Abstract (300 words)
A lack of quantitative information on the species composition of parasite communities present in fecal samples is a major limiting factor for the sensitivity, accuracy and interpretation of the diagnostic tests commonly used to assess anthelmintic efficacy and resistance. In this paper, we investigate the ability of ITS-2 rDNA nemabiome metabarcoding to enhance fecal egg count reduction testing by providing information on the effect of drug treatments on individual parasite species. Application of ITS-2 rDNA nemabiome metabarcoding to fecal samples from ewes from over 90 flocks across western Canada revealed high gastrointestinal nematode infection intensities in many flocks with Haemonchus contortus being the most abundant species followed by Teladorsagia circumcincta and then Trichostrongylus colubriformis. Integration of ITS-2 rDNA nemabiome metabarcoding with pre- and post-treatment fecal egg counting revealed consistently poor efficacy of producer-applied ivermectin and benzimidazole treatments against H. contortus, but much better efficacy against T. circumcincta and T. colubriformis, except for in a small number of flocks. Integration of nemabiome ITS-2 rDNA metabarcoding with Fecal Egg Count Reduction Tests (FECRT), undertaken on farm visits, confirmed that ivermectin and fenbendazole resistance is widespread in H. contortus but is currently less common in T. circumcincta and T. colubriformis in western Canada. FECRT/nemabiome testing did not detect moxidectin resistance in any GIN species but suggested the early emergence of levamisole resistance specifically in T. circumcincta. It also revealed that although poor efficacy to closantel was relatively common, based on total fecal egg counts, this was due to its narrow spectrum of activity rather than the emergence of anthelmintic resistance. This study illustrates the value of ITS-2 rDNA nemabiome metabarcoding to improve fecal egg count resistance testing, perform large-scale anthelmintic resistance surveillance and direct more targeted rational anthelmintic use.
Keywords: Amplicon sequencing, Molecular diagnostics, anthelmintic resistance, FECRT, Haemonchus, Benzimidazoles
Graphical abstract
Highlights
-
•
Nemabiome metabarcoding in anthelmintic resistance diagnostics and surveillance.
-
•
Producer-applied treatment results were consistent with controlled FECRT.
-
•
Widespread BZ and IVM resistance in H. contortus in western Canada.
-
•
Only sporadic BZ and IVM resistance T. circumcincta and T. colubriformis.
-
•
Early levamisole resistance in T. circumcincta, closantel resistance not prevalent.
1. Introduction
Fifty years have passed since the earliest reports of anthelmintic resistance in sheep (Drudge et al., 1964; Smeal et al., 1968) and the problem now has grown and spread to all continents, occurring in multiple nematode species and against multiple classes of drugs (Chandrawathani et al., 2003; Čerňanská et al., 2006; Domke et al., 2012; Papadopoulos et al., 2012; Falzon et al., 2013; Chandra et al., 2014; Geurden et al., 2014; Lyndal-Murphy et al., 2014; Rose et al., 2015; Learmount et al., 2016a; Gárcia et al., 2016; Salgado and Santos, 2016; Ploeger and Everts, 2018). However, the methods used to diagnose anthelmintic resistance have changed very little and predominantly rely on determining the reduction of fecal egg counts following anthelmintic treatment. One major limitation of these approaches is that coinfection commonly occurs with multiple nematode species which have strongyle-type eggs that can only be distinguished to the genus level by microscopy and with specialist expertise. Consequently, a simple percentage strongyle egg count reduction has limited sensitivity and accuracy for detection of anthelmintic resistance when the parasite populations consists of a mixture of resistant and susceptible species. The methods available to identify nematode species include coproculture combined with L3 morphology and morphometry, conventional PCR, real-time PCR, droplet digital PCR (Elmahalawy et al., 2018) and pyrosequencing (Álvarez-Sánchez et al., 2005; Roeber et al., 2011, 2017; Milhes et al., 2017). Each of these approaches have different limitations. Larval coproculture and morphological identification of L3 larvae takes 7–14 days, requires specialist expertise and is subject to inaccuracy due to species-specific culture biases. It is often not performed for routine clinical diagnosis and is very time consuming when applied to large-scale research and surveillance studies (Coles et al., 1992, 2006; Lichtenfels and Pilitt, 2000; McMurtry et al., 2000; Van Wyk et al., 2004; Amarante, 2011; Barrère et al., 2013; Ljungström et al., 2018). Real-time PCR-based and ddPCR approaches are more accurate but require careful optimisation and validation between laboratories for accurate quantitation. Further, assays need to be individually validated for each species and only those species anticipated to be present will be identified (Roeber et al., 2011, 2017).
Nemabiome metabarcoding is a recently developed “microbiome-style” approach which involves short-read next generation sequencing of the internal transcribed spacer (ITS-2) rDNA amplicons for nematode species identification and relative quantitation (Avramenko et al., 2015). It was initially developed for the relative quantitation of cattle gastrointestinal nematode (GIN) species from larval L3 coproculture, but has more recently been validated for use on eggs and L1 larvae isolated from sheep fecal samples (Redman et al., 2019). It has also been applied to bison (Avramenko et al., 2018), horses (Mitchell et al., 2019) and wildlife fecal samples (Barone et al., 2020). It has several advantages including sensitivity, specificity, versatility, scalability and high cost-efficacy for large sample sets (Avramenko et al., 2015; Redman et al., 2019). ITS-2 rDNA nemabiome metabarcoding allows hundreds of samples to be pooled and sequenced in a single sequencing run, most commonly the Illumina MiSeq (Illumina Inc., San Diego, CA, USA), reducing the individual sample cost. Another advantage is the high accuracy, generating thousands to millions reads from hundreds or thousands of eggs or larvae per sample that are compared against a reference database for species identification, without the need of species-specific primers. It can also identify a single larva in a pool of thousands, which is an advantage for surveillance and identification of rare species, that would probably be missed in conventional methods such as morphology.
In this paper, we investigate the integration of ITS-2 rDNA metabarcoding with fecal egg count reduction testing to provide specific information on parasite surveillance, anthelmintic efficacy and anthelmintic resistance for individual GIN species. We demonstrate the value of this approach using a large-scale study of anthelmintic efficacy and resistance on sheep flocks across western Canada, a region where essentially no such information has been previously reported.
2. Materials and methods
2.1. Survey of the effectiveness of producer-applied anthelmintic treatments
2.1.1. Sample collection
To evaluate the effectiveness of anthelmintic treatments undertaken by producers in western Canada, annual surveys were carried out between June and September from 2014 to 2018 in which 7 producers from British Columbia (BC), 55 from Alberta (AB), 18 from Saskatchewan (SK) and 12 from Manitoba (MB) participated. Recruitment of producers was predominantly from respondents to advertisements posted in producer newsletters asking for participants with flocks of at least 20 ewes.
Producers were requested to wait at least eight weeks from the last anthelmintic treatment in order to participate. Treatment and sampling was undertaken by producers, supplied with a sampling kit, questionnaire, gloves, plastic bags for collection of individual fecal samples and an instruction sheet. Producers were asked to collect fecal samples from 20 ewes, either per rectum or freshly voided on pasture, followed by treatment with their current drug of choice and collection of a second set of fecal samples from the same ewes two weeks later.
The samples were sealed in plastic bags, firmly pressed to remove air and prevent egg hatching and sent within 24 h of collection by prepaid courier to the laboratory. Producers were asked not to refrigerate or freeze the samples before shipping. At the laboratory, fecal samples were stored at room temperature and processed within 24 h of arrival. For farms that participated more than once between 2014 and 2018, only the results from the most recent year of participation are included here. Producers who did not send a post-treatment sample, did not followed the two weeks interval or whose samples were damaged or not sealed were not included in the treatment effectiveness study.
2.1.2. Calculation of fecal egg count reduction (FECR)
In the laboratory, individual samples were thoroughly mixed and pooled by equal weight, adding the maximum amount possible based on the smallest sample, with a minimum of 6 g. To determine the number of strongyle-type eggs per gram (EPG) of feces before and after drug treatment, we calculated the mean of four independent fecal egg counts performed on 2 g of each pooled sample, using a modified McMaster method with a sensitivity of 16.66 EP G (Paracount-EPG™, Chalex, LCC). Anthelmintic treatment effectiveness was assessed by comparing the mean EPG of the pool of 20 ewes in the pre- and post-treatment, using the following formula: . A treatment efficacy of below 95% was considered suboptimal.
2.2. Fecal egg count reduction test (FECRT)
2.2.1. Sample collection
Eleven farms in Alberta and three farms in Saskatchewan were visited twice at an interval of two weeks, between June to September from 2014 to 2018, to collect samples to perform Fecal Egg Count Reduction Tests. The criteria for inclusion of farms in this part of the study were for them to be located within a 3-h drive from University of Calgary or University of Saskatchewan, to have at least 80 ewes, to have history of high GIN infection intensities (based on an average fecal egg count higher than 400 eggs per gram on samples from 20 randomly selected ewes, sent two to three weeks before the first visit) and, in some cases, producer concern about parasite control problems. At the first visit, adult ewes were randomly assigned into treatment groups, with 20–25 animals each. The ewes were identified by their radio frequency identification (RFID) or ear tag number and weighed. Fecal samples were collected per rectum and anthelmintics were given orally according to the manufacturer's recommendations. Fecal samples were collected in accordance with an approved Animal Use Protocol (Animal Care Committee, Study #AC13-0157, University of Calgary), which is in accordance with the principles outlined in the current Guidelines of the Canadian Council on Animal Care. The anthelmintics tested included ivermectin, fenbendazole, closantel, levamisole and moxidectin (see Supplementary Table 1 for detailed information of drug classes, commercial brands, dosage and activity of anthelmintics used in this study). At the second visit, fecal samples were collected from the same ewes.
