Abstract
Ribosomes play an integral part in plant growth, development, and defence responses. We report here the role of ribosomal protein large (RPL) subunit QM/RPL10 in nonhost disease resistance. The RPL10‐silenced Nicotiana benthamiana plants showed compromised disease resistance against nonhost pathogen Pseudomonas syringae pv. tomato T1. The RNA‐sequencing analysis revealed that many genes involved in defence and protein translation mechanisms were differentially affected due to silencing of NbRPL10. Arabidopsis AtRPL10 RNAi and rpl10 mutant lines showed compromised nonhost disease resistance to P. syringae pv. tomato T1 and P. syringae pv. tabaci. Overexpression of AtRPL10A in Arabidopsis resulted in reduced susceptibility against host pathogen P. syringae pv. tomato DC3000. RPL10 interacts with the RNA recognition motif protein and ribosomal proteins RPL30, RPL23, and RPS30 in the yeast two‐hybrid assay. Silencing or mutants of genes encoding these RPL10‐interacting proteins in N. benthamiana or Arabidopsis, respectively, also showed compromised disease resistance to nonhost pathogens. These results suggest that QM/RPL10 positively regulates the defence and translation‐associated genes during nonhost pathogen infection.
Keywords: disease resistance, plant immunity, ribosome, RNA sequencing, translation regulation
Ribosomal protein QM/RPL10 and its interacting proteins RRM, RPL30, RPL23, and RPS30 have extraribosomal functions and play a role in nonhost disease resistance.
1. INTRODUCTION
Ribosomal proteins are an integral part of ribosomes and are involved in their biogenesis and assembly, thus regulating protein synthesis. It is difficult to attribute an individual function of translational activity to a single ribosomal protein. This difficulty is rooted in the highly cooperative nature of the interactions between ribosomal ribonucleic acid (rRNA) and ribosomal proteins. Ribosomal proteins regulate their own synthesis by controlling the expression of their transcripts in association with transcription factors. The ribosome consists of large and small subunits. In eukaryotes, 47 ribosomal protein large (RPL) subunits and 32 ribosomal protein small (RPS) subunits form a ribosome complex with rRNAs (Ben‐Shem et al., 2011). Interestingly, extraribosomal functions of many ribosomal proteins have been reported (Wool, 1996; Freed et al., 2010). Several ribosomal protein‐encoding genes are shown to be differentially expressed under environmental stress conditions (Saha et al., 2017; Vemanna et al., 2019).
The role of ribosomal proteins in plant immunity has not been well studied. Plants have a sophisticated and layered defence mechanism against invading pathogens. Plants can perceive pathogen‐associated molecular patterns (PAMPs) through their pattern recognition receptors and induce a general defence response, termed PAMP‐triggered immunity (PTI; Jones and Dangl, 2006). A few successful pathogens can overcome or suppress PTI by delivering virulence proteins or metabolites termed as effectors. Certain plants have the ability to detect such effectors and trigger a strong defence response called effector‐triggered immunity (ETI; Feng and Zhou, 2012). Another type of plant defence response, operating under less understood mechanisms, provides resistance against pathogens throughout all members of a species and is referred to as nonhost disease resistance (Heath, 2000; Mysore and Ryu, 2004; Senthil‐Kumar and Mysore, 2013; Ayliffe and Sorensen, 2019). A pathogen that cannot cause disease in a nonhost plant is referred to as a nonhost pathogen. Nonhost resistance can be used to confer broad and durable disease resistance in crop plants (Gill et al., 2015; Fonseca and Mysore, 2019). Although nonhost resistance mechanisms are not fully understood, a variety of preformed and inducible responses are implicated (Mysore and Ryu, 2004; Senthil‐Kumar and Mysore, 2013).
Ribosomal proteins are involved in basic cellular machinery and any abnormalities in these mechanisms may lead to compromised disease resistance. Multiple rare genetic diseases have been attributed to defects in ribosome function in humans and other mammals (Freed et al., 2010). Silencing of RPL12 and RPL19 in Nicotiana benthamiana and Arabidopsisthaliana showed compromised nonhost disease resistance against multiple bacterial pathogens (Nagaraj et al., 2016). RPL12 interacting receptor for activated C‐kinase 1 (RACK1) was identified in Arabidopsis (Kundu et al., 2013) and plays a key role in plant innate immunity (Nakashima et al., 2008; Wang et al., 2014). Differential expression of genes encoding ribosomal proteins has been reported in vanilla in response to infection by Fusarium oxysporum f. sp. vanillae (Solano‐De la Cruz et al., 2019). Similarly, induction of RPS10 in soybean in response to Phytophthora sojae (Zhang et al., 2013) and induction of RPS6, RPL19, RPL13, RPL7, and RPS2 associated with the plant response to turnip mosaic virus (TuMV), tobacco mosaic virus (TMV), and tomato bushy stunt virus (TBSV) have been reported (Yang et al., 2009). In a rice genotype resistant to Xanthomonas bacterial infection, 50 genes encoding ribosomal proteins were up‐regulated (Narsai et al., 2013). The differential responses of all the ribosomal protein‐encoding genes, including small and large subunit encoding genes, have been studied using genome‐wide studies in rice in response to multiple stress conditions (Moin et al., 2016; Saha et al., 2017). Even though many stress‐induced ribosomal protein‐encoding genes have been reported, the precise function of each ribosomal protein during stress is not known (Vemanna et al., 2019).
Ribosomal proteins also possess extraribosomal functions such as regulation of transcription, chaperone activity, and protein phosphorylation in addition to their role in protein synthesis (Kubo and Arimura, 2010). A tomato QM)‐like protein (chromosome 11 [Q26, 23]) has been shown to be involved in regulating oxidative stress (Chen et al., 2006). QM was able to rescue the Proline dehydrogenase (PUT1) mutant of Saccharomyces cerevisiae from oxidative stress (Chen et al., 2006). QM10 was initially reported as a Wilm's tumour suppressor gene in humans and has homology with RPL10 (Rivera‐Madrid et al., 1993; Shi et al., 2004; Dong et al., 2007; Chen et al., 2011). QM/RPL10 interacts with c‐Jun transcription factor in humans and has been shown to be involved in regulating reactive oxygen species (ROS) levels in prostate cancer patients (Yang et al., 2018). RPL10 is a 60S large subunit protein mainly involved in ribosome biogenesis and assembly of both large and small subunits (Eisinger et al., 1997; Loftus et al., 1997). RPL10 proteins are present at the stalk of ribosomes and are involved in the rotation of ribosomes to recognize the mRNA strand for protein synthesis (Sulima et al., 2014). Arabidopsis has three QM/RPL10 protein‐encoding gene family members that are differentially regulated by various stress stimuli. A specific set of genes is expressed in different individual rpl10 Arabidopsis mutants, signifying their role in the regulation of specific mechanisms (Falcone Ferreyra et al., 2013). Nuclear shuttle protein‐interacting kinase 1 (NIK1) phosphorylates RPL10 during viral disease signalling and translocates to the nucleus to associate with MYB transcription factor (Carvalho et al., 2008). The Arabidopsis rpl10a mutant shows reduced plant protein synthesis and reduced viral protein synthesis, resulting in improved tolerance to viral infection (Rocha et al., 2008; Zorzatto et al., 2015). The NIK1 protein is homologous to a well‐characterized plant defence protein called Botrytis‐induced kinase1 (BRI1)‐associated receptor kinase 1 (BAK1). Arabidopsis nik1/bik1 mutants exhibited resistance against Pseudomonas syringae pv. tomato DC3000 (Lal et al., 2018; Li et al., 2019).
Here we report the role of QM/RPL10 in nonhost disease resistance against bacterial pathogens. NbRPL10‐silenced N. benthamiana plants and Arabidopsis rpl10 mutants showed higher nonhost bacterial multiplication. The transcriptome profiling of NbRPL10‐silenced plants identified differential expression of several plant defence‐related genes and protein translation‐associated genes. Furthermore, we identified and characterized QM/RPL10 interacting proteins and determined their role in nonhost disease resistance. This study demonstrates that RPL10 acts as a central regulator of many ribosomal proteins and plant defence responses.
2. RESULTS
2.1. Silencing of QM10/RPL10 in N. benthamiana compromises nonhost disease resistance
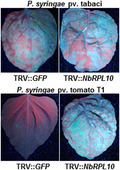
We performed a virus‐induced gene silencing (VIGS)‐based forward genetics screen in N. benthamiana to identify genes involved in nonhost disease resistance (Rojas et al., 2012; Wang et al., 2012; Senthil‐Kumar et al., 2013; Kaundal et al., 2017). One of the cDNA clones NbME23C12 (Senthil‐Kumar et al., 2018; https://vigs.noble.org/line2.php?id=NbME23C12) compromised nonhost disease resistance when silenced. The NbME23C12 sequence has homology with Arabidopsis QM/RPL10 family genes, and the domain architecture clearly shows that it belongs to the ribosomal L16_L10e superfamily of genes (Figure S1a). In Arabidopsis, RPL10 has three gene family members, A, B, and C, whereas we could find only two of them in N. benthamiana and named them NbRPL10A and NbRPL10B. To test whether silencing of a specific NbRPL10 gene in N. benthamiana compromises nonhost resistance, the VIGS constructs with different regions specifically targeting NbRPL10A or NbRPL10B or both were designed using the PssRNAit webserver tool (Ahmed et al., 2020) with minimum off‐target genes, PCR amplified, and cloned into TRV2 VIGS vector (Senthil‐Kumar and Mysore, 2014; Figure S1b). Silencing of the NbRPL10A clone showed stunted plants with variegated, mottled, and crinkled leaves (Figure 1a). The silenced plants also showed cell death starting at the petiole and midrib. Interestingly, silencing of NbRPL10B did not produce a variegated leaf phenotype when compared to NbRPL10A or NbRPL10A + NbRPL10B (NbRPL10s)‐silenced plants (Figure 1a). All silenced plants showed more than 50% down‐regulation of target transcripts (Figure S1c). To test the response of NbRPL10‐silenced plants to pathogens, silenced plants and control (TRV::GFP inoculated; the green fluorescent protein gene, GFP, has no sequence similarity to plant genomic DNA and thus will not cause gene silencing) were vacuum infiltrated with GFPuv (Wang et al., 2007)‐expressing nonhost pathogen P. syringae pv. tomato T1 and host pathogen P. syringae pv. tabaci at 104 cfu/ml concentration. All the silenced plants had a higher accumulation of nonhost bacteria P. syringae pv. tomato T1 than the control plants (Figures 1b,c and S1d,e). In contrast, the host pathogen P. syringae pv. tabaci multiplied to the same extent in both control and silenced plants. These results suggest that NbRPL10 silencing in N. benthamiana compromises nonhost resistance but not basal resistance.
