Abstract
Phaeochromocytomas and paragangliomas (PPGLs) are rare neuroendocrine tumours with a hereditary background in over one-third of patients. Mutations in succinate dehydrogenase (SDH) genes increase the risk for PPGLs and several other tumours. Mutations in subunit B (SDHB) in particular are a risk factor for metastatic disease, further highlighting the importance of identifying SDHx mutations for patient management. Genetic variants of unknown significance, where implications for the patient and family members are unclear, are a problem for interpretation. For such cases, reliable methods for evaluating protein functionality are required. Immunohistochemistry for SDHB (SDHB-IHC) is the method of choice but does not assess functionality at the enzymatic level. Liquid chromatography-mass spectrometry-based measurements of metabolite precursors and products of enzymatic reactions provide an alternative method. Here, we compare SDHB-IHC with metabolite profiling in 189 tumours from 187 PPGL patients. Besides evaluating succinate:fumarate ratios (SFRs), machine learning algorithms were developed to establish predictive models for interpreting metabolite data. Metabolite profiling showed higher diagnostic specificity compared to SDHB-IHC (99.2% versus 92.5%, p = 0.021), whereas sensitivity was comparable. Application of machine learning algorithms to metabolite profiles improved predictive ability over that of the SFR, in particular for hard-to-interpret cases of head and neck paragangliomas (AUC 0.9821 versus 0.9613, p = 0.044). Importantly, the combination of metabolite profiling with SDHB-IHC has complementary utility, as SDHB-IHC correctly classified all but one of the false negatives from metabolite profiling strategies, while metabolite profiling correctly classified all but one of the false negatives/positives from SDHB-IHC. From 186 tumours with confirmed status of SDHx variant pathogenicity, the combination of the two methods resulted in 185 correct predictions, highlighting the benefits of both strategies for patient management.
Keywords: mass spectrometry, succinate to fumarate ratio, multi-observer, Krebs cycle metabolites, linear discriminant analysis, LC–MS/MS, diagnostics, variants of unknown significance, metabolite profiling, prediction models
Introduction
Mitochondrial enzymes, such as succinate dehydrogenase (SDH), or complex II of the respiratory chain, play a central role in energy homeostasis within the cell. The complex is made up of four subunits (SDHA, SDHB, SDHC, and SDHD) and assembly is assisted by several factors, including SDHAF2. Mutations of genes encoding these proteins can result in phaeochromocytomas and paragangliomas (PPGLs), gastrointestinal stromal tumours, renal cell carcinomas, and pituitary adenomas [1]. The mutations occur almost exclusively in the germline, leaving the patient and potentially family members at lifelong risk for disease. Mutations in SDHB in particular predispose to metastatic PPGL and are associated with increased mortality [2,3]. Endocrine guidelines therefore advise that genetic testing should be offered to all patients with PPGL [4].
With advances in gene sequencing techniques and decreasing costs, genetic testing is becoming more practical and widely available, but this has also led to new challenges [5]. Variants of unknown significance, where the functional impact of the mutation has not been established, are increasingly troublesome. Also, even with advanced genetic testing, some functional variants or mutations may be missed. This includes intronic variants and epimutations, as well as mutations in other genes impacting mitochondrial energy metabolism [6–8]. Such problems can be addressed by methods assessing the functionality of involved proteins, thereby allowing classification of variants with uncertain mutational status.
For gene variants affecting SDH, the routinely applied method is immunohistochemistry for SDHB (SDHB-IHC), where staining intensity of tumoural cells is compared with that of non-tumoural cells as internal control [9]. Positively stained cells show a granular pattern, whereas negative tissue at most has a weak diffuse cytoplasmic blush. Importantly, the protein is also degraded when subunits other than SDHB are lost [10].
Another method to assess SDH functionality is based on measurements of Krebs cycle metabolites by liquid chromatography–mass spectrometry (LC—MS/MS), the same instrument also now used for biochemical diagnosis of PPGL [11,12]. Metabolite profiling assesses the functionality of SDH directly at the catalytic level by measuring the precursor succinate and the product fumarate. The ratio of these two metabolites, the tissue succinate to fumarate ratio (SFR), can predict SDHx mutations with a high sensitivity and specificity in PPGL and is also now being applied to other tumours [11,13].
So far, there has been no formal comparison of the two methods for predicting SDHx mutational status. The nature of the techniques is different, one involving functional assessment of enzyme activity versus histological information about the presence of protein, but both offering complementary potential. Based on the ability of machine learning to recognise patterns in data in a way the human mind is not trained [14,15], we also investigated whether such an approach can improve predictions from metabolite data beyond the currently used SFR [11,12]. Since one of the disadvantages of SDHB-IHC relates to the subjective nature of image interpretation, we further assessed whether local pathologists from different institutions scored slides differently from investigators experienced in the method (referred to as experts). An overall goal of the study was to explore how SDHB-IHC and metabolite profiling might be useful for streamlining diagnostic procedures for patients and their families suffering from PPGLs due to SDH impairment.
