expression of SCAMP3 was significantly correlated with HCC patients’ individual cancer stages, tumor grades, and ages. The highest mRNA expression of SCAMP3 was found in stage 3 (Figure 3A), when the highest mRNA expression of SCAMP3 was found in tumor grade 4 (Figure 3B). Furthermore, the results from a Kaplan–Meier plotter showed that higher combinatory mRNA expressions of SCAMP3 were associated with poorer overall survival (OS) in liver cancers patients with stage 1+2 or stage 3 (Supplementary A and B), when SCAMP3 mRNA expression showed the correlation with favorable OS of HCC patients with tumor grade 3 or grade 4 (Supplementary C and D). In addition, as age increased, the mRNA expression of SCAMP3 tended to be higher (Figure 3C). Similarly, a significant correlation between mRNA expression of SCAMP3 and lymph node metastasis was also found (Figure 3D). Interestingly, statistical analysis revealed the expression levels of SCAMP3 and patients’ gender was significantly correlated and lower expression of SCAMP3 in males was also associated with higher survival rate (Figures 3E and F). In a word, the results above suggest that mRNA expressions of SCAMP3 were remarkably associated with clinicopathological parameters in HCC patients.
Figure 3.
The relationship between SCAMP3 mRNA expression and tumor stages, grades, age, lymph node metastasis, and genders of HCC patients in the UALCAN database. (A) mRNA expressions of SCAMP3 was significantly correlated with patients’ individual cancer stages; *P<0.05, ***P<0.001. (B) mRNA expressions of SCAMP3 was significantly correlated with patients’ tumor grades; *P<0.05, ***P<0.001. (C) mRNA expressions of SCAMP3 was significantly correlated with patient’s age; *P<0.05, ***P<0.001. (D) mRNA expressions of SCAMP3 was significantly correlated with patients’ lymph node metastasis; *P<0.05, ***P<0.001. N0: No regional lymph node metastasis; N1: Metastases in 1 to 3 axillary lymph nodes. (E) mRNA expressions of SCAMP3 was significantly correlated with patient’s gender; *P<0.05, ***P<0.001. (F) Lower expression of SCAMP3 of male patients was also associated with higher survival rate in HCC patients by Kaplan–Meier curve.
Association Between the Expression Levels of the Interactor of SCAMP3 and Hepatocellular Carcinoma
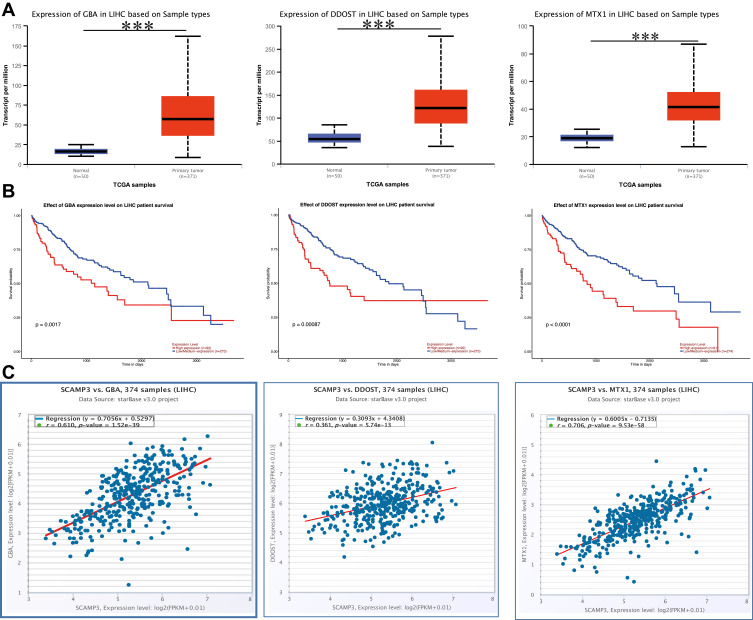
To comprehensively assess the prognostic value of SCAMP3, the association between the expression levels of the interactor of SCAMP3 and patients’ survival in hepatocellular carcinoma was analyzed. The BioGrid database displayed 73 unique interactors of SCAMP3 (Figure 4A). Combined literature and STRING database, the data showed that SOCS2 was negatively associated with SCAMP3, while GBA, MTX1, and DDOST had the most similar expression pattern and positive correlation with SCAMP3. Further analysis illustrated that SOCS2 was significantly down-regulated in hepatocellular carcinoma and associated with patients’ survival (Figures 4B and C). Moreover, co-expression analysis for the SOCS2-SCAMP3 interactions by starBase and GEPIA databases determined their negative correlation (Figure 4D), while the analysis data released that GBA, MX1, and DDOST were significantly up-regulated in hepatocellular carcinoma and their higher expression was unfavorable prognostic (Figures 5A and B). Moreover, GBA, MX1, and DDOST had higher positive correlation with SCAMP3 (Figure 5C). Therefore, we speculated that SCAMP3 might be a critical factor regulated by SOCS2 or GBA, MTX1, and DDOST to be involved in hepatocarcinogenesis.
