Abstract
Autophagy is a conserved process that recycles cellular contents to promote survival. Although nitrogen limitation is the canonical inducer of autophagy, recent studies have revealed several other nutrients important to this process. In this study, we used a quantitative, high-throughput assay to identify potassium starvation as a new and potent inducer of autophagy in the yeast Saccharomyces cerevisiae. We found that potassium-dependent autophagy requires the core pathway kinases Atg1, Atg5, and Vps34, and other components of the phosphatidylinositol 3-kinase complex. Transmission EM revealed abundant autophagosome formation in response to both stimuli. RNA-Seq indicated distinct transcriptional responses: nitrogen affects transport of ions such as copper, whereas potassium targets the organization of other cellular components. Thus, nitrogen and potassium share the ability to influence molecular supply and demand but do so in different ways. Both inputs promote catabolism through bulk autophagy, but result in distinct mechanisms of cellular remodeling and synthesis.
Keywords: autophagy, starvation, potassium, yeast, electron microscopy, RNA sequencing, phosphatidylinositide 3-kinase (PI 3-kinase), transcriptomics
Many eukaryotic organisms experience nutrient starvation, and their ability to adapt is important for their survival. Adaptation to starvation is often characterized by alterations to signaling, transcription, and metabolism (1–4). To support these changes, cellular components are recycled into useable building blocks by two distinct and complementary mechanisms. Whereas the ubiquitin proteasome system breaks down specific short-lived proteins into their constituent amino acids, autophagy targets a wider variety of cytoplasmic cargo for degradation (5).
Much of our understanding of proteostasis comes from genetic studies conducted in the budding yeast Saccharomyces cerevisiae. As first shown in yeast, and later in animals, autophagy can be induced by diverse nutritional and pharmacological signals that converge at the target of rapamycin complex 1 (TORC1) (6–10). In nutrient-rich environments, TORC1 remains active and inhibits autophagy via phosphorylation. Under pro-autophagy conditions, deactivation of TORC1 promotes assembly of autophagy-related (ATG) proteins (such as Atg1) and lipid chains. This step requires the phosphatidylinositol (PI) 3-kinase Vps34, in complex with Vps15 (regulatory kinase), Vps30 (adaptor), and either Atg14 or Vps38 (11–18). Activation of Vps34 leads to increased levels of PI 3-phosphate, which enables downstream proteins such as Atg5 to assemble into a functional complex and catalyze the conjugation of the ubiquitin-like protein Atg8 (LC3, GABARAP, and GATE-16 in animal cells) to phosphatidylethanolamine (19). This conjugate leads to membrane expansion around portions of the cytoplasm (20). This structure, known as the autophagosome, is a unique double-membrane vesicle that fuses with the vacuole (lysosome, in higher eukaryotes), resulting in the degradation and reuse of cytoplasmic contents (21–26).
In addition to removing cytoplasmic proteins, autophagy helps to replenish the cellular pool of biologically important metals, which are required in large abundance (calcium, potassium, and sodium) or in trace amounts (iron, copper, and zinc) to maintain physiological parameters such as cell volume, pH, and protein synthesis (27–29). Under basal conditions, iron is recycled through the vacuolar transport and degradation of iron-containing cargo (30). Nitrogen limitation results in the release of calcium ions from lysosomes and the subsequent induction of autophagy through the activation of the transcription factor EB (31, 32). In addition, the availability of certain ions influences basal and stress-induced autophagy. For example, zinc starvation promotes autophagic targeting of zinc-binding proteins in yeast (33). Therefore, autophagy and ion homeostasis share a complex reciprocal relationship that is only beginning to be appreciated.
Here we describe a new and important regulator of autophagy. Using a panel of independent and complementary methods, we show that short-term potassium starvation induces bulk autophagy. Using RNA-Seq, we demonstrate that nitrogen limitation and potassium starvation result in substantially different transcriptional profiles. Whereas nitrogen affects genes important to ion transport, potassium is less specific and regulates genes related to cellular organization. Taken together, our findings point to converging mechanisms for autophagic recycling of cellular materials in conjunction with distinct and complementary routes to transcriptional adaptation.
Results
Potassium starvation promotes autophagy
Nitrogen limitation has long been the canonical inducer of autophagy. However, recent reports suggest that other salts, amino acids, and micronutrients are also important (33–36). Autophagy may indeed be up-regulated to compensate for limited external availability of essential nutrients. We hypothesized that a quantitative and parallel analysis of growth medium components might reveal new pro-autophagy regulatory pathways.
To enable high-throughput comparison of autophagy responses across diverse nutritional conditions, we used Rosella, a fluorescent reporter of autophagy (Fig. 1A) (37, 38). Rosella is composed of super-ecliptic pHluorin (green, pH-sensitive) fused with DsRed.T3 (red, pH stable). Upon induction of autophagy, the reporter is transported to the lumen of the vacuole where the acidic pH lowers green fluorescence, whereas red fluorescence remains unaffected. Rosella response is presented as a ratio of red and green fluorescence. As shown in Fig. 1B, cells transferred to nitrogen limitation conditions exhibited a large increase in Rosella response as compared with control cells in standard medium.
Figure 1.
