Abstract
Potato late blight, caused by the oomycete pathogen Phytophthora infestans, significantly hampers potato production. Recently, a new Resistance to Phytophthora infestans (Rpi) gene, Rpi‐amr1, was cloned from a wild Solanum species, Solanum americanum. Identification of the corresponding recognized effector (Avirulence or Avr) genes from P. infestans is key to elucidating their naturally occurring sequence variation, which in turn informs the potential durability of the cognate late blight resistance. To identify the P. infestans effector recognized by Rpi‐amr1, we screened available RXLR effector libraries and used long read and cDNA pathogen‐enrichment sequencing (PenSeq) on four P. infestans isolates to explore the untested effectors. Using single‐molecule real‐time sequencing (SMRT) and cDNA PenSeq, we identified 47 highly expressed effectors from P. infestans, including PITG_07569, which triggers a highly specific cell death response when transiently coexpressed with Rpi‐amr1 in Nicotiana benthamiana, suggesting that PITG_07569 is Avramr1. Here we demonstrate that long read and cDNA PenSeq enables the identification of full‐length RXLR effector families and their expression profile. This study has revealed key insights into the evolution and polymorphism of a complex RXLR effector family that is associated with the recognition by Rpi‐amr1.
Keywords: Avramr1, late blight, oomycete, PenSeq, Phytophthora infestans, Rpi‐amr1, RXLR effector
Long reads and cDNA pathogen enrichment sequencing (PenSeq) are developed to reveal the RXLR effector repertoires and expression pattern of four Phytophthora infestans isolates and enable to identify a new avirulence gene Avramr1.
1. INTRODUCTION
Potato late blight, caused by the hemibiotrophic oomycete pathogen Phytophthora infestans, triggered the Irish and European famine in the late 1840s, and still causes severe losses to world potato production.
To reduce losses, breeders sought resistance genes in wild relatives of potato. Early in the 20th century, Solanum demissum, a highly resistant hexaploid (2n = 72) wild potato, was found to be a useful source of resistance to P. infestans (Rpi) genes (Salaman, 1937). Since then, many resistance traits have been transferred to cultivated potatoes by introgression breeding (Toxopeus, 1956), and many Rpi genes have been cloned from wild potatoes, for example R1, R3a, R8, Rpi‐blb1, and Rpi‐vnt1 (Ballvora et al., 2002; van der Vossen et al., 2003; Huang et al., 2005; Foster et al., 2009; Pel et al., 2009; Vossen et al., 2016). Unlike wild potatoes, Solanum nigrum and Solanum americanum have been reported to be nonhosts for P. infestans (Colon et al., 1993). Two Rpi genes encoding NLR proteins, Rpi‐amr3 and Rpi‐amr1, were cloned from S. americanum and confer late blight resistance in potato (Witek et al., 2016, 2020).
Identification of the recognized effectors for Rpi‐amr3 and Rpi‐amr1 would open the way to investigate their virulence function and distribution in P. infestans populations. Moreover, it could also help to diagnose Rpi gene repertoires in resistant plants, and individually confirm their activity in genetically modified potatoes carrying multiple Rpi genes. In oomycetes, all the cloned Avr proteins contain a signal peptide and RXLR motif (Rehmany et al., 2005), and the genomic sequencing of P. infestans revealed 563 RXLR effectors in the T30‐4 reference genome (Haas et al., 2009). This enabled a high‐throughput effectoromics approach for functional screening of the candidate effectors in plants (Vleeshouwers et al., 2008, 2011), and many Avr genes were identified by this approach, including Avrblb1, Avrblb2, and Avrvnt1 (Vleeshouwers et al., 2008; Oh et al., 2009; Pel, 2010).
However, available RXLR effector libraries do not contain recombinant clones of all P. infestans RXLR effectors, and the effector candidates were defined on the basis of expression profile, motif analysis, and distribution between P. infestans races (Vleeshouwers et al., 2008; Haas et al., 2009; Oh et al., 2009). In total, c.300/563 RXLR effectors were previously cloned into expression vectors for functional screening (Rietman, 2011).
To further explore the diversity of RXLR effectors from P. infestans, a pathogen‐enrichment sequencing (PenSeq) approach was adopted to study allelic variation of RXLR effectors and population genomics of oomycetes. A bait library of RXLR effectors and some other pathogen‐related genes was synthesized and used for enrichment prior to sequencing (Jouet et al., 2019; Thilliez et al., 2019). However, the previous PenSeq analyses used Illumina reads and genomic DNA (gDNA), making it difficult to differentiate individual effector alleles and closely related paralogs or to find out which effectors are expressed.
Here, to identify the recognized effector of the newly cloned Rpi‐amr1 protein from S. americanum (Witek et al., 2020), we screened all currently available RXLR effectors for recognition but without success. Therefore, we adapted and improved PenSeq with long read (PacBio) and cDNA sequencing, and extended the list of candidate effectors that could be screened. Amongst these additional candidate RXLR genes, we identified Avramr1 and defined orthologs and paralogs from four different isolates of P. infestans.
2. RESULTS
2.1. Available recombinant RXLR effector libraries do not contain Avramr1
To identify Avramr1, we tested 278 available RXLR effectors (Table S1) by coexpressing them with Rpi‐amr1‐2273 in Nicotiana benthamiana(Rietman, 2011). Rpi‐amr1‐2273 (hereafter Rpi‐amr1) is a functional Rpi‐amr1 homolog cloned from S. americanum SP2273 (Witek et al., 2020). However, we did not identify an effector that activated Rpi‐amr1‐dependent hypersensitive response (HR) from the available RXLR effector libraries and concluded these libraries are incomplete. Notably, Avr8 was not originally included in the core effector selection because Avr8 expression goes up earlier then 2 days postinoculation (dpi) (Jo, 2013), showing that the criteria adopted to define core effectors do not reveal all recognized effectors.
To find Avramr1, we proposed three hypotheses: (a) Avramr1 is an RXLR effector but the recognized allele is not present in the assembled version of P. infestans T30‐4 reference genome, (b) Avramr1 is an RXLR effector but it has not yet been tested in previous functional studies/libraries, and (c) Avramr1 is not a typical RXLR effector. To address the first hypothesis, we performed PacBio PenSeq to sequence the effector alleles in the four diverse P. infestans isolates, EU_13_A2 (2006_3928A), EC1_A1 (EC1_3626), EU_6_A1 (2006_3920A), and US23, all of them avirulent on potato plants carrying Rpi‐amr1. To address the second hypothesis, we performed cDNA PenSeq to try to identify other RXLR effectors that are expressed during infection but not reported or defined in previous functional studies.