2.2.2. FECR calculation
Strongyle-type eggs in each individual sample were counted using the Modified McMaster technique described above. Reduction in number of eggs was assessed by comparing fecal egg counts from the same group of animals before and after treatment, FECR was calculated using the same equation described on 2.1.2 and the confidence interval as described by Levecke et al. (2018), equation 1. All ewes with positive fecal egg counts pre-treatment were included in the calculation. We aimed to count in the pre-treatment samples a minimum of 400 eggs under the microscope per treatment group, adding an extra slide chamber and changing the multiplication factor as necessary, to increase the power of the study (Levecke et al., 2018). A pooled sample with 2 g of each individual sample was prepared for nematode species identification later.
2.3. First stage larvae isolation
First stage larvae were obtained from pre- and post-treatment samples for DNA lysate preparation, using a protocol adapted from Mes et al. (2007). A 24 g aliquot of each of the pooled samples was thoroughly mixed with 100 mL of 13% sodium chloride solution (1.06 specific gravity) and washed through a coarse kitchen sieve, allowing eggs to pass whilst retaining large particulate matter. The egg-containing filtrate was equally distributed into four 50 mL Falcon tubes and centrifuged at 3600 G for 5 min at room temperature to float the eggs. The supernatant was transferred to new 50 mL Falcon tubes and equal volume of distilled water was added, following centrifugation at the same conditions. Using vacuum pump aspiration, the supernatant was carefully removed and 5 mL of 13% sodium chloride solution was added to resuspend the eggs in the pellet before another 5 min centrifugation at 3600 G. The supernatant from the four tubes was then combined into a new 50 mL Falcon tube, an equal amount of distilled water added, and the tube centrifuged again at 3600 G for 5 min and the supernatant removed by vacuum aspiration. The pellet was resuspended with 25 mL of distilled water and washed through a pre-wetted 20 μm sieve to retain the eggs and remove remaining salt solution. The eggs retained in the sieve were washed into a Petri dish and incubated with tap water at 22 °C for 24–48 h to hatch the eggs. After incubation, L1 suspension was refrigerated at 4 °C for 3 h and then centrifuged at 4500 G at 4 °C for 10 min. Supernatant was removed with vacuum aspiration, and harvested L1 were fixed in 70% ethanol and stored at 4 °C until use. The egg hatch rate was determined by counting unhatched eggs and L1 in three 500 μL aliquots of suspension (mean 88%, range 48%–100%, median 91%, Supplementary Table 2).
2.4. Genomic DNA extraction and ITS-2 rDNA nemabiome amplicon sequencing
2.4.1. Lysate preparations for DNA extraction
Approximately 500 larvae harvested from each pooled fecal sample were used to prepare DNA lysates earlier in the study (2014–2016). However, after demonstration of the reliability of ITS-2 rDNA nemabiome metabarcoding for lower numbers of larvae (Redman et al., 2019), subsequent DNA lysates were prepared with 200 larvae, following method described by Avramenko et al. (2015). Supplementary Table 3 documents the number of larvae included in each lysate. In those samples with less than a total of 200 larvae, for example in post-treatment samples with low FECs due to high drug efficacies, DNA lysates were prepared from all larvae harvested from the sample. Ethanol fixed L1 larvae were washed three times by centrifugation at 13,000 G with made-in-house lysis buffer (50 mM KCl, 10 mM Tris pH 8.3, 2.5 mM MgCl2, 0.45% Nonidet p-40, 0.45% Tween-20, 0.01% gelatin) to remove excess ethanol. The pellet was resuspended in 50 μL lysis buffer, placed in −80 °C for 1 h and then lysed by adding 6 μL of 20 mg/mL proteinase K (Thermo Scientific) and heating at 55 °C for 120 min; followed by 95 °C for 20 min to inactivate proteinase K. A 1:10 dilution of the resulting lysate was made with molecular grade water to use as PCR template and the remaining original lysate was stored at −80 °C.
2.4.2. PCR amplification and nemabiome metabarcoding of the ITS-2 rDNA locus
Deep amplicon sequencing was performed on ITS-2 rDNA amplicons generated from L1 larvae populations harvested from pooled fecal samples. The 311–331 bp ITS-2 rDNA locus was PCR amplified using modified primers NC1 and NC2, complementary to 5.8 S and 28 S coding regions, as previously described (Gasser et al., 2008; Avramenko et al., 2015; Redman et al., 2019). A total of four forward (NC1_Adp, NC1_Adp1N, NC1_Adp2N, NC1_Adp3N) and four reverse (NC2_Adp, NC2_Adp1N, NC2_Adp2N, NC2_Adp3N) primers were used, where Adp is the Illumina Adapter sequencing tag, added to allow further annealing of sequencing primers, and N is the number of random nucleotides added between the primer sequence and the adapter to introduce diversity of amplicons to prevent fluorescence signal saturation during Illumina sequencing (Avramenko et al., 2015). The PCR reaction conditions were 5 μL 5X KAPA HiFi Buffer, 0.75 μL NC1 primer mix (10 mM), 0.75 μL NC2 primer mix (10 mM), 0.75 μL dNTPs (10 mM), 0.5 μL KAPA HiFi HotStart Polymerase (0.5 U, KAPA Biosystems, USA), 13.25 μL ddH2O and 4 μL 1:10 diluted lysate. Thermocycling parameters were 95 °C for 2 min, followed by 25 cycles of 98 °C for 20 s, 62 °C for 15 s, 72 °C for 15 s and a final extension of 72 °C for 2 min.
PCR products were purified with AMPure XP Magnetic Beads (Beckman Coulter, Inc) before Illumina barcoding indices and P5/P7 tags were added with a limited-cycle PCR amplification. In this step, 8 forward and 12 reverse barcoded primers were used to make 96 unique barcode combinations. Primer sequences were obtained from the Illumina Customer Sequencing Letter (September 7, 2012, Oligonucleotide sequences 2007–2012 Illumina, Inc. All rights reserved) as described by Avramenko et al. (2015). PCR reaction conditions were 5 μL 5X KAPA HiFi HotStart Fidelity Buffer (KAPA Biosystems, USA), 1.25 μL Forward Primer (S502–S522) (10 μM), 1.25 μL Reverse Primer (N701–N729) (10 μM), 0.75 μL dNTPs (10 mM), 0.5 μL KAPA HiFi HotStart Polymerase (0.5 U), 13.25 μL molecular grade dH2O, and 3 μL of first round purified PCR product as template. Thermocycling parameters were 98 °C for 45 s, followed by nine cycles of 98 °C for 20 s, 63 °C for 20 s, 72 °C for 2 min. Amplicons were purified with AMPure XP magnetic (Beckman Coulter, Inc). The concentration of each purified amplicon was determined using a NanoVue plus Spectrophotometer (GE) and 50 ng aliquots pooled to create a library. The final concentration of the pooled library was assessed with the KAPA qPCR Library Quantification Kit (KAPA Biosystems, USA), following the manufacturer's protocol. The library was then diluted to 4 nM and run on an Illumina Desktop Sequencer using a 500-cycle paired-end reagent kit (MiSeq Reagent Kit, v2, MS-103-2003, Illumina Inc., San Diego, CA, USA) at a final concentration of 15 pM, with the addition of 25% PhiX control v3 (Illumina, FC-110-3001).
2.5. Bioinformatic and statistical analysis
Samples were automatically de-multiplexed with the MiSeq software on the basis of their index combinations. Sequence data was analyzed using a previously described bioinformatic pipeline (Avramenko et al., 2017; Redman et al., 2019) based on Mothur software version 1.36.1 (Schloss et al., 2009). Briefly, forward and reverse sequences were aligned to make contigs which were filtered to remove reads shorter than 200 bp and longer than 450 bp. The remaining sequences were compared to a bespoke trichostrongylid nematode ITS-2 rDNA database (Avramenko et al., 2017) and filtered for a minimum similarity of 90%. Sequences were then taxonomically classified, using the k-nearest neighbor method with k = 3. Further details and protocols of the analysis can be found on https://www.nemabiome.ca/mothur_workflow.html.
Samples with total number of reads of less than 1000 were removed from the study. Classified read counts were corrected for species-specific representation biases using correction factors previously described and validated for sheep gastrointestinal nematodes (Redman et al., 2019). The relative abundance of each species was calculated based on the total number of sequence reads from each sample.
Statistical analyses were performed on GraphPad Prism version 8.1.0 for macOS, GraphPad Software, San Diego, California USA, www.graphpad.com (refer to figures for details on tests performed). Alpha diversity of each sample for the survey of species abundance was calculated using the inverse Simpson index, performed in Mothur v.1.36.1. All samples were subsampled to 1000 reads to ensure equal comparison. Samples that did not meet this threshold were removed from this analysis. To compare the inverse Simpson index between provinces, we performed a One-way ANOVA on SPSS (IBM Corp. Released 2012. IBM SPSS Statistics for Macintosh, version 26.0. Armonk, NY:IBM Corp), assuming non-equal variances and using Games-Howell post-hoc comparison test. Beta diversity between surveyed populations was estimated using MetaStats plugin in Mothur v.1.36.1, with default parameters (White et al., 2009). P < 0.05 was considered significant in all tests performed. The bar charts and scatter plots were created in GraphPad Software. The map displayed in Fig. 1 was created using package rgdal (Bivand et al., 2019) in R studio version 1.2.1335.