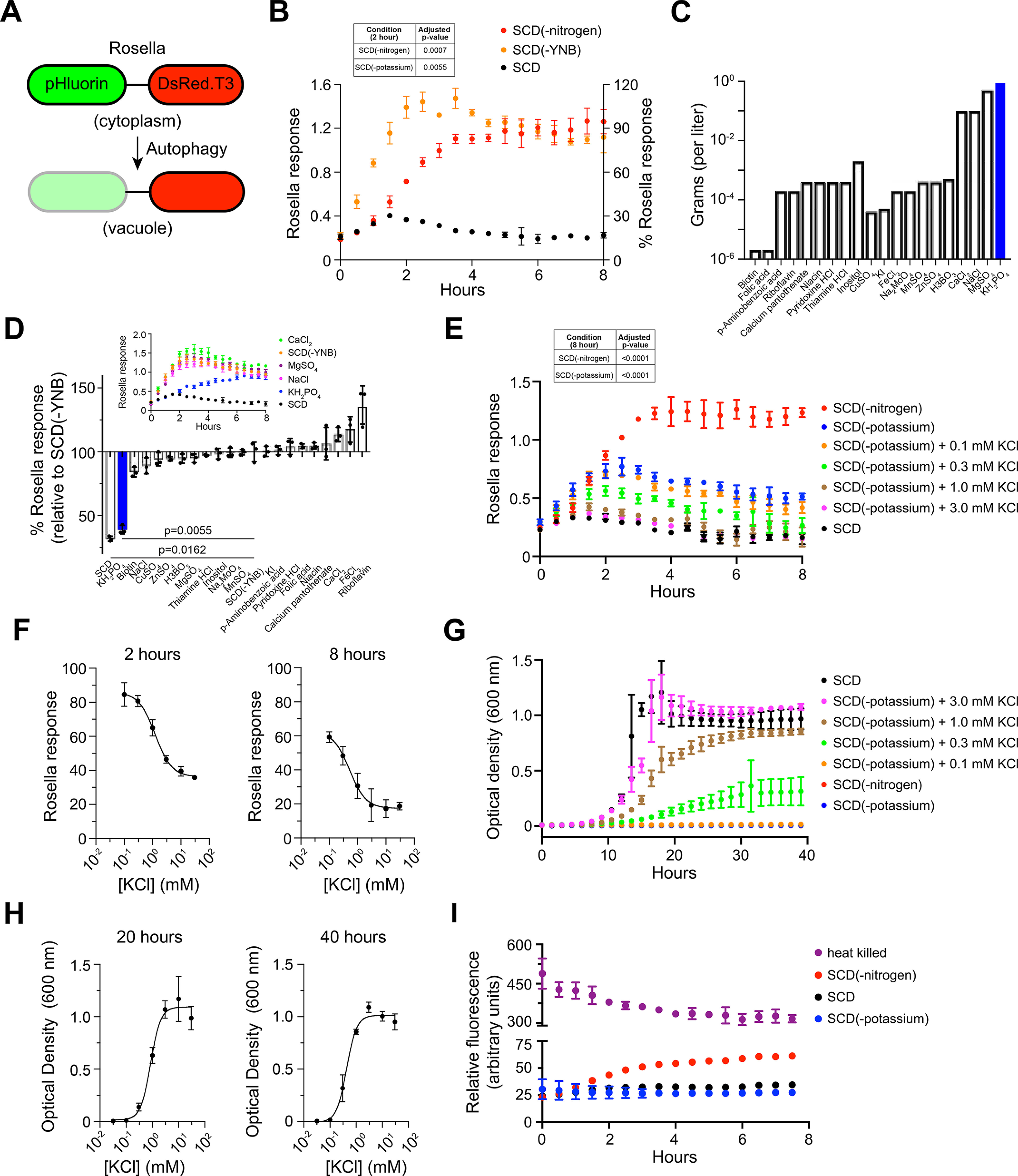
Potassium starvation promotes autophagy. A, rosella is composed of super-ecliptic pHluorin (green) and DsRed.T3 (red). Upon induction of autophagy, Rosella is transported to the vacuole where low pH results in attenuation of green fluorescence, whereas red fluorescence is unaffected. B, the cells expressing Rosella were maintained in exponential growth (A at 600 nm < 1) for 24 h prior to nitrogen limitation or starvation for YNB. Fluorescence was measured at 30-min intervals for 8 h. Response is presented as the ratio of red and green fluorescence. Inset: Table showing adjusted p-values for data at 2 h. C, components of YNB and their abundance (in grams) in 1 liter of growth medium. D, the cells were grown in SCD-leucine medium as described for (B), then transferred to SCD(-YNB) medium individually supplemented with each YNB component. Rosella response is shown as % relative to the response in SCD(-YNB) at 2 h. Inset: Time-course of response in SCD(-YNB) medium after addition of the major individual salt components. Adjusted p-values were determined for KH2PO4 addition relative to SCD(-YNB) E, time-course of response in SCD(-potassium) medium upon addition of 0–3.0 mm KCl. Inset: Table showing adjusted p-values for data at 8 h. F, dose-dependence of Rosella response shown in (E), for the 2-h and 8-h time points. G, cellular growth reported by optical density at 600 nm (A600), under the same conditions as in (E). H, dose-dependence of cell growth data shown in (G), for the 20-h and 40-h time points. I, time-course of cell viability measured as fluorescence from propidium iodide. Rosella experiments were performed at least three times, and data are presented as ± S.D. (error bars) for four technical replicates. Growth and viability data are reported for three biological replicates. Data were analyzed in Microsoft Excel and GraphPad Prism. Adjusted p-values were determined with one-way ANOVA relative to untreated cells (SCD).
Next, we used Rosella to test other nutrients important for proper cellular growth and homeostasis. As described in Table 1, synthetic complete medium with 2% dextrose (SCD) is composed of a nitrogen source (ammonium sulfate), sugar (dextrose), amino acids, nucleotides, and a base mixture (yeast nitrogen base, or YNB) composed of vitamins, trace elements, and salts. YNB does not provide nitrogen, which is included separately as ammonium sulfate. Other investigators have shown that sugar starvation does not lead to completion of autophagy (39, 40). Therefore, we turned our attention to other nutrients, and in particular, components of yeast nitrogen base. As shown in Fig. 1B, in the absence of YNB, cells displayed an increase in response that was similar in magnitude and substantially faster (t1/2 ∼ 1.25 h) than that observed in nitrogen limitation media (t1/2 ∼ 4 h). To identify the component(s) of YNB that individually contributed to the Rosella response (Fig. 1B) we supplemented SCD(-YNB) medium with each individual vitamin, trace element, or salt (Table 2 and Fig. 1C) and monitored fluorescence over time. As shown in Fig. 1D, Rosella response was significantly diminished upon addition of potassium phosphate (+KH2PO4) alone, whereas re-addition of other salts had no effect (Fig. 1D, inset). We note the comparatively small response to zinc sulfate. Depletion for Zn2+ ions was reported to induce autophagy, but over longer timescales of >16 h (33). We conclude that, by this measure, potassium is the primary driver of autophagy observed in the absence of YNB.
Table 1.
Components of synthetic complete medium with 2% dextrose (SCD)
| Nutrient | Name | Amount (g/l, unless specified otherwise) |
|---|---|---|
| Sugar | Dextrose | 20 |
| Nitrogen | Ammonium sulfate | 5 |
| Nucleotides + Amino acids | CSM dropout mixture (CSM-Ura) | 0.69 |
| Uracil | 20 mg | |
| Adenine | 30 mg | |
| NaOH | 1 pellet | |
| Vitamins, compounds supplying trace elements, and salts | YNB without amino acids and without ammonium sulfate | 1.7 |
CSM, Complete Supplement Mixture.
Table 2.
Components of yeast nitrogen base (without amino acids and without ammonium sulfate)
| Nutrient | Name | Amount (μg/l) |
|---|---|---|
| Vitamins | Biotin | 2 |
| Calcium pantothenate | 400 | |
| Folic acid | 2 | |
| Inositol | 2000 | |
| Niacin | 400 | |
| p-Aminobenzoic acid | 200 | |
| Pyridoxine hydrochloride | 400 | |
| Riboflavin | 200 | |
| Thiamine hydrochloride | 400 | |
| Compounds supplying trace elements | Boric acid | 500 |
| Copper sulfate | 40 | |
| Potassium iodide | 100 | |
| Ferric chloride | 200 | |
| Manganese sulfate | 400 | |
| Sodium molybdate | 200 | |
| Zinc sulfate | 400 | |
| Salts | Potassium phosphate | 1000 |
| Magnesium sulfate | 500 | |
| Sodium chloride | 100 | |
| Calcium chloride | 100 |
To distinguish between the contributions of the constituent ions (K+ and H2PO4−), we measured autophagy in cells exposed to growth medium in which potassium phosphate (in the YNB) was replaced with ammonium phosphate SCD(-potassium). As shown in Fig. 1E, Rosella signal was elevated in these conditions, indicating that autophagy is specifically regulated by potassium cations. The response was recovered by the addition of 0.1–3 mm KCl, which has been used by others to replenish extracellular K+ ions (41, 42). In complete SCD medium, K+ ions are present at a concentration of ∼7 mm. As shown in the time profiles in Fig. 1E, the response at 2 h was similar for both nitrogen and potassium but diverged substantially at later time points. The Rosella response at 2 h showed an EC50 of 1.2 mm KCl (Fig. 1F), which was substantially lower than that required to activate the Hog1 kinase (>50 mm), which is crucial for cellular adaptation to external osmotic changes (43, 44).
