Abstract
The recently emerged severe acute respiratory syndrome coronavirus-2 (SARS–CoV-2) is the causative agent of the devastating COVID-19 lung disease pandemic. Here, we tested the inhibitory activities of the antiviral interferons of type I (IFN-α) and type III (IFN-λ) against SARS–CoV-2 and compared them with those against SARS–CoV-1, which emerged in 2003. Using two mammalian epithelial cell lines (human Calu-3 and simian Vero E6), we found that both IFNs dose-dependently inhibit SARS–CoV-2. In contrast, SARS–CoV-1 was restricted only by IFN-α in these cell lines. SARS–CoV-2 generally exhibited a broader IFN sensitivity than SARS–CoV-1. Moreover, ruxolitinib, an inhibitor of IFN-triggered Janus kinase/signal transducer and activator of transcription signaling, boosted SARS–CoV-2 replication in the IFN-competent Calu-3 cells. We conclude that SARS–CoV-2 is sensitive to exogenously added IFNs. This finding suggests that type I and especially the less adverse effect–prone type III IFN are good candidates for the management of COVID-19.
Keywords: virology, infection, innate immunity, interferon, virus, antiviral agent, cytokine action, COVID-19, interferon-alpha/beta, interferon-lambda, ruxolitinib, SARS–CoV-2
The massive pandemic caused by coronavirus SARS–CoV-2 (1, 2) is calling for rapid evaluation of potential therapeutics through repurposing of drugs already in clinical use. Interferons of type I (IFN-α/β) and type III (IFN-λ) constitute an important branch of the mammalian innate immune response. These cytokines are produced by virus-infected cells and are able to establish an antiviral state in target cells by triggering the so-called JAK/STAT signaling pathway (3–5). Both type I and type III IFNs are clinically used or being tested, respectively, against a range of ailments that include viral diseases (6, 7). Previously, we and others have demonstrated the potential of IFNs to inhibit the two related, previously emerged pathogenic coronaviruses SARS–CoV-1 and MERS-CoV (8–15). Here, we investigated the potential of type I and type III IFNs against the newly emerged SARS–CoV-2.
Results
Type I IFN
We tested the effect of type I IFN against SARS–CoV-2 compared with the SARS–CoV-1 from 2003. Two different cell lines were employed, namely the human bronchial epithelial Calu-3 and the primate kidney epithelial Vero E6. The cells were first treated for 16 h with 100, 500, or 1000 units/ml of recombinant human IFN-α(B/D) and then infected with the viruses at a multiplicity of infection (MOI) of 0.01 plaque forming units (PFU)/cell to obtain multistep growth. Virus titers in supernatants were determined 24 h later, when titers are reaching a plateau (see below). The data of three biological replicates are shown in Fig. 1. Because several titers were below the detection limit of our plaque assay, a rank correlation test (Spearman's exact rank correlation test) was used for statistical dose-response correlation analysis. For SARS–CoV-2 (dark gray bars), statistically significant negative correlation coefficients (CC) were obtained for both cell lines, indicating that viral replication is increasingly inhibited by IFN-α. For SARS–CoV-1 (light gray bars), titers were also affected. However, at least in Vero E6 cells, the reduction of SARS–CoV-1 appears to be weaker than the reduction of SARS–CoV-2 (Fig. 1B). Observations were similar when the input MOI was reduced to 0.001 (Fig. S1), except that titers of SARS–CoV-1 in Calu-3 cells were already very low in the absence of any IFN-α, resulting in a nonsignificant effect of additional IFN. These data may suggest that the potency of IFN to reduce viral titers may be stronger and more consistent against SARS–CoV-2 than against SARS–CoV-1.
Figure 1.
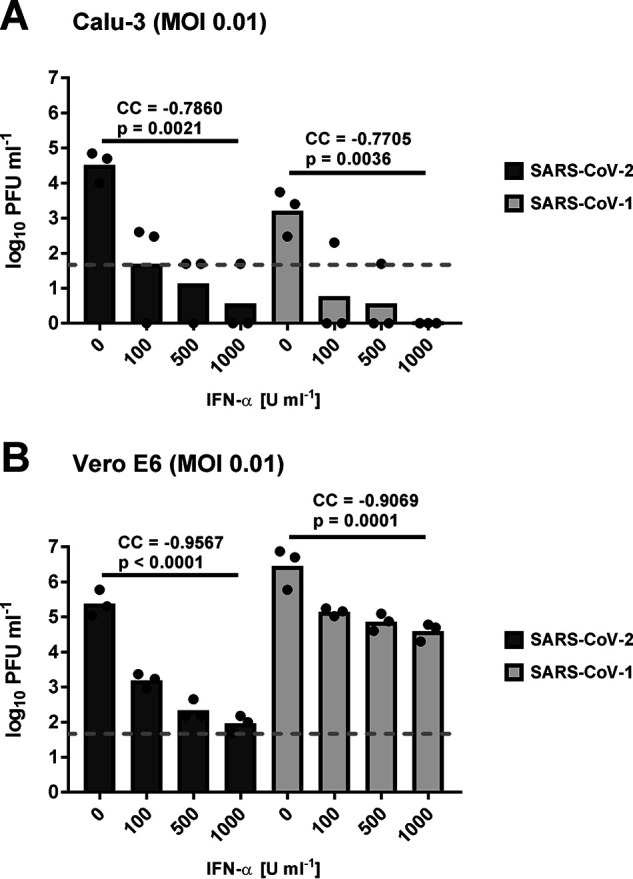
Sensitivity of SARS–CoV-2 and SARS–CoV-1 to type I IFN dose escalation. Calu-3 (A) and Vero E6 cells (B) were pretreated with recombinant human IFN-α and infected at an MOI of 0.01. Titers were measured at 24 h postinfection by plaque assay. Individual titers (dots) and geometric mean values (bars) from three biological replicates are shown. Log-transformed titers of each virus dose-response experiment were analyzed by Spearman's exact rank correlation test. CC and exact one-sided p values are provided. Note that titer values that were below the plaque assay detection level (50 PFU/ml; indicated by the dashed line) were set to 1 PFU/ml.