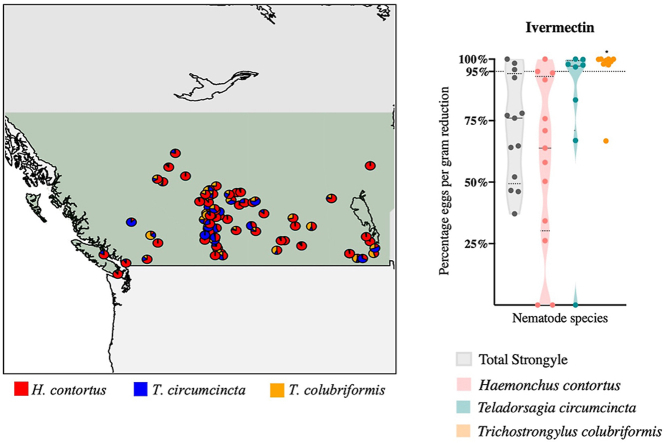
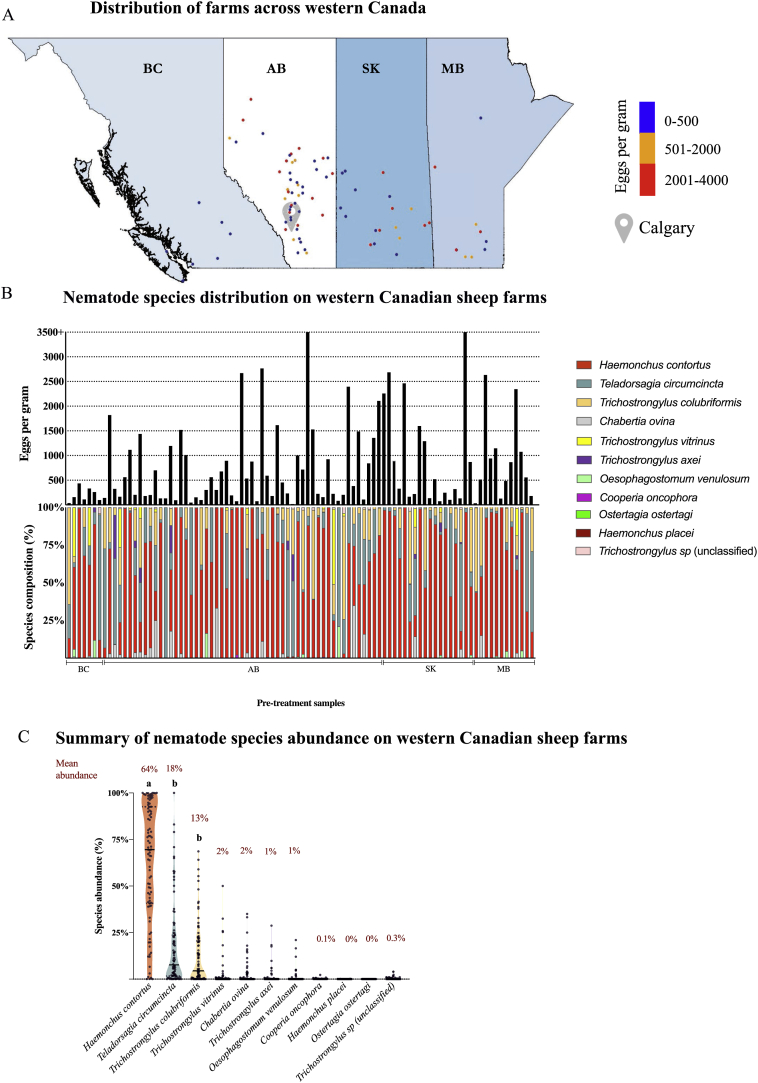
Figure 1.
ITS-2 rDNA nemabiome metabarcoding enabled survey of gastrointestinal nematode species infection intensities in western Canada. Data shown in Panels A, B and C is from 92 sheep flocks based on fecal egg counts (Modified McMaster) and ITS-2 rDNA nemabiome metabarcoding applied to pooled samples from 20 ewes/flock taken between 2014 and 2018 (7 farms from British Columbia, 55 farms from Alberta, 18 farms from Saskatchewan and 12 farms from Manitoba).
Panel A. Distribution of gastrointestinal nematode infection intensities in ewes from sheep flocks across western Canada. BC – British Columbia, AB – Alberta, SK – Saskatchewan, MB – Manitoba. Calgary location is shown as the grey shaded pin. To preserve data privacy, approximate location of each farm based on Post office coordinates were transformed into a Special Points data frame and plotted using package rgdal on RStudio Software Version 1.2.1335 using boundary shape files of Canada available on Statistics Canada. Pre-treatment fecal egg counts (eggs per gram) were determined by Modified McMaster technique and flocks were categorized according to their fecal egg counts as blue (0–500 epg), orange (501–2000 epg) and red (2001–4000 epg).
Panel B. Relative abundance of gastrointestinal nematode species in ewes from 92 western Canadian sheep flocks determined by fecal egg counting and ITS-2 rDNA nemabiome metabarcoding in pre-treatment samples. Pre-treatment fecal egg counts for each flock are represented as black bars on the upper histogram. Relative species composition (%), determined by ITS-2 rDNA nemabiome metabarcoding of at least 200 L1 larvae in each pre-treatment sample and are represented by the lower coloured stacked bars. Province location is indicated below the chart (BC-British Columbia, AB-Alberta, SK-Saskatchewan, MB-Manitoba).
Panel C. Summary of gastrointestinal nematode species abundance in ewes from western Canadian sheep flocks. The violin plot summarizes the relative abundance of the different gastrointestinal nematode species on each of 92 sheep flocks determined by ITS-2 rDNA nemabiome metabarcoding of pre-treatment samples. Beta diversity analysis was performed using the command MetaStats on mothur software v.1.36.1. Abundance of H. contortus was significantly higher compared to all other species (a, p < 0.0001). Teladorsagia circumcincta and Trichostrongylus colubriformis abundance was significantly different (b, p < 0.0001) than all species identified, except for each other.
3. Results
3.1. ITS-2 rDNA nemabiome metabarcoding reveals high gastrointestinal nematode infection intensities and a predominance of H. contortus in western Canadian sheep flocks
A total of eleven operational taxonomic units (OTUs) were identified on the dataset: Chabertia ovina, Oesophagostomum venulosum, Cooperia oncophora, Haemonchus contortus, Haemonchus placei, Ostertagia ostertagi, Teladorsagia circumcincta, Trichostrongylus axei, Trichostrongylus colubriformis, Trichostrongylus vitrinus and Trichostrongylus sp (unclassified). We first investigated the infection intensities of different species of gastrointestinal strongylid nematodes in ewes for 92 sheep flocks across western Canada using a combination of fecal egg counting and ITS-2 rDNA nemabiome metabarcoding. The strongyle fecal egg count ranged from 17 epg to 3915 epg (mean 785, median 446, standard deviation 864). Eggs from Nematodirus sp, Trichuris sp and Moniezia sp, as well as Eimeria oocysts, were only occasionally seen in small number, and are not discussed in this paper.
Haemonchus contortus was the most abundant strongylid nematode species overall (p < 0.0001, Kruskal-Wallis, followed by Dunn's multiple comparison) and was detected in all but 5 flocks in Alberta. Teladorsagia circumcincta was the next most abundant species followed by T. colubriformis. Teladorsagia circumcincta and T. colubriformis abundance was significantly different (p < 0.0001) from the abundance of all the other species identified (p values ranged from <0.0001 to > 0.9999). There was no significant difference in abundance between T. circumcincta and T. colubriformis (p > 0.9999, Fig. 1B and C). Of the 92 ewe flocks surveyed, H. contortus, T. circumcincta and T. colubriformis were the most abundant species in 69, 14 and 7 flocks respectively. In two flocks, Trichostrongylus vitrinus was the most abundant species (Farm 5 with 26% and Farm 90 with 50%, both in AB). Other species such as Chabertia ovina, Oesophagostomum venulosum and Trichostrongylus axei also had a high relative abundance in a few flocks (Fig. 1C). There was significant difference in beta-diversity of T. colubriformis between BC and AB (p = 0.02). Supplementary Table 6 shows comparison of beta diversity of H. contortus, T. circumcincta and T. colubriformis between provinces.
3.2. ITS-2 rDNA nemabiome metabarcoding as a tool to assess the effectiveness of producer-applied anthelmintic treatment against specific gastrointestinal nematode species
From the 92 farms that participated in the nematode species abundance survey, we investigated the effectiveness of producer-applied anthelmintic treatments for 56 western Canadian sheep flocks. Fecal egg counts were determined for pooled samples collected by producers. The percentage post-treatment reduction in total strongyle fecal egg counts was less than 95% in 32/32 flocks, 11/11 and 10/13 for ivermectin, fenbendazole and albendazole respectively (Fig. 2A, grey violin plots and Supplementary Fig. 1, upper histogram bars). The overall mean strongyle fecal egg count reduction after treatment was 41% (median 38%, range 0–91%), 41% (median 60% range 0–85%) and 59% (median 65% range 0–100%) for ivermectin, fenbendazole and albendazole respectively (Supplementary Table 5A).
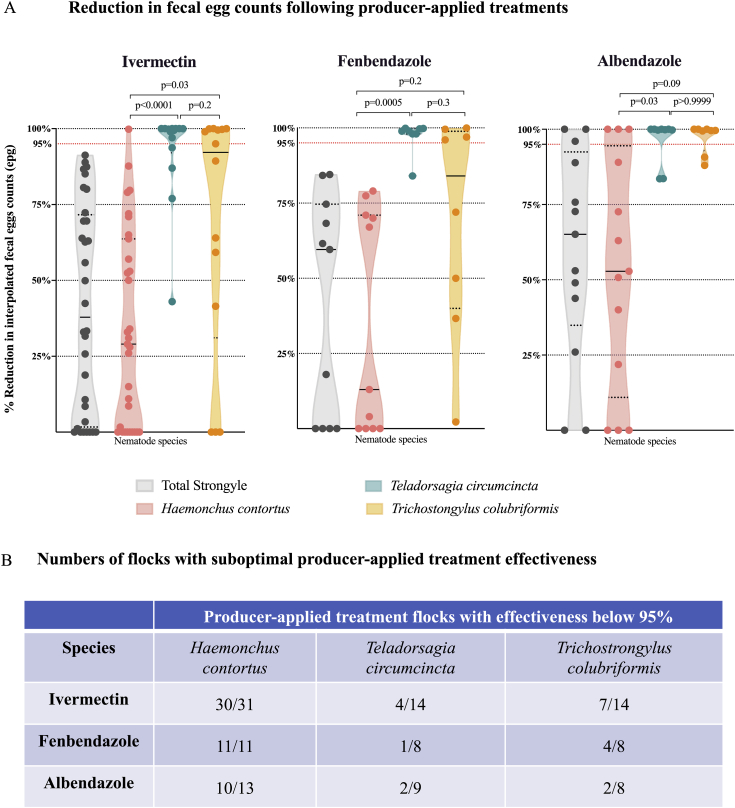
Figure 2.