FIGURE 1.
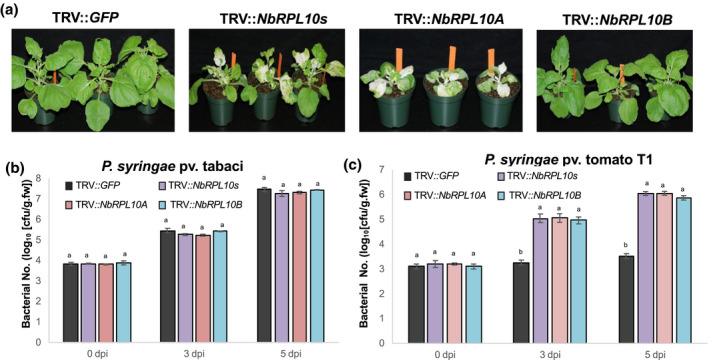
Silencing NbQM/NbRPL10 in Nicotiana benthamiana enhances multiplication of nonhost pathogen Pseudmonas syringae pv. tomato T1. (a) Visualization of developmental changes in N. benthamiana plants individually inoculated with TRV::NbRPL10s (silences both NbRPL10A and NbRPL10B), TRV::NbRPL10A, TRV::NbRPL10B, and TRV::GFP (control; GFP does not have any sequence similarity to plant DNA and therefore will not cause gene silencing). Three weeks after TRV inoculation, N. benthamiana plants were vacuum‐infiltrated with host pathogen P. syringae pv. tabaci or nonhost pathogen P. syringae pv. tomato T1 at 104 cfu/ml concentration. Photographs were taken 3 days postinoculation (dpi). (b) and (c) Quantification of host and nonhost bacterial multiplication in TRV::NbRPL10‐silenced and TRV::GFP inoculated plants at 0, 3, and 5 dpi. Bars represent average values of three biological replicates and experiments were repeated three times with similar results. Error bars indicate standard error. Different letters above the bars indicate a significant difference from two‐way analysis of variance at p < .05 with Tukey's HSD means separation test (α = .05) within a time point among respective control and silenced plants.
2.2. NbQM/NbRPL10‐silenced N. benthamiana plants have fewer differentially expressed genes after nonhost pathogen treatment
To understand the molecular mechanisms involved in nonhost disease resistance by NbQM/NbRPL10 silencing in N. benthamiana, we generated a transcriptome profile of NbRPL10A‐silenced and control (TRV::GFP) plants by RNA‐Seq after inoculation with the nonhost pathogen P. syringae pv. tomato T1 at 104 cfu/ml concentration. The differentially expressed gene (DEG) analysis of RNA‐Seq data identified a diverse list of genes that play a defence role in QM/RPL10‐mediated nonhost resistance. When NbRPL10A was silenced in N. benthamiana, molecular‐function associated genes were highly represented in the DEGs in both control and nonhost pathogen treatment. Following this group, the genes that encode proteins having catalytic, biosynthetic, and transferase activities were highly represented among DEGs. Interestingly, the expression of genes associated with morphology, cell proliferation, nuclear chromosomes, and embryonic development were not affected (Figure S2a,b).
In the NbRPL10A‐silenced plants, 2,928 and 3,134 genes were up‐ or down‐regulated, respectively, when compared to the control plant without any pathogen inoculation. On nonhost pathogen infection (12 hr postinoculation, hpi), 2,844 and 2,708 genes were up‐ or down‐regulated, respectively, in NbRPL10‐silenced plants when compared to control. There were 619 and 577 genes that were up‐ or down‐regulated commonly between 0 and 12 hpi samples (Figure 2a). We compared the DEGs to assess the differences in control and NbRPL10A‐silenced N. benthamiana plants in response to nonhost pathogen P. syringae pv. tomato T1. Interestingly, we observed that more genes were differentially regulated in response to the nonhost pathogen in control plants than the NbRPL10‐silenced plants (Figure 2b). These results suggest that NbQM/NbRPL10A‐silenced plants were muted to some extent in inducing defence responses when compared to control. The data also suggest that the expression of these genes could be directly or indirectly regulated by NbQM/NbRPL10.
FIGURE 2.
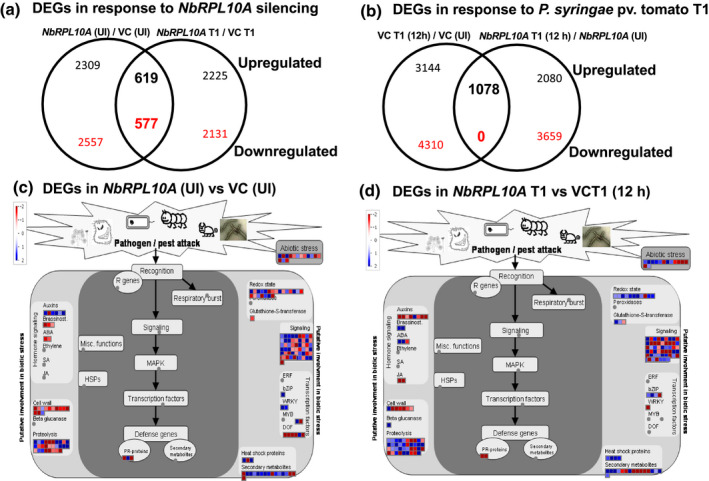
Transcriptome analysis of NbRPL10A‐silenced Nicotiana benthamiana plants treated with nonhost pathogen. Two‐week‐old N. benthamiana plants were inoculated with TRV::GFP (vector control‐VC, UI‐ uninfected) or TRV::NbRPL0A constructs. Three weeks after TRV inoculation, the plants were vacuum‐infiltrated with nonhost pathogen Pseudomonas syringae pv. tomato T1 at 104 cfu/ml concentration. Leaf samples were collected 12 hr after pathogen inoculation, RNA was isolated and subjected to RNA‐Seq experiment. (a) Venn diagram showing up‐ and down‐regulated genes in NbRPL10A‐silenced plants over vector control uninfected (VC UI) samples and 12 hr after pathogen treatment. (b) Venn diagram showing up‐ and down‐regulation of genes in NbRPL10A‐silenced or VC plants 12 hr after pathogen treatment compared to uninfected. (c) MapMan analysis of differentially regulated genes in uninfected NbRPL10A silenced plants compared to uninfected VC. (d) Differential expression of genes in NbRPL10A‐silenced plants compared to VC 12 hr after nonhost pathogen inoculation
2.3. Genes involved in pathogen signalling and protein translational processes were differentially expressed in NbQM/NbRPL10A‐silenced N. benthamiana plants
The DEGs from the transcriptome profiling experiment described above were mapped using MapMan analysis to identify the key pathways that are differentially regulated due to NbQM/NbRPL10A silencing in N. benthamiana and to identify the pathways responsible for compromised disease resistance against nonhost pathogen P. syringae pv. tomato T1 (Figure 2c,d). Many genes associated with abiotic stress response, redox state, signalling, proteolysis, and cell wall mechanisms were up‐regulated, indicating that the plants were experiencing stress in NbRPL10A‐silenced plants without the nonhost pathogen infection compared to nonsilenced control plants (Figure 2c). The redox genes like Thioredoxin, Ascorbate peroxidase, and Glutaredoxin were up‐regulated more than 2.5‐fold, and others like Dehydroascorbate reductase, 2‐oxoglutarate dependent dioxygenase, and so on were up‐regulated more than 2‐fold. Furthermore, the transcription factor‐encoding genes WRKY41, WRKY53, MYB, and Dof Zinc finger were up‐regulated more than 3‐fold. In addition, leucine‐rich repeat (LRR) genes, MAP kinase‐encoding genes, and genes involved in calcium signalling were up‐regulated more than 2‐fold (Data S1). Interestingly, when the NbRPL10A‐silenced plants were infected with a nonhost pathogen, the transcripts of genes that are associated with abiotic stress (early responsive to dehydration [ERD3, ERD15], Dehydration‐responsive element‐binding [DREB], basic zipper [bZIP], and WRKY41), redox state (Glutaredoxin), proteolytic mechanisms (genes encoding C3HC4‐type RING finger family and LysM domain containing proteins), phytohormone signalling (Lipoxygenase LOX3, Allene oxide synthase‐ AOS, Biogenic amine synthase BAS1), pathogen defence (genes encoding LRR, trypsin and protease inhibitor family proteins) and cell‐wall‐associated genes (UDP‐gluconic acid deacetylase, Epimerase, Lyase and a gene‐encoding polygalactorunase inhibiting protein) were significantly less when compared to nonsilenced control (Data S1). These results suggest that, unlike the nonsilenced control, NbRPL10A‐silenced plants were not able to induce expression of several stress‐ or defence‐associated genes upon infection with a nonhost pathogen. This could be due to the regulatory role of QM/RPL10 in controlling the expression of many genes associated with stress or defence response.
Furthermore, we identified 17 up‐regulated and 13 down‐regulated genes associated with plant defence responses in uninfected NbRPL10A‐silenced plants compared to nonsilenced control plants. Upon nonhost pathogen infection, nine defence‐related genes were up‐regulated and 17 defence‐related genes were down‐regulated in NbRPL10A‐silenced plants compared to nonsilenced control (Figure 3a and Data S2). As expected, many DEGs between the NbRPL10A‐silenced plant and the nonsilenced control plant were associated with protein translation mechanisms (Figure 3 and Table 1), with 238 up‐regulated and 855 genes down‐regulated in uninfected NbRPL10A‐silenced plants compared to the control. Upon nonhost pathogen infection, there were 148 translation‐associated genes up‐regulated and 286 genes down‐regulated in NbRPL10A‐silenced plants compared to the control (Table 1 and Data S2).
FIGURE 3.