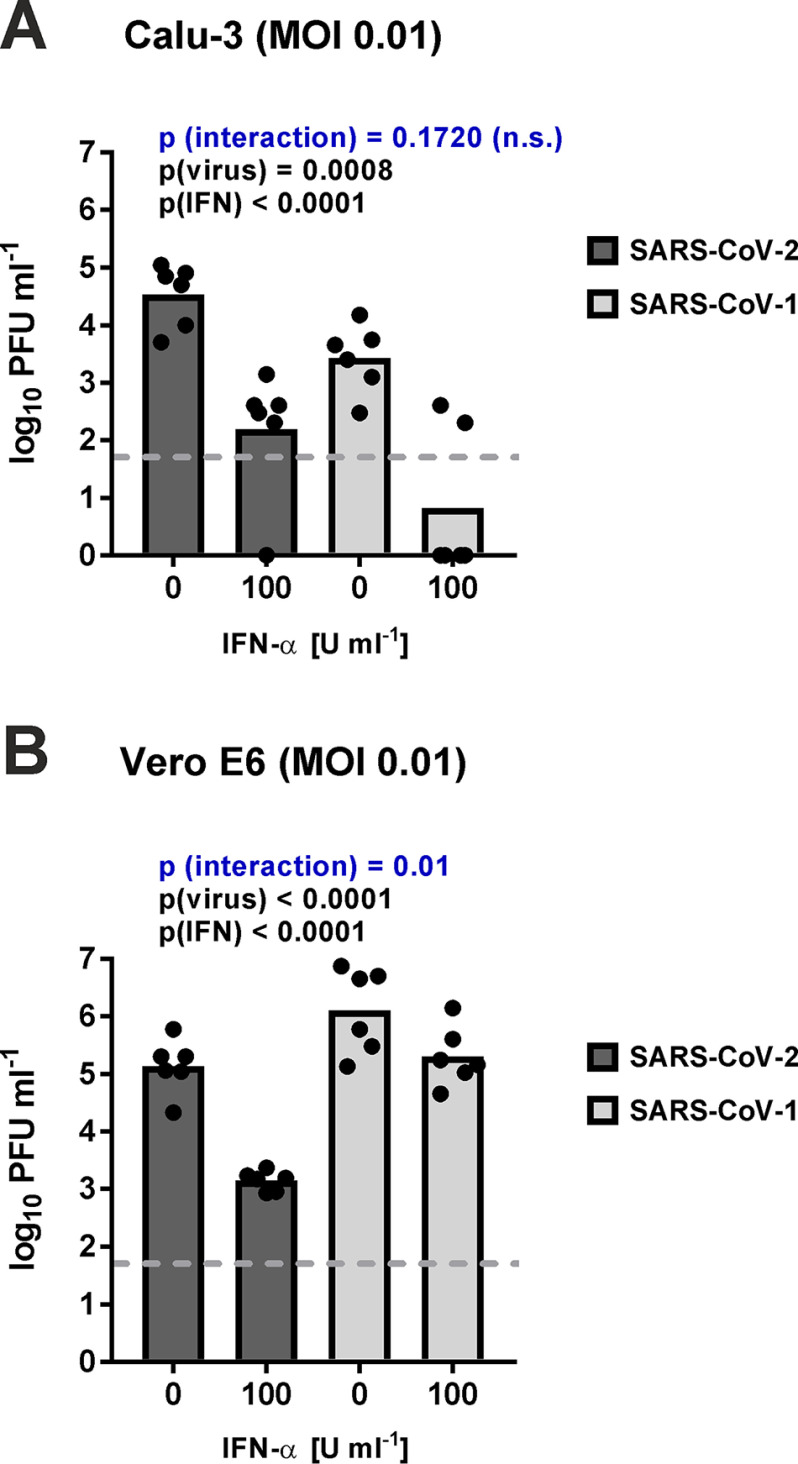
To further investigate the potential differences between the viruses, we repeated the experiment three times more with the intermediate dose of 100 units/ml and analyzed the data statistically after pooling them with the previous three replicates. Two-way ANOVA was used to simultaneously evaluate the influence of both IFN-α and virus species on virus reduction. This analysis (Fig. 2, A and B) showed again that (i) both viruses are reduced by IFN (comparison of 0 versus 100 units/ml IFN-α, p(IFN)) and (ii) there are differences between the SARS–CoV species (comparison of the virus experiments, p(virus)). Moreover, the “interaction” p value showed that, at least in Vero cells, the degree of IFN sensitivity depends on the virus species, again indicating that SARS–CoV-2 is more IFN-sensitive than SARS–CoV-1.