Use of ITS-2 rDNA nemabiome metabarcoding to determine effectiveness of producer-applied anthelmintic treatments against specific gastrointestinal nematode species in western Canada.
Panel A. Reductions in fecal egg counts following producer-applied treatments interpolated from ITS-2 rDNA nemabiome data for specific gastrointestinal nematode species.
Relative species abundance, determined by ITS-2 rDNA nemabiome metabarcoding in each sample pre- and post-treatment, was used to interpolate species-specific eggs counts and treatment effectiveness for each group in 56 producer-applied treatment samples. Violin plots show drug effectiveness based on the reduction of total strongyle-type fecal egg counts (grey) and the effectiveness against Haemonchus contortus (pink), Teladorsagia circumcincta (teal) and Trichostrongylus colubriformis (orange) for ivermectin, fenbendazole and albendazole, respectively. Dotted lines represent quartiles and solid lines represent median of each violin plot. Statistical significance of the differences between the groups was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test using GraphPad Prism version 8.1.0 for macOS, GraphPad Software, San Diego, California USA, www.graphpad.com.
Panel B. Numbers of flocks with suboptimal producer-applied treatment effectiveness . Each cell in the table shows the number of flocks with specific treatment effectiveness below 95%/total number of flocks tested with ivermectin, fenbendazole and albendazole against Haemonchus contortus, Teladorsagia circumcincta and Trichostrongylus colubriformis respectively.
We used the pre- and post-treatment ITS-2 rDNA nemabiome metabarcoding data to interpolate species-specific fecal egg count values and percentage fecal egg count reductions for each of the three major GIN species for producer-applied ivermectin, fenbendazole and albendazole treatments (Fig. 2A and B, and Supplementary Table 5A). All samples with at least 5% abundance of each species pre-treatment were included, which equates to at least 10 larvae in each 200 larvae pooled sample or 25 in samples where 500 larvae were used being taxonomically classified as H. contortus, T. circumcincta, or T. colubriformis. In the case of ivermectin, the effectiveness of producer-applied treatments was below 95% in 30/31 flocks, 4/14 flocks and 7/14 flocks for H. contortus, T. circumcincta and T. colubriformis respectively. In the case of fenbendazole, the effectiveness of producer-applied treatments was below 95% in 11/11 flocks, 1/8 flocks and 4/8 flocks for H. contortus, T. circumcincta and T. colubriformis respectively. In the case of albendazole, the effectiveness of producer-applied treatments was below 95% in 10/13 flocks, 2/9 flocks and 2/8 flocks for H. contortus, T. circumcincta and T. colubriformis respectively (Fig. 2B).
Ivermectin was less effective against H. contortus compared to the other two species (p < 0.0001 and p < 0.03 for T. circumcincta and T. colubriformis, respectively, Kruskal-Wallis followed by Dunn's multiple comparison test) (Fig. 2). Treatment effectiveness with fenbendazole was significantly higher against T. circumcincta compared to H. contortus (p < 0.0005) (Fig. 2). Treatment effectiveness with albendazole was significantly lower against H. contortus compared to T. circumcincta (p < 0.03) (Fig. 2). The total strongyle mean % effectiveness and standard deviation of these drugs against the three species are summarized in Fig. 2A and B and Supplementary Table 5A. Other nematode species were also present after treatment with ivermectin and benzimidazoles at a lower abundance, such as O. venulosum, T. vitrinus and T. axei (Supplementary Fig. 1).
3.3. Integration of ITS-2 rDNA nemabiome metabarcoding and fecal egg count reduction testing (FECRT) for the diagnosis of anthelmintic resistance
We investigated the presence of anthelmintic resistant gastrointestinal trichostrongylid nematodes against several different anthelmintic drugs by performing FECRT on fecal samples from visited farms. Based on the WAAVP criteria, resistance is present if the percentage reduction is less than <95% and the lower 95% confidence interval is less than 90%. Confidence intervals were calculated according to equation 1 in Levecke et al. (2018). Anthelmintic resistant strongylid nematodes were determined to be present against ivermectin, fenbendazole, moxidectin and levamisole on 10/13 flocks, 9/13 flocks, 0/6 flocks and 1/4 flocks, respectively (Fig. 3A and Supplementary Fig. 2).
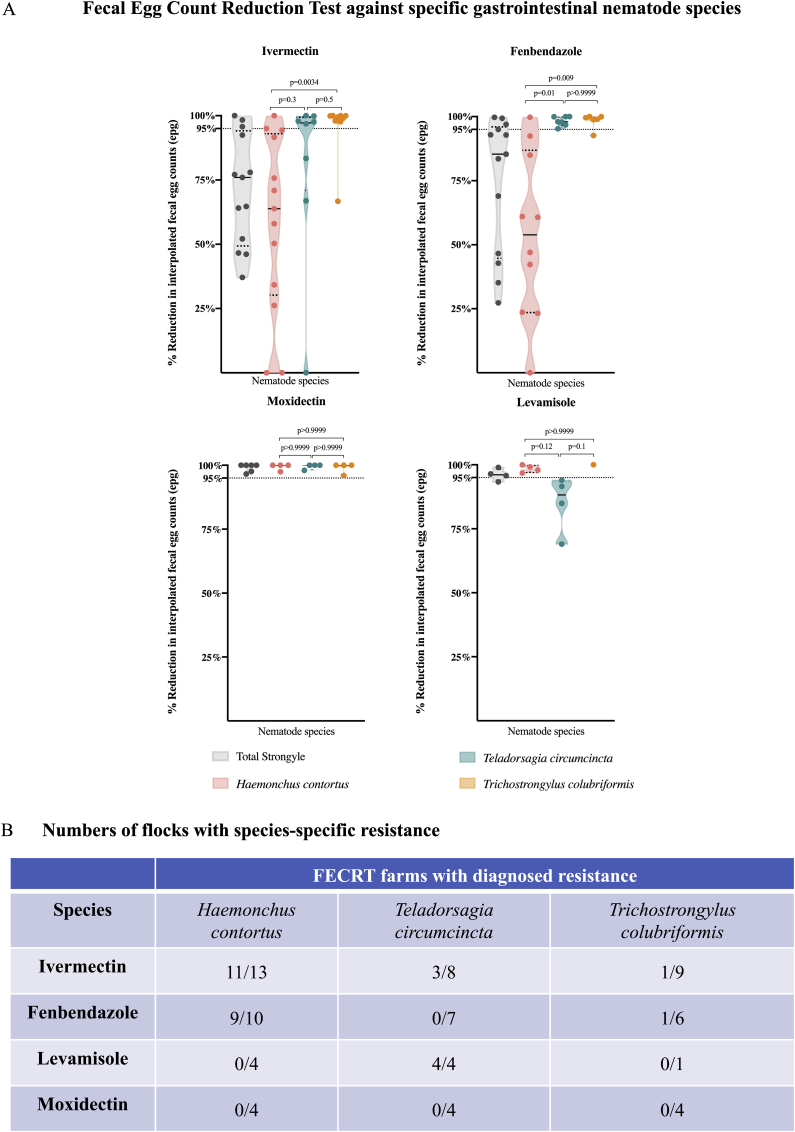
Fig. 3.
Integration of ITS-2 rDNA nemabiome metabarcoding with formal Fecal Egg Count Reduction Tests (FECRT) to investigate anthelmintic resistance against specific gastrointestinal nematode species in western Canada.
Panel A. ITS-2 rDNA nemabiome data for specific gastrointestinal nematode species. Relative species abundance in each sample pre- and post-treatment, as determined by ITS-2 rDNA nemabiome metabarcoding, was used to interpolate species-specific fecal egg counts and to show Fecal Egg Count Reduction Test treatment efficacy data for individual gastrointestinal nematode species. Violin plots show drug efficacy based on the reduction of total strongyle-type fecal egg counts (grey), and efficacy against Haemonchus contortus (pink), Teladorsagia circumcincta (teal) and Trichostrongylus colubriformis (orange) for ivermectin, fenbendazole, levamisole and moxidectin, respectively. Not all anthelmintics were tested in all farms. Dotted lines represent quartiles and solid lines represent median of each violin plot. Statistical significance of the differences between the groups was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test using GraphPad Prism version 8.1.0 for macOS, GraphPad Software, San Diego, California USA, www.graphpad.com.
Panel B. Numbers of flocks with species-specific efficacy below 95% from Fecal Egg Count Reduction Test (FECRT) data. Each cell in the table shows number of flocks with specific treatment effectiveness below 95%/total number of flocks tested with ivermectin, fenbendazole, levamisole and moxidectin against Haemonchus contortus, Teladorsagia circumcincta and Trichostrongylus colubriformis respectively.
We used ITS-2 rDNA nemabiome metabarcoding data from the FECRT pre- and post-treatment samples to interpolate species-specific fecal egg counts and fecal egg count reductions for H. contortus, T. circumcincta and T. colubriformis (Fig. 3A and B and Supplementary Table 5B). All samples with at least 5% abundance of each species in the pre-treatment were included, which equates to at least 10 larvae in each 200 larvae or 25 larvae in each 500 larvae pooled sample being taxonomically classified as H. contortus, T. circumcincta, or T. colubriformis (Supplementary Table 3 shows the number of larvae included in each lysate preparation, samples not included in the specific treatment efficacy dataset are marked with “*“). In the case of ivermectin, 11/13 flocks, 3/8 flocks and 1/9 had a fecal egg count reduction less than 95% for H. contortus, T. circumcincta and T. colubriformis respectively. In the case of fenbendazole, 9/10 flocks, 0/7 flocks and 1/6 had a less than 95% fecal egg count reduction for H. contortus, T. circumcincta and T. colubriformis respectively. In the case of levamisole, 0/4 flocks, 4/4 flocks and 0/1 had a less than 95% fecal egg count reduction for H. contortus, T. circumcincta and T. colubriformis respectively. In the case of moxidectin, no flock had a less than 95% fecal egg count reduction for any of the species studied (Fig. 3B). Ivermectin treatment efficacy was significantly higher (p < 0.0034, Kruskal-Wallis, followed by Dunn's multiple comparison test) against T. colubriformis compared to H. contortus. Efficacy of fenbendazole treatment was significantly lower against H. contortus, compared to the other two species (p < 0.01 and p = 0.009). There was no difference between treatment efficacy with levamisole and moxidectin against the three species.