2.2. PacBio PenSeq of four P. infestans isolates, EU_13_A2, EC1_A1, EU_6_A1, and US23
PacBio gDNA PenSeq was performed on four P. infestans isolates of genotypes EU_13_A2, EC1_A1, EU_6_A1, and US23 (Figure 1a). To evaluate the enrichment efficiency, quantitative polymerase chain reaction (qPCR) was performed with the DNA pre‐ and postcapture. In general, the targeted genes of different length were well enriched at concentration × time (Cot) value <20, while the untargeted genes were almost undetectable, with Cot > 27 (Peterson et al., 2002) (Figure S1). Furthermore, we found that the capture efficiency was increased by including a 10‐fold molar excess of nonadaptor‐ligated fragmented P. infestans DNA (500–1,000 bp) in the reannealing reaction to reduce the extent to which sequences were recovered due to concatenation of transposon‐containing sequences adjacent to RXLR genes. After sequence capture, enrichment of most effector genes was more efficient when nonadaptor‐ligated P. infestans DNA was included (Figure S1).
FIGURE 1.
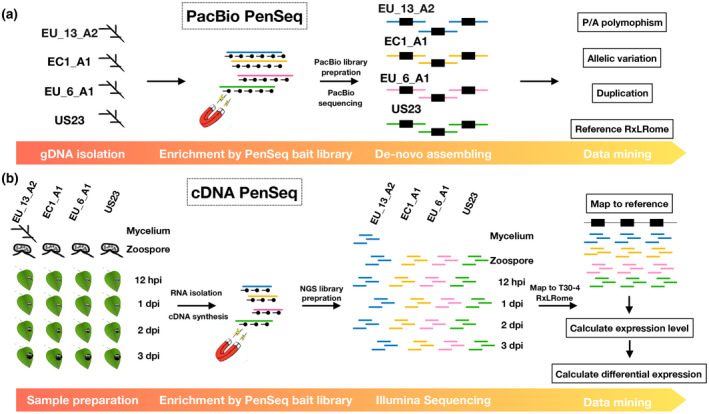
The pipelines of PacBio and cDNA pathogen‐enrichment sequencing (PenSeq). (a) The pipeline of PacBio gDNA PenSeq. Briefly, the gDNA isolated from various Phytophthora infestans was enriched for RXLR effectors, sequenced by PacBio and de novo assembled for data mining. (b) The pipeline of cDNA PenSeq. The cDNA was synthetized using RNA sampled from various P. infestans at different stages (mycelium, zoospore, 12 hr postinoculation, 1, 2, and 3 days postinoculation). The libraries enriched for RXLR effectors were sequenced, reads were mapped to the RXLRome of the reference P. infestans genome T30‐4 and the expression levels of samples were calculated and compared. Black lines with dots represent the baits, the enriched fragments are depicted in blue (EU_13_A2), yellow (EC1_A1), pink (EU_6_A1), and green (US23). The black boxes indicate RXLR effectors. EU_13_A2, EC1_A1, EU_6_A1 and US23 are P. infestans isolates.
Following the enrichment sequencing, circular consensus sequencing reads were assembled (Figure 1a) and contigs with fewer than 10 reads were removed. The average length of the contigs of coverage >10 reads was 7 kb (Table 1), and the size of the largest contig was over 50 kb. This suggests that the PacBio PenSeq successfully captured the target effector genes and the adjacent flanking DNA sequences. In total, 1,137, 1,054, 1,283, and 925 contigs were obtained from EU_13_A2, EC1_A1, EU_6_A1, and US23, respectively, of which 687, 650, 741, and 571 contigs contain RXLR effectors (Table 1 and Notes [Link], [Link], [Link], [Link]). The remaining contigs contained non‐RXLR effectors, which were included in the bait library design for other purposes (Thilliez et al., 2019).
TABLE 1.
PacBio PenSeq for EU_13_A2, EC1_A1, EU_6_A1, and US23
| Phytophthora infestans isolate | Total contigs | RXLR effector | Non_RXLR | Average length (bp) | Minimum length (bp) | Maximum length (bp) |
|---|---|---|---|---|---|---|
| EU_13_A2 | 1,137 | 687 | 450 | 7,732 | 2,617 | 30,571 |
| EC1_A1 | 1,054 | 650 | 404 | 7,663 | 2,696 | 33,203 |
| EU_6_A1 | 1,283 | 741 | 542 | 7,463 | 3,052 | 50,275 |
| US23 | 925 | 571 | 354 | 7,598 | 2,648 | 29,901 |
The PacBio PenSeq data allowed us to detect new RXLR effector alleles from different haplotypes of various P. infestans isolates and even in polyploid genotypes like EU_13_A2 (Li et al., 2017). This data set can also be used to extensively study allelic variation, presence/absence (P/A) polymorphism, and effector evolution. For example, Avr1 (PITG_16663) and a paralogous Avr1‐like gene (PITG_06432) are located on supercontigs 1.51 and 1.8 of the reference T30‐4 genome, respectively. The R1‐breaking clonal lineage EU_13_A2 was reported to have an 18 kb deletion comprising the Avr1 locus (Cooke et al., 2012). Also, the Illumina PenSeq data showed that the Avr1 locus is missing in EU_13_A2, EC1_A1, and US23 (Thilliez et al., 2019). We mapped the four Avr1 contigs from EU_13_A2 (contigs 192, 261, 296, and 329) to supercontig 1.51 and 1.8, and found that all four contigs map to the Avr1‐like supercontig 1.8. Two contigs (contig 261 and 286) mapped to the Avr1‐like locus, and two other contigs (contig 192 and 329) mapped to a locus next to Avr1‐like that was not previously annotated (Figure S2), although the genes in those two contigs might be pseudogenes as the signal peptide is missing in both of them. Additionally, in EU_6_A1 and US23, two Avr1 contigs did not map to Avr1 or Avr1‐like loci of T30‐4. Thus, our PacBio PenSeq data set can provide the means to detect novel RXLR effector paralogs absent from the reference genome.
As another example, our data set carries in total 504 of the 563 predicted RXLR effectors from the reference genome T30‐4 (Haas et al., 2009). To investigate P/A polymorphism of RXLR effectors in the four sequenced isolates, we performed a basic local alignment search (BLAST) of the 504 effectors against the PacBio contigs, with hits with <50% coverage defined as absent (Table S2). We found that 17, 28, 15, and 33 RXLR effectors out of the 504 are missing in EU_13_A2, EC1_A1, EU_6_A1, and US23, respectively.
Taken together, we have generated a rich data set that could help to define full‐length RXLR effector genes, deliver robust information on alleles and paralogs, and reveal conserved or race‐specific effectors from different isolates. The data set is available in full in Notes [Link], [Link], [Link], [Link].