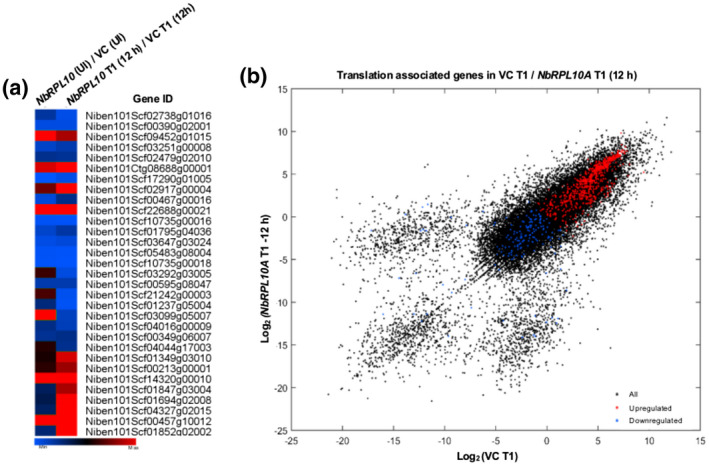
Plant defence and protein translation‐associated genes that are differentially expressed in NbRPL10A‐silenced Nicotiana benthamiana plants. A select number of genes from the data presented in Figure 2 is shown here. (a) Heat map showing differentially expressed genes that are known to play a role in plant defence. The map was generated using multiple expression viewer (MeV). (b) Distribution of differentially expressed protein translation‐associated genes from the whole genome RNA‐Seq data. A total of 6,817 genes (black dots) were used for analysis, out of which 419 genes were up‐regulated (red) and 494 genes were down‐regulated (blue) in NbRPL10A‐silenced plants
TABLE 1.
Differentially expressed genes related to defence and translation mechanisms
| Processes | NbRPL10 (UI)/VC (UI) | NbRPL10 A T1/VC T1 (12h) | VC T1 (12h)/VC (UI) | NbRPL10A T1 (12h)/NbRPL10 (UI) | |
|---|---|---|---|---|---|
| Defence response | Up‐regulated | 17 | 9 | 23 | 31 |
| Down‐regulated | 13 | 17 | 13 | 16 | |
| Translation | Up‐regulate | 238 | 148 | 201 | 208 |
| Down‐regulated | 855 | 286 | 527 | 33 | |
| Total | Up‐regulated | 2,927 | 2,843 | 4,221 | 3,157 |
| Down‐regulated | 3,133 | 2,707 | 4,310 | 3,659 | |
In NbRPL10A‐silenced plants without any pathogen infection, the transcripts that are involved in organelle organization, cellular metabolic process, oxidation reduction, small molecule metabolic process, response to diverse stimulus, and signalling genes were highly represented in the down‐regulated gene set compared to the control (Figure S3). Similarly, genes that are involved in cell death, regulation of immune system process, positive regulation of biological organellar process, ribonucleoprotein complex, protein complex biogenesis, photosynthesis, secondary metabolite synthesis, and so on were up‐regulated in uninfected NbRPL10A‐silenced plants (Figure S4). Upon inoculation with nonhost pathogen P. syringae pv. tomato T1, genes involved in cell death, regulation of immune response, signalling, response to diverse stresses, flavonoid synthesis, redox mechanisms, and salicylic acid (SA) and jasmonic acid (JA) metabolism were highly represented in the down‐regulated set in NbRPL10A‐silenced plants when compared to the control (Figure S5). The genes that are involved in systemic acquired resistance and redox metabolism were also represented in the up‐regulated gene set in NbRPL10A‐silenced plants on nonhost pathogen infection compared to the control, but were fewer in number compared to down‐regulated genes (Figure S6). The transcriptome data clearly suggest that QM/RPL10 regulates several genes involved in plant defence signalling and translation mechanisms (Figure S7). The transcriptome data further revealed down‐regulation of Phytoene synthase and Phytoene desaturase genes in NbRPL10A‐silenced plants when compared to the control, which correlates with the photobleaching phenotype observed in silenced plants (Figure 1a).
2.4. Overexpression of AtRPL10A in Arabidopsis reduces susceptibility to host bacteria and RNAi, and mutants show compromised resistance to nonhost pathogens
To determine if the function of QM/RPL10 in nonhost disease resistance is conserved in other plant species, we identified qm/rpl10 Arabidopsis mutants and generated AtRPL10A overexpression lines in Arabidopsis. We also generated RNAi lines that can silence all the family members (RPLs) and lines that can specifically silence AtRPL10A or AtRPL10B. Homozygous lines were made for all transgenic and mutant lines, and gene overexpression or down‐regulation was confirmed by quantitative reverse transcription PCR (RT‐qPCR) (Figure S8). Arabidopsis Atrpl10b mutants were embryo‐lethal. Therefore, AtRPL10B RNAi lines were generated. All the Arabidopsis mutants and transgenic lines mentioned above along with wild‐type (Col‐0) were inoculated with nonhost pathogens P. syringae pv. tabaci, P. syringae pv. tomato T1, and host pathogen P. syringae pv. tomato DC3000 by flood inoculation using 105 cfu/ml bacterial concentration. Overexpression of AtRPL10A resulted in less accumulation of host pathogen P. syringae pv. tomato DC3000 and produced fewer disease symptoms (Figure 4a,b). Similar to N. benthamiana results, AtRPL10A, AtRPL10B, and AtRPLs RNAi lines were susceptible to nonhost pathogen (Figure 4a,c,d). In addition, Atrpl10a and Atrpl10c mutants were also susceptible to nonhost pathogens P. syringae pv. tabaci and P. syringae pv. tomato T1 (Figure 4a,c,d). These results suggest that QM/RPL10 also plays a positive role in Arabidopsis nonhost resistance against bacterial pathogens.
FIGURE 4.
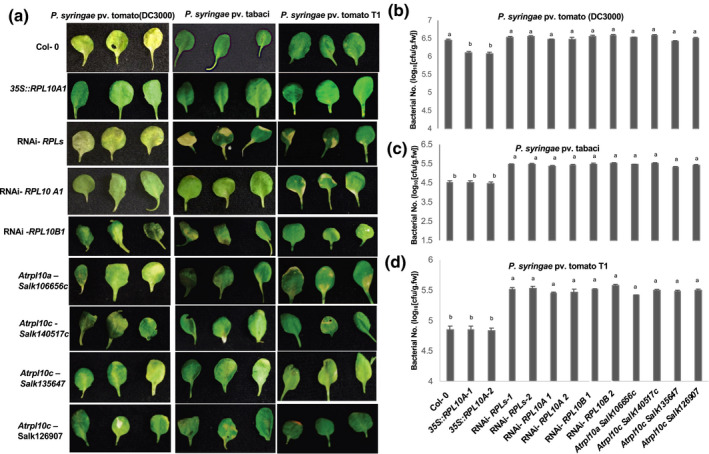
AtRPL10 RNAi and Atrpl10 mutant plants compromise nonhost disease resistance, and AtRPL10A overexpression plants show less susceptibility to host pathogen. Four‐week‐old Arabidopsis AtRPL10A overexpressors (35S::AtRPL10A‐1 and 35S::AtRPL10A‐2), AtRPL10s RNAi (RNAi‐AtRPL10s‐1and RNAi‐AtRPL10s‐2), AtRPL10A RNAi (RNAi‐ AtRPL10A‐1 and RNAi‐AtRPL10A‐2), AtRPL10B RNAi (RNAi‐AtRPL10B‐1 and RNAi‐AtRPL10B‐2), rpl10a mutant (Salk106656c), rpl10c mutants (Salk140517c, Salk135647, and Salk126907), and wild‐type Col‐0 were flood‐inoculated with host pathogen Pseudomonas syringae pv. tomato DC3000 or nonhost pathogens P. syringae pv. tabaci or P. syringae pv. tomato T1 at 105 cfu/ml concentration. (a) Photographs of a few representative lines showing disease symptoms after inoculation with host and nonhost pathogens. Photographs were taken 3 days postinoculation (dpi). (b)–(d) Bacterial titre was assessed at 3 dpi from leaves of seedlings that were flood‐inoculated with host and nonhost pathogens. Error bars represent the standard error for three biological replicates in three independent experiments. Different letters above the bars indicate a significant difference from two‐way analysis of variance at p < .05 with Tukey's HSD means separation test (α = .05)
2.5. RPL10 interacts with other ribosomal proteins involved in translation mechanisms
To understand the molecular mechanism of QM/RPL10‐mediated nonhost resistance, the Arabidopsis AtRPL10A was cloned into yeast two‐hybrid (Y2H) bait vector pDEST32 and screened against the Arabidopsis stress‐induced library (Lee et al., 2017). Several putative RPL10A‐interacting proteins were identified from this screen (Table S2). Among them, chloroplast, cytoplasmic‐intracellular‐component‐associated, and ribosomal proteins were identified. Many of these proteins have binding function, enzymatic activity, and other molecular functions (Figure S9). The ribosomal proteins such as AtRPL23, AtRPL30, AtRPS30, and RNA‐recognition motif (AtRRM) were found to interact with AtRPL10A in Y2H assay. Furthermore, to understand the role of protein translational mechanisms in plant defence, Arabidopsis ribosomal protein‐encoding full‐length genes of AtRPL23, AtRPL30, AtRPS30, and AtRRM were cloned in pDEST22 and we confirmed their encoded protein interaction with AtRPL10A by Y2H assay. The X‐Gal assay on triple‐dropout media further confirms their interaction (Figure 5a). Furthermore, the Arabidopsis proteins interacting with AtRPL10A were predicted by in silico analysis using STRING protein–protein interaction networks by functional enrichment analysis (https://string‐db.org). The analysis indicated that several ribosomal proteins potentially interact with AtRPL10A along with the identified proteins from Y2H assay (Figure 5b).
FIGURE 5.