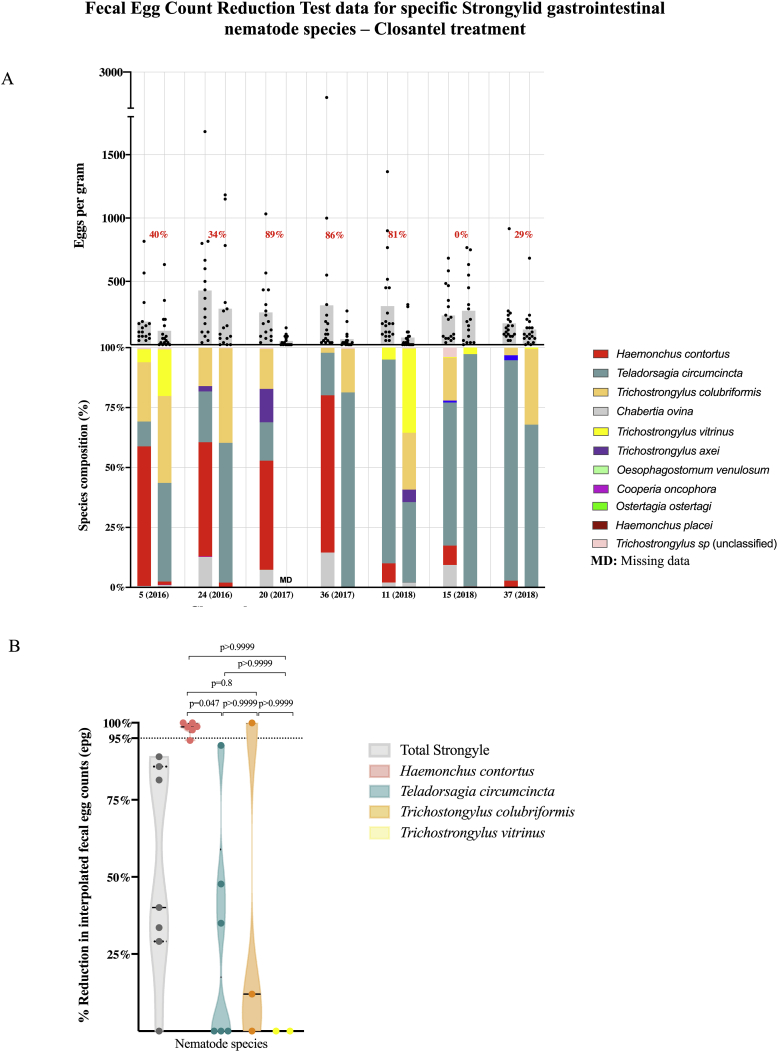
We also undertook FECRT for closantel, on seven flocks (Fig. 4A and B). Based on total fecal egg count data, there was a <95% reduction of fecal egg counts (<90% lower 95% confidence interval) on 7/7 flocks. We used ITS-2 rDNA nemabiome metabarcoding data from the pre- and post-treatment FECRT samples from six flocks to interpolate species-specific fecal egg counts and fecal egg count reductions for H. contortus, T. circumcincta, T. colubriformis and T. vitrinus. In 5/6 flocks there was a greater than 95% reduction in interpolated fecal egg counts for H. contortus (Fig. 4, panels A and B). In contrast, 6/6 flocks, 2/3 flocks and 2/2 had a less than 95% fecal egg count reduction for T. circumcincta, T. colubriformis and T. vitrinus respectively. Closantel treatment efficacy was significantly higher (p < 0.047, Kruskal-Wallis, followed by Dunn's multiple comparison test) against H. contortus compared to T. circumcincta.
Fig. 4.
Integration of ITS-2 rDNA nemabiome metabarcoding with formal Fecal Egg Count Reduction Test to investigate closantel efficacy against specific gastrointestinal nematode species.
Panel A. Fecal egg count and ITS-2 rDNA nemabiome data for Fecal Egg Count Reductions Tests (FECRT) conducted to assess closantel efficacy. Pre- and post-treatment fecal egg counts, from Fecal Egg Count Reduction Tests (FECRT), are represented as scatter dot plots (grey). Individual dots represent egg counts of individual animals in each group, pre- and post-treatment. Grey bars represent mean egg counts of each group of animals, pre- and post-treatment. Numbers above scatter dot plots are the overall percentage fecal egg count reduction comparing pre and post treatment egg counts with closantel, calculated according to Levecke et al. (2018) (green: susceptible, red: resistant). Relative species composition (%) were determined by ITS-2 rDNA nemabiome metabarcoding of at least 200 first stage larvae in each pre- and post-treatment sample, represented at the bottom stacked bars. Panel B. Fecal Egg Count Reduction Test (FECRT) data for closantel interpolated from ITS-2 rDNA nemabiome data for specific gastrointestinal nematode species.
Relative species abundance in each sample pre- and post-treatment, as determined by ITS-2 rDNA nemabiome metabarcoding, was used to interpolate species-specific fecal egg counts and treatment efficacy for closantel groups from Fecal Egg Count Reduction Test data. Violin plots show drug efficacy based on the reduction of total strongyle-type egg counts (grey) and the efficacy against Haemonchus contortus (pink), Teladorsagia circumcincta (teal), Trichostrongylus colubriformis (orange) and Trichostrongylus vitrinus (yellow) for Closantel. Dotted lines represent quartiles and solid lines represent median of each violin plot. Statistical significance of the differences between the groups was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test using GraphPad Prism version 8.1.0 for macOS, GraphPad Software, San Diego, California USA, www.graphpad.com.
4. Discussion
4.1. ITS-2 rDNA nemabiome metabarcoding reveals mixed gastrointestinal nematode species infections with a predominance of H. contortus in western Canada
There has been no published data on gastrointestinal species prevalence, infection intensities and anthelmintic resistance status on sheep farms in western Canada for over 15 years, with only two small scale studies published in the last 40 years (Moteane et al., 1979; Colwell et al., 2002). Over the last 10–15 years there have been an increasing number of anecdotal reports from sheep producers and veterinarians of clinical haemonchosis in spite of widespread use of both ivermectin and benzimidazole drugs. Consequently, the situation in western Canada provided us with an opportunity to apply ITS-2 rDNA nemabiome metabarcoding to produce a precise contemporary picture of infection intensities and anthelmintic resistance status of specific ovine GIN species. Of the 92 ewe flocks surveyed between 2014 and 2018, infection intensities were high in many flocks, with 44 flocks (48%) having FECs of >500 epg pre-treatment.
Haemonchus contortus, T. circumcincta and T. colubriformis were the most abundant species with a mean overall abundance of 64%, 18% and 13% respectively. Haemonchus contortus was the species with highest infection intensity in 69 of the flocks, T. circumcincta in 14 of the flocks and T. colubriformis in 7 of the flocks. The high infection intensity and predominance of H. contortus was consistent with the frequent anecdotal reports of clinical haemonchosis but is in marked contrast to the previously available data. In the two previous necropsy-based studies, H. contortus infection intensities were generally low with Teladorsagia, Trichostrongylus and Nematodirus being the predominant genera reported (Moteane et al., 1979; Colwell et al., 2002). H. contortus was not reported by Colwell et al. (2002), and Moteane et al. (1979) reported only very low infection intensities with mean abomasal adult worm counts of 62 (range 4–600). Although these previous studies were small, the results support the suggestion that H. contortus prevalence and infection intensities have increased in the region in the last 15–20 years. The high prevalence and infection intensities of H. contortus that we have found in western Canada is similar to recent work from other regions with temperate climates and cold winters such as eastern Canada (Barrère et al., 2013; Falzon et al., 2013). The ability of H. contortus to thrive in these regions, despite originally being a tropical/sub-tropical parasite (O'Connor et al., 2006), is likely due to its ability to undergo arrested larval development and survive inside the host during the cold dry winters (Waller et al., 2004, 2006; O'Connor et al., 2006; Abbott et al., 2012; Falzon et al., 2014; Westers et al., 2016). The reason for an increase in H. contortus in western Canada over the last 15–20 years could be due to several factors, including introduction and spread by imported sheep, anthelmintic resistance compromising control and climate change. Further research will be needed to determine the relative importance of these and other factors.
Although H. contortus was found to be the predominant species in western Canada, T. circumcincta and T. colubriformis were also present at relatively high abundance on several flocks in the region. This suggests that the risk of diarrheal disease associated with parasitic gastroenteritis should also be considered. It is worth contrasting these results with a previous study in eastern Canada where H. contortus comprised over 80% species abundance in 18/18 flocks pre-anthelmintic treatment (Falzon et al., 2013). Whether the higher levels of T. circumcincta and T. colubriformis reflects regional differences in species abundance or just different methodological or sampling strategies remains to be determined.