We observed substantial differences when comparing the response to SCD(-YNB) (Fig. 1B) with SCD(-potassium) (Fig. 1E). Although KH2PO4 appears to have a predominant role in mediating the autophagic response in SCD(-YNB), we infer that the lack of additional components in SCD(-YNB) contributes in some way, perhaps in a synergistic or cooperative fashion, to the kinetics and magnitude of the differences when comparing these conditions. The differences are not due to specific anions, given that autophagy is absent in SCD(-potassium) medium supplemented with KCl (Fig. 1E) or SCD(-YNB) medium supplemented with KH2PO4 (Fig. 1D, inset).
Potassium ions are highly abundant within yeast cells (∼150–300 mm) and regulate ionic strength, turgor pressure, and enzyme function (28, 45, 46). In accordance with these requirements and as shown previously, cell growth is slowed by the lack of extracellular potassium (47). Addition of 0.3 mm KCl to SCD(-potassium) medium resulted in partial recovery of growth (Fig. 1G). At 1.0 and 3.0 mm KCl, long-term recovery of growth was near complete. The growth response at 20 h showed an EC50 of 0.9 mm KCl (Fig. 1H). To ensure that cells remained viable, we monitored propidium iodide fluorescence (48) from individual cells in microplate wells exposed to the same growth media as in Fig. 1G. As shown in Fig. 1I, propidium iodide fluorescence remained low under potassium starvation conditions and was elevated upon nitrogen limitation. Thus, potassium is required for proper cell growth and autophagy. The autophagy response to potassium starvation is ∼30% of that seen in response to nitrogen limitation. In contrast to nitrogen, however, potassium is not required to maintain cell viability, at least in the short term.
Potassium-dependent autophagy is mediated by autophagosomes
Our findings presented in Fig. 1 indicate that the potassium-dependent autophagy response is lower than that for nitrogen. The magnitude of autophagy is regulated by many factors that control the number or size of autophagosomes (49, 50). To further compare potassium starvation and nitrogen limitation, we examined the accumulation of autophagosomes in individual cells using transmission EM (TEM). For these experiments, we used cells lacking the vacuolar protease Pep4, which is required for breakdown of autophagosomes within the vacuole (6). We first grew the cells for 6 h in SCD(-nitrogen) or SCD(-potassium) medium, harvested the cells by centrifugation, and prepared the cells for TEM imaging (fixed, dehydrated, embedded, cut, and stained). As expected, nitrogen limitation resulted in the formation of multiple autophagosomes in ∼95% of the cells. On average, we observed 6.9 ± 0.9 autophagosomes/cell with a cross-sectional area of 0.102 ± 0.042 μm2 (Fig. 2, A and B). In potassium-starved samples, autophagosomes were evident in only ∼10% of the cells, far fewer than that observed in nitrogen-limited cells. However, when present, the number (6.5 ± 1 per cell) and size (0.089 ± 0.037 μm2) of autophagosomes were similar under both conditions. The presence of autophagosomes is a hallmark of macroautophagy, as opposed to other types of autophagy, in particular microautophagy. In both conditions, we observed accumulation of glycogen granules (G) and lipid droplets (L) in the cytoplasm (Fig. 2, A and C). These features are characteristic of bulk autophagy (6). Although most of the autophagosomes observed in our TEM images were already engulfed by the vacuole, we also observed some that were fused to the vacuolar membrane (Fig. 2C) prior to engulfment. From these results, we conclude that potassium starvation leads to autophagosome formation but does so in only a subset of cells.
Figure 2.
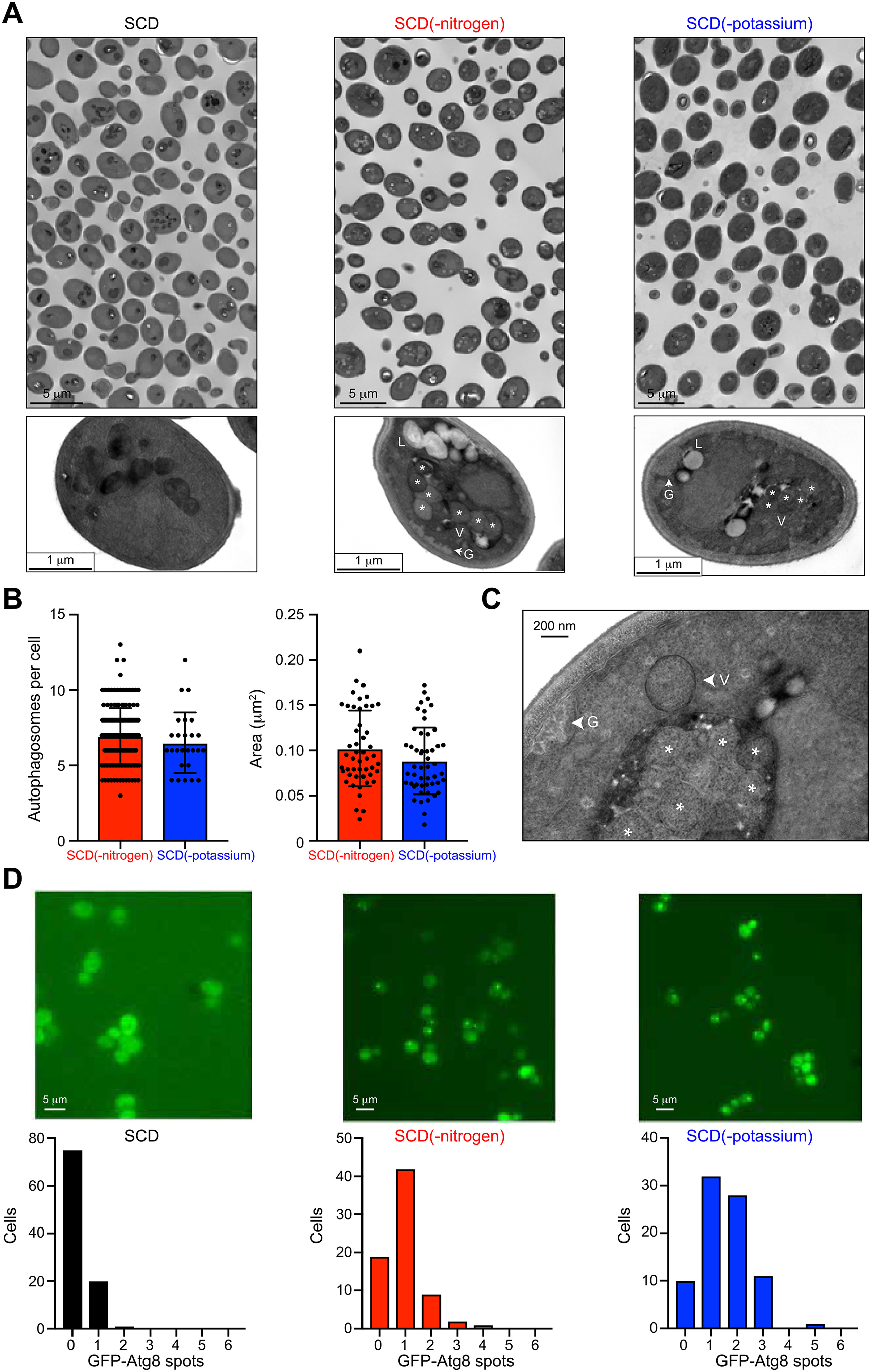
Potassium-dependent autophagy is mediated by autophagosomes. A, representative images of individual cell sections after 6 h of nitrogen limitation or potassium starvation. Vacuole (V), autophagosomes (*), lipid droplet (L), glycogen (G). B, distribution of the abundance (per cell) and size (area in µm2) of autophagosomes (n = 50). C, high magnification image (80,000×) showing an autophagosome (arrowhead) fused to the vacuolar membrane. D, representative single-cell images and distributions of GFP-Atg8 spots formed in response to 2 h of nitrogen limitation or potassium starvation (n = 85 cells).