Figure 4.
An interactor of SCAMP3 was negative correlation with SCAMP3 and involved in hepatocellular carcinoma. (A) Network of SCAMP3 was analyzed by BioGrid database. (B) Box plot showing the correlation between SOCS2 and Hepatocellular carcinoma in the UALCAN database. Blue: normal samples; Red: tumor samples. ***P<0.001. (C) Kaplan–Meier survival curves revealing SOCS2 expression and survival probability in hepatocellular carcinoma. (D) starBase database analyses the co-expression of the RNA-RNA interactions.
Figure 5.
SCAMP3 interactors were positive correlation with SCAMP3 and involved in hepatocellular carcinoma. (A) Box plot showing the mRNA expression of GBA, MTX1, DDOST in HCC in UALCAN database. Blue: normal samples; Red: tumor samples. ***P<0.001. (B) Kaplan–Meier survival curves revealing the expression of GBA, MTX1, DDOST, and survival probability in HCC. (C) Starbase database analyses the co-expression of the RNA-RNA interactions.
Correlation Between ncRNA Targets of SCAMP3 and Hepatocellular Carcinoma
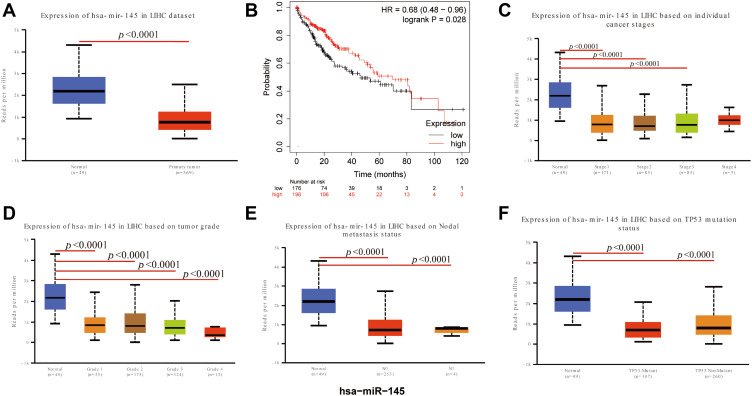
Noncoding RNAs (ncRNAs) have been demonstrated to play a central role in transcription regulation, posttranscriptional regulation, and translational regulation, as well as several biological processes and disease development, including cancer. Therefore, to reveal the lncRNA targets of SCAMP3, LncRNA2Target, and Starbase were utilized to uncover lncRNA interactor of SCAMP3, the result showed that SBF2-AS1 and LINC00922 might act as lncRNA interactors of SCAMP3 to regulate hepatocellular carcinoma metastasis and progression. In addition, the LncRNA-miRNA-mRNA network was analyzed though the ENCORI database and found that SBF2-AS1 and LINC00922 might serve as a competing endogenous RNA of has-miR-1321 that targets SCAMP3 (Figure 6A). Consistently, SBF2-AS1 was significantly up-regulated in hepatocellular carcinoma (Figure 6B) and Kaplan–Meier plotter analysis determined that higher expression of SBF2-AS1 revealed significantly poorer survival and progression than those in the lower expression group (P=0.00015, Figure 6C). Furthermore, co-expression analysis illustrated that there was a positive interaction between SCAMP3 and SBF2-AS1 (Figure 6D), suggesting that SBF2-AS1 might interact with SCAMP3 to regulate hepatocarcinogenesis. Furthermore, in order to uncover whether SCAMP3 also acts as a target gene of miRNAs, four databases (miRWalk, miRDB, microRNA.org and TargetScanHuman) were used to identify miRNAs targeting SCAMP3. The predicted results showed that has-miR-128-3p, has-miR-145-5p, has-miR-5195-3p, and has-miR-216a-3p might target the SCAMP3 gene, but only has-miR-145-5p expression in hepatocellular carcinoma was down-regulated (Figure 7A) and was significantly associated with the patients’ survival in hepatocellular carcinoma, revealing that low expression of has-miR-145-5p was correlated with poor survival of patients (Figure 7B). Moreover, the mRNA expression of has-miR-145-5p was significantly related to HCC patients’ individual cancer stages, tumor grades, nodal metastasis status, and TP53 mutation tumor grades, and, as tumor grade increased, the mRNA expression of CBXs tended to be higher (Figures 7C–F), indicating that has-miR-145-5p might target SCAMP3 to regulate migration and invasion in HCC.