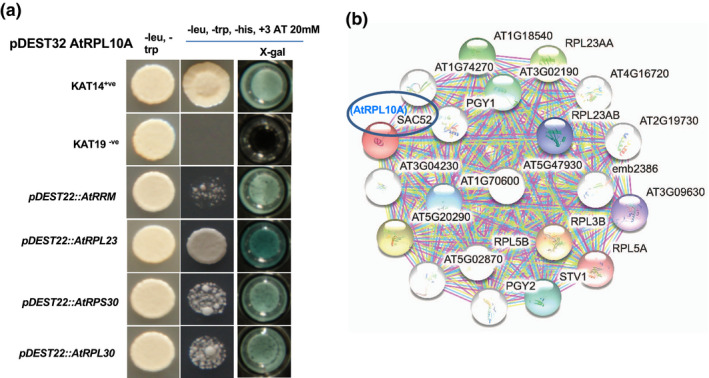
Identification of AtRPL10A‐interacting proteins. (a) Yeast two‐hybrid (Y2H) assay to confirm the protein–protein interactions of a few selected proteins that were identified from the Y2H library screen. The AtRPL10A full‐length gene cloned in pDEST32 and AtRPL23, AtRPL30, AtRPS30, and AtRRM genes were cloned in pDEST22 and cotransformed into MaV203 yeast cells, then were grown on double (−leu, −trp) or triple (−leu, −trp, −his with 20 mM 3‐AT) drop‐out media. Krev1 RalGDS and Krev1 RalGDS‐m1 were used as positive and negative controls, respectively. The X‐Gal staining was done to confirm the interaction of the two proteins. (b) Representative image showing in silico prediction of AtRPL10A‐interacting proteins using STRING online tool (https://string‐db.org)
The AtRPL10B‐ and AtRPL10C‐interacting proteins were also predicted (Figure S10a,b), and we found that 13 common proteins, including AtRPL23, interact with all the RPL10s, that is, 10A, 10B, and 10C (Figure S10c and Table S3). Three proteins are common between RPL10A and 10B, seven are common with 10A and 10C, and eight are common with 10B and 10C. A few unique proteins were also predicted to specifically interact with individual RPL10 proteins (Figure S10 and Table S3), suggesting their independent functions. These observations are consistent with a peptidyl transferase centre in the ribosome structure where RPL10 is found to play a role in assembly and rotation of the ribosome and therefore has to interact with many proteins. Based on the ribosome structure, several proteins, such L3, L5, L12, L20, L21, L23, L40, NMD3, and a few others, are in close proximity to RPL10. Some of these proteins were identified in our in silico and Y2H analyses (Tables S2 and S3).
2.6. Silencing of QM/RPL10‐interacting ribosomal protein‐encoding genes in N. benthamiana showed varied response to nonhost pathogen infection
To evaluate if the N. benthamiana homologs of Arabidopsis QM/RPL10‐interacting proteins play a role in nonhost disease resistance, we identified N. benthamiana cDNA clones from VIGS phenomics and the functional genomics database (Senthil‐Kumar et al., 2018). NbTI01C11 (NbRPL30), NbME01F11 (NbRPL23), NbME14G06 (NbRPS30), and NbME39D07 (NbRRM) clones were recovered from the library, and VIGS was performed in 4‐week‐old N. benthamiana plants. Silencing of NbRPL23 caused a variegated phenotype, and other selected genes when silenced caused stunted growth (Figure 6a). The silenced plants showed more than 50% down‐regulation of their respective transcripts (Figure 6b). These plants were vacuum‐infiltrated with host pathogen P. syringae pv. tabaci or with the nonhost pathogen P. syringae pv. tomato T1 at 104 cfu/ml concentration (Figure 6b). The N. benthamiana‐silenced plants did not show any difference in host bacterial multiplication when compared to control plants (Figure 6c). However, all the silenced plants showed significantly higher multiplication of nonhost pathogen P. syringae pv. tomato T1 when compared to the nonsilenced control (Figure 6d). These results suggest that most of the ribosomal proteins that potentially interact with QM/RPL10 also play a role in nonhost resistance against a bacterial pathogen in N. benthamiana.
FIGURE 6.
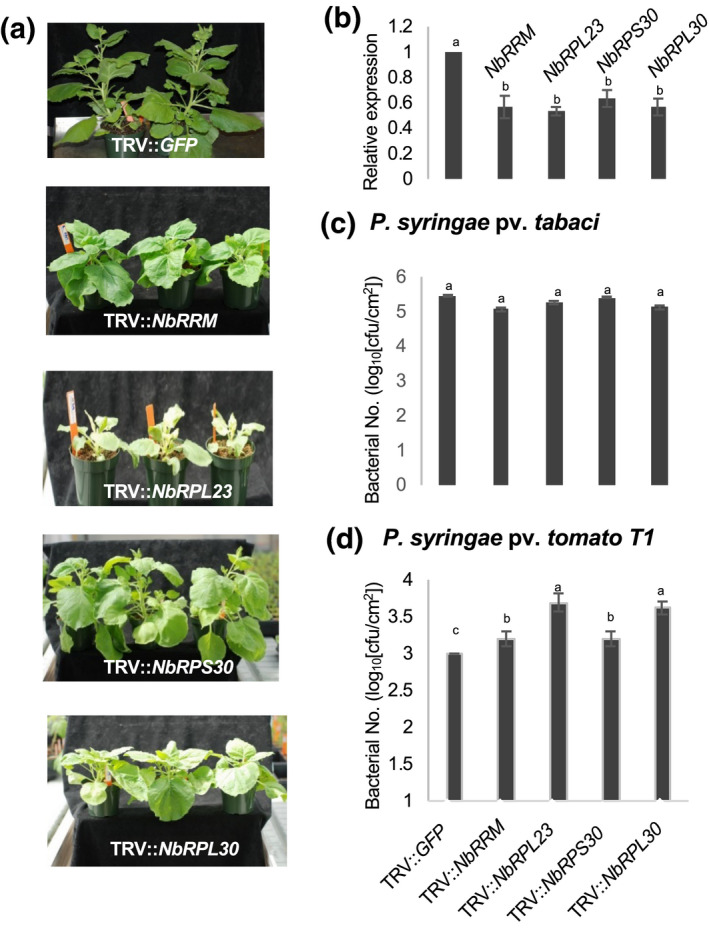
Silencing of RPL10A‐interacting ribosomal protein‐encoding genes in Nicotiana benthamiana compromises nonhost disease resistance. (a) Altered developmental phenotypes (dwarf, variegated, etc.) of N. benthamiana 3 weeks after inoculation with TRV::NbRPL23 or TRV::NbRPL30 or TRV::NbRPS30 or TRV::NbRRM or TRV::GFP (control). (b) Expression of respective genes in specific virus‐induced gene silenced N. benthamiana plants using quantitative reverse transcription PCR (c) Bacterial multiplication rate of host pathogen and (d) nonhost pathogen at 3 days postinoculation (dpi) on silenced plants. Three weeks after TRV inoculation, N. benthamiana plants were vacuum‐infiltrated with host pathogen Pseudomonas syringae pv. tabaci and nonhost pathogen P. syringae pv. tomato T1 at 104 cfu/ml concentration. Bars represent the average of three biological replicates. Experiments were repeated three times with similar results. Error bars indicate standard error. Different letters above the bars indicate a significant difference from two‐way analysis of variance at p < .05 with Tukey's HSD means separation test (α = .05)
2.7. Arabidopsis QM/RPL10‐interacting ribosomal protein‐encoding mutants show compromised resistance to nonhost pathogens
The Arabidopsis mutant lines Atrpl23, Atrpl30, and Atrrm were obtained from the Arabidopsis Biological Resource Center and were made homozygous. These mutants, along with the wild type (Col‐0), were flood‐inoculated with host pathogen P. syringae pv. tomato DC3000 and nonhost pathogen P. syringae pv. tomato T1 or P. syringae pv. tabaci at 105 cfu/ml concentrations. Arabidopsis rpl23c, rrm, and rpl30 mutants showed slightly reduced disease symptoms when treated with host pathogen P. syringae pv. tomato DC3000 (Figure 7a). However, the bacterial multiplication assay did not show any significant difference when compared to the wild type (Figure 7b). On the contrary, all the tested mutants showed compromised nonhost resistance against P. syringae pv. tomato T1 and P. syringae pv. tabaci (Figure 7a). The mutants showed increased bacterial multiplication with both the nonhost pathogens tested when compared to the wild type (Figure 7c,d). Taken together, these results suggest that in addition to QM/RPL10, some of their interacting proteins are also required for nonhost disease resistance against bacterial pathogens in N. benthamiana and Arabidopsis.
FIGURE 7.
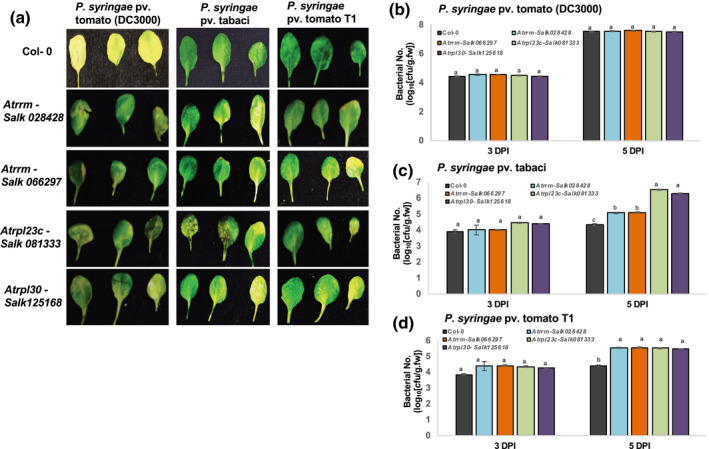
Response of Arabidopsis RPL10A‐interacting ribosomal protein‐encoding gene mutants to host and nonhost pathogens. Four‐week‐old Arabidopsis mutants Atrrm, Atrpl23, and Atrpl30 were flood inoculated with nonhost pathogens Pseudomonas syringae pv. tabaci, P. syringae pv. tomato T1, and host pathogen P. syringae pv. tomato DC3000 at 105 cfu/ml concentration. (a) Leaves of host and nonhost pathogen inoculated Arabidopsis mutants and wild‐type (Col‐0) were detached 3 days postinoculation (dpi) and photographs were taken. (b)–(d) Bacterial accumulation at 3 and 5 dpi was measured from leaves that were floodinoculated with P. syringae pv. tomato DC3000 or P. syringae pv. tabaci or P. syringae pv. tomato T1. Bars represent the average of three biological replicates in three independent experiments. Error bars represent the standard error. Different letters above the bars indicate a significant difference from two‐way analysis of variance at p < .05 with Tukey's HSD means separation test (α = .05) within a time point among respective wild‐type and mutant lines
3. DISCUSSION
Plant immunity against pathogens relies on the capacity of plant cells to detect pathogens and activate defence responses. Plants do not possess specialized defence cells, as most animals do. Some bacterial pathogens have evolved to evade plant defence by hijacking host cellular machinery to cause disease (Wang et al., 2007; Jones et al., 2016). However, the majority of potential plant pathogens (nonhost pathogens) cannot infect most plants due to a broad‐spectrum nonhost disease resistance mechanism (Heath, 2000; Mysore and Ryu, 2004). Using a VIGS‐based forward genetics screen, we identified and characterized several genes involved in nonhost disease resistance (Rojas et al., 2012; Senthil‐Kumar and Mysore, 2012; Wang et al., 2012; Kaundal et al., 2017; Lee et al., 2017). Previously, we reported that when silenced, two ribosomal genes NbRPL12 and NbRPL19 compromised nonhost disease resistance against P. syringae pv. tomato T1 (Nagaraj et al., 2016). In addition, many ribosomal protein‐encoding genes have been shown to be important for gene‐for‐gene mediated resistance using VIGS‐mediated forward genetic screens (Lu et al., 2003; Gabriels et al., 2006). Many plant defence‐related transcriptome profiles show differential regulation of ribosomal protein‐encoding genes (Vemanna et al., 2019; Solano‐De La Cruz et al., 2019). However, the role of ribosomal proteins in plant defence was not further characterized.