Figure 2.
Sensitivity of SARS–CoV-2 and SARS–CoV-1 to intermediate-dose type I IFN. Calu-3 (A) and Vero E6 cells (B) were pretreated with 100 units/ml IFN-α, infected at an MOI of 0.01, and titrated 24 h later. Log-transformed data were analyzed by two-way ANOVA with factors “IFN” and “virus,” for each of which the specific p values are indicated. p (interaction) designates the probability that IFN sensitivity depends on the virus species. Data points and geometric mean values from six independent experiments are shown. Note that three of the six biological repeats are repeats from Fig. 1.
Type III IFN
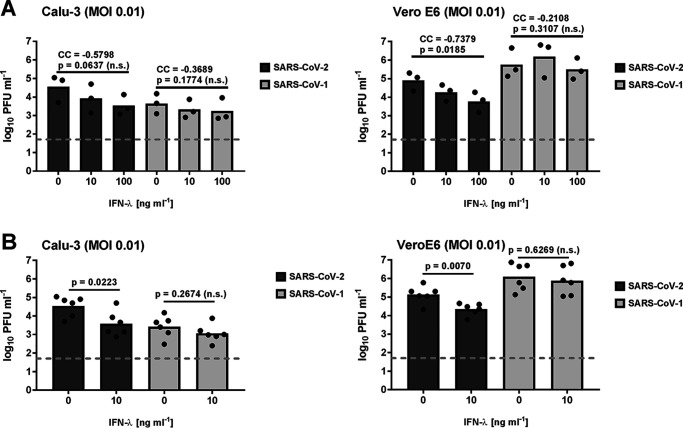
The primary tropism of coronaviruses typically involves epithelia of the respiratory and gastrointestinal tracts (16). On such mucosal barriers, type III IFNs rather than type I IFNs are the predominant antiviral cytokine (4, 5). Although the IFN induction as well as signaling and up-regulation of IFN-stimulated genes (ISGs) are very similar, type III IFNs engage a different receptor that is restricted to epithelial cells, and generate a weaker but longer-lasting antiviral response (5, 17). IFN-λ was previously shown to have activity against coronaviruses (11, 18, 19) and proposed as potential COVID-19 treatment (20). Hence, we compared the sensitivity of the two SARS–coronaviruses also to recombinant human IFN-λ. As shown in Fig. 3A, pretreatment with 10 or 100 ng/ml IFN-λ exhibited only in Vero E6 cells a dose-dependent inhibitory effect on SARS–CoV-2. For SARS–CoV-1, by contrast, no significant inhibition was noted in any of the cell lines. To further investigate the difference between the viruses, we repeated the IFN-λ experiment three times more with the intermediate dose of 10 ng/ml and analyzed the data after pooling with the previous 10 ng/ml IFN-λ experiment (Fig. 3B). Conventional statistical analysis (one-tailed Student's t test, because none of the values was below the detection limit) again revealed a significant impact of IFN-λ on SARS–CoV-2 and the lack of an effect for SARS–CoV-1. Our data thus show that IFN-λ can inhibit SARS–CoV-2 but not SARS–CoV-1.
Figure 3.
Sensitivity of SARS–CoV-2 and SARS–CoV-1 to type III IFN. A, experiments were performed as described for Fig. 1, except that recombinant human IFN-λ was used. Log-transformed titers of each virus dose-response experiment with concentrations of 10 and 100 ng/ml IFN-λ were analyzed by Spearman's exact rank correlation test. CCs and exact one-sided p values are provided. B, three additional biological replicates of the 10 ng/ml IFN-λ were performed, and the resulting titer data were pooled with the 10 ng/ml IFN-λ data from A. Log-transformed titers were analyzed by unpaired one-tailed Student's t test. n.s., nonsignificant.
Blocking JAK/STAT signaling by ruxolitinib
A recent study on the host cell interactome of SARS–CoV-2 identified a number of human proteins for which Food and Drug Administration–approved drugs are available (21). Ruxolitinib, a compound known to target the type I and type III IFN-triggered JAK/STAT signaling pathway (22), was among the proposed inhibitors of virus–host cell interactions (21). Because virus inhibition by an IFN inhibitor seems counterintuitive, we aimed to clarify the influence of this compound on SARS–CoV-2 replication. Cells were pretreated with 1 μm ruxolitinib for 16 h and infected at the two different MOIs, and titers were measured 24 or 48 h later. As shown in Fig. 4, with this setting titers in nontreated controls are already reaching a plateau at the 24-h time point. In Calu-3 cells, ruxolitinib had a clear boosting effect on SARS–CoV-2 replication, mostly at 48 h postinfection, and at both MOI 0.01 and 0.001 (Fig. 4A and Fig. S2A). By contrast, in Vero E6 there was neither a positive nor a negative effect discernible (Fig. 4B and Fig. S2B). Of note, Calu-3 cells are capable of inducing IFN in response to virus infection, whereas Vero cells are not (15). Our data thus indicate that (i) if anything, ruxolitinib is an enhancer rather than an inhibitor of SARS–CoV-2 multiplication, and (ii) the boosting effect is most likely due to inhibition of the antiviral JAK/STAT signaling pathway, because it is not present in the IFN induction–deficient Vero E6 cells.
Figure 4.