Materials and methods
Patient cohorts and tumour procurement
Tumour collections were approved under Intramural Review Board protocols with informed consent signed at each participating centre. A total of 397 patients with 401 different tumours were included in this study (supplementary material, Table S1). The present report builds on our previously reported data on metabolite profiles in 391 of these patients [12] by additional comparisons to immunohistopathological data and introduction of machine learning for interpretation of metabolite profiles. Tumour material was collected as freshly frozen (FF) and/or formalin-fixed and paraffin-embedded (FFPE) samples. Patients were divided into two separate cohorts (Table 1 and supplementary material, Figure S1): cohort 1 with 187 patients and 189 PPGLs included results for matched metabolite profiling and SDHB-IHC, whereas for cohort 2 (210 patients with 212 PPGLs) data from metabolite profiles, without availability of SDHB-IHC, were included and used for training the machine learning algorithms.
Table 1.
Diagnostic performance of SDHB-IHC compared to LC-MS/MS based measurements of succinate:fumarate [SFR] in cohort 1.
| SDHB-IHC* | SFR | p-value | |||
| Sensitivity [%] | 85.2 [46/54] | 88.1 [52/59] | 0.774 | ||
| Specificity [%] | 92.5 [111/120] | 99.2 [126/127] | 0.021 | ||
| Accuracy [%] | 90.2 [157/174] | 95.7 [178/186] | |||
| AUC | 0.88 [0.82,0.91] | 0.96 [0.89,0.98] | 0.048 | ||
|
SFR |
|||||
| FP | TN | FN | TP | ||
| SDHB-IHC | FP [n = 9] | 0 | 9 | – | – |
| FN [n = 8] | – | – | 1 | 7 | |
| inc. [n =12] | 0 | 7 | 1 | 4 | |
Inconclusive samples (12) not included
inc. = inconclusive IHC results, FP: false positive; FN: false negative; TP: true positives; TN: true negatives. Sensitivity, specificity and accuracy are given as percentages with absolute numbers in brackets. AUC is given as a ratio with CI in brackets.
Genetic characteristics
Genetic testing, accomplished as previously described [12], yielded findings of germline or somatic variants in 18 genes in 49.1% (195/397) of patients (for simplicity, only 11 are displayed in Table 1). Eleven patients had variants of unknown significance in SDH genes, classified as variants of unknown significance according to the standards and guidelines of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology [16]. In silico prediction of mutation significance was performed on variants of unknown significance using Mutation Taster [17], Polyphen [18], and SIFT [19].
Immunohistochemistry
FFPE tissue was sectioned and stained for SDHB using rabbit polyclonal anti-SDHB (HPA002868, 1:400 dilution; Sigma-Aldrich, St Louis, MO, USA) according to the local procedures of six different centres (Dresden, Bethesda, Madrid, Florence, Nijmegen, and Rotterdam) [9]. For 23% of tumours (44 samples), a tissue microarray was constructed with three cores of 1.0 mm per sample. Local pathologists evaluated SDHB staining in one slide per tumour and gave the results in four categories: as positive, for the typical granular staining pattern; as negative, for completely negative or weak diffuse staining; as inconclusive, when both patterns were present; or as non-informative, when tissue or staining artefacts were observed. For 50 samples, the SDHB-IHC interpretations were as established previously, from combined interpretations of seven expert pathologists [20]; for these cases, results were rated as inconclusive when fewer than five pathologists agreed. All pathologists were blinded to the genetic status.
Interpretation of immunohistochemistry by experienced pathologists
In a subset of cohort 1, termed subcohort 1b (see supplementary material, Figure S1), SDHB-IHC slides from the local centres were scanned and high-resolution images were provided to a panel of three experts in SDHB-IHC. Experts rated the staining according to the four categories described above.
Metabolite measurements
Seven carboxylic acids of the Krebs cycle (succinate, fumarate, malate, citrate, isocitrate, cis-aconitate, α-ketoglutarate), 2-hydroxyglutarate, pyruvate, and lactate were measured in methanol extracts of FF or FFPE tissue by LC—MS/MS as detailed in supplementary material, Supplementary materials and methods and with resulting data provided in supplementary material, Table S2. The cut-off for succinate:fumarate-based interpretation was 97.7, as previously established [11].
Machine learning-assisted interpretation of metabolite data
Tissue metabolite concentrations (ng/mg tissue) were normalised to natural logarithmic values. These and their ratios were used for formulating predictive models. To establish the need for batch corrections (according to measurement dates), a principal component analysis was generated with the normalised metabolite values of SDHx-mutated or SDHx-wild-type patients (supplementary material, Figure S2). There was a clear distinction between the groups and none of the 27 different batches showed any bias towards SDHx-mutated or SDHx-wild type, allowing formulation of models without the need for batch correction.
Feature selection for models was performed using either logarithmically transformed values or ratios of all metabolites against each other. The results of genetic testing were used to separate patients into the categories SDHx-mutated or SDHx-wild type. The ‘LDA MATLAB’ function (MATLAB; MathWorks, Natick, MA, USA) with application of a cross validation was used to train the algorithm and generate models based on linear discriminant analysis (LDA) [21]. Patients from cohort 2 (excluding SDHx variants of unknown significance and FFPE only tissue) were used to develop the models and were randomly divided into training and internal validation sets in ratios ranging from 50/50 to 90/10 in steps of 10%. This randomisation and model generation was performed ten times and an average model was generated. In total, this resulted in five LDA models for absolute values and five models for metabolite ratios. The predictive models were applied to the PPGLs of cohort 1 to calculate the likelihood of SDHx mutations (external validation). Performance scores for the different models on the training, internal validation, and external validation sets were calculated (supplementary material, Figure S3) and the two best models selected according to performance (supplementary material, Figure S4). Selected models are referred to as LDA A, using absolute values, and LDA B for metabolite ratios. Since the models were built on values only from FF tissues, ten samples with metabolite values for FF and FFPE tissue were used to assess the suitability of LDA B for PPGLs stored as FFPE tissues.