2.3. cDNA PenSeq enables effector expression detection in early stages of infection
To clarify whether the untested effectors might be putative Avr genes, we performed cDNA PenSeq for the four P. infestans isolates EU_13_A2, EC1_A1, EU_6_A1, and US23, at different time points after infection (12 hr postinoculation [hpi], 1, 2, and 3 dpi) and in mycelium and zoospores (Figure 1b). To analyse and visualize the cDNA PenSeq data, we built an artificial DNA sequence contig (“RXLRome”) for the RXLR effectors. In addition, nine non‐RXLR genes from the bait library were included as controls (Jouet et al., 2019; Figure 2). The cDNA PenSeq reads were mapped to the RXLRome and gene expression compared over time (Figure 1b).
FIGURE 2.
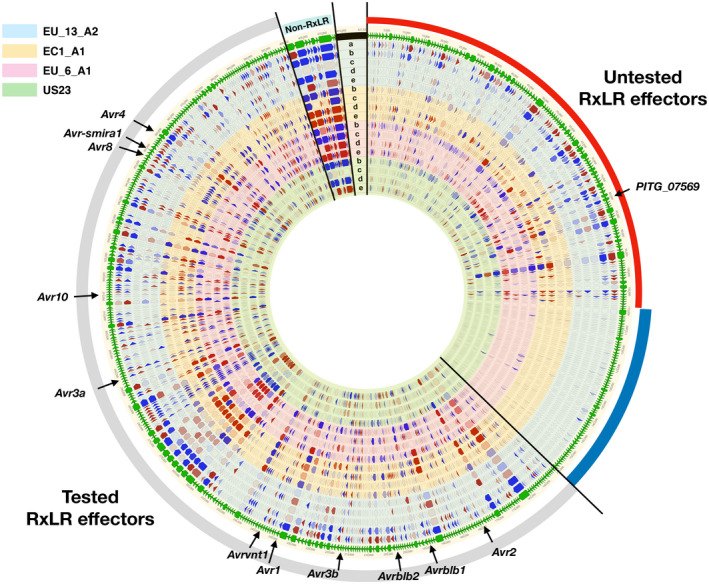
cDNA pathogen‐enrichment sequencing (PenSeq) of RXLR effectors from EU_13_A2, EC1_A1, EU_6_A1, and US23. The cDNA PenSeq data for the RXLR effectors from four Phytophthora infestans at different stages were mapped to an artificial contig (RXLRome) of 499 RXLR effectors and nine non‐RXLR genes, demarcated by bright green arrows on the outer edge of the diagram. Black lines separate the previously tested RXLR effectors (grey bar), new effector candidates with differential expression (red bar), unexpressed effectors (blue bar) and non‐RXLR controls (cyan). The concentric circles in blue, yellow, pink, and green represent data from P. infestans EU_13_A2, EC1_A1, EU_6_A1, and US23, respectively. The arrows on them indicate differential expression (red, up‐regulation; blue, down‐regulation; no fill, no difference), where the more intense the colour, the bigger the difference. The data are plotted as follows: a, mycelium vs. zoospores (for EU_13_A2 only); b, zoospores vs. 12 hr postinoculation (hpi); c, 12 hpi vs. 1 day postinoculation (dpi); d, 1 dpi vs. 2 dpi; e, 2 dpi vs. 3 dpi. Eleven known Avr genes, namely Avr4, AvrSmira1, Avr8, Avr10, Avr3a, Avrvnt1, Avr1, Avr3b, Avrblb2, Avrblb1, and Avr2, are indicated by black arrows. PITG_07569 is indicated by a black arrow
Most of the RXLR effectors that were included in previous effector libraries show an up‐regulation of expression in the early stages of infection (Figure 2). Eleven known Avr genes from P. infestans, namely Avr4, Avr‐smira1, Avr8, Avr10, Avr3a, Avrvnt1, Avr1, Avr3b, Avrblb2, Avrblb1, and Avr2, are shown in Figure 2. Some of the untested RXLR effectors show a similar pattern of expression and might also represent additional potential Avr genes, while others are poorly expressed in some isolates. The details of the cDNA PenSeq are available in Table S3.
2.4. Identification of Avramr1
To test if Avramr1 is among the untested effectors, we selected 47 highly expressed RXLR effectors (Figure 3) present in all tested lineages that had not previously been investigated. The effectors were synthesized, cloned into an expression vector with CaMV 35S promoter, and transformed into Agrobacterium GV3101‐pMP90 for agroinfiltration in N. benthamiana (Figure 4a). All the effectors were infiltrated alone or coinfiltrated with Rpi‐amr1 (Witek et al., 2020). Among the 47 effectors, PITG_07569 was the only effector that triggered an HR when coexpressed with Rpi‐amr1. Hence, we concluded PITG_07569 is Avramr1. To verify if both proteins were expressed in planta, we cloned Avramr1 and Rpi‐ amr1 with C‐terminal green fluorescent protein (GFP) and His‐FLAG (HF), respectively. Both recombinant proteins were expressed and detected in N. benthamiana by western blot (Figure 4c) The same constructs were used for agroinfiltration in N. benthamiana, and HR was observed specifically after coexpression of Avramr1‐GFP and Rpi‐amr1‐HF (Figure 4b).
FIGURE 3.
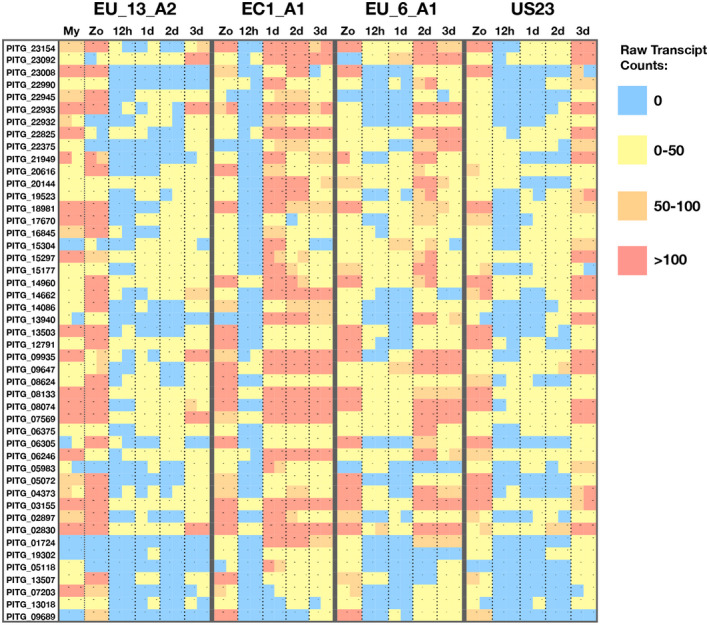
Raw transcript counts for the new candidate RXLR effectors. The 47 most differentially expressed RXLR effectors from the previously untested set were selected, and the raw transcript counts were visualized as a heat map across time points and treatments. Each square indicates a single data point derived from two independent biological replicates. The colours red, orange, yellow, and blue represent >100, 50–100, 0–50, or 0 raw transcripts, respectively
FIGURE 4.