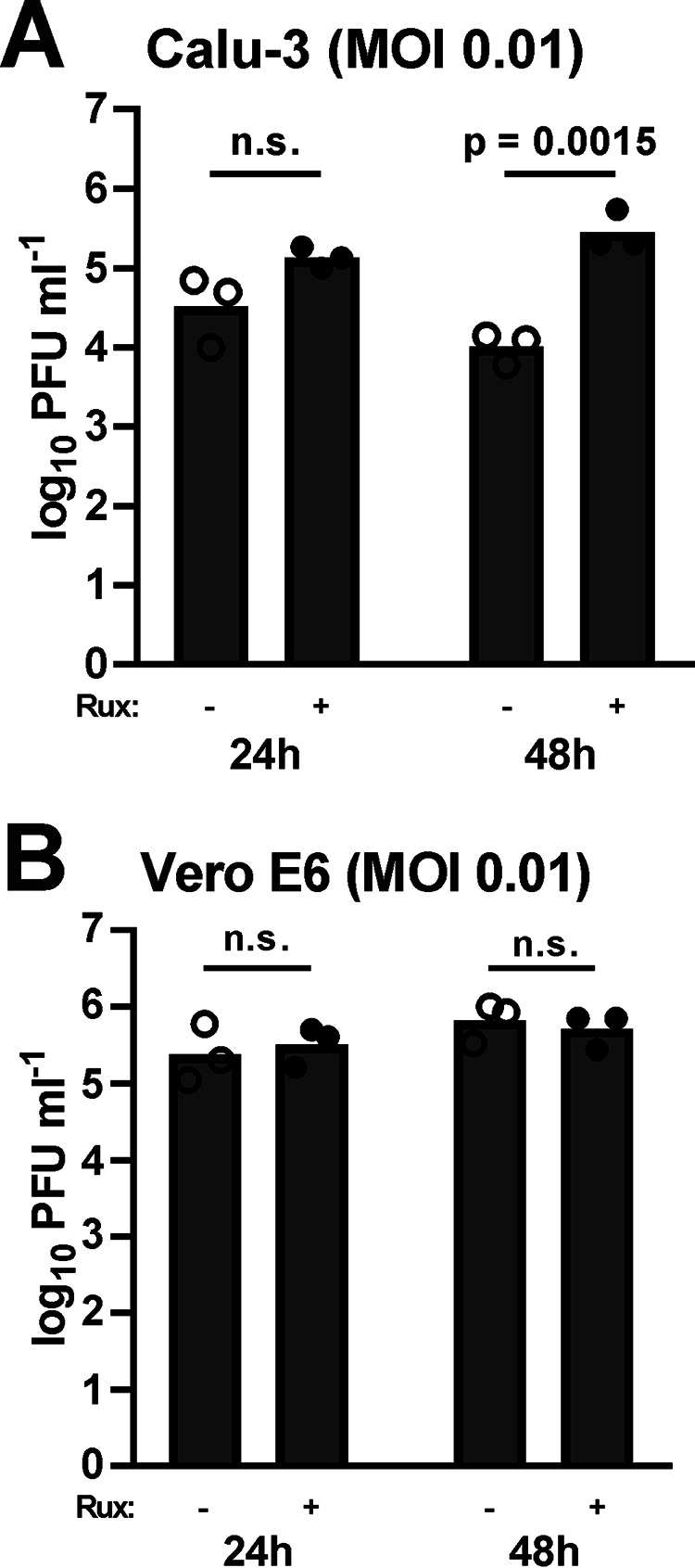
Effect of the JAK/STAT inhibitor ruxolitinib on SARS–CoV-2 replication. Calu-3 (A) and Vero E6 (B) cells were pretreated with 1 μm ruxolitinib (Rux) and infected with SARS–CoV-2 at an MOI of 0.01, and titers were determined at 24 and 48 h postinfection. Individual titers (dots) and geometric mean values (bars) from three biological replicates are shown. Log-transformed titers were analyzed by unpaired two-tailed Student's t test. n.s., nonsignificant.