Calculation of diagnostic performance
Based on confusion matrices of genetically determined versus predicted mutational status for SDHx, diagnostic performance was assessed from estimates of sensitivity, specificity, accuracy, and precision as detailed below:
sensitivity = TP/(TP + FN),
specificity = TN/(TN + FP),
accuracy = (TN + TP)/(TN + TP + FN + FP),
precision/positive predictive value = TP/(TP + FP),
F1-Score = 2 * [(precision * sensitivity)/(precision + sensitivity)], where TP represents true positives, FN false negatives, TN true negatives, and FP false positives.
Statistics
Receiver operating characteristics (ROC) curves were produced using logistic regression. Areas under ROC curves (AUCs) of the SDHB-IHC, SFR, LDA A, and LDA B were compared using the Model Comparison tool in JMP Pro (version 14; SAS, Cary, NC, USA). Logistic regression models for combinations of SDHB-IHC with any of the other models were produced in JMP and these combined models’ AUCs were compared with those from using only SFR, LDA Model A or LDA Model B. As there were different numbers of tumours available for the different models, the AUC comparisons were performed with all available tumours (186 versus 185 versus 186 versus 174) and with equal numbers (173 versus 173 versus 173 versus 173). Comparisons of sensitivity and specificity between different predictive methods in the same patient group utilised McNemar’s test for matched pairs data, while comparisons of predictive methods in different patient groups utilised Fisher’s exact test. Differences were considered significant for P values below 0.05.
Results
Diagnostic performance of SDHB-IHC and metabolite profiling by SFR
Among a total of 186 PPGLs (cohort 1 excluding three variants of unknown significance), SDHB-IHC incorrectly predicted the SDH status in nine cases (false positives) and missed SDH impairment in eight cases (false negatives) (Table 1). In 12 tumours, the results were deemed inconclusive according to heterogeneous staining patterns or observer disagreements (the latter applied to five cases with SDHB-IHC results taken from a previously published study). SFR-based metabolite profiling correctly predicted all nine false positives and seven of eight false negatives, and all but one inconclusive case were predicted correctly by the SFR. Diagnostic specificity (p = 0.021) and AUC (p = 0.048) were higher for SFR-based metabolite profiling than for SDHB-IHC.
Machine learning-assisted interpretation of metabolite profiling
Two different methods for machine learning-assisted interpretation of metabolite profiles were compared with the SFR (Table 2). Model LDA A uses four metabolites and is restricted to measurements from frozen tissue, where weight normalisation is possible. LDA B requires input of ratios of ten metabolites and is applicable to both FF and FFPE specimens (supplementary material, Table S3; supplementary material, Table S4 to produce the ratios). Both models are provided as supplementary material, MATLAB File S1 and MATLAB File S2 (for MATLAB Model LDA A and B, respectively).
Table 2.
Comparison of different predictive models for SDHx impairment based on metabolite profiling.
| LDA-Model A | LDA-Model B | SFR | ||||||||||||||||
| Model Parameters | Tissue amount normalized values (ng/mg) of: | Ratlos of Metabolites: | Ratios of metabolites: | |||||||||||||||
| • Succinate | • Succinate | • Succinate | ||||||||||||||||
| • Citrate | • Fumarate | • Fumarate | ||||||||||||||||
| • Malate | • Citrate | |||||||||||||||||
| • Pyruvate | • Malate | |||||||||||||||||
| • Pyruvate | ||||||||||||||||||
| • Cis-aconitate | ||||||||||||||||||
| • Isocitrate | ||||||||||||||||||
| • Lactate | ||||||||||||||||||
| • 2-hydroxyglutarate | ||||||||||||||||||
| • α-ketoglutarate | ||||||||||||||||||
| Tissue Type | FF | FF + FFPE | FF + FFPE | |||||||||||||||
| Confusion Matrix | Predicted | Predicted | Predicted | |||||||||||||||
| n=186 | NO | YES | n=185 | NO | YES | n=186 | NO | YES | ||||||||||
| Actual | NO | 126 | 1 | Actual | NO | 126 | 1 | Actual | NO | 126 | 1 | |||||||
| YES | 4 | 55 | YES | 6 | 52 | YES | 7 | 52 | ||||||||||
| Diagnostic Performance | p-value | p-value | p-value | |||||||||||||||
| Sensitivity | 93.2% | 0.344 | Sensitivity | 89.7% | 0.774 | Sensitivity | 88.1% | 0.774 | ||||||||||
| Specificity | 99.2% | 0.022 | Specificity | 99.2% | 0.021 | Specificity | 99.2% | 0.021 | ||||||||||
| Accuracy | 97.3% | Accuracy | 96.2% | Accuracy | 95.7% | |||||||||||||
| AUC | 0.982 | 0.004 | AUC | 0.977 | 0.012 | AUC | 0.961 | 0.048 | ||||||||||
| SDHB-IHC | SDHB-IHC | SDHB-IHC | ||||||||||||||||
| FP | TN | FN | TP | inc | FP | TN | FN | TP | inc | FP | TN | FN | TP | inc | ||||
| FP(1) | 0 | 1 | – | – | 0 | FP(1) | 0 | 1 | – | – | 0 | FP(1) | 0 | 1 | – | – | 0 | |
| FN(4) | – | – | 1 | 3 | 0 | FN(6) | – | – | 1 | 5 | 0 | FN(7) | – | – | 1 | 5 | 1 | |
FP: false positive; TN: True Negative; FN: False Negative; TP: True Positive; inc: inconclusive results in IHC.