Whereas Rosella reports on the transport of cytoplasmic cargo to the vacuole, other methods directly monitor components of the autophagy machinery such as autophagosomes and vacuolar proteins. To confirm the pro-autophagy effect of potassium starvation, we monitored the spatial localization of the ubiquitin-like protein Atg8. Upon induction of autophagy, Atg8 conjugates with phosphatidylethanolamine to form autophagosomes, which deliver cytoplasmic contents to the vacuole (19, 51). During this process, Atg8 assembles into autophagosomal clusters, which are observed as bright puncta within the cell (52). As shown in Fig. 2D, we observed a substantial increase in GFP-Atg8 clusters under potassium starvation. This response was greater than that for nitrogen-limited cells, as is evident from the frequency distribution plots. These results indicate that autophagy is initiated in a majority of the cells but that the process is completed in only a subset of those cells.
Potassium-dependent autophagy requires core ATG kinases and the PI 3-kinase complex
After being internalized by the vacuole, the autophagosomal cargo is released for degradation and recycling. This process can be monitored by cleavage of GFP-Atg8 (53). As shown in Fig. 3A, GFP was observed after 3 h of potassium starvation and the response increased further at 6 h, indicating a sustained effect. Because GFP-Atg8 processing is typically more variable than the Rosella response, we validated our findings using the Pho8Δ60 enzymatic assay (54). The alkaline phosphatase Pho8 is translocated to the vacuole using the secretory pathway. Although the truncated form is maintained in the cytosol under basal conditions, it is transported to the vacuole under nitrogen-limited conditions and subsequently activated via proteolytic processing. The increase in enzyme activity is a quantitative indicator of autophagy and is measured as the conversion of a substrate to a fluorescent product. As shown in Fig. 3B and in agreement with our findings from the other assays, potassium starvation resulted in an increase in activity that was ∼38% of that observed with nitrogen limitation.
Figure 3.
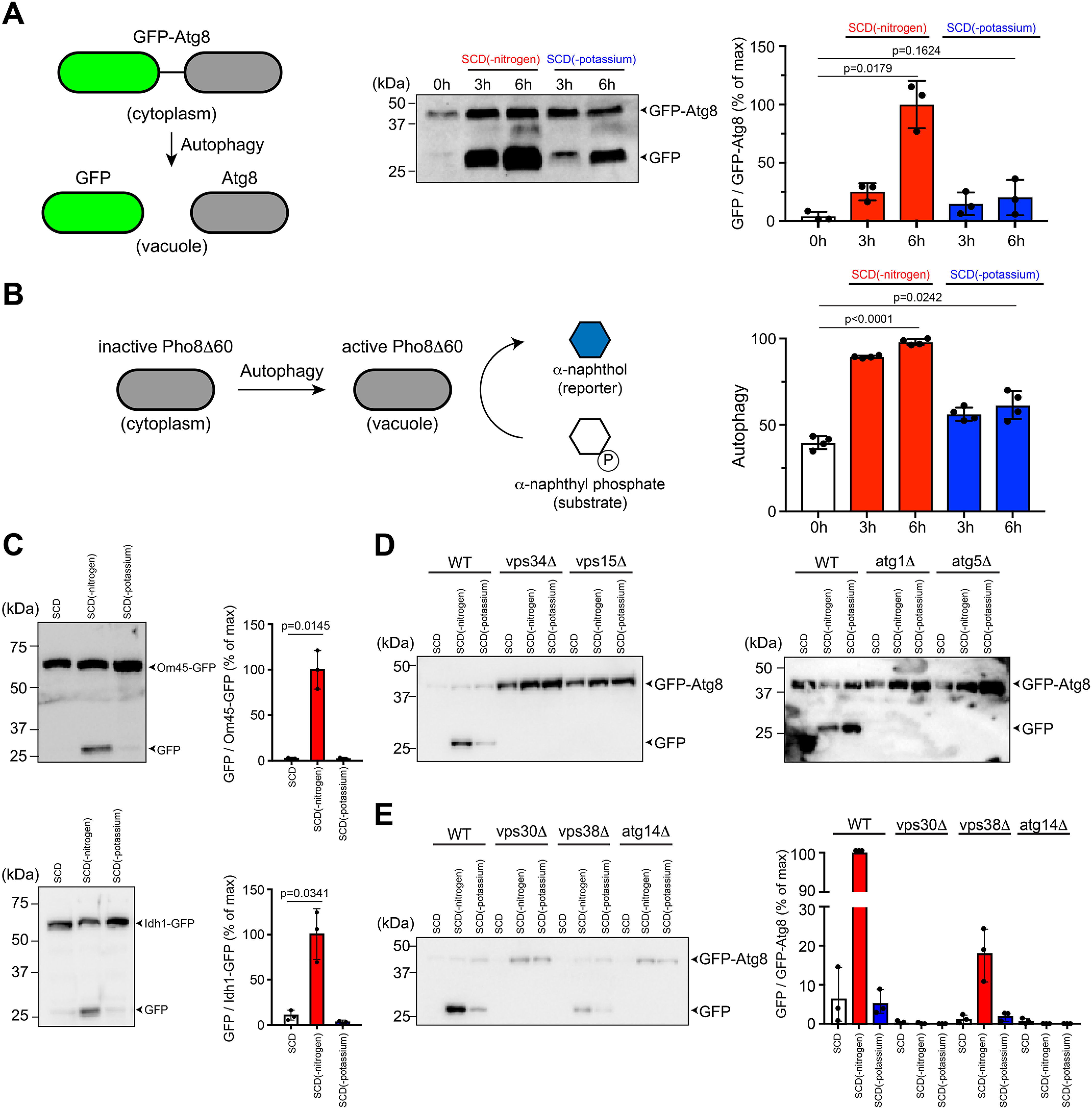
Potassium-dependent autophagy requires autophagy-related kinases Atg1 and Atg5 and the PI 3-kinase complex. A, in pro-autophagy conditions, the GFP-Atg8 reporter is transported to the vacuole and enzymatically processed to release GFP. GFP-Atg8 processing was monitored in WT BY4741 cells after 0, 3, and 6 h of nitrogen limitation or potassium starvation. Protein bands were quantified using ImageLab and presented as the ratio of GFP and GFP-Atg8 (% of maximum). B, induction of autophagy results in activation of the Pho8Δ60 enzyme, which converts a fluorogenic substrate (α-naphthyl phosphate) into a fluorescent reporter (α-naphthol). Shown are data for Pho8Δ60 activity in the same conditions as (A). C, processing of mitophagy reporters Om45-GFP and Idh1-GFP in WT BY4741 after 6 h of nitrogen limitation or potassium starvation. Data are presented as the ratio of GFP and Om45-GFP/Idh1-GFP (% of maximum). D, processing of autophagy reporter GFP-Atg8 after 6 h of nitrogen limitation or potassium starvation in BY4741 cells lacking core components of the PI 3-kinase complex (Vps34 or Vps15) or the autophagy pathway (Atg1 or Atg5). E, processing of GFP-Atg8 after 6 h of nitrogen limitation or potassium starvation in BY4741 cells lacking regulatory components of the PI 3-kinase complex (Vps30, Vps38, or Atg14). All experiments were repeated three times and data are ± S.D. (error bars) for three or four biological replicates. Adjusted p-values were determined by one-way ANOVA relative to untreated cells (SCD).