Comparison of the cell lines
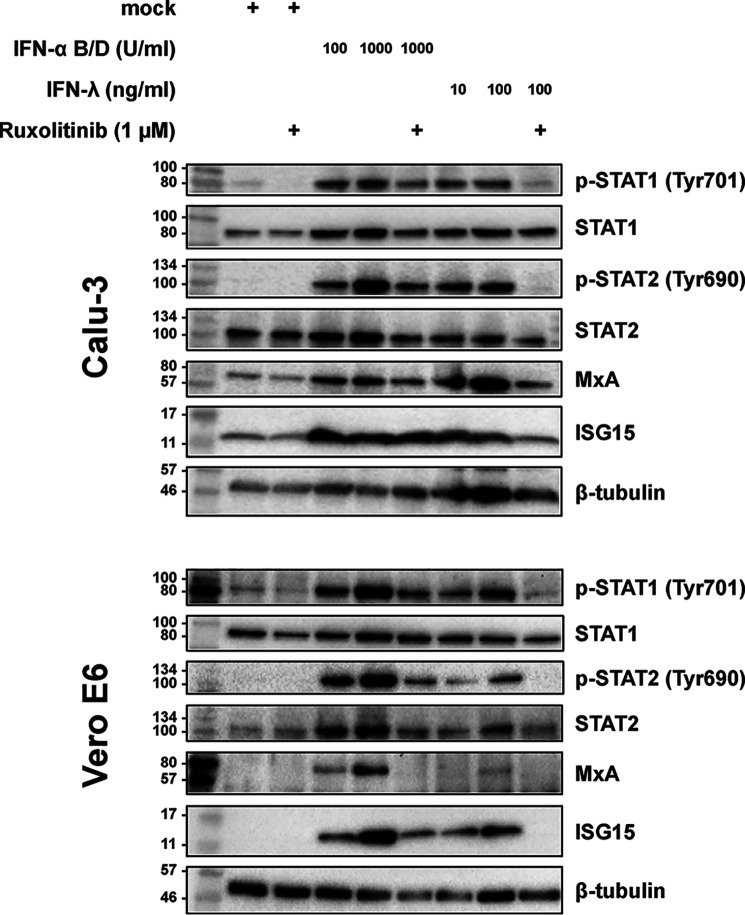
Our observations so far suggest that SARS–CoV-2 is consistently more sensitive to IFNs than SARS–CoV-1. Moreover, type I IFN seems to have a more profound effect than type III IFN. To see whether principal differences in signaling or subsequent gene expression could account for these phenomena, we tested the ability of the cell lines to respond to the IFNs. The immunoblot analysis (Fig. 5) shows that Calu-3 cells have a very similar reaction to both types of IFN concerning phosphorylation of STAT1 and STAT2 and expression of the IFN-stimulated MxA and ISG15. Vero E6 cells also responded to IFN-λ as expected (23), but the ISG response was lower than to IFN-α. Moreover, in nontreated Calu-3 cells, there was already a background ISG expression, which was not observed in Vero cells. Ruxolitinib was in principle able to influence these ISG responses, as expected, but it was more potent against IFN-λ than against IFN-α, and its effects on IFN-stimulated genes were more evident in the Vero E6 compared with the Calu-3 (Fig. 5). Thus, both cell lines are capable to respond to the different types of IFN, but IFN-λ was less potent, which is in agreement with our observations on SARS–CoV sensitivity, as well as with previous studies (5, 17).
Figure 5.
Effect of IFNs and ruxolitinib on Calu-3 and Vero E6 cells. Calu-3 and Vero E6 cells were incubated with the indicated amounts of IFNs and ruxolitinib (added 1 h before IFN) and 24 h later were analyzed for the indicated antigens using immunoblotting. The data are representative of three independent experiments. Molecular markers are shown on the left sides of the blots.
Discussion
The recently emerged SARS–CoV-2 is responsible for major health crises all over the world. Here, we show that type I and type III IFNs are able to inhibit SARS–CoV-2 replication, with effects that in our hands were consistently more profound than against the SARS–CoV-1 from 2003. It should be noted however, that the differences between the viruses could be due to the cell types used or due to the observed differences in virus replication (which could result in higher production of IFN antagonists). Thus, the question whether SARS–CoV-2 is intrinsically more sensitive to IFNs remains to be solved.
PEGylated IFN-α was the standard of care against chronic infection with hepatitis C virus until the recent introduction of other, directly acting antiviral drugs (24). Although associated with some side effects, IFN-α is well-characterized, has been used to treat millions of patients, is considered safe, and is available immediately. IFN-λ has undergone phase I and II clinical trials with hepatitis C virus (25). It exhibited excellent tolerance as well as efficacy, but the phase III trials were abandoned because of the availability of effective direct antivirals. IFN-λ holds promise as having fewer side effects because of its restriction to mucosal tissue and the less sudden but more prolonged antiviral response it triggers (5, 17). In line with our results, a series of preprints show that also others found type I and type III IFNs to be effective against SARS–CoV-2 replication in cell culture (26–28). Also in earlier in vivo studies with SARS–CoV-1, both type I and type III IFNs were shown to be important for the control of infection or the associated disease (29–33). Clinical data on the usage of type I IFN against SARS–CoV-1 or the related MERS-CoV, however, are limited, are not always conclusive, or did not show a clear benefit (34–38). Thus, type III IFN-λ rather than the side effect–prone type I IFNs (39) might be considered for clinical testing against SARS–CoV-2.
Ruxolitinib was proposed as a potential treatment against SARS–CoV-2 (21, 40), and a small clinical trial is underway (clinicaltrials.gov, NCT04334044), although case reports were discouraging (41). The replication boost obtained with ruxolitinib on the IFN-competent Calu-3 cells indicates that ruxolitinib is not at all inhibiting SARS–CoV-2 replication. Thus, drugs that interfere with viral host interactors may not necessarily be antiviral but rather boost the infection.
Experimental procedures
Cells and viruses
Calu-3 and Vero E6 cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) in a 5% CO2 atmosphere at 37 °C. SARS–CoV-2 (strain SARS–CoV-2 /München-1.2/2020/984, p.2) (42) and SARS–CoV-1 (strain SARS-FRA1, p.2) (43) were grown on Vero E6 cells and purified via VivaSpin columns (Sartorius Stedim Biotech). The viruses were titrated on Vero E6 cells. Infection experiments were done under biosafety level 3 conditions with enhanced respiratory personal protection equipment. Of note, all cells were tested mycoplasma-negative.