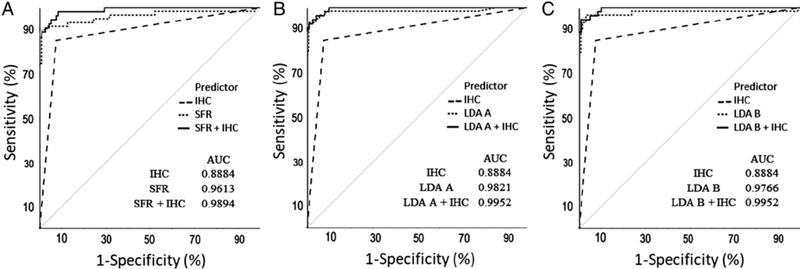
Our metabolite profiling-based models (LDA A, LDA B, and SFR) were applied to 186 tumours (cohort 1). In the case of LDA B, only 185 tumours of the 186 could be used because measurement of one metabolite failed in one tumour. Comparisons of metabolite profiling predictions showed that the performance of LDA A was improved over that of the SFR (p = 0.044), while there were no significant differences between LDA B and SFR or LDA A and LDA B (Figure 1 and Table 3). SFR-based interpretation of metabolite profiles resulted in seven false negatives, whereas LDA B produced six and LDA A four false negatives (Table 2). The LDA models calculate probabilities for the likelihood of an SDHx mutation based on the metabolite inputs; for the samples differently rated in LDA A and LDA B, the probabilities were 57% and 99% for LDA A, and 42% and 32% for LDA B, respectively (supplementary material, Table S5).
Figure 1.
ROC curve comparisons for SDHB-IHC, metabolite profiling, and their combination. (A) ROC curves of SFR, SDHB-IHC, and their combination. (B) ROC curves of LDA A, SDHB-IHC, and their combination. (C) ROC curves of LDA B, SDHB-IHC, and their combination.
Table 3.
Statistical comparisons of AUC differences.
| Predictor | versus Predictor | AUC Difference | p-value* |
|---|---|---|---|
| IHC | SFR | −0.070 | 0.048 |
| IHC | LDA A | −0.092 | 0.004 |
| IHC | LDA B | −0.086 | 0.012 |
| SFR | LDA A | −0.023 | 0.044 |
| SFR | LDA B | −0.016 | 0.533 |
| LDA A | LDA B | 0.006 | 0.775 |
| SFR | SFR + IHC | −0.031 | 0.113 |
| LDA B | LDA B + IHC | −0.021 | 0.235 |
| LDA A | LDA A +IHC | −0.014 | 0.301 |
p<0.05 considered significant.
It should also be noted that of the eight tumours where SFR had an erroneous prediction, six were head and neck paragangliomas with all seven false negatives carrying an SDHB, SDHC or SDHD mutation. When LDA A and LDA B were applied to these samples, LDA A correctly predicted three of the head and neck paragangliomas, while LDA B correctly predicted two of them (supplementary material, Table S5).
Since model generation of LDA B was performed exclusively on data from freshly frozen (FF) samples, we determined if the model could be applied to both FF and FFPE tissues. For this purpose, LDA B was applied to ten tumours where both FF and FFPE tissues were available. LDA B correctly predicted the mutational status of all ten FFPE and nine of ten FF tissues (supplementary material, Figure S5).
Combining metabolite profiling and SDHB-IHC for best possible predictions on SDHx mutational status
We observed a trend towards improved predictions when SDHB-IHC results were combined with the results from metabolite profiling compared with the latter alone, supporting complementary utility of SDHB-IHC and LC—MS/MS approaches (Figure 1). Metabolite profiling complemented SDHB-IHC by correctly predicting the SDHx mutational status of all but one case where SDHB-IHC incorrectly predicted the mutational status. In turn, SDHB-IHC correctly identified all but one of the false-negative cases from metabolite profiling (supplementary material, Table S5). With the available number of samples, statistical significance was not reached (Table 3).
As it was observed that paragangliomas of the head and neck region were classified more often as false negatives with metabolite profiling, we compared the diagnostic sensitivity and specificity between phaeochromocytomas, paragangliomas of the thorax or abdomen, and head and neck paragangliomas (Table 4). Although statistical significance was not reached, it was apparent that the sensitivity was lowest for head and neck paragangliomas with all four methods, SDHB-IHC, SFR, LDA A, and LDA B. Specificity, on the other hand, showed no regional bias with metabolite-based methods, but was slightly lower for all paragangliomas compared with phaeochromocytomas using SDHB-IHC.
Table 4.