In addition to the bulk cytoplasm, organelles such as mitochondria, peroxisomes, and ribosomes are also recognized as specific cargo for autophagic degradation. These selective forms of autophagy employ many of the core pathway components and cargo-specific receptors (55–57). Previously, it was shown that pharmacological alterations to potassium ion homeostasis can lead to the production of reactive oxygen species, mitochondrial damage, and autophagic degradation of mitochondria (mitophagy) (58). Given these findings, we sought to determine the effects of our potassium starvation conditions on mitophagy. To that end, we monitored two GFP-tagged proteins, the mitochondrial outer membrane protein Om45 and the matrix protein Idh1 (59–61). As shown in Fig. 3C, whereas nitrogen limitation resulted in the release of GFP from these proteins, potassium starvation had a negligible effect. Thus, bulk autophagy is regulated by nitrogen or potassium whereas mitophagy depends on nitrogen alone.
Vps34 is the sole PI 3-kinase in yeast and is essential for autophagy. PI 3-phosphate, generated by Vps34, is localized to autophagosomal membranes and is required for recruitment of other autophagy-related (ATG) proteins. Complex I is composed of Vps34, a regulatory kinase Vps15, Vps30 (Beclin-1 in animal cells), and Atg14. Complex II contains Vps38 (UVRAG) in place of Atg14 (11–13). Whereas complex I mediates autophagy in nutrient-limiting conditions, complex II is essential for proper sorting of vacuolar enzymes under nutrient-rich conditions. To test the role of the effector complexes in potassium-dependent autophagy, we analyzed individual gene deletions using the GFP-Atg8 immunoblotting assay. As anticipated, we observed no processing of GFP-Atg8 in cells lacking Vps34 and Vps15 (Fig. 3D). Potassium-dependent autophagy required the canonical components Atg1 and Atg5, in a manner similar to the nitrogen response (Fig. 3D). Moreover, autophagy was completely abrogated in cells lacking Atg14 (complex I) or the common adapter Vps30, and reduced by deletion of Vps38 (complex II) (Fig. 3E). This is consistent with recent work suggesting reciprocal regulation of potassium signaling and TORC1 activity, which inhibits autophagy by phosphorylating Atg13, a component of the Atg1 kinase complex (41, 62). Thus, both potassium- and nitrogen-dependent autophagy are mediated by Atg1, Atg5, and the PI 3-kinase complex.
Potassium starvation and nitrogen limitation exhibit distinct transcriptional profiles
Our data indicate that potassium and nitrogen regulate a common autophagy response to supply raw materials for cellular biosynthesis. To determine how this information is interpreted, we first considered phosphorylation-mediated activation of the mitogen-activated protein kinase (MAPK) Kss1. In addition to its effects on autophagy, nitrogen limitation promotes cellular remodeling events such as filamentous growth (63) and the induction of genes involved in these processes, such as FUS1 (64–67). Notably, filamentation requires elements of the MAPK pathway (68), and Kss1 activation is important for the transcription of filamentation genes under nitrogen limitation conditions (69). Preliminary evidence from others suggests that potassium starvation also induces FUS1 transcription (42). However, the effect of nitrogen limitation or potassium starvation on Kss1 activation has not been investigated, and more generally, the regulatory mechanisms shared by the autophagy and filamentous growth pathways are not fully understood (70, 71). To address this question, we used Phos-tag gel electrophoresis to determine whether either stimulus led to phosphorylation of Kss1. As expected, Kss1 was activated upon nitrogen limitation, although the effect was substantially less than that reported previously for addition of mating pheromone (Fig. S1) (72). In contrast, Kss1 was unaffected by potassium starvation.
Another way to understand the consequences of autophagy is to measure global changes in gene transcription. To that end, we sequenced the transcriptome of cells exposed to nitrogen limitation or potassium starvation (in duplicate) prior to RNA extraction (Fig. 4A). To compare the effects of the two stimuli, we carried out differential expression analysis, which revealed a dramatic impact on the transcriptome. Nitrogen limitation produced a highly dispersed response that included a large number of differentially expressed genes (DEGs) (log2(fold change) > 1 and adjusted p-value < 0.05) (Fig. 4B). In comparison, potassium starvation resulted in fewer DEGs with substantially smaller fold-change values. Using this approach, we identified 107 DEGs common to the two conditions (Fig. 4C). In addition to these shared targets, we found 68 and 626 unique DEGs for potassium and nitrogen, respectively. Principal component analysis revealed that nitrogen limitation and potassium starvation lead to distinct transcriptional profiles (Fig. 4D). To understand the functional processes targeted by these stimuli, we performed Gene Ontology (GO) enrichment analysis using the DEGs independently for each condition. Whereas potassium targets processes that organize and modify cellular components and organelles, nitrogen affects transport of copper and transition metal ions (Fig. 4E). We conclude that potassium and nitrogen mediate their responses through distinct cellular mechanisms, as indicated by the appearance of differentially expressed genes (Fig. 4, B and C) and distinct GO annotations (Fig. 4E).