Inhibitor assays
The cells were pretreated for 16 h with the indicated amounts of pan-species IFN-α(B/D) (PBL Assay Science) (44), purified recombinant IFN-λ3 (18, 45), or with 1 μm ruxolitinib (Selleckchem). Infections were performed at a MOI of 0.01 and 0.001. At the indicated times postinfection, cell supernatants were collected and titrated by plaque assay on Vero E6 cells.
Immunoblot analysis
The cells were treated for 24 h with the indicated amounts of IFNs or ruxolitinib (added 1 h before IFN) and lysed in T-PER protein extraction reagent (Thermo Fisher) supplemented containing 1× Protease inhibitor mixture (c0mplete, Roche), 1× phosphatase Inhibitor mixture set II (Calbiochem), and sample buffer (35.8 mm Tris-HCl, pH 6.8, 7.15% glycerol, 1.43% SDS, 1.08 mm bromphenol blue). Protein samples were run on 12% acrylamide gels and transferred to polyvinylidene fluoride membranes (Millipore) via semidry blotting. After blocking in TBS with 5% BSA (for detection of phospho-STATs, MxA, and total STAT2) or milk powder (all other detections), primary antibody staining was performed overnight at 4 °C. The membranes were washed in TBS with 0.1% Tween 20, stained with secondary antibodies for 45 min, and washed again in TBS with 0.1% Tween 20 and once in TBS. Finally, the membranes were developed with a SuperSignal West Femto kit (Pierce), and the bands were visualized using a ChemiDoc imaging system (Bio-Rad).
The primary antibodies used were phospho-STAT1, Tyr701 (7649S, Cell Signaling) (1:1000); phospho-STAT2, Tyr690 (88410S, Cell Signaling) (1:1000); STAT1 (610186, BD Biosciences) (1:1000); STAT2 (610188, BD Biosciences) (1:1000); ISG15 (sc-166755, Santa Cruz) (1:4000); MXA (MABF938, Sigma–Aldrich) (1:1000); and β-tubulin (ab6046, Abcam) (1:2000). The secondary antibodies used were peroxidase-conjugated goat anti-mouse IgG (31430; Thermo Fisher) (1:10,000) and peroxidase-conjugated goat anti-rabbit IgG (31460; Thermo Fisher) (1:10,000).
Statistical analyses
The statistical analysis of the data were done by means of the statistical program packages BMDP (46) and StatXact® (version 9.0.0). For the statistical testing of the dose-response effect of IFN (type I and III) against SARS–coronaviruses, the typical regression procedures were not applicable because of several values below the detection limit and some ties in the data. Instead of this, the nonparametric Spearman rank CC was used in the exact version (software StatXact). Because the scientific question was clearly one-sided formed (only PFU reduction under application of IFN), one-sided p values were given.
If only two IFN concentrations were to compare with no data below the detection limit, then the t test for independent samples was used (program BMDP3D). For testing the effect of IFN and virus type simultaneously, the two-way ANOVA (program BMDP7D) was applied especially considering a possible interaction between the two tested factors.
In the parametric statistical analyses as well as the graphical representations, the response variable PFU was logarithmically transformed because of its right skewed statistical distribution. In all cases a statistical significance level of α = 0.05 was applied.
Data availability
All data presented and discussed are contained within the article.
Supplementary Material
This article contains supporting information.
Author contributions—U. F., A. S., R. H., A. R. S., K. F., C. D., and F. W. conceptualization; U. F., C. D., and F. W. validation; U. F., A. S., A. R. S., and K. F. investigation; U. F. and F. W. visualization; U. F., A. S., H. H. G., R. H., A. R. S., K. F., C. D., and F. W. methodology; H. H. G., R. H., and F. W. resources; R. H. and F. W. writing-review and editing; C. D. software; C. D. and F. W. supervision; C. D. and F. W. funding acquisition; F. W. formal analysis; F. W. writing-original draft; F. W. project administration.
Funding and additional information—This work was supported by Deutsche Forschungsgemeinschaft Grants SFB 1021 (project number 197785619), We 2616/7-2 (to F. W.) and DR 772/10-2 (to C. D.), by the RAPID (Risk assessment in pre-pandemic respiratory infectious diseases) Consortium of the Bundesministerium für Bildung und Forschung under Grants 01KI1723E (to F. W.) and 01KI1723A (to C. D.), and by the European Union's Horizon 2020 Research and Innovation Program under Grant Agreement 101003666 (OPENCORONA) (to F. W.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ANOVA
- analysis of variance
- CC
- correlation coefficient
- CoV
- coronavirus
- COVID-19
- coronavirus disease 2019
- IFN
- interferon
- JAK
- Janus kinase
- MERS
- Middle East respiratory syndrome
- PFU
- plaque forming units
- Rux
- ruxolitinib
- SARS
- severe acute respiratory syndrome
- STAT
- signal transducer and activator of transcription
- MOI
- multiplicity of infection
- ISG
- IFN-stimulated gene.