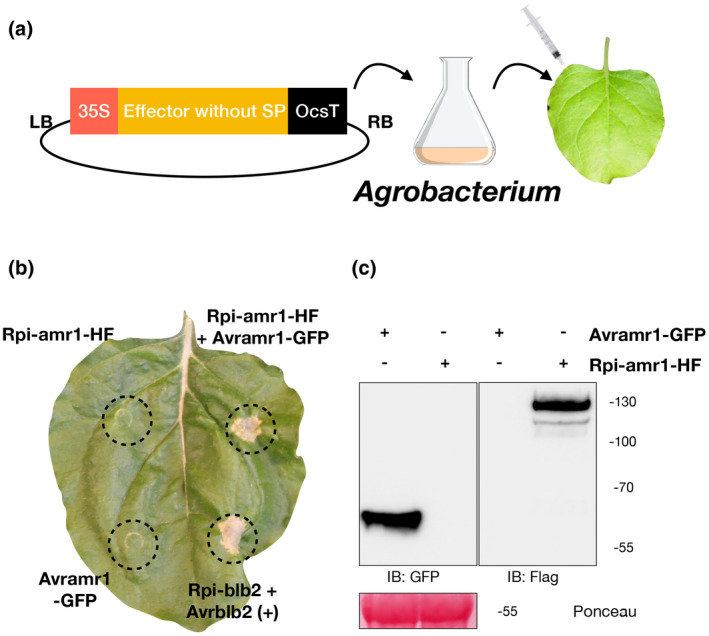
Identification of Avramr1. (a) All 47 selected effectors without signal peptides (SP) were synthesized and cloned into an expression vector under a CaMV35S promoter for Agrobacterium‐mediated transient expression. (b) Transient expression of Avramr1‐green fluorescent protein (GFP), Rpi‐amr1‐His‐FLAG (HF) on their own coexpression in Nicotiana benthamiana. Dashed circles demarcate the infiltration sites. Hypersensitive response (HR) was observed when coexpressed Avramr1‐GFP with Rpi‐amr1‐HF. Rpi‐blb2 and Avrblb2 were used as positive control (+). This experiment was repeated three times with the same results. (c) Expression of Avramr1‐GFP and Rpi‐amr1‐HF fusion proteins in N. benthamiana leaves. The expression of Avramr1‐GFP was confirmed by immunoblot (IB) using anti‐GFP antibody, and expression of Rpi‐amr1‐HF was confirmed by anti‐FLAG antibody. Size markers are indicated in kDa and Ponceau stain was used to show the protein loading
2.5. Avramr1 homologs in different P. infestans isolates and other Phytophthora species
Avramr1 is a canonical RXLR effector with RYLR and EER motifs and an N‐terminal signal peptide (Figure 5b). Avramr1 locates on supercontig 1.11 of the P. infestans reference genome T30‐4. Avramr1‐like (hereafter Avramr1L), a truncated paralog (PITG_07566), maps adjacent to Avramr1 (Figure 5a,b). Two known Avr effectors, Avr8 (PITG_07558) and Avrsmira1 (PITG_07550), are physically close to the Avramr1 locus in the T30‐4 genome (Figure 5a) (Rietman et al., 2012).
FIGURE 5.
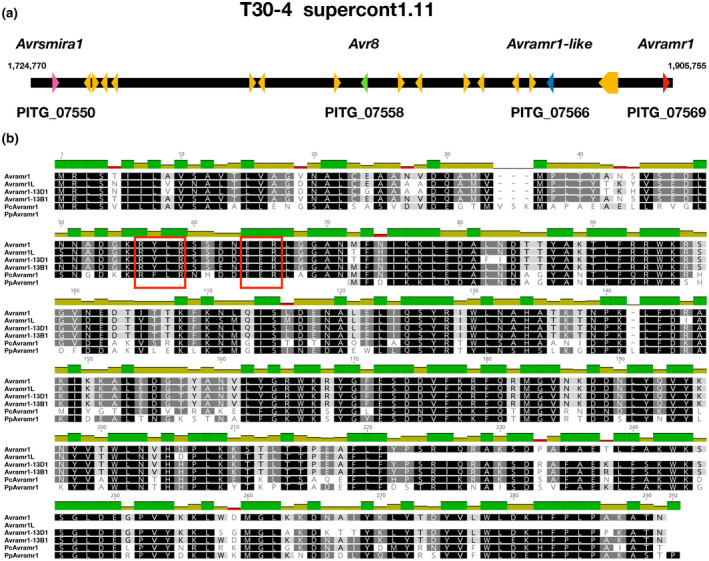
Genomic localization and amino acid alignment of Avramr1. (a) The localization of Avramr1 (PITG_07569, red arrow) on supercontig 1.11 of the reference Phytophthora infestans T30‐4 genome. A paralog Avramr1L gene (PITG_07566, blue arrow) is located close to Avramr1. The supercontig contains two other known Avr genes, Avrsmira1 (PITG_07550, pink arrow) and Avr8 (PITG_07558, green arrow). (b) The alignment of protein sequences of Avramr1 and selected homologs from P. infestans, Phytophthora cactorum (Pc), and Phytophthora parasitica (Pp). The dark green bars on top of the alignment indicate 100% identity while olive green and red bars indicate various degrees of polymorphism between the sequences. RXLR and EER motifs are highlighted by red boxes
To study the sequence polymorphism of Avramr1 homologs in P. infestans, we used BLAST to search for Avramr1 homologs in the PacBio PenSeq assemblies generated in this study. This revealed that EU_13_A2, EC1_A1, EU_6_A1, and US23 carry six, four, three, and six Avramr1 homologs, respectively (Note S5). Next, we aligned the corresponding Avramr1 amino acid sequences and generated a neighbour‐joining (NJ) tree for phylogenetic analysis (Figure 6a). Two Avramr1 homologs from Phytophthora parasitica and Phytophthora cactorum were identified from public databases, and they were used as an out‐group (Figures 5b and 6a). Based on the phylogenetic tree, we distinguished four Avramr1 clades, clade A (containing Avramr1 from T30‐4) and clade C (with Avramr1L from T30‐4), and two more clades, B and D (Figure 5a). For a more detailed analysis, we selected one Avramr1 homolog from clade B and one from D (Avramr1‐13B1 and Avramr1‐13D1 from EU_13_A2) and aligned them with Avramr1 homologs from clade A and C, and with P. parasitica and P. cactorum homologs. Significant sequence polymorphisms were observed between effectors from different clades (Figure 5b). Meanwhile, the Avramr1 homologs within the same clade were almost identical (Figure 6a).