Figure 4.
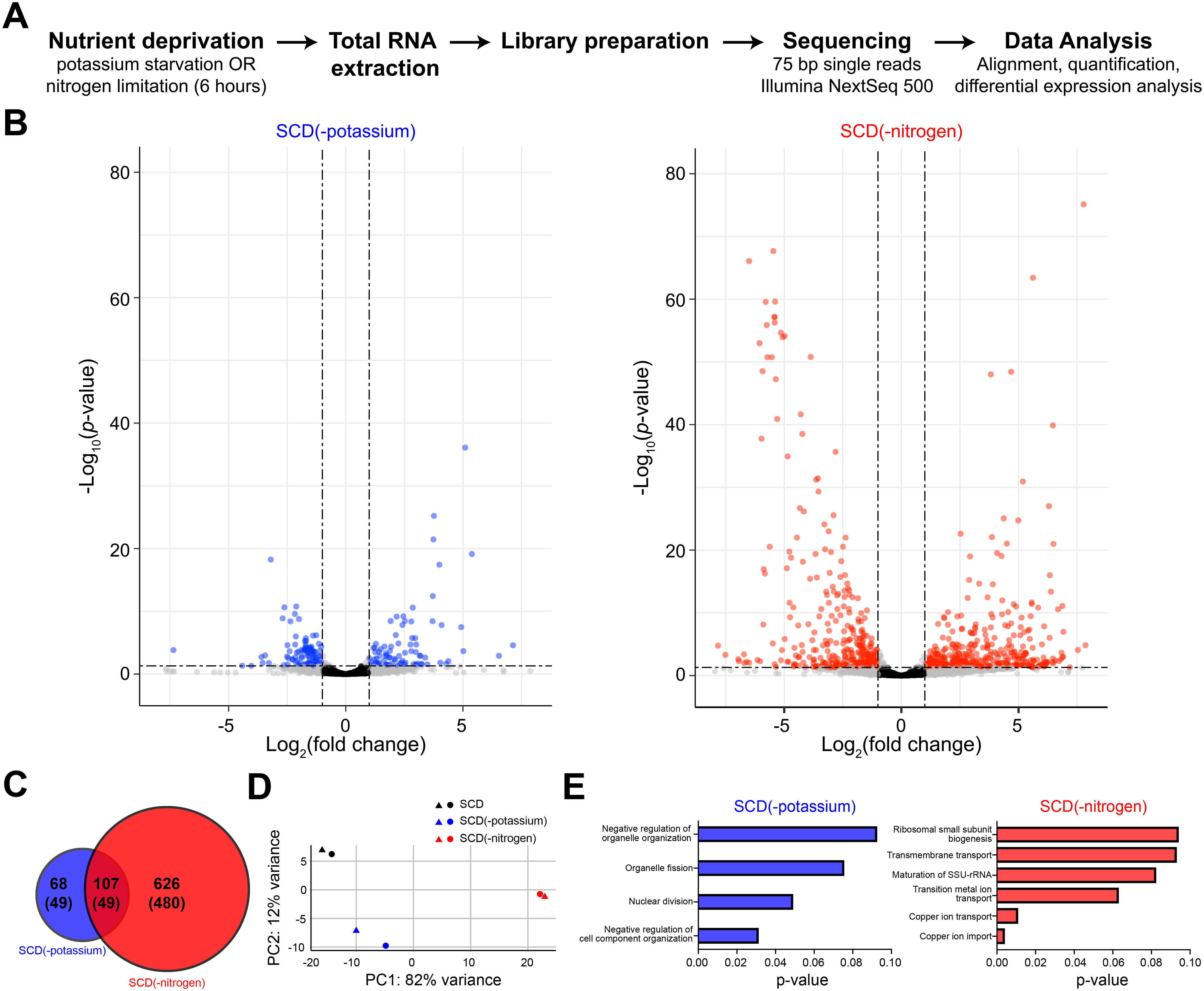
Potassium starvation and nitrogen limitation exhibit distinct transcriptional profiles. A, schematic representation of the experimental approach to obtain RNA-Seq data for potassium starvation or nitrogen limitation. B, differential expression analysis reveals statistically significant changes in RNA abundance. Genes with adjusted p-value < 0.05 and log2(fold change) > 1 are highlighted in blue (potassium starvation) and red (nitrogen limitation). C, venn diagram showing overlap between DEGs for potassium starvation and nitrogen limitation. Numbers in parentheses represent genes with a known GO annotation. D, principal component analysis of sequencing data showing the first two principal components (PC1 and PC2). Colors represent treatment conditions and shapes denote biological replicates. Axis labels describe the percentage of variance explained by each principal component. Together, principal components 1 and 2 explain 94% variance of the data. For all conditions, replicates are closely spaced, which indicates that variance arises primarily from the nutritional treatments. E, GO process terms enriched among the DEGs for each condition. RNA abundance was quantified for two biological replicates for each condition.
Discussion
Most pioneering studies of autophagy used nitrogen limitation as the stimulus, primarily because of its large and rapid response. Here we demonstrate, using a panel of reporter assays in individual cells and populations, that potassium starvation is a new and potent inducer of autophagy. The magnitude of the initial response is comparable with that observed with nitrogen limitation. Both stimuli transmit their effects via the canonical pathway components Atg1, Vps34, Vps15, Vps30, Atg14, and Atg5, which suggests a common route to cellular recycling. In contrast to these shared features, transcriptional responses to nitrogen limitation and potassium starvation are substantially different. Collectively, our data indicate that nitrogen and potassium share the ability to supply metabolic building blocks but adopt distinct ways to reallocate these resources for future use.
Traditional methods to study autophagy require extensive sample preparation prior to readout and are not conducive to high-throughput analysis. In comparison, the Rosella plate reader assay provides a convenient, automated platform for quantitative measurements in diverse genetic backgrounds and growth conditions. Our comprehensive analysis showed that whereas nitrogen transmits the largest autophagy effects, potassium is also very effective, particularly in its induction of GFP-Atg8 clusters and the early Rosella response (<2 h). By integrating the Rosella reporter with other independent and well-established methods (EM, GFP-Atg8 immunoblotting and imaging, and the Pho8Δ60 alkaline phosphatase assay), we were able to compare nearly 30 conditions and multiple time points, thereby accelerating the discovery process.
Our findings add to a growing list of novel regulators of autophagy such as amino acids, sulfate, glucose and, most recently, zinc ions (6, 36). The potassium response is robust and mirrors that of nitrogen limitation, particularly at early time points (<2 h). This is in contrast with zinc, which induces a response that is comparatively small and is evident only after prolonged treatment (>16 h) (Fig. 1) (33). Another class of inducers of autophagy operates through G protein–coupled receptors. Prominent among these are the mammalian muscarinic cholinergic and β-adrenergic receptors, which bind to acetylcholine and epinephrine, respectively (73–78). Likewise, we have shown that the pheromone G protein–coupled receptor in yeast promotes vacuolar delivery of cytoplasmic contents (38, 79). Whereas the response to nitrogen and potassium depends on both complex I and II of the yeast PI 3-kinase, mating pheromone requires only complex II. Collectively, these studies demonstrate that multiple biochemically and physiologically distinct inputs converge on a common cellular process, one that is needed to support survival during changing growth conditions.