Comparison of predictive methods in the different subcategories of tumours.
| PHEO n = 112 | PGL n = 30 | HNP n = 44 | HNP versus (PHEO + PGL) p-value* | ||
|---|---|---|---|---|---|
| IHC | Sensitivity (%) | 88.8 [8/9] | 90.9 [20/22] | 78.3 [18/23] | 0.264 |
| Specificity (%) | 93.8 [91/97] | 87.5 [7/8] | 86.7 [13/15] | 0.313 | |
| inconclusive (n) | 6 | 0 | 6 | ||
| SFR | Sensitivity (%) | 100 [9/9] | 95.5 [21/22] | 78.6 [22/28] | 0.130 |
| Specificity (%) | 99 [102/103] | 100 [8/8] | 100 [16/16] | 1.000 | |
| LDA A | Sensitivity (%) | 100 [9/9] | 95.5 [21/22] | 89.3 [25/28] | 0.337 |
| Specificity (%) | 99 [102/103] | 100 [8/8] | 100 [16/16] | 1.000 | |
| LDA B | Sensitivity (%) | 100 [8/8] | 95.5 [21/22] | 82.1 [23/28] | 0.097 |
| Specificity (%) | 99 [102/103] | 100 [8/8] | 100 [16/16] | 1.000 |
calculated using Fisher’s exact test.
HNP = Head and neck paraganglioma, PGL = paraganglioma, PHEO = Phaeochromocytomas.
SDHB-IHC interpretations by local pathologists and a panel of experienced experts
To address the question of whether expertise in SDHB-IHC influences the interpretation of SDHB-IHC results, we utilised a subset of samples in which interpretations from local pathologists were compared with those of experts (see supplementary material, Figure S1). Accuracy of predictions of SDHx mutations according to SDHB-IHC indicated no significant differences between local pathologists and experts, suggesting that SDHB-IHC does not require specialised training (Table 5). In 71% of cases, all pathologists agreed and when at least two experts on the panel agreed on the prediction (100 of 105 cases), they in turn agreed with the local pathologist in 83 of these cases (79%) (supplementary material, Figure S6). Agreement was higher in non-SDHx cases (86%) than in SDHx-mutated cases (70%) (Table 5).
Table 5.
SDHB-IHC interpretations of a panel of experienced researchers versus local pathologists in cohort 1b.
| Local | Panel# | p-value | ||
| Sensitivity [%] | 80.0 [16/20] | 65.0 [13/20] | 0.250 | |
| Specificity [%] | 92.4 [61/66] | 98.5 [65/66] | 0.125 | |
| Accuracy [%] | 89.5 [77/86] | 90.7 [78/86] | ||
| # Inconclusive/non-informative cases * | 9 | 11 | ||
|
Agreement amongst panel |
Agreement of panel [>2/3] with local interpretations | |||
| 3 | 2 | 0 | ||
| non-SDHx (n = 73) | 62 | 9 | 2 | 63 [86.3%] |
| SDHx (n = 30) | 21 | 6 | 3 | 19 [70.0%] |
| SDHx VUS (n = 2) | 2 | 0 | 0 | 1 [50.0%] |
| Total (n = 105) | 85 | 15 | 5 | 83 [79.0%] |
Inconclusive cases were removed from the analysis.
For the panel of experienced researchers, interpretations where at least two out of three researchers agreed, were used.
VUS, Variant of Unknown Significance.
Amongst the samples with full agreement between all pathologists, there were six cases (out of 75) with incorrect predictions. These six cases comprised five head and neck paragangliomas with SDHx mutations and one adrenal PPGL without an SDHx mutation, indicating again that head and neck paragangliomas are the most challenging specimens to interpret (supplementary material, Table S5). Local pathologists rated nine slides as inconclusive, whereas experts rated five slides as inconclusive and six as non-informative. Only one inconclusive case overlapped between the two groups, indicating some variable subjectivity of interpretations. Non-informative cases arose due to technical problems with scanned images or staining of slides, such as related to uneven staining or high background (example images in supplementary material, Figure S7).
Re-evaluation of variants of unknown significance in SDHx
Amongst the tumours evaluated, there were 11 patients (three in cohort 1 and eight in cohort 2) with a variant of unknown significance in one of the SDHx genes. Both LDA models and the SFR were applied to the metabolite profiles and compared with in silico predictions for protein changes (supplementary material, Table S6). Both LDA models agreed in all four cases where the SFR predicted SDH impairment. In two of these cases, SDHB-IHC was available, but did not support the metabolite-based interpretations. LDA Model B predicted SDH impairment in two further PPGLs, a splice site variant in SDHA (NM_004168.3:c.457–1G>A) and an indel variant in SDHC (NM_003001.3:c.256_257insTTT, p. (Gly86delinsValCys). For the former, the same variant was found in a second unrelated patient, where SDHB-IHC showed negative staining, supporting the classification as ‘likely pathogenic’. The missense variant of SDHA, NM_004168.3:c.1772C>T, p.(Ala591Val), was predicted to have no functional impact based on metabolite profiling and SDHB-IHC as interpreted by a local pathologist. Experts, however, all agreed on a negative staining pattern for SDHB. Two out of three in silico protein prediction tools rated the variant as ‘disease causing’ or ‘possibly damaging’.
Discussion
This study establishes for the first time that SDHB-IHC and metabolite profiling provide complementary diagnostic tools for the prediction of SDH impairment in PPGL tumour tissue. Moreover, we show that diagnostic performance of metabolite profiling can be improved by machine learning-assisted interpretation of metabolite data and that there is a trend towards further improvement by inclusion of findings from SDHB-IHC. We therefore propose an approach that combines metabolite profiling and SDHB-IHC to better facilitate identifying or excluding SDH impairment when tumour material is available, particularly for selected patients in whom there is a suspicion of the presence of SDH mutation and where genetic testing yields equivocal or negative results or is unavailable. First, the high specificity of metabolite profiling (99%) translates to high positive predictive value of a positive result, strongly indicating a mutation in an SDHx gene. If the genetic change identified is a variant of unknown significance, the predictive models will, together with in silico prediction tools, aid in determining whether the variant is pathogenic and whether the patient and affected family members require life-long surveillance. If metabolite profiling predicts no SDH impairment, then SDHB-IHC provides utility to exclude false negatives by metabolite profiling. Applying this approach to cohort 1 (excluding three variants of unknown significance), we would have correctly predicted impairments of SDH in 185 of the 186 PPGLs, providing an advantage over either method alone.