FIGURE 6.
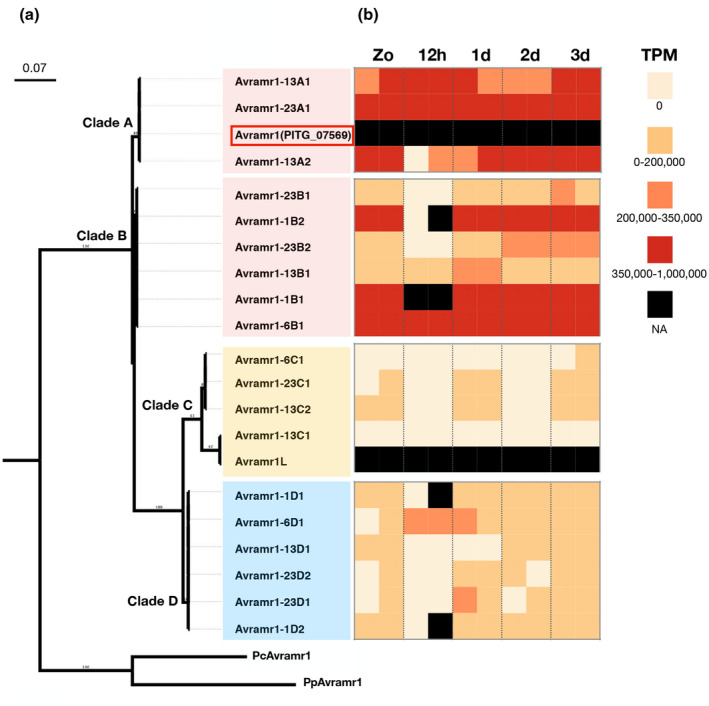
Phylogeny and expression profile of Avramr1 homologs from EU_13_A2, EC1_A1, EU_6_A1, and US23. (a) Maximum‐likelihood phylogeny of the protein sequences of the Avramr1 homologs were made by IQ‐TREE (Minh et al., 2020). PcAvramr1 and PpAvramr1 are Avramr1 homologs from Phytophthora cactorum and Phytophthora parasitica, and they were used as out‐groups. (b) The expression profile of Avramr1 homologs at different stages and time points (zoospores, 12 hr postinoculation, 1, 2, and 3 days postinoculation). Transcripts per kilobase million (TPM) for each effector homologs were visualized as follows: black, data not available; red, 350,000–1,000,000 TPM; orange, 200,000–350,000 TPM; yellow, 0–200,000 TPM; beige, 0 TPM. Avramr1 and Avramr1L are from the reference genome T30‐4, which was not included in the cDNA PenSeq
2.6. Differential expression of Avramr1 homologs in different P. infestans isolates
To investigate the expression patterns of Avramr1 homologs defined in the PacBio PenSeq data, we mapped the corresponding cDNA PenSeq reads to the PacBio PenSeq contigs from EU_13_A2, EC1_A1, EU_6_A1, and US23. The transcripts per kilobase per million (TPM) values for each time point are visualized in Figure 6b. The clade A homologs Avramr1‐23A1, Avramr1‐13A1, and Avramr1‐13A2 were highly expressed at almost all stages, and the Avramr1 homologs from clade B showed a similar expression pattern. For clade C, some homologs, like Avramr1‐13C, Avramr1‐6C1 or Avramr1L gene from T30‐4, were weakly expressed at all stages. However, two other Avramr1L homologs, Avramr1‐23C1 and Avramr1‐13C2, showed moderately elevated expression in zoospores, and at 1 and 3 dpi. Interestingly, the Avramr1 homologs from clade D, which are missing in the reference genome T30‐4, showed an intermediate expression level compared to clades A, B, and C, and most Avramr1 homologs in clade D showed an increase in expression at the zoospore stage, and at 1, 2, and 3 dpi.
In summary, our PacBio PenSeq analysis created a rich data set to reveal new Avr variants from different P. infestans isolates, and to quantify their expression profile individually. This facilitates the analysis of the polymorphism of pathogen effectors and their potential differential recognition patterns with the corresponding Rpi genes (Witek et al., 2020).
3. DISCUSSION
The availability of the P. infestans genome sequence enabled a step‐change in the rate of investigation of this pathogen, accelerating the discovery of recognized effectors, and of new Rpi genes (Vleeshouwers et al., 2008, 2011; Haas et al., 2009). However, some questions remain open. For example, how different are the effector repertoires in different P.infestans isolates? To what extent do they show differential expression between races? The study of plant NLR gene repertoires faces similar challenges, and sequence capture, combined with long‐read sequencing technologies, has enabled the refinement of tools to cost‐effectively investigate diversity, such as RenSeq, SMRT RenSeq, RLP/KSeq, and AgRenSeq (Jupe et al., 2013; Witek et al., 2016; Arora et al., 2019; Lin et al., 2020). Recently, the pan‐NLRome of 65 diverse Arabidopsis thaliana accessions was determined by a similar strategy, revealing that any one accession lacks many of the NLRs found in the species pan‐NLRome (Van de Weyer et al., 2019).
PenSeq was developed to facilitate cost‐effective investigation of pathogen diversity on infected plants, and polymorphism of pathogen effectors (Jouet et al., 2019; Thilliez et al., 2019). The first PenSeq studies, however, were conducted using Illumina short reads. This significantly limited their resolving power as many oomycete genomes are highly heterozygous, and some effectors belong to large gene families with multiple sequence‐related paralogs that can lead to false assemblies (Gilroy et al., 2011; Oliva et al., 2015).
In this study, we combined long read and cDNA Penseq, enabling a detailed analysis of the RXLR genes and their expression patterns in different P. infestans isolates. The cDNA PenSeq data set allowed us to define an additional set of 47 RXLR genes expressed during infection that were not previously investigated. Amongst these, we identified Avramr1, which encodes the cognate recognized effector for Rpi‐amr1 from S. americanum (Witek et al., 2020). It is noteworthy that PITG_07569 (Avramr1) was identified by an alternative splicing reporter system as a splicing regulatory effector; furthermore, it was shown to promote the colonization of P. infestans (Huang et al., 2020).