The involvement of protein translation machinery in plant defence is poorly studied. Some reports have shown that many ribosomal proteins are known to play extraribosomal functions, such as the involvement in plant–bacteria or plant–virus interactions (Gabriels et al., 2006; Yang et al., 2009). We identified NbME23C12, encoding QM/RPL10, in N. benthamiana that when silenced showed compromised nonhost disease resistance against P. syringae pv. tomato T1. The VIGS fragment predicted for NbRPL10A using newly designed webserver PssRNAit has identified fewer and precise off‐targets (Ahmed et al., 2020). The RNA‐Seq analysis revealed several defence‐related DEGs in NbRPL10‐silenced plants in the response to the nonhost pathogen P. syringae pv. tomato T1. For example, Flagellin sensitive 2 (Fls2) homolog was down‐regulated in NbRPL10‐silenced plants when compared to nonsilenced control. Arabidopsis fls‐2 mutant is hypersusceptible to the host pathogen P. syringae pv. tomato DC3000 (Zheng and He 2010). Arabidopsis fls‐2 mutant has also been shown to compromise nonhost resistance against several bacterial pathogens (Ishiga et al., 2011). We speculate that PAMP‐mediated plant defence is compromised in NbRPL10A‐silenced plants. RNA‐Seq data also indicated that many Ca2+ signalling‐associated genes were differentially regulated in NbRPL10‐silenced plants when compared to the control. Genes such as Calcium‐dependent protein kinase (CDPK or CPK)6, CPK3, and genes encoding the calmodulin binding EF motif family were up‐regulated, and genes like CPK13, CDPK19, Salt overlay sensitive (SOS3), Receptor lectin kinase (RLK), Wall associated kinase (WAK2), and a gene‐encoding calmodulin‐related binding protein were significantly down‐regulated in NbRPL10‐silenced plants. The PAMP signalling genes were induced at early stages of infection and subsequently down‐regulated in NbRPL10‐silenced plants with respect to nonhost pathogen. This is consistent with calcium signalling, where it is up‐regulated in response to PAMPs at early stages of infections and subsequently declines to a steady state (Blume et al., 2000; Ranf et al., 2008). We speculate that PAMP‐signalling mediated nonhost disease response is compromised in the NbRPL10‐silenced plants. However, it is intriguing that despite down‐regulation of PAMP signalling genes, the NbRPL10‐silenced plants did not compromise basal resistance to the host pathogen. Perhaps silencing of these genes is not enough to compromise basal resistance against the host pathogen but sufficient to compromise nonhost resistance.
The genes encoding AAA+‐ATPase family proteins were up‐regulated 10‐fold in NbRPL10‐silenced plants, which could lead to dissociation of macromolecular complexes such as H+‐ATPase involved in defence response (Kaundal et al., 2017). Multiple MAP kinases were down‐regulated in NbRPL10‐silenced plants, and transcription factor‐encoding genes like WRKY41 and C3H4‐Zinc finger family genes were also down‐regulated. Down‐regulation of Egg binding receptor (EBR1) or C3H4 type ring finger E3 ligase led to a hypersensitive response against bacterial leaf blight caused by Xanthomonas oryzae pv. oryzae (You et al., 2016). Down‐regulation of Suppressor of acaulis (SAC) 51 encoding bHLH transcription factor in sac52/rpl10 mutants resulted in a dwarf phenotype in Arabidopsis (Imai et al., 2008), indicating that RPL10 has an extraribosomal role in the regulation of transcription factors.
The phytohormone JA precursor enzyme LOX3‐ and AOS‐encoding genes are highly down‐regulated in response to nonhost pathogen P. syringae pv. tomato T1. We expect that if JA is low, SA could accumulate to provide resistance against biotrophic and hemibiotrophic pathogens (Zhang et al., 2018). However, NbRPL10‐silenced plants showed compromised nonhost disease resistance against the hemibiotrophic pathogen P. syringae pv. tomato T1, suggesting that the phytohormone‐independent defence pathway is regulated by NbRPL10. In addition, we did not find down‐regulation of SA biosynthetic or signalling genes in NbRPL10‐silenced plants. Furthermore, down‐regulation of many genes involved in the response to wounding also correlates with the expression of genes in association with systemic acquired resistance or SA‐mediated resistance (Park et al., 2007). Interestingly, the Proline dehydrogenase 2 (PDH2) gene was down‐regulated in NbRPL10‐silenced plants. Down‐regulation of proline biosynthesis genes in Arabidopsis and N. benthamiana causes compromised nonhost disease resistance (Senthil‐Kumar et al., 2013). The pdh yeast mutants were rescued by QM overexpression under paraquat stress (Chen et al., 2006). The role of QM/RPL10 in the regulation of proline biosynthesis or metabolic pathway in plants needs further study.
NbRPL10‐silenced plants showed a variegated or photobleaching phenotype, which could be due to the down‐regulation of Phytoene synthase (Psy1) and Phytoene desaturase (PDS) genes (Figure S6). The silencing of PDS through VIGS results in a photobleaching phenotype (Senthil‐Kumar and Mysore, 2011). Similarly, Psy1‐RNAi lines of oncidium hybrid orchid showed reduced chlorophyll content (Liu et al., 2014). The reduced expression of pyrabactin 1 (PYR1), an ABA receptor, in response to nonhost pathogen P. syringae pv. tomato T1 correlates with the expression of PDS1 and PSY1 in NbRPL10‐silenced plants. The down‐regulation of PYR1 in NbRPL10‐silenced plants may disrupt immune responses associated with ABA signal transduction and could occur at the level of Ca2+ signalling (Kim et al., 2010). Genes involved in Ca2+ signalling were also down‐regulated in NbRPL10‐silenced plants. Other defence‐associated genes such as MLO‐like protein 6, MLP‐like 43, and Defensin genes were also down‐regulated in NbRPL10‐silenced plants in response to nonhost pathogen. In contrast, genes encoding peptidyl‐prolyl‐cis‐trans isomerases were up‐regulated in NbRPL10‐silenced plants. In Arabidopsis, elevated expression of Peptidyl‐prolyl‐cis‐trans isomerases showed defence against Xanthomonas campestris infection (Mokryakova et al., 2014).
Approximately 900 genes associated with protein translation mechanisms were differentially regulated in NbRPL10‐silenced plants, indicating that QM/RPL10 could be a central regulator of gene expression as well as of protein translation mechanisms. Eukaryotic elongation, initiation factors, ribosomal proteins L‐29, L‐30, L‐23, L‐3, S‐4, S‐3, L‐38, EIF‐3C, L‐5, S‐4, S‐11, S‐30, S‐17, S‐13, L‐5, L‐17, L‐2, L‐24, S‐13, and so on were highly down‐regulated with or without pathogen responses in NbRPL10‐silenced plants. Similar observations were reported in F. oxysporum f. sp. vanillae infection in vanilla (Solano‐De La Cruz et al., 2019), X. oryzae pv. oryzae infection in rice (Moin et al., 2016; Saha et al., 2017; Vemanna et al., 2019), and P. syringae pv. tomato T1 infection in tomato (Mysore et al., 2002) wherein genes associated with ribosomal proteins were up‐regulated. These data substantiate that ribosomal proteins play an intrinsic role in plant defence mechanisms. It is possible that QM/RPL10 plays a transcriptional role in regulating translational mechanisms and defence‐associated genes. The interaction of QM/RPL10 with MYB transcription factor and regulation of defence genes against virus infection (Carvalho et al., 2008; Zorzatto et al., 2015) substantiates this theory. The Arabidopsis rpl10a mutant plants showed reduced translation efficiency, and also specific protein translation is affected in individual Atrpl10 mutants (Falcon‐Ferreyra et al., 2010). Based on this report we can hypothesize that the DEGs in the NbRPL10A‐silenced plants were due to reduced RPL10A protein and not due to an unusual ribosome. The gene expression reprogramming in RPL10‐R98S mutation has shown profound structural, biochemical, and translational fidelity defects that cause cancer in humans (Sulima et al., 2014). Similarly, RPL12 and RPL19 proteins are known to be required for proper functioning of a number of factors involved in ribosome biogenesis and protein synthesis (Plafker and Macara, 2002).