Figure 6.
SBF2-AS1 expression was associated with SCAMP3 expression and patients’ survivals. (A) ENCORI database predicts the LncRNA-miRNA-SCAMP3 network that was visualized by cytoscape. (B) Box plot showing the correlation between SBF2-AS1 and Hepatocellular carcinoma. Purple: normal samples; Orange: tumor samples. ***P<0.001. (C) Kaplan–Meier survival curves revealing the expression of SBF2-AS1 and survival probability in hepatocellular carcinoma. (D) starBase database analyses the co-expression of the RNA-RNA interactions.
Figure 7.
miR-145-5p expression was associated with SCAMP3 expression and patients’ survivals. (A) Box plot showing the correlation between miR-145-5p and Hepatocellular carcinoma. Blue: normal samples; Red: tumor samples. (B) Kaplan–Meier survival curves revealing the expression of miR-145-5p and survival probability in hepatocellular carcinoma. (C) The expressions of miR-145-5p was significantly correlated with patients’ individual cancer stages. (D) The expressions of miR-145-5p was significantly correlated with patients’ individual cancer grades. (E) The expressions of miR-145-5p was significantly correlated with patients’ lymph node metastasis; N0: No regional lymph node metastasis; N1: Metastases in 1 to 3 axillary lymph nodes. (F) The expressions of miR-145-5p was significantly correlated with TP53 mutation status in patients.
Discussion
Secretory Carrier Membrane Protein 3(SCAMP3) is an endocytosis-associated protein that contributes to the regulation of EGFR trafficking and ubiquitin-mediated receptor degradation as well as endosomal sorting via interaction with ESCRT proteins.4 Moreover, SCAMP3 plays a role in the process of multivesicular endosome biogenesis. Previous studies have released the role of SCAMP3 as a tumor-related protein in inflammatory breast cancer,6 glioblastoma multiforme.5,21 In this study, we comprehensively evaluated the diagnostic and prognostic values of SCAMP3 in hepatocellular carcinoma patients by comprehensive bioinformatic analysis. Our results demonstrate that SCAMP3 is significantly highly expressed, and the most dramatic difference in hepatocellular carcinoma specimens compared with normal liver tissues and others tumors. Simultaneously, the expression of SCAMP3 is also correlated with age, gender, stage, and grade of hepatocellular carcinoma patients. Importantly, higher SCAMP3 expression associated with poorer survival and progression, indicating that SCAMP3 may be a statistically significant prognostic biomarker. Interaction regulatory network analysis shows that some interactors of SCAMP3, such as SOCS2, GAB, MTX1, and DDOST also correlated with poorer survival and progression, suggesting that incorporation of SCAMP3 expression with these interactors could improve the predictive accuracy for survival in hepatocellular carcinoma patients. Previous global gene expression profiling of HCC tissue samples showed that SCAMP3 mRNA expression level was significantly up-regulated when compared to normal tissues.1 Therefore, how high expression of SCAMP3 affects hepatocarcinogenesis urgently needs to be understood.