References
- 1. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020) The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS–CoV-2. Nat. Microbiol. 5, 536–544 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., et al. (2020) Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lazear H. M., Schoggins J. W., and Diamond M. S. (2019) Shared and distinct functions of type I and type III interferons. Immunity 50, 907–923 10.1016/j.immuni.2019.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wack A., Terczyńska-Dyla E., and Hartmann R. (2015) Guarding the frontiers: the biology of type III interferons. Nat. Immunol. 16, 802–809 10.1038/ni.3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ye L., Schnepf D., and Staeheli P. (2019) Interferon-λ orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 19, 614–625 10.1038/s41577-019-0182-z [DOI] [PubMed] [Google Scholar]
- 6. O'Brien T. R., Young H. A., Donnelly R. P., and Prokunina-Olsson L. (2019) Interferon lambda: disease impact and therapeutic potential. J. Interferon Cytokine Res. 39, 586–591 10.1089/jir.2019.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Snell L. M., McGaha T. L., and Brooks D. G. (2017) Type I interferon in chronic virus infection and cancer. Trends Immunol. 38, 542–557 10.1016/j.it.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan R. W. Y., Chan M. C. W., Agnihothram S., Chan L. L. Y., Kuok D. I. T., Fong J. H. M., Guan Y., Poon L. L. M., Baric R. S., Nicholls J. M., and Peiris J. S. M. (2013) Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 87, 6604–6614 10.1128/JVI.00009-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., and Doerr H. W. (2003) Treatment of SARS with human interferons. Lancet 362, 293–294 10.1016/S0140-6736(03)13973-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Falzarano D., de Wit E., Martellaro C., Callison J., Munster V. J., and Feldmann H. (2013) Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci. Rep. 3, 1686 10.1038/srep01686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kindler E., Jonsdottir H. R., Muth D., Hamming O. J., Hartmann R., Rodriguez R., Geffers R., Fouchier R. A., Drosten C., Muller M. A., Dijkman R., and Thiel V. (2013) Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4, e00611–12 10.1128/mBio.00611-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kindler E., Thiel V., and Weber F. (2016) Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv. Virus Res. 96, 219–243 10.1016/bs.aivir.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spiegel M., Pichlmair A., Mühlberger E., Haller O., and Weber F. (2004) The antiviral effect of interferon-beta against SARS–coronavirus is not mediated by MxA protein. J. Clin. Virol. 30, 211–213 10.1016/j.jcv.2003.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ströher U., DiCaro A., Li Y., Strong J. E., Aoki F., Plummer F., Jones S. M., and Feldmann H. (2004) Severe acute respiratory syndrome related coronavirus is inhibited by interferon-alpha. J. Infect. Dis. 189, 1164–1167 10.1086/382597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A. M., Matrosovich M., Becker S., and Weber F. (2013) Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 87, 5300–5304 10.1128/JVI.03496-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hulswit R. J. G., de Haan C. A. M., and Bosch B. J. (2016) Coronavirus spike protein and tropism changes. Adv. Virus Res. 96, 29–57 10.1016/bs.aivir.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pervolaraki K., Rastgou Talemi S. R., Albrecht D., Bormann F., Bamford C., Mendoza J. L., Garcia K. C., McLauchlan J., Höfer T., Stanifer M. L., and Boulant S. (2018) Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 14, e1007420 10.1371/journal.ppat.1007420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamming O. J., Terczyńska-Dyla E., Vieyres G., Dijkman R., Jørgensen S. E., Akhtar H., Siupka P., Pietschmann T., Thiel V., and Hartmann R. (2013) Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 32, 3055–3065 10.1038/emboj.2013.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G., Schwemmle M., Günther S., Drosten C., Michiels T., and Staeheli P. (2010) Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 84, 5670–5677 10.1128/JVI.00272-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prokunina-Olsson L., Alphonse N., Dickenson R. E., Durbin J. E., Glenn J. S., Hartmann R., Kotenko S. V., Lazear H. M., O'Brien T. R., Odendall C., Onabajo O. O., Piontkivska H., Santer D. M., Reich N. C., Wack A., et al. (2020) COVID-19 and emerging viral infections: The case for interferon lambda. J. Exp. Med. 217, e20200653 10.1084/jem.20200653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., O'Meara M. J., Rezelj V. V., Guo J. Z., Swaney D. L., Tummino T. A., Huettenhain R., Kaake R. M., Richards A. L., Tutuncuoglu B., et al. (2020) A SARS–CoV-2 protein interaction map reveals targets for drug repurposing. Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davis M. I., Hunt J. P., Herrgard S., Ciceri P., Wodicka L. M., Pallares G., Hocker M., Treiber D. K., and Zarrinkar P. P. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 10.1038/nbt.1990 [DOI] [PubMed] [Google Scholar]
- 23. Stoltz M., and Klingström J. (2010) Alpha/Beta interferon (IFN-alpha/beta)–independent induction of IFN-lambda 1 (interleukin-29) in response to Hantaan virus infection. J. Virol. 84, 9140–9148 10.1128/JVI.00717-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fried M. W., Shiffman M. L., Reddy K. R., Smith C., Marinos G., Gonçales F. L. Jr., Häussinger D., Diago M., Carosi G., Dhumeaux D., Craxi A., Lin A., Hoffman J., and Yu J. (2002) Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347, 975–982 10.1056/NEJMoa020047 [DOI] [PubMed] [Google Scholar]