SDHB-IHC requires a simple setup and can be easily incorporated into pathology workflows. Since our preliminary evidence suggests that interpretation does not require expert review, the technique is readily adoptable anywhere. LC—MS/MS, on the other hand, requires specialised instrumentation and expertise, but is becoming more and more available in clinical laboratories where the instruments are used for many routine diagnostic tests. While SDHB-IHC assesses presence of the protein, LC—MS/MS-based metabolite profiling provides information about functionality of the succinate complex through measurements of precursor and product metabolites. There is also the added benefit of measuring a panel of metabolites to identify impaired function of other enzymes, such as fumarate hydratase and isocitrate dehydrogenase [12,22]. Metabolite measurements also address some of the limitations of SDHB-IHC: in particular, there is no subjective bias of interpretation and there is always a numerical result rather than inconclusive interpretations.
On the other hand, there are limitations of metabolite profiling. Generating cut-offs or machine learning models requires large numbers of samples and whether such data are transferrable among laboratories (i.e. method harmonisation) using LC—MS/MS is not yet established. Another limitation of metabolite profiling is tissue selection. False negatives can occur due to excessive amounts of non-tumour tissue in the sample. We suspect that this is also the reason for different predictions (probabilities for SDH impairment) produced by LDA A and LDA B, since depending on the type of stromal contaminant, metabolite levels will differ. One possible solution, requiring interdisciplinary connections between anatomic and chemical diagnostic laboratories, is to assess tumour content first by haematoxylin and eosin staining, perform macro-dissection of tumour areas, and use this material for metabolite extraction.
Inter-observer variability of SDHB-IHC interpretations could be addressed by applying deep learning to establish a pipeline for automated image interpretation, as has been done for immunohistochemistry directed to other purposes [14,23]. Machine learning was also demonstrated to be suitable for cancer diagnosis on whole-slide images [24]. Nevertheless, not only histology but also biomarker interpretation and analysis of omics data (transcriptomics, proteomics, and metabolomics) can benefit from machine learning approaches [15]. A recent example is the identification of PPGL-specific long intergenic noncoding RNAs and their use for molecular subtyping of PPGL patients [25].
In this study, we used pattern recognition and multidimensional strategies from the field of artificial intelligence for analysing metabolite data to gain information beyond simple ratios such as the SFR. In this way, machine learning offered improved diagnostic sensitivity. This was especially useful for identifying functional impairment of SDH in head and neck paragangliomas, for which false negatives can be a problem when relying on the SFR [11]. We advise that the tumour content of the input material for head and neck paragangliomas be evaluated carefully and that further available methods to test protein status, such as immunohistochemistry for SDHB, SDHA or SDHD, be used [9,26,27].
While the predictive models generated in this study were targeted towards identifying SDHx mutations, other models could be generated based on measurements of metabolites in the same panel to predict mutations and functional deficiencies impacting other enzymes. Application of the generated models (supplied as supplementary material, MATLAB Files S1 and S2) is relatively straightforward since it only requires tissue concentrations of measured metabolites or ratios of metabolites (to calculate the matrices needed for input of this data into the LDA models, see supplementary material, Table S4). Results are provided as per cent probabilities of the tumour harbouring an SDHx mutation. The interpretation of ten metabolites is thereby converted into a simplified single output variable to guide clinical decision-making.
With ongoing data collection for PPGLs and other tumour entities, machine learning-assisted data interpretation can be used to further stratify patients according to mutational background or other clinically relevant features as new models can be generated as more data become available. This approach was also suggested in the context of steroid metabolomics for the diagnosis of adrenal cortical tumours [28].
A challenge of next-generation panel sequencing in genetic testing is the interpretation of variants of unknown significance. In our combined cohorts, a total of 11 variants of unknown significance in SDHx genes were identified. From those, four had elevated SFR indicating loss of functionality; however, in silico predictions and SDHB-IHC showed varying agreement. Such discrepancies between SDHB-IHC and in silico predictions were reported previously [29]. In two more tumours, where SFR failed to indicate SDH impairment, the new machine learning-based LDA B predicted loss of SDH functionality. At least in one of these cases, SDHA NM_004168.3: c.457–1G>A, evidence suggests a true mutation.
Limitations of the present study are that SDHB-IHC data were not available for all patients and that immunohistochemistry was performed in different centres to varying standards and quality. The latter may partly explain the lower level of agreement among pathologists for interpreting SDHB-IHC than found in a previous study [20]. Despite this limitation, diagnostic sensitivity and specificity for SDHB-IHC were in the range of former reports [9,20].
Although machine learning was only applied to metabolite data and not to images from SDHB-IHC, we could show that there is potential for this approach to improve diagnostic and prognostic workflows, especially when data complexity is high. Our study also highlights the benefit of interdisciplinary connections between physicians, pathologists, clinical chemists, geneticists, and data scientists. By working together, advanced genotypic stratification of PPGLs can be expected to better facilitate targeted therapies with increased efficacy and improved patient outcomes [30–33].