It has been demonstrated that receptor-mediated endocytosis is important for tumorigenesis and progression that needs receptor-mediated signals cascades for survivability, invasion, and metastasis. Membranes receptor tyrosine kinases, such as EGFR, are critical for the activation of cellular signaling pathways and tumor progression.22 As an endocytosis-associated protein, SCAMP3 has been explored to regulate EGFR degradation4 and its signaling system via regulation of multivesicular endosomes pathway to control tumorigenic progression.
Results from our study showed that SCAMP3 expression can be regulated by multiple regulators, including ncRNA. SOCS2 may act as a negative regulator of SCAMP3.23 Low expression of SOCS2 is significantly associated with the presence of intrahepatic metastasis and poor survival in hepatocellular carcinoma patients.24,25 In addition, SOCS2 is an E3 ubiquitin ligase to be involved in ubiquitination and proteasomal degradation.26 Based on the above, we speculate that hepatocellular carcinoma may inhibit the expression of SOCS2 to block SCAMP3 ubiquitylation for degradation. Furthermore, ncRNA data elucidates that SCAMP3 can interact with LncRNA SBF2-AS1 and miRNA miR-145-5p, all whose expression are significantly correlated with survival,27–30 suggesting that SCAMP3, SBF2-AS1, and miR-145-5p may construct a regulatory network to regulate the migration and invasion of hepatocellular carcinoma cells, however, further experimental authentication needs to be done for explaining the competing endogenous RNA (ceRNA) regulatory mechanism.
Conclusion
By comprehensive bioinformatics analysis, this study expands to evaluate the diagnostic and prognostic values of SCAMP3 in hepatocellular carcinoma. High expression of SCAMP3 is closely correlated with poorer survival, and SCAMP3 may act as a central downstream factor regulated by SOCS2, SBF2-AS1, and miR-145-5p to affect tumor invasion and migration. These results suggest that SCAMP3 may act as a specific prognostic biomarker in hepatocellular carcinoma. However, the mechanism of SCAMP3 as tumor-related protein in hepatocellular carcinoma and how SCAMP3 is regulated by ubiquitylation and ncRNA is needed to further elucidate.
Acknowledgments
This study was supported by Key Lab of Process Analysis and Control of Sichuan Universities (No.2018001), The Project-sponsored by Sichuan Province for ROCS (0903/00021728). A. Zhou is supported by Southwest Medical University (0903/00030699, 0903/00030827).
Author Contributions
A. Zhou designed this project and did most of the experiments, analyzed and interpreted data, and contributed to the writing of the manuscript; A. Zhou and H. J. Liu analyzed and interpreted data; B. Tang provided financial support to A. Zhou; A. Zhou and B. Tang designed the experiments, supervised the research, and wrote the manuscript.
Disclosure
The authors have declared that they have no competing interests for this work.