4.2. Integration of ITS-2 rDNA nemabiome metabarcoding with fecal egg count reduction testing reveals benzimidazole and ivermectin resistance to be widespread in H. contortus but less frequent in T. circumcincta and T. colubriformis in western Canada
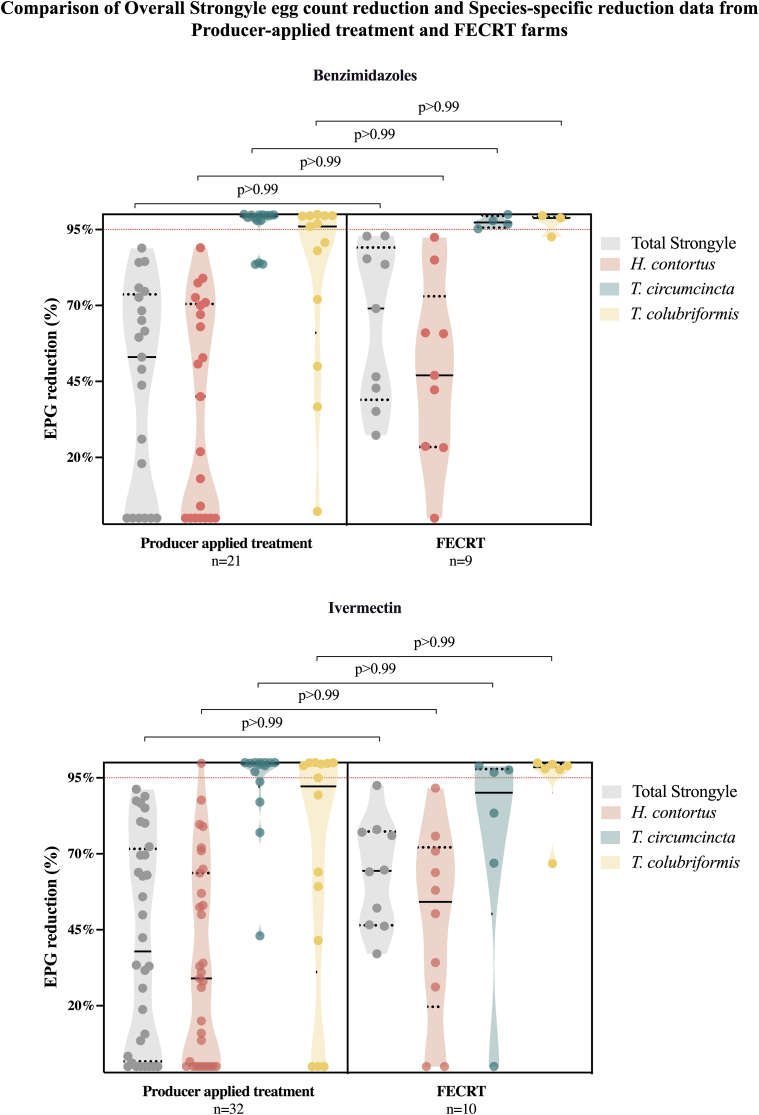
Both producer-applied anthelmintic treatment efficacy testing and formal Fecal Egg Count Reduction Testing (FECRT) shows that ivermectin and benzimidazole resistance is widespread in strongylid nematode species in western Canada. The integration of ITS-2 rDNA metabarcoding with these methods provided an additional layer of information revealing that resistance to both these drugs is essentially ubiquitous in H. contortus but still relatively rare for the second two most important GIN parasite species T. circumcincta and T. colubriformis. Caution is needed when interpreting the results of fecal egg count reduction data from producer-applied anthelmintic treatments, since treatments may be sub-optimal not just due to anthelmintic resistance but also to other factors such as incorrect dosing of animals, use of expired drug, failure to collect samples from the same animals and other factors. However, it is interesting to note that there was no statistically significant difference between mean percentage fecal egg count reduction values for total strongyle eggs or the species-specific interpolated egg counts when the two data sets were compared (Fig. 5). This argues that the lack of efficacy in the larger dataset of producer-applied treatments (56 farms) is predominantly due to anthelmintic resistance rather than other methodological factors. This in turn suggests that ivermectin and fenbendazole resistance in H. contortus are truly widespread in the region. Nevertheless, there is a general trend for the mean fecal egg count reductions to be lower for producer-applied treatments than for the FECRT data and so caution is still needed when interpreting the details of producer applied treatment data (Fig. 5).
Fig. 5.
Comparison of producer-applied treatment effectiveness and Fecal Egg Count Reduction Test data . Relative species abundance determined by ITS-2 rDNA nemabiome metabarcoding in each sample pre- and post-treatment was used to interpolate species-specific fecal egg count reduction of Haemonchus contortus (pink), Teladorsagia circumcincta (teal), and Trichostrongylus colubriformis (orange), in producer-applied treatment and Fecal Egg Count Reduction Test flocks, for both benzimidazole and ivermectin treatments. Solid lines represent the mean fecal egg count reduction, dotted red lines mark the 95% threshold. Statistical significance of the differences between the groups was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test using GraphPad Prism version 8.1.0 for macOS, GraphPad Software, San Diego, California USA, www.graphpad.com. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The widespread and, in many cases high level, of resistance to both ivermectin and benzimidazole in H. contortus is both interesting and concerning given the nature of anthelmintic use in western Canada. Although these two drug classes have been the mainstay of ovine parasite control in western Canada for over 30 years, ivermectin is the only licensed anthelmintic for use in sheep in Canada, while fenbendazole and albendazole are licensed for use in cattle and often used in sheep extra-label (Supplementary Table 1). In addition, most producers only treat ewes and lambs once a year and yet the levels of resistance we have detected are as high, or higher, than in many countries with more frequent anthelmintic use. For example, in a United Kingdom study published in 2012 from 188 farms surveyed, only 22 had confirmed anthelmintic resistance and none had resistance to ivermectin (Burgess et al., 2012). There are a number of potential reasons for this high level of benzimidazole and ivermectin resistance in western Canada in spite of mainly once yearly treatments of ewes and lambs. Since H. contortus does not overwinter on pastures in regions with cold winters, the early spring is a time when most of the parasite population is within the host and refugia is low (Waller et al., 2004; Troell et al., 2005; Grosz et al., 2013; Falzon et al., 2014). Treatment around lambing time has been previously shown to be associated with high selective pressure in the UK (Leathwick et al., 1995). This is particularly the case when the whole flock is treated with anthelmintic at the same time which has been relatively common in western Canada. The role of low refugia during early spring treatments of ewes may also, at least partially, explain why anthelmintic resistance is less advanced in T. circumcincta and T. colubriformis than H. contortus. The former two parasites have been shown to overwinter on pastures to some extent in eastern Canada which would increase refugia and reduce selection (Falzon et al., 2014). There are also a variety of other potential risk factors relevant to western Canadian sheep flocks including animal movement and importing stock from places such as the United States, where there are high levels of both benzimidazole and ivermectin resistance in H. contortus together with the lack of rigorous quarantine procedures for recently introduced animals. This discussion underlines how the factors driving anthelmintic resistance are much more complex than simply the frequency of anthelmintic treatment (Learmount et al., 2016b).
4.3. Integration of ITS-2 rDNA nemabiome metabarcoding with fecal egg count reduction testing to assess moxidectin, levamisole and closantel resistance in western Canada
Given the high level of resistance to ivermectin and benzimidazole drugs in H. contortus and emerging resistance in T. circumcincta and T. colubriformis in western Canada, there is a need for judicious use of other drugs against which resistance is less common. Consequently, we also used FECRT/nemabiome metabarcoding to assess resistance to several other anthelmintic that are less commonly used in western Canada. Moxidectin has been used extensively in the United States but less so in western Canada to date. We found moxidectin to be highly effective against all the major trichostrongylid species present in all four flocks in which it was tested (Fig. 3). We found levamisole efficacy, based on total fecal egg counts, high for all four flocks tested. However, although the species-specific fecal egg count reductions, interpolated from the nemabiome data, indicated high efficacy against H. contortus, the efficacy against T. circumcincta was less than 95% on all four flocks. Although the total strongyle fecal egg count reductions were 93%, 99%, 96% and 96% (Supplementary Table 5), the interpolated efficacy for T. circumcincta was only 69%, 92%, 85% and 94% efficacy. Levamisole was originally reported to have high efficacy against both H. contortus and T. circumcincta, including against inhibited L4 larvae (McKenna, 1974; Andrews, 2000) and so this sub-optimal efficacy specifically against T. circumcincta may suggest the early emergence of resistance in this species. Evidence of T. circumcincta resistant to levamisole have been reported in T. circumcincta from multiple countries, including Greece, New Zealand, Brazil, Ireland and Spain (Cezar et al., 2010; Martínez-Valladares et al., 2012; Geurden et al., 2014; Keegan et al., 2015; Leathwick et al., 2015). Levamisole was licensed for use in sheep in Canada until 2005 and is still used to some extent in eastern Canada, and so some drug selection with levamisole has occurred. The integration of ITS-2 nemabiome metabarcoding into the FECRT suggests the early emergence of levamisole resistance in T. circumcincta which was not detected as an overall loss of efficacy in the FECRT data alone. Sequencing of the T. circumcincta acr-8 genre from these and other populations would be useful to help confirm levamisole resistance and monitor further development.
In 2016, closantel (Flukiver©) was licensed for sheep in Canada providing an important new option for parasite control. The efficacy of this drug, based on the FECRT data on western Canadian sheep farms, varied between 0% and 89%. Since the spectrum of activity of this drug is restricted to blood-feeding parasites such as H. contortus, it is not possible to assess whether this variable efficacy is due to closantel resistance using the FECRT based on total strongyle egg counts alone because many of the farms comprise mixed species infections. Integration of the ITS-2 rDNA nemabiome data revealed that the drug was still highly effective against its target parasite H. contortus in all cases and the apparent low efficacy was due to the narrow spectrum of activity of the drug and the mixed species composition of the parasite communities. This provides a good illustration of how the integration of nemabiome metabarcoding into the FECRT can provide critical information to the diagnosis and confirmation of anthelmintic resistance.