Supplementary Material
MATLAB File S1. Matlab_Model_LDA_A.mat – a MATLAB model for use with absolute metabolite concentrations
MATLAB File S2. Matlab_Model_LDA_B.mat – a MATLAB model for use with relative metabolite concentrations
Table S4. Excel file to produce matrices for use in LDA models
Table S2. Tumour identifiers, clinical data, IHC results, and metabolite concentrations
Table S1. Characterisation of patient/tumour cohorts
Table S3. Input ratios for LDA model B in the order they need to be supplied in MATLAB
Table S5. Results of all predictive methods where the tumour is misclassified by at least one of the methods
Table S6. Interpretation of metabolite profiles for SDHx VUS based on predictive models generated with linear discriminant analysis (LDA)
Table S7. Concentrations of pre-calibrators Pre-Cal 1–8
Table S8. Concentrations of internal standards in the Internal Standard Mix (IS-Mix)
Table S9. Concentrations of calibrators Cal 0–8
Table S10. Assay precision estimated using two quality control (QC) samples
Table S11. Multiple reaction monitoring transitions, fragmentation parameters,m and quantifiers
Figure S1. Flow diagram of patient and tumour sample numbers used in this study
Figure S2. PCA plot of all tumours showing grouping of SDHx mutations (black triangles) and SDHx wild types (open circles) based on the normalised values of all ten measured metabolites
Figure S3. Performance of the models generated based on different ratios of how to separate cohort 2 into learning set and validation set
Figure S4. Performance of the two chosen models LDA A and LDA B
Figure S5. Comparison of predictions generated with LDA Model B from FFPE and FF tissue extracted metabolites
Figure S6. Distribution of true negatives (TN), true positives (TP), false positives (FP), and false negatives (FN) in cases of agreement between the panel of researchers and local pathologists
Figure S7. Example images for SDHB-IHC scored by local pathologists and experts in the field of SDHB-IHC
Acknowledgements
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Projektnummer: 314061271 — TRR 205; RI 2684/1-1; KL 2541/2-1, the AES PI17/01796, co-financed by Fondo Europeo de Desarrollo Regional (FEDER), the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement No 259735, the Paradifference Foundation, and the Intramural Research Program of the NIH, NICHD.
References
- 1.Gill AJ. Succinate dehydrogenase (SDH)-deficient neoplasia. Histopathology 2018; 72: 106–116. [DOI] [PubMed] [Google Scholar]
- 2.Amar L, Baudin E, Burnichon N, et al. Succinate dehydrogenase B gene mutations predict survival in patients with malignant pheochromocytomas or paragangliomas. J Clin Endoncrinol Metab 2007; 92: 3822–3828. [DOI] [PubMed] [Google Scholar]
- 3.Rijken JA, van Hulsteijn LT, Dekkers OM, et al. Increased mortality in SDHB but not in SDHD pathogenic variant carriers. Cancers (Basel) 2019; 11: el03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenders JW, Duh QY, Eisenhofer G, et al. Pheochromocytoma and paraganglioma: an Endocrine Society Clinical Practice Guideline. J Clin Endocrinol Metab 2014; 99: 1915–1942. [DOI] [PubMed] [Google Scholar]
- 5.Toledo RA, Burnichon N, Cascon A, et al. Consensus statement on next-generation-sequencing-based diagnostic testing of hereditary phaeochromocytomas and paragangliomas. Nat Rev Endocrinol 2017; 13: 233–247. [DOI] [PubMed] [Google Scholar]
- 6.Curras-Freixes M, Pineiro-Yanez E, Montero-Conde C, et al. Pheo-Seq: a targeted next-generation sequencing assay for pheochromocytoma and paraganglioma diagnostics. J Mol Diagn 2017; 19: 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter S, Klink B, Nacke B, et al. Epigenetic mutation of the succinate dehydrogenase C promoter in a patient with two paragangliomas. J Clin Endocrinol Metab 2016; 101: 359–363. [DOI] [PubMed] [Google Scholar]
- 8.Remacha L, Comino-Mendez I, Richter S, et al. Targeted exome sequencing of Krebs cycle genes reveals candidate cancer-predisposing mutations in pheochromocytomas and paragangliomas. Clin Cancer Res 2017; 23: 6315–6324. [DOI] [PubMed] [Google Scholar]
- 9.van Nederveen FH, Gaal J, Favier J, et al. An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis. Lancet Oncol 2009; 10: 764–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill AJ. Succinate dehydrogenase (SDH) and mitochondrial driven neoplasia. Pathology 2012; 44: 285–292. [DOI] [PubMed] [Google Scholar]
- 11.Richter S, Peitzsch M, Rapizzi E, et al. Krebs cycle metabolite profiling for identification and stratification of pheochromocytomas/paragangliomas iomas due to succinate dehydrogenase deficiency. J Clin Endocrinol Metab 2014; 99: 3903–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter S, Gieldon L, Pang Y, et al. Metabolome-guided genomics to identify pathogenic variants in isocitrate dehydrogenase, fumarate hydratase, and succinate dehydrogenase genes in pheochromocytoma and paraganglioma. Genet Med 2019; 21: 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E, Wright MJ, Sioson L, et al. Utility of the succinate:fumarate ratio for assessing SDH dysfunction in different tumor types. Mol Genet Metab Rep 2016; 10: 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abels E, Pantanowitz L, Aeffner F, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol 2019; 249: 286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grapov D, Fahrmann J, Wanichthanarak K, et al. Rise of deep learning for genomic, proteomic, and metabolomic data integration in precision medicine. OMICS 2018; 22: 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz JM, Cooper DN, Schuelke M, et al. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–362. [DOI] [PubMed] [Google Scholar]