The Arabidopsis plants overexpressing AtRPL10A showed reduced susceptibility to host pathogen P. syringae pv. tomato DC3000 when compared to wild‐type Col‐0. Because the AtRPL10A overexpression lines did not confer complete resistance, the infected plants will eventually die but can survive longer than wild‐type plants. In contrast, the mutants or RNAi lines of AtRPL10A, B, and C compromised resistance against nonhost pathogens P. syringae pv. tabaci and P. syringae pv. tomato T1. This further confirms that QM/RPL10 family proteins have conserved mechanisms to confer nonhost resistance against bacterial pathogens in N. benthamiana and Arabidopsis. We identified RPL30, RPS30, RPL23, and RRM proteins that interact with AtRPL10A. Silencing the homologs of these genes in N. benthamiana or Arabidopsis rpl23c, rpl30, and rrm mutants showed higher nonhost pathogen growth when compared to the wild type, suggesting that these ribosomal proteins may have an extraribosomal function, including a role in plant immunity. The nonhost bacterial growth in NbRPL10‐silenced or Atrpl10 mutants was higher than in other ribosomal gene‐silenced plants. This further confirms that RPL10 is a central regulator of defence and the RPL10A‐interacting proteins studied have a minor role in nonhost resistance. Induction of RPL23 with other ribosomal proteins was reported in spinach and resurrection plant Tortula ruralis under desiccation stress (Thomas et al., 1988; Wood et al., 2000). RPL23 promotes multidrug resistance in gastric cancer cells by suppressing drug‐induced apoptosis, and overexpression of RPL23 induces Glutathione‐S transferase in yeast (Shi et al., 2004). The eukaryotic RPS30 that is conserved from yeast to humans showed antimicrobial activity within the cytosolic fraction. RPS30 peptide fragments kill mycobacteria through autophagy mechanisms (Ponpuak et al., 2010). Posttranslation modification (sumoylation) of RPL30 by SUMO protease protein, suppressor of mat3‐7 (SMT7), regulates the cell size checkpoint function, supporting the importance of the extraribosomal functions of ribosomal proteins (Lin et al., 2020). Glycine‐rich protein 7 (GRP7) belonging to the RRM protein family is ADP‐ribosylated by the P. syringae pv. tomato DC3000 secreted effector protein, ADP‐ribosyl transferase hopU1. The Arabidopsis grp7 mutant plants were more susceptible than the wild type against host pathogen P. syringae pv. tomato DC3000 (Fu et al., 2007). Overall, our study clearly demonstrates that QM/RPL10 is involved in the regulation of many genes that are important for bacterial defence and translation mechanisms. Our results provide a novel strategy by altering the expression of ribosomal protein‐encoding genes to confer disease resistance in crop plants.
4. EXPERIMENTAL PROCEDURES
4.1. Gene constructs, bacterial strains, and plant materials
Agrobacterium tumefaciens GV3101 containing TRV2::23C12 (NbME23C12 http://vigs.noble.org) was grown at 28°C in a Luria Bertani (LB) medium supplemented with rifampicin (10 µg/ml) and kanamycin (50 µg/ml). VIGS was carried out by mixing a 1:1 ratio of Agrobacterium strains containing TRV1 and TRV2 as described (Senthil‐Kumar and Mysore, 2014). P. syringae strains were grown in King's B medium at 30°C supplemented with rifampicin (10 µg/ml), kanamycin (50 µg/ml), or streptomycin (50 µg/ml) when needed. The TRV2::RPL10s, TRV2::RPL10A, TRV2::RPL10B, TRV2::RRM (NbME39D07), TRV2::RPL23 (NbME01F11), TRV2::RPL30 (NbTI01C11), TRV2::RPS30 (NbME14G06), or TRV2::GFP constructs were developed and used for VIGS as described (Senthil‐Kumar and Mysore, 2014).
Arabidopsis T‐DNA insertion mutants rpl10c‐Salk140517c, Salk135647, Salk126907‐1, rpl23‐Salk 081333, rpl30‐Salk 099025, rpl30‐Salk 12246, rrm‐Salk 028428, and Salk 066297 were obtained from the Arabidopsis Biological Resource Center and identified homozygous lines for further analysis. Full‐length AtRPL10A cDNA was cloned into a pMDC32 binary vector under the CaMV 35S promoter and RNAi constructs for AtRPL10A and AtRPL10B were developed in pH7GWIW G2 (II) vector (Karimi et al., 2007) and mobilized into A. tumefaciens GV3101. The floral dip method of transformation (Clough and Bent, 1998) was used to develop transgenic lines that were screened on hygromycin (25 μg/ml) and Basta (25 μM).
4.2. Disease assays in N. benthamiana and Arabidopsis
Three weeks after TRV inoculation, the silenced and nonsilenced plants were vacuum‐infiltrated with GFPuv (Wang et al., 2007) expressing host (P. syringae pv. tabaci) or nonhost pathogen (P. syringae pv. tomato T1) and tissue was collected 8 hr postinoculation (hpi). To determine the bacterial titre, leaf samples at 0, 3, and 5 dpi from four biological replicates were collected using a 1 cm2 core borer. Leaf samples were ground, subjected to serial dilution, plated on King's B agar supplemented with appropriate antibiotics and incubated at 28°C for 2 days for bacterial colony counting.
Flood inoculation assay was used to infect Arabidopsis plants. Four‐week‐old Arabidopsis plants grown in Murashige and Skoog (MS) plates were incubated for 1 min with 40 ml of bacterial suspension as described (Ishiga et al., 2011). Symptoms were observed after 5 days. To quantify bacteria, the entire rosette was harvested, ground, and serially diluted as described previously (Ishiga et al., 2017). Leaf samples from three biological replicates were collected using a core borer and the bacteria were quantified in a similar fashion to that described above for N. benthamiana.
4.3. Differential gene expression analysis by RNA‐sequencing
The leaf samples from three biological replicates from 4‐week‐old NbRPL10A‐silenced N. benthamiana were harvested at different times after infection with P. syringae pv. tomato T1 and immediately frozen in liquid nitrogen. The total RNA was extracted and RNA samples with high quality (RIN > 7.5 assessed by Agilent 2100 Bioanalyzer) were used for Illumina library preparation following the manufacturer's instructions (Illumina). Paired‐end reads were generated from each sample having a minimum of two replicate (eight) libraries sequenced with Illumina 70 HiSeq 2000. After adapter sequences and low‐quality reads were removed, the remaining reads were aligned to the annotated N. benthamiana reference transcriptome (https://solgenomics.net/organism/1490/view) and the gene‐wise raw counts were calculated. The differential analysis was carried out using DE seq analysis. Genes with more than 2‐fold change in expression and a p value of 0.05 were considered DEGs. Data from all the samples have been uploaded into our GEA universal gene expression atlas platform (https://bioinfo.noble.org/vigs/) for data normalization, visualization, and differential expression analysis using RNA‐Seq analysis software. To predict the pathways and functions of DEGs, the N. benthamiana RNA‐Seq information was BLAST analysed against the Arabidopsis genome and the Arabidopsis IDs were used for agriGO analysis (http://bioinfo.cau.edu.cn/agriGO/) and MapMan analysis (https://mapman.gabipd.org) to predict the pathways and functions of DEGs.
4.4. RT‐qPCR
Tissue was collected 3 weeks after TRV inoculation to test the down‐regulation of NbRPL10 and other ribosomal protein‐encoding gene transcripts in N. benthamiana‐silenced plants. Leaf tissue was collected 3 weeks after germination to determine the expression of AtRPL10A, AtRPL10B, and AtRPL10C in Arabidopsis wild type (Col‐0), AtRPL10A overexpression, and RNAi or mutant plants. Total RNA was extracted according to the manufacturer's instructions using a Qiagen total RNA extraction kit. The cDNA was synthesized by oligo (dT) primers using Molony murine leukaemia virus reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer's instructions. The real‐time RT‐qPCR was performed with a Sigma KicQStart SYBR green kit. The conditions for PCR were as follows: 95°C for 2 min, 25 cycles of denaturation at 94°C for 45 s, annealing for 30 s at 58°C, polymerization for 45 s (72°C), followed by plate reading at 72°C for 5 min, estimation of melting curve from 50°C to 95°C, and incubation at 72°C for 4 min. The primers used in the study are given in Table S1.
4.5. Yeast two‐hybrid assay
Yeast two‐hybrid assays were performed following the manufacturer's protocol using the ProQuest Two‐Hybrid System (Thermo Fisher Scientific). AtRPL10A was fused to the GAL4 DNA‐g binding domain in pDEST32 as the bait construct. The Arabidopsis mixed elicitor treated cDNA library was used for screening (Lee et al., 2018). Based on the screening the full‐length genes of AtRRM1, AtRPL30, AtRPL23, and AtRPS30 were used as prey proteins and fused to the GAL4 activation domain in pDEST22. Bait and prey constructs were cotransformed into yeast MaV203 competent cells. Positive clones were identified by their ability to grow on synthetic defined medium minus leucine/tryptophan/histidine (triple‐dropout medium) or leucine/tryptophan/histidine/uracil (quadruple‐dropout medium) containing 20 mM 3‐aminotriazole. Liquid media contained X‐Gal was used to confirm the interactions.
CONFLICT OF INTEREST
Authors declare no conflict of interest.
Supporting information
FIGURE S1 RPL10 protein architecture and its response to nonhost pathogen when silenced in Nicotiana benthamiana
FIGURE S2 Classification of differentially expressed genes from the NbRPL10‐silenced plant
FIGURE S3 Gene ontology analysis of down‐regulated genes in NbRPL10‐silenced samples over vector control samples
FIGURE S4 Gene ontology analysis of up‐regulated genes in NbRPL10‐silenced samples over vector control samples
FIGURE S5 Gene ontology analysis of down‐regulated genes in nonhost pathogen Pseudomonas syringae pv. tomato T1 treated to NbRPL10‐silenced Nicotiana benthamiana samples over pathogen‐treated control samples
FIGURE S6 Gene ontology analysis of up‐regulated genes with response to nonhost pathogen Pseudomonas syringae pv. tomato T1 treated to NbRPL10‐silenced Nicotiana benthamiana samples over pathogen‐treated control samples
FIGURE S7 Differentially expressed genes in NbRPL10‐silenced plants under normal conditions
FIGURE S8 Genetic map of AtRPL10 mutants and expression analysis in overexpression, RNAi, and mutant Arabidopsis plants using qRT‐PCR
FIGURE S9 Classification of different proteins that are interacted with RPL10A protein in yeast two hybrid screen
FIGURE S10 Prediction of AtRPL10A, AtRPL10B, and AtRPL10C interacting proteins
TABLE S1 Primers used in the study
TABLE S2 AtRPL10A interacting proteins from yeast two hybrid screening
TABLE S3 Prediction of RPL10 interacting proteins using the STRING bioinformatics tool based on text mining, experiments, and database parameters
DATA S1 Differentially regulated genes in NbRPL10A silenced and vector control after infection with nonhost pathogen Pseudomonas syringae pv. tomato T1. The list was generated from the MapMan analysis
DATA S2 Differentially regulated genes belonging to proline, defence, and protein translation related pathways in NbRPL10A silenced and vector control after infection with nonhost pathogen Pseudomonas syringae pv. tomato T1
ACKNOWLEDGMENTS
This work was supported by the Noble Research Institute, LLC, and the Ramanujan Fellowship, SERB to V.S.R., and Regional Centre for Biotechnology core funding. V.S.R. acknowledges a Fulbright‐Nehru postdoctoral fellowship from USIEF and a Ramanujan Fellowship, SERB, India (SB/S2/RJN‐046/2016). K.S.M. acknowledges the Fulbright‐Nehru Academic and Professional Excellence Award.