5. The value of nemabiome-metabarcoding enhanced anthelmintic efficacy testing
This work illustrates the power of integrating ITS-2 rDNA nemabiome metabarcoding into both routine pre- and post-treatment fecal egg count efficacy testing and the Fecal Egg Count Reduction Test (FECRT). An example of the benefit is in how the approach revealed benzimidazole and ivermectin resistance to be at an advanced stage in H. contortus but a much earlier stage in T. circumcincta and T. colubriformis in western Canada. This opens up the possibility of a more targeted use of these older drugs whilst reserving newer, often more expensive, drugs for when they are specifically needed. For example, effective treatment outcomes could be achieved on some farms by closantel alone and on other farms by a combination of closantel with fenbendazole or ivermectin. However, on some farms, other drugs such as moxidectin or abamectin/derquantel would be required. Nemabiome metabarcoding-enhanced anthelmintic resistance testing could be used to direct such rational drug choices in a way not possible by fecal egg count-based efficacy testing alone. The ITS-2 nemabiome metabarcoding also revealed evidence for the early emergence of levamisole resistant in T. circumcincta which was not apparent by the FECRT data alone. It also enabled the status of closantel resistance to be assessed, which again was not possible using the FECRT based on total fecal egg counts alone. The merits of nemabiome metabarcoding, relative to other methods used to quantify relative species abundance, has been discussed in detail elsewhere (Avramenko et al., 2015, 2017; Redman et al., 2019). Important key benefits include greater precision (assignment to species level), flexibility (no need to anticipate the species present or have pre-validated protocols for each individual species) and its scalability for large surveillance and research projects. The current main limitation for routine diagnostic use on individual farms is the need to batch large numbers of samples on a single sequencing run to make the cost per sample economically viable. In spite of this limitation, the approach has significant potential for routine diagnostic use in larger diagnostic facilities processing large numbers of samples. Also, sequencing platforms and costs are rapidly changing and so we suggest that nemabiome metabarcoding methodology will continue to evolve for more flexible smaller scale use in the future.
Declaration of competing interest
None.
Acknowledgements
We thank summer students Amanda Kuzyk, Amelia Whitelaw, Colby Morris, Elspeth Yates, Emily Dorey, Erica Ward, Erika Brandson, Jeff Lees, Jesse Pawlak and Keeley Haight, and the laboratory technician Bruna Palmeira, for collaborating with sample collection and processing. We also thank the dedicated sheep producers who volunteered to participate in this project. This work was supported by an Alberta Innovates Technology Futures scholarship to (CQ), NSERC Discovery grant RGPIN-2015-03976 (JSG) and Alberta Agriculture and Forestry (AAF) grants 2015R029R and 2017R021R(JSG and ML), University of Calgary, Canada, NSERC-CREATE Host-Parasite Interactions (HPI) graduate training program and the Faculty of Veterinary Medicine Summer Undergraduate Research Program (SURE). Many thanks to Brent Wagner, University of Saskatchewan for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2020.09.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abbott K., Taylor M., Stubbings L. a. Sustainable worm control strategies for sheep - a technical manual for veterinary surgeons and advisors. Scops. 2012:27–32. [Google Scholar]
- Álvarez-Sánchez M.A., Pérez-García J., Cruz-Rojo M.A., Rojo-Vázquez F.A. Real time PCR for the diagnosis of benzimidazole resistance in trichostrongylids of sheep. Vet. Parasitol. 2005;129:291–298. doi: 10.1016/j.vetpar.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Amarante A.F.T. Why is it important to correctly identify Haemonchus species? Rev. Bras. Parasitol. Vet. 2011;20:263–268. doi: 10.1590/s1984-29612011000400002. [DOI] [PubMed] [Google Scholar]
- Andrews S.J. The efficacy of levamisole, and a mixture of oxfendazole and levamisole, against the arrested stages of benzimidazole-resistant Haemonchus contortus and Ostertagia circumcincta in sheep. Vet. Parasitol. 2000;88:139–146. doi: 10.1016/S0304-4017(99)00195-8. [DOI] [PubMed] [Google Scholar]
- Avramenko R.W., Bras A., Redman E.M., Woodbury M.R., Wagner B., Shury T., Liccioli S., Windeyer M.C., Gilleard J.S. High species diversity of trichostrongyle parasite communities within and between Western Canadian commercial and conservation bison herds revealed by nemabiome metabarcoding. Parasites Vectors. 2018;11:1–13. doi: 10.1186/s13071-018-2880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramenko R.W., Redman E.M., Lewis R., Bichuette M.A., Palmeira B.M., Yazwinski T.A., Gilleard J.S. The use of nemabiome metabarcoding to explore gastro-intestinal nematode species diversity and anthelmintic treatment effectiveness in beef calves. Int. J. Parasitol. 2017;47:893–902. doi: 10.1016/j.ijpara.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Avramenko R.W., Redman E.M., Lewis R., Yazwinski T.A., Wasmuth J.D., Gilleard J.S. Exploring the gastrointestinal “nemabiome”: deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PloS One. 2015;10:1–18. doi: 10.1371/journal.pone.0143559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone C.D., Wit J., Hoberg E.P., Gilleard J.S., Zarlenga D.S. Wild ruminants as reservoirs of domestic livestock gastrointestinal nematodes. Vet. Parasitol. 2020;279 doi: 10.1016/j.vetpar.2020.109041. [DOI] [PubMed] [Google Scholar]
- Barrère V., Falzon L.C., Shakya K.P., Menzies P.I., Peregrine A.S., Prichard R.K. Assessment of benzimidazole resistance in Haemonchus contortus in sheep flocks in Ontario, Canada: comparison of detection methods for drug resistance. Vet. Parasitol. 2013;198:159–165. doi: 10.1016/j.vetpar.2013.07.040. [DOI] [PubMed] [Google Scholar]
- Bivand R., Keitt T., Rowlingson B. Abstraction Library; 2019. Rgdal: Bindings for the “Geospatial” Data. [Google Scholar]
- Burgess C.G.S., Bartley Y., Redman E., Skuce P.J., Nath M., Whitelaw F., Tait A., Gilleard J.S., Jackson F. A survey of the trichostrongylid nematode species present on UK sheep farms and associated anthelmintic control practices. Vet. Parasitol. 2012;189:299–307. doi: 10.1016/j.vetpar.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Čerňanská D., Várady M., Čorba J. A survey on anthelmintic resistance in nematode parasites of sheep in the Slovak Republic. Vet. Parasitol. 2006;135:39–45. doi: 10.1016/j.vetpar.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Cezar A.S., Toscan G., Camillo G., Sangioni L.A., Ribas H.O., Vogel F.S.F. Multiple resistance of gastrointestinal nematodes to nine different drugs in a sheep flock in southern Brazil. Vet. Parasitol. 2010;173:157–160. doi: 10.1016/j.vetpar.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Chandra S., Prasad A., Sankar M., Yadav N., Dalal S. Molecular diagnosis of benzimidazole resistance in Haemonchus contortus in sheep from different geographic regions of North India. Vet. 2014;7:337–341. doi: 10.14202/vetworld.2014.337-341. World. [DOI] [Google Scholar]
- Chandrawathani P., Waller P.J., Adnan M., Höglund J. Evolution of high-level, multiple anthelmintic resistance on a sheep farm in Malaysia. Trop. Anim. Health Prod. 2003;35:17–25. doi: 10.1023/a:1022023620599. [DOI] [PubMed] [Google Scholar]
- Coles G.C., Bauer C., Borgsteede F.H.M., Geerts S., Klei T.R., Taylor M.A., Waller P.J. World Association for the Advancement of Veterinary Parasitology (W.A.A.V.P.) methods for the detection of anthelmintic resistance in nematodes of veterinary importance. Vet. Parasitol. 1992;44:35–44. doi: 10.1016/0304-4017(92)90141-U. [DOI] [PubMed] [Google Scholar]
- Coles G.C., Jackson F., Pomroy W.E., Prichard R.K., von Samson-Himmelstjerna G., Silvestre A., Taylor M.A., Vercruysse J. The detection of anthelmintic resistance in nematodes of veterinary importance. Vet. Parasitol. 2006;136:167–185. doi: 10.1016/j.vetpar.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Colwell D.D., Goater C.P., Jacobson K.M. Prevalence and intensity of gastrointestinal nematodes in slaughter lambs from central Alberta. Can. Vet. J. = La Rev. Vet. Can. 2002;43:775–777. [PMC free article] [PubMed] [Google Scholar]
- Domke A.V.M., Chartier C., Gjerde B., Höglund J., Leine N., Vatn S., Stuen S. Prevalence of anthelmintic resistance in gastrointestinal nematodes of sheep and goats in Norway. Parasitol. Res. 2012;111:185–193. doi: 10.1007/s00436-012-2817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drudge J.H., Szanto J., Wyant Z.N., Elam G. Field studies on parasite control in sheep: comparison of thiabendazole, ruelene and phenothiazine. Am. J. Vet. Res. 1964;25:1512–1518. [PubMed] [Google Scholar]
- Elmahalawy S.T., Halvarsson P., Skarin M., Höglund J. Droplet digital polymerase chain reaction (ddPCR) as a novel method for absolute quantification of major gastrointestinal nematodes in sheep. Vet. Parasitol. 2018;261:1–8. doi: 10.1016/j.vetpar.2018.07.008. [DOI] [PubMed] [Google Scholar]
- Falzon L.C., Menzies P.I., Shakya K.P., Jones-Bitton A., Vanleeuwen J., Avula J., Stewart H., Jansen J.T., Taylor M.A., Learmount J., Peregrine A.S. Anthelmintic resistance in sheep flocks in Ontario, Canada. Vet. Parasitol. 2013;193:150–162. doi: 10.1016/j.vetpar.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Falzon L.C., Menzies P.I., VanLeeuwen J., Shakya K.P., Jones-Bitton A., Avula J., Jansen J.T., Peregrine A.S. Pilot project to investigate over-wintering of free-living gastrointestinal nematode larvae of sheep in Ontario, Canada. Can. Vet. J. 2014;55:749–756. [PMC free article] [PubMed] [Google Scholar]