- 18.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002; 30: 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim NL, Kumar P, Hu J, et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012; 40: W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papathomas TG, Oudijk L, Persu A, et al. SDHB/SDHA immunohistochemistry in pheochromocytomas and paragangliomas: a multicenter interobserver variation analysis using virtual microscopy: a Multinational Study of the European Network for the Study of Adrenal Tumors (ENS@T). Mod Pathol 2015; 28: 807–821. [DOI] [PubMed] [Google Scholar]
- 21.Fischer RA. The use of multiple measurements in taxonomic problems. Ann Eugen 1936; 7: 179–188. [Google Scholar]
- 22.Dwight T, Kim E, Novos T, et al. Metabolomics in the diagnosis of pheochromocytoma and paraganglioma. Horm Metab Res 2019; 51: 443–450. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Yang DM, Rong R, et al. Pathology image analysis using segmentation deep learning algorithms. Am J Pathol 2019; 189: 1686–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Chen P, McGough M, et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat Mach Intell 2019; 1: 236–245. [Google Scholar]
- 25.Ghosal S, Das S, Pang Y, et al. Long intergenic non-coding RNA profiles of pheochromocytoma and paraganglioma: a novel prognostic biomarker. Int J Cancer 2020; 146: 2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korpershoek E, Favier J, Gaal J, et al. SDHA immunohistochemistry detects germline SDHA gene mutations in apparently sporadic paragangliomas and pheochromocytomas. J Clin Endocrinol Metab 2011; 96: E1472–E1476. [DOI] [PubMed] [Google Scholar]
- 27.Menara M, Oudijk L, Badoual C, et al. SDHD immunohistochemistry: a new tool to validate SDHx mutations in pheochromocytoma/paraganglioma. J Clin Endocrinol Metab 2015; 100: E287–E291. [DOI] [PubMed] [Google Scholar]
- 28.Eisenhofer G, Durán C, Chavakis T, et al. Steroid metabolomics: machine learning and multidimensional diagnostics for adrenal cortical tumors, hyperplasias, and related disorders. Curr Opin Endocrinol Metab Res 2019; 8: 40–49. [Google Scholar]
- 29.Evenepoel L, Papathomas TG, Krol N, et al. Toward an improved definition of the genetic and tumor spectrum associated with SDH germline mutations. Genet Med 2015; 17: 610–620. [DOI] [PubMed] [Google Scholar]
- 30.Pang Y, Liu Y, Pacak K, et al. Pheochromocytomas and paragangliomas: from genetic diversity to targeted therapies. Cancers (Basel) 2019; 11: e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang Y, Lu Y, Caisova V, et al. Targeting NAD+/PARP DNA repair pathway as a novel therapeutic approach to SDHB-mutated cluster I pheochromocytoma and paraganglioma. Clin Cancer Res 2018; 24: 3423–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacak K, Taieb D. Pheochromocytoma (PHEO) and paraganglioma (PGL). Cancers (Basel) 2019; 11: e1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Pang Y, Caisova V, et al. Targeting NRF2-governed glutathione synthesis for SDHB-mutated pheochromocytoma and paraganglioma. Cancers (Basel) 2020; 12: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MATLAB File S1. Matlab_Model_LDA_A.mat – a MATLAB model for use with absolute metabolite concentrations
MATLAB File S2. Matlab_Model_LDA_B.mat – a MATLAB model for use with relative metabolite concentrations
Table S4. Excel file to produce matrices for use in LDA models
Table S2. Tumour identifiers, clinical data, IHC results, and metabolite concentrations
Table S1. Characterisation of patient/tumour cohorts
Table S3. Input ratios for LDA model B in the order they need to be supplied in MATLAB
Table S5. Results of all predictive methods where the tumour is misclassified by at least one of the methods
Table S6. Interpretation of metabolite profiles for SDHx VUS based on predictive models generated with linear discriminant analysis (LDA)
Table S7. Concentrations of pre-calibrators Pre-Cal 1–8
Table S8. Concentrations of internal standards in the Internal Standard Mix (IS-Mix)
Table S9. Concentrations of calibrators Cal 0–8
Table S10. Assay precision estimated using two quality control (QC) samples
Table S11. Multiple reaction monitoring transitions, fragmentation parameters,m and quantifiers
Figure S1. Flow diagram of patient and tumour sample numbers used in this study
Figure S2. PCA plot of all tumours showing grouping of SDHx mutations (black triangles) and SDHx wild types (open circles) based on the normalised values of all ten measured metabolites
Figure S3. Performance of the models generated based on different ratios of how to separate cohort 2 into learning set and validation set
Figure S4. Performance of the two chosen models LDA A and LDA B
Figure S5. Comparison of predictions generated with LDA Model B from FFPE and FF tissue extracted metabolites
Figure S6. Distribution of true negatives (TN), true positives (TP), false positives (FP), and false negatives (FN) in cases of agreement between the panel of researchers and local pathologists
Figure S7. Example images for SDHB-IHC scored by local pathologists and experts in the field of SDHB-IHC