References
- 1.Naboulsi W, Bracht T, Megger DA, et al. Quantitative proteome analysis reveals the correlation between endocytosis-associated proteins and hepatocellular carcinoma dedifferentiation. Biochim Biophys Acta. 2016;1864(11):1579–1585. doi: 10.1016/j.bbapap.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 2.Skawran B, Steinemann D, Weigmann A, et al. Gene expression profiling in hepatocellular carcinoma: upregulation of genes in amplified chromosome regions. Mod Pathol. 2008;21(5):505–516. doi: 10.1038/modpathol.3800998 [DOI] [PubMed] [Google Scholar]
- 3.Thomas P, Wohlford D, Aoh QL. SCAMP 3 is a novel regulator of endosomal morphology and composition. Biochem Biophys Res Commun. 2016;478(3):1028–1034. doi: 10.1016/j.bbrc.2016.08.012 [DOI] [PubMed] [Google Scholar]
- 4.Aoh QL, Castle AM, Hubbard CH, Katsumata O, Castle JD. SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling. Mol Biol Cell. 2009;20(6):1816–1832. doi: 10.1091/mbc.E08-09-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chunliu L, Zhang Z, Peng L, Zhan Y, Zhong Q. SCAMP3 promotes glioma proliferation and indicates unfavorable prognosis via multiple pathways. Onco Targets Ther. 2020;13:3677–3687. doi: 10.2147/OTT.S242462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suárez-Arroyo IJ, Feliz-Mosquea YR, Pérez-Laspiur J, et al. The proteome signature of the inflammatory breast cancer plasma membrane identifies novel molecular markers of disease. Am J Cancer Res. 2016;6(8):1720–1740. [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Rodriguez IP. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Z, Kang B, Chenwei L, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from argonaute CLIP-Seq and degradome-seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menyhart O, Nagy A, Gyorffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5(122):181006. doi: 10.1098/rsos.181006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oughtred R, Stark C, Breitkreutz BJ, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng L, Wang P, Tian R, et al. Lncrna2target V2.0: a comprehensive database for target genes of lncrnas in human and mouse. Nucleic Acids Res. 2018;gky1051–gky51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 17.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152. doi: 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. doi: 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucl Acids Res. 2016;gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh D, Ulasov IV, Chen L, et al. TGFβ-responsive HMOX1 expression is associated with stemness and invasion in glioblastoma multiforme. Stem Cells. 2016;34(9):2276–2289. doi: 10.1002/stem.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366(1):2–16. doi: 10.1016/j.gene.2005.10.018 [DOI] [PubMed] [Google Scholar]
- 23.Pilling C, Cooper JA. SOCS2 binds to and regulates EphA2 through multiple mechanisms. Sci Rep. 2017;7(1):10838. doi: 10.1038/s41598-017-11040-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu X, Zheng J, Guo X, et al. Reduced expression of SOCS2 and SOCS6 in hepatocellular carcinoma correlates with aggressive tumor progression and poor prognosis. Mol Cell Biochem. 2013;378(1–2):99–106. doi: 10.1007/s11010-013-1599-5 [DOI] [PubMed] [Google Scholar]
- 25.Cui M, Sun J, Hou J, et al. The suppressor of cytokine signaling 2 (SOCS2) inhibits tumor metastasis in hepatocellular carcinoma. Tumour Biol. 2016;37(10):13521–13531. doi: 10.1007/s13277-016-5215-7 [DOI] [PubMed] [Google Scholar]
- 26.Vesterlund M, Zadjali F, Persson T, et al. The SOCS2 ubiquitin ligase complex regulates growth hormone receptor levels. PLoS One. 2011;6(9):e25358. doi: 10.1371/journal.pone.0025358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Liu G, Li X, Dong H, Xiao W, Lu S. Long non-coding RNA SBF2-AS1 promotes hepatocellular carcinoma progression through regulation of miR-140-5p-TGFBR1 pathway. Biochem Biophys Res Commun. 2018;503(4):2826–2832. doi: 10.1016/j.bbrc.2018.08.047 [DOI] [PubMed] [Google Scholar]
- 28.Zhang YT, Li BP, Zhang B, et al. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22(19):6333–6341. doi: 10.26355/eurrev_201810_16044 [DOI] [PubMed] [Google Scholar]
- 29.Noh JH, Chang YG, Kim MG, et al. MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013;335(2):455–462. doi: 10.1016/j.canlet.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 30.Duan X, Hu J, Wang Y, Gao J, Peng D, Xia L. MicroRNA-145: a promising biomarker for hepatocellular carcinoma (HCC). Gene. 2014;541(1):67–68. doi: 10.1016/j.gene.2014.03.018 [DOI] [PubMed] [Google Scholar]