- Gárcia C.M.B., Sprenger L.K., Ortiz E.B., Molento M.B. First report of multiple anthelmintic resistance in nematodes of sheep in Colombia. An. Acad. Bras. Cienc. 2016;88:397–402. doi: 10.1590/0001-3765201620140360. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., Bott N.J., Chilton N.B., Hunt P., Beveridge I. Toward practical, DNA-based diagnostic methods for parasitic nematodes of livestock — bionomic and biotechnological implications. Biotechnol. Adv. 2008;26:325–334. doi: 10.1016/j.biotechadv.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Geurden T., Hoste H., Jacquiet P., Traversa D., Sotiraki S., Frangipane di Regalbono A., Tzanidakis N., Kostopoulou D., Gaillac C., Privat S., Giangaspero A., Zanardello C., Noé L., Vanimisetti B., Bartram D. Anthelmintic resistance and multidrug resistance in sheep gastro-intestinal nematodes in France, Greece and Italy. Vet. Parasitol. 2014;201:59–66. doi: 10.1016/j.vetpar.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Grosz D.D., Eljaki A.A., Holler L.D., Petersen D.J., Holler S.W., Hildreth M.B. Overwintering strategies of a population of anthelmintic-resistant Haemonchus contortus within a sheep flock from the United States Northern Great Plains. Vet. Parasitol. 2013;196:143–152. doi: 10.1016/j.vetpar.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Keegan J.D., Keane O.M., Farrell L., Byrne W., De Waal T., Good B. Characterisation of ivermectin and multi-drug resistance in two field isolates of Teladorsagia circumcincta from Irish sheep flocks. Vet. Parasitol. Reg. Stud. Reports. 2015;1(2):3–9. doi: 10.1016/j.vprsr.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Learmount J., Stephens N., Boughtflower V., Barrecheguren A., Rickell K. The development of anthelmintic resistance with best practice control of nematodes on commercial sheep farms in the UK. Vet. Parasitol. 2016;229:9–14. doi: 10.1016/j.vetpar.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Learmount J., Stephens N., Boughtflower V., Barrecheguren A., Rickell K., Massei G., Taylor M. Three-year evaluation of best practice guidelines for nematode control on commercial sheep farms in the UK. Vet. Parasitol. 2016;226:116–123. doi: 10.1016/j.vetpar.2016.06.037. [DOI] [PubMed] [Google Scholar]
- Leathwick D.M., Ganesh S., Waghorn T.S. Evidence for reversion towards anthelmintic susceptibility in Teladorsagia circumcincta in response to resistance management programmes. Int. J. Parasitol. Drugs Drug Resist. 2015;5:9–15. doi: 10.1016/j.ijpddr.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leathwick D.M., Vlassofft A., Agricultural F.H., Zealand N., Animal W., Hutt U., Zealand N., Agriculture C., Centre S., Bag P., Zealand N. A model for nematodiasis in New Zealand Lambs : the effect of drenching regime and grazing management on the development of anthelmintic resistance. Int. J. Parasitol. 1995;25:1479–1490. doi: 10.1016/0020-7519(95)00059-3. [DOI] [PubMed] [Google Scholar]
- Levecke B., Kaplan R.M., Thamsborg S.M., Torgerson P.R., Vercruysse J., Dobson R.J. How to improve the standardization and the diagnostic performance of the fecal egg count reduction test? Vet. Parasitol. 2018;253:71–78. doi: 10.1016/j.vetpar.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Lichtenfels J.R., Pilitt P.A. Synlophe patterns of the haemonchinae of ruminants (nematoda: trichostrongyloidea) J. Parasitol. 2000;86:1093. doi: 10.2307/3284828. [DOI] [PubMed] [Google Scholar]
- Ljungström S., Melville L., Skuce P.J., Höglund J. Comparison of four diagnostic methods for detection and relative quantification of Haemonchus contortus eggs in feces samples. Front. Vet. Sci. 2018;4 doi: 10.3389/fvets.2017.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndal-Murphy M., Ehrlich W., Mayer D. Anthelmintic resistance in ovine gastrointestinal nematodes in inland southern Queensland. Aust. Vet. J. 2014;92:415–420. doi: 10.1111/avj.12250. [DOI] [PubMed] [Google Scholar]
- Martínez-Valladares M., Famularo M.R., Fernández-Pato N., Cordero-Pérez C., Castañón-Ordóñez L., Rojo-Vázquez F.A. Characterization of a multidrug resistant Teladorsagia circumcincta isolate from Spain. Parasitol. Res. 2012;110:2083–2087. doi: 10.1007/s00436-011-2753-1. [DOI] [PubMed] [Google Scholar]
- McKenna P.B. The anthelmintic efficacy of thiabendazole and levamisole against inhibited Haemonchus contortus larvae in sheep. N. Z. Vet. J. 1974;22:163–166. doi: 10.1080/00480169.1974.34157. [DOI] [PubMed] [Google Scholar]
- McMurtry L.W., Donaghy M.J., Vlassoff A., Douch P.G.C. Distinguishing morphological features of the third larval stage of ovine Trichostrongylus spp. Vet. Parasitol. 2000;90:73–81. doi: 10.1016/S0304-4017(00)00230-2. [DOI] [PubMed] [Google Scholar]
- Mes T.H.M., Eysker M., Ploeger H.W. A simple, robust and semi-automated parasite egg isolation protocol. Nat. Protoc. 2007;2:486–489. doi: 10.1038/nprot.2007.56. [DOI] [PubMed] [Google Scholar]
- Milhes M., Guillerm M., Robin M., Eichstadt M., Roy C., Grisez C., Prévot F., Liénard E., Bouhsira E., Franc M., Jacquiet P. A real-time PCR approach to identify anthelmintic-resistant nematodes in sheep farms. Parasitol. Res. 2017;116:909–920. doi: 10.1007/s00436-016-5364-z. [DOI] [PubMed] [Google Scholar]
- Mitchell C.J., O'Sullivan C.M., Pinloche E., Wilkinson T., Morphew R.M., McEwan N.R. Using next-generation sequencing to determine diversity of horse intestinal worms: identifying the equine ʼnemabiome’. J. Equine Sci. 2019;30:1–5. doi: 10.1294/jes.30.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moteane M., Middleton D.M., Polley L.R. A survey of disease conditions in adult and feeder sheep in Saskatchewan. Can. Vet. J. 1979;20:2–7. [PMC free article] [PubMed] [Google Scholar]
- O'Connor L.J., Walkden-Brown S.W., Kahn L.P. Ecology of the free-living stages of major trichostrongylid parasites of sheep. Vet. Parasitol. 2006;142:1–15. doi: 10.1016/j.vetpar.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Papadopoulos E., Gallidis E., Ptochos S. Anthelmintic resistance in sheep in Europe: a selected review. Vet. Parasitol. 2012;189:85–88. doi: 10.1016/j.vetpar.2012.03.036. [DOI] [PubMed] [Google Scholar]
- Ploeger H.W., Everts R.R. Alarming levels of anthelmintic resistance against gastrointestinal nematodes in sheep in The Netherlands. Vet. Parasitol. 2018;262:11–15. doi: 10.1016/j.vetpar.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Redman E., Queiroz C., Bartley D.J., Levy M., Avramenko R.W., Gilleard J.S. Validation of ITS-2 rDNA nemabiome sequencing for ovine gastrointestinal nematodes and its application to a large scale survey of UK sheep farms. Vet. Parasitol. 2019;275:108933. doi: 10.1016/j.vetpar.2019.108933. [DOI] [PubMed] [Google Scholar]
- Roeber F., Jex A.R., Campbell A.J.D., Campbell B.E., Anderson G.A., Gasser R.B. Evaluation and application of a molecular method to assess the composition of strongylid nematode populations in sheep with naturally acquired infections. Infect. Genet. Evol. 2011;11:849–854. doi: 10.1016/j.meegid.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Roeber F., Morrison A., Casaert S., Smith L., Claerebout E., Skuce P. Multiplexed-tandem PCR for the specific diagnosis of gastrointestinal nematode infections in sheep: an European validation study. Parasites Vectors. 2017;10:226. doi: 10.1186/s13071-017-2165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose H., Rinaldi L., Bosco A., Mavrot F., de Waal T., Skuce P., Charlier J., Torgerson P.R., Hertzberg H., Hendrickx G., Vercruysse J., Morgan E.R. Widespread anthelmintic resistance in European farmed ruminants: a systematic review. Vet. Rec. 2015;176:546. doi: 10.1136/vr.102982. 546. [DOI] [PubMed] [Google Scholar]
- Salgado J.A., Santos C. de P. Overview of anthelmintic resistance of gastrointestinal nematodes of small ruminants in Brazil. Rev. Bras. Parasitol. Vet. 2016;25:3–17. doi: 10.1590/s1984-29612016008. [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeal M.G., Gough P.A., Jackson A.R., Hotson I.K. The occurrence of strains of Haemonchus contortus resistant to thiabendazole. Aust. Vet. J. 1968;44:108–109. doi: 10.1111/j.1751-0813.1968.tb09033.x. [DOI] [PubMed] [Google Scholar]
- Troell K., Waller P., Höglund J. The development and overwintering survival of free-living larvae of Haemonchus contortus in Sweden. J. Helminthol. 2005;79:373–379. doi: 10.1079/joh2005286. [DOI] [PubMed] [Google Scholar]
- Van Wyk J.A., Cabaret J., Michael L.M. Morphological identification of nematode larvae of small ruminants and cattle simplified. Vet. Parasitol. 2004;119:277–306. doi: 10.1016/j.vetpar.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Waller P.J., Rudby-Martin L., Ljungström B.L., Rydzik A. The epidemiology of abomasal nematodes of sheep in Sweden, with particular reference to over-winter survival strategies. Vet. Parasitol. 2004;122:207–220. doi: 10.1016/j.vetpar.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Waller P.J., Rydzik A., Ljungström B.L., Törnquist M. Towards the eradication of Haemonchus contortus from sheep flocks in Sweden. Vet. Parasitol. 2006;136:367–372. doi: 10.1016/j.vetpar.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Westers T., Jones-Bitton A., Menzies P., VanLeeuwen J., Poljak Z., Peregrine A.S. Identification of effective treatment criteria for use in targeted selective treatment programs to control haemonchosis in periparturient ewes in Ontario, Canada. Prev. Vet. Med. 2016;134:49–57. doi: 10.1016/j.prevetmed.2016.09.021. [DOI] [PubMed] [Google Scholar]
- White J.R., Nagarajan N., Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000352. e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.