Ramu VS, Dawane A, Lee S, et al. Ribosomal protein QM/RPL10 positively regulates defence and protein translation mechanisms during nonhost disease resistance. Molecular Plant Pathology. 2020;21:1481–1494. 10.1111/mpp.12991
Contributor Information
Vemanna S. Ramu, Email: ramu.vemanna@rcb.res.in.
Kirankumar S. Mysore, Email: ksmysore@noble.org.
DATA AVAILABILITY STATEMENT
VIGS clones are available at the Noble Research Institute VIGS Database at https://vigs.noble.org/line2.php?id=NbME23C12 as NbME23C12. RNA‐Seq data are available at https://bioinfo.noble.org/vigs/ as VIGS: NicotianaBenthamiana_Niben101 Gene Expression Atlas and Network Analysis.
REFERENCES
- Ahmed, F. , Senthil‐Kumar, M. , Dai, X. , Ramu, S.V. , Lee, S. , Mysore, K.S. et al (2020) pssRNAit: a web server for designing highly effective and specific plant siRNAs with in silico genome‐wide off‐target gene assessment. Plant Physiology, 184, 65–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayliffe, M. and Sørensen, C.K. (2019) Plant nonhost resistance: paradigms and new environments. Current Opinion in Plant Biology, 50, 104–113. [DOI] [PubMed] [Google Scholar]
- Ben‐Shem, A. , Loubresse, N.G.D. , Melnikov, S. , Jenner, L. , Yusupova, G. and Yusupov, M. (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science, 334, 1524–1529. [DOI] [PubMed] [Google Scholar]
- Blume, B. , Nurnberger, T. , Nass, N. and Scheel, D. (2000) Receptor‐mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. The Plant Cell, 12, 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, C.M. , Santos, A.A. , Pires, S.R. , Rocha, C.S. , Saraiva, D.I. , Machado, J.P.B. et al (2008) Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS Pathogens, 4, e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Wanduragala, S. , Becker, D.F. and Dickman, M.B. (2006) Tomato QM‐like protein protects Saccharomyces cerevisiae cells against oxidative stress by regulating intracellular proline levels. Applied and Environment Microbiology, 72, 4001–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.Y. , Clark, A.J. , Chan, D.C. , Ware, J.L. , Holt, S.E. , Chidambaram, A. et al (2011) Wilms’ tumor 1 silencing decreases the viability and chemoresistance of glioblastoma cells in vitro: A potential role for IGF‐1R de‐repression. Journal of Neuro‐oncology, 103, 87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Eisinger, D.P. , Dick, F.A. and Trumpower, B.L. (1997) Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Molecular and Cellular Biology, 17, 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, G. , Ni, Z. , Yao, Y. , Nie, X. and Sun, Q. (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha‐gliadin gene during wheat seed development. Plant Molecular Biology, 63, 73–84. [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra, M.L. , Pezza, A. , Biarc, J. , Burlingame, A.L. and Casati, P. (2010) Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet‐B stress. Plant Physiology, 153, 1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra, M.L. , Casadevall, R. , Luciani, M.D. , Pezza, A. and Casati, P. (2013) New evidence for differential roles of L10 ribosomal proteins from Arabidopsis. Plant Physiology, 163, 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, F. and Zhou, J.M. (2012) Plant–bacterial pathogen interactions mediated by type III effectors. Current Opinion in Plant Biology, 15, 469–476. [DOI] [PubMed] [Google Scholar]
- Fonseca, J.P. and Mysore, K.S. (2019) Genes involved in nonhost disease resistance as a key to engineer durable resistance in crops. Plant Science, 279, 108–116. [DOI] [PubMed] [Google Scholar]
- Freed, E.F. , Bleichert, F. , Dutca, L.M. and Baserga, S.J. (2010) When ribosomes go bad: diseases of ribosome biogenesis. Molecular BioSystems, 6, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z.Q. , Guo, M. , Jeong, B.R. , Tian, F. , Elthon, T.E. , Cerny, R.L. et al (2007) A type III effector ADP‐ribosylates RNA‐binding proteins and quells plant immunity. Nature, 447, 284–288. [DOI] [PubMed] [Google Scholar]
- Gabriëls, S.H.E.J. , Takken, F.L.W. , Vossen, J.H. et al (2006) cDNA‐AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Molecular Plant‐Microbe Interactions, 19, 567–576. [DOI] [PubMed] [Google Scholar]
- Gill, U.S. , Lee, S. and Mysore, K.S. (2015) Host versus nonhost resistance: distinct wars with similar arsenals. Phytopathology, 105, 580–587. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000) Nonhost resistance and nonspecific plant defenses. Current Opinion in Plant Biology, 3, 315–319. [DOI] [PubMed] [Google Scholar]
- Imai, A. , Komura, M. , Kawano, E. , Kuwashiro, Y. and Takahashi, T. (2008) A semi‐dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana . The Plant Journal, 56, 881–890. [DOI] [PubMed] [Google Scholar]
- Ishiga, Y. , Ishiga, T. , Uppalapati, S.R. and Mysore, K.S. (2011) Arabidopsis seedling flood‐inoculation technique: A rapid and reliable assay for studying plant–bacterial interactions. Plant Methods, 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga, Y. , Ishiga, T. , Ichinose, Y. and Mysore, K. (2017) Pseudomonas syringae flood‐inoculation method in Arabidopsis. Bio‐Protocol, 7, e2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. , Vance, R.E. and Dangl, J.L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science, 354, 1117. [DOI] [PubMed] [Google Scholar]
- Karimi, M. , Depicker, A. and Hilson, P. (2007) Recombinational cloning with plant gateway vectors. Plant Physiology, 145, 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal, A. , Ramu, V.S. , Oh, S. , Lee, S. , Pant, B. , Lee, H.K. et al (2017) General control nonrepressible4 degrades 14‐3‐3 and the RIN4 complex to regulate stomatal aperture with implications on nonhost disease resistance and drought tolerance. The Plant Cell, 29, 2233–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.H. , Böhmer, M. , Hu, H. , Nishimura, N. and Schroeder, J.I. (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Review of Plant Biology, 61, 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, N. and Arimura, S.I. (2010) Discovery of the rpl10 gene in diverse plant mitochondrial genomes and its probable replacement by the nuclear gene for chloroplast RPL10 in two lineages of angiosperms. DNA Research, 17, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu, N. , Dozier, U. , Deslandes, L. , Somssich, I.E. and Ullah, H. (2013) Arabidopsis scaffold protein RACK1A interacts with diverse environmental stress and photosynthesis related proteins. Plant Signaling & Behavior, 8, e24012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, N.K. , Nagalakshmi, U. , Hurlburt, N.K. , Flores, R. , Bak, A. , Sone, P. et al (2018) The receptor‐like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host & Microbe, 23, 485–497.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Senthil‐Kumar, M. , Kang, M. , Rojas, C.M. , Tang, Y. , Oh, S. et al (2017) The small GTPase, nucleolar GTP‐binding protein 1 (NOG1), has a novel role in plant innate immunity. Scientific Reports, 7, 9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Rojas, C.M. , Oh, S. , Kang, M. , Choudhury, S.R. , Lee, H.K. et al (2018) Nucleolar GTP‐binding protein 1–2 (NOG1‐2) interacts with jasmonate‐ZIMDomain protein 9 (JAZ9) to regulate stomatal aperture during plant immunity. International Journal of Molecular Sciences, 19, 1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Ferreira, M.A. , Huang, M. , Camargos, L.F. , Yu, X. , Teixeira, R.M. et al (2019) The receptor‐like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nature Communications, 10, 4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.L. , Chung, C.L. , Chen, M.H. , Chen, C.H. and Fang, S.C. (2020) SUMO protease SMT7 modulates ribosomal protein L30 and regulates cell‐size checkpoint function. The Plant Cell, 32, 1285–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.X. , Chiou, C.Y. , Shen, C.H. , Chen, P.J. , Liu, Y.C. , Jian, C.D. et al (2014) RNA interference‐based gene silencing of phytoene synthase impairs growth, carotenoids, and plastid phenotype in Oncidium hybrid orchid. Springerplus, 3, 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus, T.M. , Nguyen, Y.H. and Stanbridge, E.J. (1997) The QM protein associates with ribosomes in the rough endoplasmic reticulum. Biochemistry, 36, 8224–8230. [DOI] [PubMed] [Google Scholar]
- Lu, R. , Martin‐Hernandez, A.M. , Peart, J.R. , Malcuit, I. and Baulcombe, D.C. (2003) Virus‐induced gene silencing in plants. Methods, 30, 296–303. [DOI] [PubMed] [Google Scholar]
- Moin, M. , Bakshi, A. , Saha, A. , Dutta, M. , Madhav, S.M. and Kirti, P.B. (2016) Rice ribosomal protein large subunit genes and their spatio‐temporal and stress regulation. Frontiers in Plant Science, 7, 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokryakova, M.V. , Pogorelko, G.V. , Bruskin, S.A. , Piruzian, E.S. and Abdeeva, I.A. (2014) The role of peptidyl‐prolyl cis/trans isomerase genes of Arabidopsis thaliana in plant defense during the course of Xanthomonas campestris infection. Russian Journal of Genetics (Translation of Genetika (Moscow, Russian Federation)), 50, 140–148. [PubMed] [Google Scholar]
- Mysore, K.S. and Ryu, C.M. (2004) Nonhost resistance: how much do we know? Trends in Plant Science, 9, 97–104. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S. , Crasta, O.R. , Tuori, R.P. , Folkerts, O. , Swirsky, P.B. and Martin, G.B. (2002) Comprehensive transcript profiling of Pto‐ and Prf‐mediated host defense responses to infection by Pseudomonas syringae pv. tomato . The Plant Journal, 32, 299–315. [DOI] [PubMed] [Google Scholar]