The long read PenSeq data helped us to obtain full‐length RXLR effector haplotypes with their flanking sequences. This allowed us to distinguish individual alleles from polyploid isolates like EU_13_A2, and also distinct effector paralogs. The sequences flanking the RXLR genes enabled us to understand the possible translocation events and identify new RXLR loci. We were also able to identify multiple new Avramr1 homologs from different isolates, and identified a new Avramr1 clade D that is not present in T30‐4. This data set allows us to study the differential recognition pattern of Rpi‐amr1 and Avramr1 homologs. Indeed, different Rpi‐amr1 homologs could recognize different sets of Avramr1 homologs, including the Avramr1 homolog from the newly identified clade D (Witek et al., 2020). So far, no Rpi‐amr1‐breaking P. infestans isolates have been found (Witek et al., 2020), and therefore we propose that Avramr1 might be crucial for the virulence of P. infestans. The identification of Avramr1 will enable us to study its virulence function and its polymorphism in the P. infestans population, and study the effector‐triggered immunity mediated by Rpi‐amr1.
Collectively, the PenSeq data set constitutes a valuable community resource for investigating the allelic and expression diversity of multiple recognized effectors. The long reads and cDNA PenSeq methods will contribute to understanding this fast‐evolving and destructive oomycete pathogen, and to achieving durable late blight resistance in potato.
4. EXPERIMENTAL PROCEDURES
4.1. Sample preparation
To collect the mycelium of P. infestans for DNA extraction, P. infestans strains were grown on rye sucrose agar (RSA) for 7 days and then moved to Plich liquid medium for 14 days. Mycelia were washed and harvested, freeze‐dried using a vacuum pump, and stored at −80°C until DNA or RNA extraction.
To collect the infection samples or zoospores for RNA extraction, P. infestans strains were cultured for 10 days on RSA. Grown mycelia were covered with cold (4°C) sterile water and then incubated at 4–6°C for 2–3 hr. The concentration of the inoculum was adjusted to about 50,000 zoospores/ml and 10 μl drops of inoculum were placed on the detached leaves of potato plants. Detached leaf assays were incubated at 20°C in high humidity for a required time after inoculation. Leaf discs of the infection area were collected and stored at −80°C until DNA or RNA extraction.
4.2. DNA and RNA extraction
DNA was extracted using phenol/chloroform. P. infestans mycelium samples or infected leaf discs were ground into powder in liquid nitrogen. Ground material was resuspended in 500 μl of Shorty buffer (0.2 M Tris.HCl pH 9, 0.4 M LiCl, 25 mM EDTA, 1% SDS) and one volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added. The upper aqueous phase containing DNA was mixed with one volume of 100% ice‐cold isopropanol to precipitate DNA. The pellet was washed twice using 70% ethanol, heated at 70°C for 2–5 min to completely remove ethanol, and resuspended in sterile water. Resuspended DNA was then heated at 65°C for 20 min to inactivate DNases before RNase treatment was performed (2 μl of 10 mg/ml RNase A, 37°C, 1 hr) and RNase A removed by chloroform precipitation. Genomic DNA was resuspended in water and sheared into 3–5 kb fragments using the S220 Focused‐ultrasonicator (Covaris Inc.).
RNA samples were extracted with Direct‐zol RNA MiniPrep kit (Zymo Research) according to the manufacturer's instructions.
4.3. PacBio and Illumina PenSeq capture
The biotinylated RNA bait library of 120 nucleotides (nt) was designed for enriching RXLR effectors from P. infestans and some other genes of interest. The library contains 18,348 baits, as described previously (Jouet et al., 2019; Thilliez et al., 2019).
PacBio library was constructed with DNA samples from the mycelium of four P. infestans strains, EU_13_A2 (2006_3928A), EC1_A1 (EC1_3626), EU_6_A1 (2006_3920A), and US23. The library construction and target DNA sequence capture were performed according to Witek et al. (2016), with minor modifications. A Qubit Fluorometer (ThermoFisher) was used to quantify the barcoded DNA library from each isolate. Equimolar amounts of DNA from the four individually barcoded samples were pooled to obtain 250 ng of total DNA and then subjected to sequence capture. A 10× excess of nonadaptor‐ligated P. infestans DNA at about 500–1,000 bp was added for the hybridization. The final mixture of the amplicons of the captured library was further size selected by SageELF electrophoresis system (Sage Science) according to the manufacturer’s instructions.
The Illumina library was constructed with RNA samples from zoospores of the four P. infestans strains, from the corresponding infected leaf discs harvested at 12 hpi, 1, 2, and 3 dpi, and from mycelium of EU_13_A2. An Illumina library for each sample was constructed with KAPA mRNA HyperPrep Kit for Illumina Platforms (KR1352 v. 5.17) following the manufacturer's instructions. mRNA was fragmented to 300–400 bp. The barcoded libraries were mixed together at a ratio of 16:8:4:1:1 for 12 hpi, 1, 2, and 3 dpi and zoospores samples, respectively.
Before and after sequence capture, quantitative PCR was performed on a Bio‐Rad CFX96 real‐time detection system with an input of 1 ng DNA to assess the efficiency of capture.
4.4. Sequencing
PacBio PenSeq libraries were sequenced at the Earlham Institute (Norwich, UK) using the Sequel platform. Illumina PenSeq cDNA libraries were sequenced at Novogene (Hong Kong, China) using HiSeq, PE250.
4.5. gDNA PenSeq assembly
PacBio raw reads were processed as described in Witek et al. (2016) to generate ROI reads and demultiplexed using custom script (Van de Weyer et al., 2019). Demultiplexed ROI were assembled using Geneious R8 (http://www.geneious.com/) using the settings in Witek et al. (2016).
4.6. Analysis of cDNA PenSeq
All RXLR effectors from the P. infestans reference genome T30‐4 were used to generate an artificial “RXLRome” contig, where RXLR effectors’ sequences were separated by stretches of 500 “Ns”. The contig also contained nine non‐RXLR control genes (Jouet et al., 2019). The cDNA PenSeq reads from all treatments were mapped to the T30‐4 RXLRome, and the expression analyses were performed and visualized using Geneious R10 (Kearse et al., 2012).
4.7. New candidate RXLR effectors
For the previously untested RXLR effectors, we first selected the effectors showing differential expression at different stages and ranked them based on the raw transcript counts. Next, local alignment searches (BLAST) were performed against the 563 predicted RXLR effectors (Haas et al., 2009) to remove the previously tested effectors. This analysis revealed 47 candidate RXLR effectors that were not included in the previous functional study. The 47 RXLR effectors were synthesized by Twist Bioscience. The signal peptides were removed, the sequences were domesticated for Golden Gate cloning, and overhangs containing BsaI restriction sites were added to both ends of all effector sequences.
All the effectors were cloned into vector pICSL86977 (TSL SynBio) with CaMV 35S promoter and OCS terminator. To further verify the expression Avramr1 and Rpi‐amr1, they were fused with C‐terminal GFP and His‐FLAG (HF) tags, respectively. Agrobacterium strain GV3101‐pMP90 was transformed with the constructs for agroinfiltration.