- 25. Muir A. J., Arora S., Everson G., Flisiak R., George J., Ghalib R., Gordon S. C., Gray T., Greenbloom S., Hassanein T., Hillson J., Horga M. A., Jacobson I. M., Jeffers L., Kowdley K. V., et al. (2014) A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol. 61, 1238–1246 10.1016/j.jhep.2014.07.022 [DOI] [PubMed] [Google Scholar]
- 26. Lokugamage K. G., Hage A., Schindewolf C., Rajsbaum R., and Menachery V. D. (2020) SARS–CoV-2 is sensitive to type I interferon pretreatment. bioRxiv 10.1101/2020.03.07.982264 [DOI] [PMC free article] [PubMed]
- 27. Mantlo E. K., Bukreyeva N., Maruyama J., Paessler S., and Huang C. (2020) Potent antiviral activities of type I interferons to SARS–CoV-2 infection. bioRxiv 10.1101/2020.04.02.022764 [DOI] [PMC free article] [PubMed]
- 28. Stanifer M. L., Kee C., Cortese M., Triana S., Mukenhirn M., Kraeusslich H.-G., Alexandrov T., Bartenschlager R., and Boulant S. (2020) Critical role of type III interferon in controlling SARS–CoV-2 infection, replication and spread in primary human intestinal epithelial cells. bioRxiv 10.1101/2020.04.24.059667 [DOI] [PMC free article] [PubMed]
- 29. Channappanavar R., Fehr A. R., Vijay R., Mack M., Zhao J., Meyerholz D. K., and Perlman S. (2016) Dysregulated type I interferon and inflammatory monocyte–macrophage responses cause lethal pneumonia in SARS–CoV–infected mice. Cell Host Microbe 19, 181–193 10.1016/j.chom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frieman M. B., Chen J., Morrison T. E., Whitmore A., Funkhouser W., Ward J. M., Lamirande E. W., Roberts A., Heise M., Subbarao K., and Baric R. S. (2010) SARS–CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 6, e1000849 10.1371/journal.ppat.1000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haagmans B. L., Kuiken T., Martina B. E., Fouchier R. A., Rimmelzwaan G. F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K. H., Tashiro M., and Osterhaus A. D. (2004) Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10, 290–293 10.1038/nm1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mahlakõiv T., Ritz D., Mordstein M., DeDiego M. L., Enjuanes L., Müller M. A., Drosten C., and Staeheli P. (2012) Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J. Gen. Virol. 93, 2601–2605 10.1099/vir.0.046284-0 [DOI] [PubMed] [Google Scholar]
- 33. Mordstein M., Kochs G., Dumoutier L., Renauld J. C., Paludan S. R., Klucher K., and Staeheli P. (2008) Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4, e1000151 10.1371/journal.ppat.1000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arabi Y. M., Shalhoub S., Mandourah Y., Al-Hameed F., Al-Omari A., Al Qasim E., Jose J., Alraddadi B., Almotairi A., Al Khatib K., Abdulmomen A., Qushmaq I., Sindi A. A., Mady A., Solaiman O., et al. (2020) Ribavirin and interferon therapy for critically ill patients with middle east respiratory syndrome: a multicenter observational study. Clin. Infect. Dis. 70, 1837–1844 10.1093/cid/ciz544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loutfy M. R., Blatt L. M., Siminovitch K. A., Ward S., Wolff B., Lho H., Pham D. H., Deif H., LaMere E. A., Chang M., Kain K. C., Farcas G. A., Ferguson P., Latchford M., Levy G., et al. (2003) Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290, 3222–3228 10.1001/jama.290.24.3222 [DOI] [PubMed] [Google Scholar]
- 36. Omrani A. S., Saad M. M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A. Y., Almakhlafi G. A., Albarrak M. M., Memish Z. A., and Albarrak A. M. (2014) Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 14, 1090–1095 10.1016/S1473-3099(14)70920-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strayer D. R., Dickey R., and Carter W. A. (2014) Sensitivity of SARS/MERS CoV to interferons and other drugs based on achievable serum concentrations in humans. Infect. Disord. Drug Targets 14, 37–43 10.2174/1871526514666140713152858 [DOI] [PubMed] [Google Scholar]
- 38. Stockman L. J., Bellamy R., and Garner P. (2006) SARS: systematic review of treatment effects. PLoS Med. 3, e343 10.1371/journal.pmed.0030343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davidson S., Maini M. K., and Wack A. (2015) Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J. Interferon Cytokine Res. 35, 252–264 10.1089/jir.2014.0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., and Richardson P. (2020) COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 20, 400–402 10.1016/S1473-3099(20)30132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gaspari V., Zengarini C., Greco S., Vangeli V., and Mastroianni A. (2020) Side effects of ruxolitinib in patients with SARS–CoV-2 infection: two case reports. Int. J. Antimicrob. Agents, in press 10.1016/j.ijantimicag.2020.106023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., et al. (2020) Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382, 970–971 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H. R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A., Berger A., Burguiere A. M., Cinatl J., Eickmann M., Escriou N., et al. (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- 44. Horisberger M. A., and de Staritzky K. (1987) A recombinant human interferon-alpha B/D hybrid with a broad host-range. J. Gen. Virol. 68, 945–948 10.1099/0022-1317-68-3-945 [DOI] [PubMed] [Google Scholar]
- 45. Møhlenberg M., Gad H. H., and Hartmann R. (2019) The influence of the rs30461 single nucleotide polymorphism on IFN-λ1 activity and secretion. J. Interferon Cytokine Res. 39, 661–667 10.1089/jir.2019.0051 [DOI] [PubMed] [Google Scholar]
- 46. Dixon W. J. (1993) BMDP Statistical Software Manual, University of California Press, Berkeley, CA, USA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented and discussed are contained within the article.