Although nitrogen and potassium induce a similar autophagy response, they are likely to serve distinct cellular needs. Nitrogen provides raw materials for synthesis of amino acids and nucleotides that enable the formation of proteins and nucleic acids. Potassium is accumulated within yeast and other eukaryotes to maintain important cellular parameters such as electroneutrality, pH, turgor pressure, and cell volume. Although yeast is not well-suited for direct (electrophysiological) analysis of potassium channel function, future studies could examine autophagy in mutants deficient in potassium transporter expression and activity. We observed that nitrogen targets genes that regulate transport of ions such as copper. In contrast, potassium impacts genes involved in a broader set of cellular processes, which overlap only partially with the nitrogen response. These data reveal that seemingly “redundant” input signals can nevertheless diverge and initiate distinct and complementary processes that prepare the cell for prolonged periods of nutrient deprivation.
In summary, our systematic and comprehensive studies highlight the power of high-throughput methods in hypothesis testing and discovery. Many important cellular pathways, including those mediated by the autophagy machinery, were first discovered in yeast and subsequently confirmed in more complex organisms. Given the conservation of pathways governing autophagy and ion homeostasis, we expect that potassium likewise regulates autophagy-related processes in humans. Such mechanisms may be important in understanding the functional consequences of hypokalemia as occurs in chronic kidney disease, alcohol abuse, and diabetic ketoacidosis.
Experimental procedures
Strains, plasmids, and growth media
Yeast S. cerevisiae strains used in this study were BY4741 (MATa leu2Δ met15Δ his3Δ ura3Δ), BY4741-derived gene deletion mutants obtained from the Yeast Knockout Collection (Invitrogen) or remade by homologous recombination of PCR-amplified drug resistance genes with flanking homology to the gene of interest (Table S1), and BY4741-derived GFP-gene fusions obtained from the Yeast GFP Clone Collection (Thermo Fisher Scientific) (80). PHO8Δ60 (gift from Mara Duncan, University of Michigan) was stably integrated into the genome, replacing the native PHO8 gene. Plasmid pRS416-GFP-ATG8 (Addgene, 49425) was a gift from Daniel Klionsky (81), and the GFP-ATG8 construct was subsequently introduced into the pRS415 vector (82). Rosella plasmid pAS1NB-DsRed.T3–super-ecliptic pHluorin (SEP) (2μ, ampR, LEU2+) was a gift from Mark Prescott and Rodney Devenish (37). The Kss1–9xMyc-tagged strain was generated by homologous recombination of a PCR-amplified 9xMyc cassette harboring a resistance gene to hygromycin B from plasmid pYM20 (pYM-9xMyc-hphNT1) at the C terminus of the KSS1 ORF (83, 84).
Cells were grown in rich medium containing yeast extract (10 g/liter), peptone (20 g/liter), and 2% (w/v) dextrose, or SCD containing 2% (w/v) dextrose, ammonium sulfate, nucleotides, amino acids, and a base mixture of vitamins, trace elements, and salts (YNB) (Table 1). Plasmid selection was maintained by antibiotic supplementation or exclusion of appropriate nutrients. For nutrient starvation analysis, SCD(-nitrogen) medium was prepared by excluding ammonium sulfate, the major source of nitrogen. SCD(-YNB) medium was prepared by omitting yeast nitrogen base. To study individual YNB ingredients, SCD(-YNB) medium was supplemented with each component as listed in Table 2. The cells were cultured overnight with shaking at 30 °C. Saturated cultures were diluted with fresh medium to optical density at 600 nm (A600) = 0.1 and cultured for 6 h, then further diluted to A600 = 0.001 and cultured for 18 h to maintain A600 < 1 prior to use. For potassium starvation, the cells were transferred to SCD(-potassium) medium, which was prepared using a modified YNB containing ammonium sulfate instead of potassium sulfate (translucent K+ free medium containing ∼15 μm K+; ForMedium, CYN7501). As a benchmark for autophagy, cells were transferred to nitrogen limitation medium SCD(-nitrogen) lacking ammonium sulfate.
Rosella microplate-reader assay
The cells were transformed with pAS1NB-DsRed.T3-SEP and cultured in SCD-leucine medium. To start the starvation time-course, 1 ml of cells at A600 ∼ 1 were transferred to SCD(-potassium) medium after centrifugation at 13,000 × g for 1 min, washing, and resuspension. 200 μl of cells were then added to individual wells in black, clear-bottom 96-well microplates (Greiner Bio-One, 655087 or Corning, 3631). Nitrogen-limited cells served as a reference treatment for autophagy. Untreated “control” cells in SCD-leucine medium were included in separate wells to enable measurement of basal response. Microplates were sealed to reduce evaporation (adhesive PCR plate seal; Thermo Fisher Scientific, AB0558) and placed in a microplate reader (Molecular Devices, SpectraMax i3x) for 8 h at 30 °C. At each time point, the samples were shaken and fluorescence was measured for SEP (488-nm excitation and 530-nm emission) and DsRed.T3 (543-nm excitation and 587-nm emission). Background signal was measured using clear, cell-free medium. All experiments were performed in duplicate with three technical replicates. For each time point, Rosella response was calculated as the ratio of background-corrected dsRed.T3 and SEP fluorescence. Starvation-induced response was normalized with basal response observed for cells maintained in SCD-leucine medium. Dose-response profiles were calculated for the 2- or 8-h time points using a variable slope (four parameters) nonlinear regression with least squares fit (GraphPad Prism).
Statistical analysis
Statistical analyses of autophagy data were performed in Graphpad Prism with one-way analysis of variance (ANOVA) followed by Dunnett''s test for multiple comparisons. Adjusted p-values were calculated relative to the untreated control (SCD) for Rosella response (Fig. 1, B, D, and E), GFP-Atg8 cleavage (Fig. 3A), Pho8Δ60 activity (Fig. 3B), and Om45-GFP/Idh1-GFP cleavage (Fig. 3C).
Growth and viability measurements
For cell growth measurements, WT BY4741 cells were cultured as described above to A600 ∼ 1, then diluted to A600 = 0.05 into either SCD, SCD(-potassium), or SCD(-nitrogen) medium. 200 μl of cells were transferred to a 96-well microplate and A600 was monitored in a SpectraMax i3x microplate reader at 30-min intervals for 48 h.