4.8. Cell death assay
Transient expression of RXLR effectors and Rpi‐amr1 in N. benthamiana was performed as described previously (Bos et al., 2006). Agrobacterium was infiltrated at OD600 = 1, and each effector was coinfiltrated with Rpi‐amr1 (Witek et al., 2020). The cell death phenotype was observed at 4 dpi. p35S‐Rpi‐blb2 and p35S‐Avrblb2 were coexpressed as positive control in the HR assay (Oh et al., 2009).
4.9. Protein extraction and immunoblot analysis
The p35S‐Rpi‐amr1‐HF and p35S‐Avramr1‐GFP constructs were used to transiently express the fusion proteins in N. benthamiana. The leaf tissue was harvested 2 days after infiltration and proteins were extracted as described in Guo et al. (2020). The expression of recombinant Avramr1‐GFP and Rpi‐amr1‐HF was determined by SDS‐PAGE as described in Guo et al. (2020). Horseradish peroxidase‐conjugated antibodies (anti‐FLAG M2, 1:10,000 dilution, Sigma; anti‐GFP, 1:10,000 dilution, Santa Cruz Biotechnology) were used for the immunoblot. The chemiluminescence was detected by ImageQuant LAS 4,000 (Life Sciences) after chemiluminescent substrate incubation (SuperSignal West Pico & West Femto).
4.10. Sequence and phylogenetic analysis
All sequences were analysed in Geneious R10 (Kearse et al., 2012), MAFFT was used for sequence alignment (Katoh and Standley, 2013), and the signal peptides of Avramr1 homologs were removed manually for the phylogenetic analysis. IQ‐TREE was used for the phylogenetic analysis and the JTTDCMut model was selected as best‐fit model by IQ‐TREE (Minh et al., 2020).
Supporting information
FIGURE S1 Enrichment efficiency with/without non‐adaptor‐ligated DNA
FIGURE S2 Comparison of EU_13_A2 Avr1 contigs and the T30‐4 reference genome
TABLE S1 278 RxLR effectors from previously available effector libraries
TABLE S2 P/A polymorphism of RxLR effectors from EU_13_A2, EC1_A1, EU_6_A1, and US23
TABLE S3 cDNA PenSeq for EU_13_A2, EC1_A1, EU_6_A1, and US23. Raw data of differential expression in Figure 2. Raw transcript counts in Figure 3
NOTES S1 PacBio PenSeq contigs of EU_13_A2
NOTES S2 PacBio PenSeq contigs of EC1_A1
NOTES S3 PacBio PenSeq contigs of EU_6_A1
NOTES S4 PacBio PenSeq contigs of US23
NOTES S5 Protein sequences of Avramr1 homologs
ACKNOWLEDGMENTS
This research was financed from BBSRC grant BB/P021646/1. We would like to thank Sophien Kamoun and Joe Win for valuable discussion, and the TSL Bioinformatics Team, the SynBio Team, and the horticultural team for their support. The authors have no conflict of interest to declare.
Lin X, Song T, Fairhead S, et al. Identification of Avramr1 from Phytophthora infestans using long read and cDNA pathogen‐enrichment sequencing (PenSeq). Molecular Plant Pathology. 2020;21:1502–1512. 10.1111/mpp.12987
DATA AVAILABILITY STATEMENT
Raw PacBio and cDNA PenSeq read sequences have been deposited in the Sequence Read Archive (SRA) at https://www.ncbi.nlm.nih.gov/sra under BioProject IDs PRJNA623167 and PRJNA598824.
REFERENCES
- Arora, S. , Steuernagel, B. , Gaurav, K. , Chandramohan, S. , Long, Y. , Matny, O. , et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nature Biotechnology, 37, 139–143. [DOI] [PubMed] [Google Scholar]
- Ballvora, A. , Ercolano, M.R. , Weiß, J. , Meksem, K. , Bormann, C.A. , Oberhagemann, P. et al (2002) The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. The Plant Journal, 30, 361–371. [DOI] [PubMed] [Google Scholar]
- Bos, J.I.B. , Kanneganti, T.D. , Young, C. , Cakir, C. , Huitema, E. , Win, J. et al (2006) The C‐terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana . The Plant Journal, 48, 165–176. [DOI] [PubMed] [Google Scholar]
- Colon, I.T. , Eijlander, R. , Budding, D.J. , Van Ijzendoorn, M.T. , Pieters, M.M.J. and Hoogendoorn, J. (1993) Resistance to potato late blight (Phytophthora infestans (Mont.) de Bary) in Solanum nigrum, S. villosum and their sexual hybrids with S. tuberosum and S. demissum . Euphytica, 66, 55–64. [Google Scholar]
- Cooke, D.E.L. , Cano, L.M. , Raffaele, S. , Bain, R.A. , Cooke, L.R. , Etherington, G.J. et al (2012) Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathogens, 8, e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, S.J. , Park, T.‐H. , Pel, M. , Brigneti, G. , Śliwka, J. , Jagger, L. et al (2009) Rpi‐vnt1.1, a Tm‐22 homolog from Solanum venturii, confers resistance to potato late blight. Molecular Plant‐Microbe Interactions, 22, 589–600. [DOI] [PubMed] [Google Scholar]
- Gilroy, E.M. , Breen, S. , Whisson, S.C. , Squires, J. , Hein, I. , Kaczmarek, M. et al (2011) Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2‐like in Phytophthora infestans determine virulence on R2 plants. New Phytologist, 191, 763–776. [DOI] [PubMed] [Google Scholar]
- Guo, H. , Ahn, H.K. , Sklenar, J. , Huang, J. , Ma, Y. , Ding, P. et al (2020) Phosphorylation‐regulated activation of the Arabidopsis RRS1‐R/RPS4 immune receptor complex reveals two distinct effector recognition mechanisms. Cell Host & Microbe, 27, 769–781.e6. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. et al (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Huang, S. , van der Vossen, E.A.G. , Kuang, H. , Vleeshouwers, V.G.A.A. , Zhang, N. , Borm, T.J.A. et al (2005) Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. The Plant Journal, 42, 251–261. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Lu, X. , Wu, H. , Xie, Y. , Peng, Q. , Gu, L. et al (2020) Phytophthora effectors modulate genome‐wide alternative splicing of host mRNAs to reprogram plant immunity. Molecular Plant, 10.1016/j.molp.2020.07.007 [DOI] [PubMed] [Google Scholar]
- Jo, K.R. (2013) Unveiling and Deploying Durability of Late Blight Resistance in Potato. PhD thesis, Wageningen University and Research. [Google Scholar]
- Jouet, A. , Saunders, D.G.O. , McMullan, M. , Ward, B. , Furzer, O. , Jupe, F. et al (2019) Albugo candida race diversity, ploidy and host‐associated microbes revealed using DNA sequence capture on diseased plants in the field. New Phytologist, 221, 1529–1543. [DOI] [PubMed] [Google Scholar]