- Nagaraj, S. , Senthil‐Kumar, M. , Ramu, V.S. , Wang, K. and Mysore, K.S. (2016) Plant ribosomal proteins, RPL12 and RPL19, play a role in nonhost disease resistance against bacterial pathogens. Frontiers in Plant Science, 6, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, A. , Chen, L. , Nguyen, P.T. , Fujiwara, M. , Wong, H.L. , Kuwano, M. et al (2008) RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. The Plant Cell, 20, 2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai, R. , Wang, C. , Chen, J. , Wu, J. , Shou, H. and Whelan, J. (2013) Antagonistic, overlapping and distinct responses to biotic stress in rice (Oryza sativa) and interactions with abiotic stress. BMC Genomics, 14, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.W. , Kaimoyo, E. , Kumar, D. , Mosher, S. and Klessig, D.F. (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science, 318, 113–116. [DOI] [PubMed] [Google Scholar]
- Plafker, S.M. and Macara, I.G. (2002) Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Molecular and Cellular Biology, 22, 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak, M. , Davis, A.S. , Roberts, E.A. , Delgado, M.A. , Dinkins, C. , Zhao, Z. et al (2010) Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity, 32, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf, S. , Wünnenberg, P. , Lee, J. , Becker, D. , Dunkel, M. , Hedrich, R. et al (2008) Loss of the vacuolar cation channel, AtTPC1, does not impair Ca 2+ signals induced by abiotic and biotic stresses. The Plant Journal, 53, 287–299. [DOI] [PubMed] [Google Scholar]
- Rivera‐Madrid, R. , Marinho, P. , Chartier, Y. and Meyer, Y. (1993) Nucleotide sequence of an Arabidopsis thaliana cDNA clone encoding a homolog to a suppressor of Wilms’ tumor. Plant Physiology, 102, 329–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, C.S. , Santos, A.A. , Machado, J.P.B. and Fontes, E.P.B. (2008) The ribosomal protein L10/QM‐like protein is a component of the NIK‐mediated antiviral signaling. Virology, 380, 165–169. [DOI] [PubMed] [Google Scholar]
- Rojas, C.M. , Senthil‐Kumar, M. , Wang, K. , Ryu, C.M. , Kaundal, A. and Mysore, K.S. (2012) Glycolate oxidase modulates reactive oxygen species‐mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. The Plant Cell, 24, 336–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, A. , Das, S. , Moin, M. , Dutta, M. , Bakshi, A. , Madhav, M.S. et al (2017) Genome‐wide identification and comprehensive expression profiling of ribosomal protein small subunit (RPS) genes and their comparative analysis with the large subunit (RPL) genes in rice. Frontiers in Plant Science, 8, 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2011) New dimensions for VIGS in plant functional genomics. Trends in Plant Science, 16, 656–665. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2012) Ornithine‐delta‐aminotransferase and proline dehydrogenase genes play a role in non‐host disease resistance by regulating pyrroline‐5‐carboxylate metabolism‐induced hypersensitive response. Plant, Cell and Environment, 35, 1329–1343. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2013) Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annual Review of Phytopathology, 51, 407–427. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2014) Tobacco rattle virus‐based virus‐induced gene silencing in Nicotiana benthamiana . Nature Protocols, 9, 1549–1562. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. , Lee, H.K. and Mysore, K.S. (2013) VIGS‐mediated forward genetics screening for identification of genes involved in nonhost resistance. Journal of Visualized Experiments, 78, e51033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. , Wang, M. , Chang, J. , Ramegowda, V. , Del Pozo, O. , Liu, Y. et al (2018) Virus‐induced gene silencing database for phenomics and functional genomics in Nicotiana benthamiana . Plant Direct, 2, e00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Zhai, H. , Wang, X. , Han, Z. , Liu, C. , Lan, M. et al (2004) Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug‐induced apoptosis. Experimental Cell Research, 296, 337–346. [DOI] [PubMed] [Google Scholar]
- Solano‐De La Cruz, M.T. , Adame‐García, J. , Gregorio‐Jorge, J. , Jiménez‐Jacinto, V. , Vega‐Alvarado, L. , Iglesias‐Andreu, L.G. et al (2019) Functional categorization of de novo transcriptome assembly of Vanilla planifolia Jacks. potentially points to a translational regulation during early stages of infection by Fusarium oxysporum f. sp. vanillae . BMC Genomics, 20, 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima, S.O. , Gülay, S.P. , Anjos, M. , Patchett, S. , Meskauskas, A. , Johnson, A.W. et al (2014) Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Research, 42, 2049–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, F. , Massenet, O. , Dome, A.M. , Briat, J.F. and Mache, R. (1988) Expression of the rpl23, rpl2 and rps19 genes in spinach chloroplasts. Nucleic Acids Research, 16, 2461–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemanna, R.S. , Bakade, R. , Bharti, P. , Kumar, M.K.P. , Sreeman, S.M. , Senthil‐Kumar, M. et al (2019) Cross‐talk signaling in rice during combined drought and bacterial blight stress. Frontiers in Plant Science, 10, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Kang, L. , Anand, A. , Lazarovits, G. and Mysore, K.S. (2007) Monitoring in planta bacterial infection at both cellular and whole‐plant levels using the green fluorescent protein variant GFPuv: methods. New Phytologist, 174, 212–223. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Senthil‐Kumar, M. , Ryu, C.M. , Kang, L. and Mysore, K.S. (2012) Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiology, 158, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Yu, J. , Zhu, D. , Chang, Y. and Zhao, Q. (2014) Maize ZmRACK1 is involved in the plant response to fungal phytopathogens. International Journal of Molecular Sciences, 15, 9343–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, A.J. , Duff, R.J. and Oliver, M.J. (2000) The translational apparatus of Tortula ruralis: polysomal retention of transcripts encoding the ribosomal proteins RPS14, RPS16 and RPL23 in desiccated and rehydrated gametophytes. Journal of Experimental Botany, 51, 1655–1662. [DOI] [PubMed] [Google Scholar]
- Wool, I.G. (1996) Extraribosomal functions of ribosomal proteins. Trends in Biochemical Sciences, 21, 164–165. [PubMed] [Google Scholar]
- Yang, C. , Zhang, C. , Dittman, J.D. and Whitham, S.A. (2009) Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology, 390, 163–173. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Chen, Z. , Liu, N. and Chen, Y. (2018) Ribosomal protein L10 in mitochondria serves as a regulator for ROS level in pancreatic cancer cells. Redox Biology, 19, 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Q. , Zhai, K. , Yang, D. , Yang, W. , Wu, J. , Liu, J. et al (2016) An E3 ubiquitin ligase‐BAG protein module controls plant innate immunity and broad‐spectrum disease resistance. Cell Host & Microbe, 20, 758–769. [DOI] [PubMed] [Google Scholar]
- Zeng, W. and He, S.Y. (2010) A prominent role of the flagellin receptor FLAGELLIN‐SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiology, 153, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Xia, C. , Duan, C. , Sun, S. , Wang, X. , Wu, X. et al (2013) Identification and candidate gene analysis of a novel Phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PLoS One, 8, e69799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Zhao, F. , Jiang, L. , Chen, C. , Wu, L. and Liu, Z. (2018) Different pathogen defense strategies in Arabidopsis: more than pathogen recognition. Cells, 7, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzatto, C. , MacHado, J.P.B. , Lopes, K.V.G. , Nascimento, K.J.T. , Pereira, W.A. , Brustolini, O.J.B. et al (2015) NIK1‐mediated translation suppression functions as a plant antiviral immunity mechanism. Nature, 520, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 RPL10 protein architecture and its response to nonhost pathogen when silenced in Nicotiana benthamiana
FIGURE S2 Classification of differentially expressed genes from the NbRPL10‐silenced plant
FIGURE S3 Gene ontology analysis of down‐regulated genes in NbRPL10‐silenced samples over vector control samples
FIGURE S4 Gene ontology analysis of up‐regulated genes in NbRPL10‐silenced samples over vector control samples
FIGURE S5 Gene ontology analysis of down‐regulated genes in nonhost pathogen Pseudomonas syringae pv. tomato T1 treated to NbRPL10‐silenced Nicotiana benthamiana samples over pathogen‐treated control samples
FIGURE S6 Gene ontology analysis of up‐regulated genes with response to nonhost pathogen Pseudomonas syringae pv. tomato T1 treated to NbRPL10‐silenced Nicotiana benthamiana samples over pathogen‐treated control samples
FIGURE S7 Differentially expressed genes in NbRPL10‐silenced plants under normal conditions
FIGURE S8 Genetic map of AtRPL10 mutants and expression analysis in overexpression, RNAi, and mutant Arabidopsis plants using qRT‐PCR
FIGURE S9 Classification of different proteins that are interacted with RPL10A protein in yeast two hybrid screen
FIGURE S10 Prediction of AtRPL10A, AtRPL10B, and AtRPL10C interacting proteins
TABLE S1 Primers used in the study
TABLE S2 AtRPL10A interacting proteins from yeast two hybrid screening
TABLE S3 Prediction of RPL10 interacting proteins using the STRING bioinformatics tool based on text mining, experiments, and database parameters
DATA S1 Differentially regulated genes in NbRPL10A silenced and vector control after infection with nonhost pathogen Pseudomonas syringae pv. tomato T1. The list was generated from the MapMan analysis
DATA S2 Differentially regulated genes belonging to proline, defence, and protein translation related pathways in NbRPL10A silenced and vector control after infection with nonhost pathogen Pseudomonas syringae pv. tomato T1
Data Availability Statement
VIGS clones are available at the Noble Research Institute VIGS Database at https://vigs.noble.org/line2.php?id=NbME23C12 as NbME23C12. RNA‐Seq data are available at https://bioinfo.noble.org/vigs/ as VIGS: NicotianaBenthamiana_Niben101 Gene Expression Atlas and Network Analysis.