- Jupe, F. , Witek, K. , Verweij, W. , Śliwka, J. , Pritchard, L. , Etherington, G.J. , et al. (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB‐LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. The Plant Journal, 76, 530–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. and Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Shen, H. , Zhou, Q. , Qian, K. , van der Lee, T. and Huang, S. (2017) Changing ploidy as a strategy: the Irish potato famine pathogen shifts ploidy in relation to its sexuality. Molecular Plant‐Microbe Interactions, 30, 45–52. [DOI] [PubMed] [Google Scholar]
- Lin, X. , Armstrong, M. , Baker, K. , Wouters, D. , Visser, R.G.F. , Wolters, P.J. et al (2020) RLP/K enrichment sequencing; a novel method to identify receptor‐like protein (RLP) and receptor‐like kinase (RLK) genes. New Phytologist, 227, 1264–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh, B.Q. , Schmidt, H.A. , Chernomor, O. , Schrempf, D. , Woodhams, M.D. , von Haeseler, A. et al (2020) IQ‐TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.K. , Young, C. , Lee, M. , Oliva, R. , Bozkurt, T.O. , Cano, L.M. et al (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi‐blb2 . The Plant Cell, 21, 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva, R.F. , Cano, L.M. , Raffaele, S. , Win, J. , Bozkur, T.O. , Belhaj, K. et al (2015) A recent expansion of the RXLR effector gene Avrblb2 is maintained in global populations of Phytophthora infestans indicating different contributions to virulence. Molecular Plant‐Microbe Interactions, 28, 901–912. [DOI] [PubMed] [Google Scholar]
- Pel, M. (2010) Mapping, Isolation and Characterization of Genes Responsible for Late Blight Resistance in Potato. PhD thesis, Wageningen University and Research. [Google Scholar]
- Pel, M.A. , Foster, S.J. , Park, T.H. , Rietman, H. , van Arkel, G. , Jones, J.D. et al (2009) Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Molecular Plant‐Microbe Interactions, 22, 601–615. [DOI] [PubMed] [Google Scholar]
- Peterson, D.G. , Wessler, S.R. and Andrew, H.P. (2002) Efficient capture of unique sequences from eukaryotic genomes. Journal of Integrative Agriculture, 18, 547–550. [DOI] [PubMed] [Google Scholar]
- Rehmany, A.P. , Gordon, A. , Rose, L.E. , Allen, R.L. , Armstrong, M.R. , Whisson, S.C. et al (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. The Plant Cell, 17, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietman, H. (2011) Putting the Phytophthora infestans Genome Sequence at Work: Multiple Novel Avirulence and Potato Resistance Gene Candidates Revealed. PhD thesis, Wageningen University and Research. [Google Scholar]
- Rietman, H. , Bijsterbosch, G. , Cano, L.M. , Lee, H.‐R. , Vossen, J.H. , Jacobsen, E. et al (2012) Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Molecular Plant‐Microbe Interactions, 25, 910–919. [DOI] [PubMed] [Google Scholar]
- Salaman, R. (1937) Potato variety production: a new departure. Gardeners Chronicle, 102, 326–327. [Google Scholar]
- Thilliez, G.J.A. , Armstrong, M.R. , Lim, T.Y. , Baker, K. , Jouet, A. , Ward, B. et al (2019) Pathogen enrichment sequencing (PenSeq) enables population genomic studies in oomycetes. New Phytologist, 221, 1634–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toxopeus, H.J. (1956) Reflections on the origin of new physiologic races in Phytophthora infestans and the breeding for resistance in potatoes. Euphytica, 5, 221–356. [Google Scholar]
- Van de Weyer, A.‐L. , Monteiro, F. , Furzer, O.J. , Nishimura, M.T. , Cevik, V. , Witek, K. et al (2019) A species‐wide inventory of NLR genes and alleles in Arabidopsis thaliana . Cell, 178, 1260–1272.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.‐K. et al (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE, 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A. , Raffaele, S. , Vossen, J.H. , Champouret, N. , Oliva, R. , Segretin, M.E. et al (2011) Understanding and exploiting late blight resistance in the age of effectors. Annual Review of Phytopathology, 49, 507–531. [DOI] [PubMed] [Google Scholar]
- van der Vossen, E. , Sikkema, A. , Hekkert, B.T.L. , Gros, J. , Stevens, P. , Muskens, M. et al (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad‐spectrum resistance to Phytophthora infestans in cultivated potato and tomato. The Plant Journal, 36, 867–882. [DOI] [PubMed] [Google Scholar]
- Vossen, J.H. , van Arkel, G. , Bergervoet, M. , Jo, K.R. , Jacobsen, E. and Visser, R.G.F. (2016) The Solanum demissum R8 late blight resistance gene is an Sw‐5 homologue that has been deployed worldwide in late blight resistant varieties. Theoretical and Applied Genetics, 129, 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witek, K. , Jupe, F. , Witek, A.I. , Baker, D. , Clark, M.D. and Jones, J.D.G. (2016) Accelerated cloning of a potato late blight–resistance gene using RenSeq and SMRT sequencing. Nature Biotechnology, 34, 656–660. [DOI] [PubMed] [Google Scholar]
- Witek, K. , Lin, X. , Karki, H.S. , Jupe, F. , Witek, A.I. , Steuernagel, B. et al (2020) A complex non‐host resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. bioRxiv, [preprint]. 10.1101/2020.05.15.095497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Enrichment efficiency with/without non‐adaptor‐ligated DNA
FIGURE S2 Comparison of EU_13_A2 Avr1 contigs and the T30‐4 reference genome
TABLE S1 278 RxLR effectors from previously available effector libraries
TABLE S2 P/A polymorphism of RxLR effectors from EU_13_A2, EC1_A1, EU_6_A1, and US23
TABLE S3 cDNA PenSeq for EU_13_A2, EC1_A1, EU_6_A1, and US23. Raw data of differential expression in Figure 2. Raw transcript counts in Figure 3
NOTES S1 PacBio PenSeq contigs of EU_13_A2
NOTES S2 PacBio PenSeq contigs of EC1_A1
NOTES S3 PacBio PenSeq contigs of EU_6_A1
NOTES S4 PacBio PenSeq contigs of US23
NOTES S5 Protein sequences of Avramr1 homologs
Data Availability Statement
Raw PacBio and cDNA PenSeq read sequences have been deposited in the Sequence Read Archive (SRA) at https://www.ncbi.nlm.nih.gov/sra under BioProject IDs PRJNA623167 and PRJNA598824.