Cell viability was measured using a Celigo S Imaging Cytometer (Nexcelom Bioscience) as described previously (83). Briefly, 200 μl of cells at A600 ∼ 0.05 were mixed with 20 μm propidium iodide (Molecular Probes, P3566) and plated in half-area, black, clear-bottom 96-well plates (Greiner Bio-One, CELLSTAR) by centrifugation at 500 × g for 5 min at 4 °C. The cells were imaged at 30-min intervals for 8 h at room temperature using the “Target 1+Mask” settings. Green fluorescence from propidium iodide was measured with the inbuilt GFP channel, whereas brightfield images were acquired simultaneously and used as masks for cell segmentation using Celigo's native algorithm. Debris and cell clumps were excluded from the analysis by gating analysis based on the mean intensity and aspect ratio of propidium iodide. Background correction was performed by subtracting residual intensity from cell-free regions on the microplate. For data presentation, the mean propidium iodide intensity was averaged across 10,000 cells for each condition.
GFP-Atg8 immunoblotting
For the GFP-Atg8 processivity assay, cells were propagated in SCD-leucine medium prior to nitrogen limitation or potassium starvation (85). Cell extracts were obtained after 0, 3, and 6 h of starvation. The cells were lysed with Trichloroacetic Acid (TCA) buffer (10 mm Tris-HCl, pH 8.0, 10% (w/v) TCA, 25 mm NH4OAc, and 1 mm Na2EDTA). Protein extracts were reconstituted in resuspension buffer (100 mm Tris-HCl and 3% (w/v) SDS, pH 11.0), and protein concentration was determined using the Bio-Rad detergent compatible assay. The samples were normalized to 0.5–1 μg/μl with resuspension buffer and sample buffer (500 mm Tris-HCl, 20% (v/v) glycerol, 2% (w/v) SDS, 200 mm DTT, and 0.01% (w/v) bromphenol blue, pH 8.5). Proteins were resolved on 10% SDS-PAGE gels, transferred to nitrocellulose membranes, and detected by first incubating with blocking buffer (5% nonfat milk in Tris-buffered saline with Tween 20 with 10 mm sodium azide) and, subsequently, immunoblotting with GFP antibodies (sc-999c, clone B-2, Santa Cruz Biotechnology) at 1:1,000 dilution in blocking buffer or glucose 6-phosphate dehydrogenase antibodies (8866, Cell Signaling Technology, 1:1,000). Immunoreactive species were detected with antibodies conjugated with horseradish peroxidase (715-035-150, Jackson ImmunoResearch Laboratories) at 1:10,000 using ECL-plus reagent (Life Technologies). Protein bands were quantified on a Bio-Rad Chemidoc Touch Imaging System using Image Laboratory software version 6.0.1 (Bio-Rad).
GFP-Atg8 microscopy
Clustering of GFP-Atg8 was measured using brightfield illumination on a Nikon Ti 2000 inverted microscope. Agar pads were prepared by dissolving 2% (w/v) agar in SCD-leucine, SCD(-potassium) or SCD(-nitrogen) medium and spotting (200 μl) onto a clear glass slide. Another slide was quickly placed on top and the agarose was allowed to solidify. After removing the top slide, 5 μl of starved cells were mounted on the pad and sealed with a clean glass coverslip as described previously (86). GFP-Atg8 within single cells was imaged by excitation at 488 nm and emission at 500–550 nm. GFP-Atg8 spots were counted in 85–100 individual cells for each condition using FIJI image analysis software, and graphs were plotted with Graphpad Prism (87).
Om45-GFP and Idh1-GFP immunoblotting
BY4741 cells expressing Om45-GFP or Idh1-GFP were sourced from the Yeast GFP Clone Collection (Thermo Fisher Scientific) and maintained A600 < 1 in rich medium containing yeast extract (10 g/liter), peptone (20 g/liter), and 2% (w/v) dextrose for ∼24 h. To promote increased expression of the GFP-tagged proteins, the cells were then diluted 100-fold with YPL medium (2% lactate instead of dextrose) and cultured for 12–16 h. Nitrogen limitation or potassium starvation was carried out for 6 h prior to protein extraction and immunoblotting analysis, as described above for GFP-Atg8.
Pho8Δ60 enzymatic assay
The Pho8Δ60 assay was performed as described previously (54). The cells were cultured for 24 h in SCD medium to A600 ∼ 1 prior to transfer to SCD(-nitrogen) or SCD(-potassium) medium. After 0, 3, or 6 h of starvation, 5 ml of cells were collected by centrifugation at 3000 × g for 5 min. Cell pellets were resuspended in 200 μl of cold assay buffer (250 mm Tris-HCl, pH 9.0, 10 mm MgSO4, and 10 μm ZnSO4), mixed with ∼100 μl of acid-washed glass beads (500-μm diameter), and lysed on an automatic vortex mixer (5 min at 4 °C). The suspension was diluted with 50 μl of assay buffer, and cell debris were removed by centrifugation at 13,000 × g for 5 min. Protein concentration in the supernatant solution was determined with the Bio-Rad detergent compatible protein assay. To measure enzyme activity, cell lysates were diluted 10-fold and incubated with substrate (5 mm α-naphthyl phosphate, Sigma-Aldrich, N7255, dissolved in assay buffer) at 30 °C for 20 min. The reaction was stopped by adding 1 ml of 2 m glycine-NaOH (pH 11.0) solution. Fluorescence was measured on a SpectraMax i3x microplate reader (345-nm excitation and 472-nm emission), and enzyme activity was calculated as emission/unit protein (mg) in the reaction.
Transmission EM
BY4741 cells lacking the Pep4 vacuolar protease were maintained at A600 < 1 for ∼24 h prior to nitrogen limitation or potassium starvation for 6 h. The cells were collected by centrifugation (1000 × g for 2 min) and the samples were prepared for transmission EM according to standard methods (88, 89). The cells were fixed with 2% (w/v) glutaraldehyde in 0.1 m PIPES buffer (pH 6.8 and containing 1 mm MgCl2, 1 mm CaCl2, and 0.1 m sorbitol) at room temperature for 1 h and stored at 4 °C for several days. The cell wall material was permeabilized by mixing with 1% (w/v) sodium metaperiodate. The pellets were post-fixed with 1% osmium tetroxide/1.25% potassium ferrocyanide (w/v)/0.1 m PIPES buffer (pH 6.8) for 1 h and stained en bloc in 2% (v/v) aqueous uranyl acetate for 20 min. Cell pellets were dehydrated using a series of increasing ethanol concentrations, rinsed with 100% propylene oxide, and embedded in Spurr's epoxy resin (Electron Microscopy Sciences). Ultrathin sections (70–80 nm) were cut with a diamond knife, mounted on 200 mesh copper grids, and stained with 4% (v/v) aqueous uranyl acetate for 12 min and with Reynold's lead citrate for 8 min (90). Samples were observed using a JEOL JEM-1230 transmission electron microscope operating at 80 kV (JEOL USA), and images were acquired with a Gatan Orius SC1000 CCD Digital Camera and Gatan Microscopy Suite 3.0 software (Gatan).
TEM images were analyzed using FIJI image processing software (87). Autophagosome frequency (number/cell) was estimated by manual counting for 50 representative cells. To measure autophagosome size, outlines of individual autophagosomes were traced using the freehand selection tool. Next, the image was spatially calibrated using the “set scale” tool to convert pixels to micrometers. Area (in square micrometers) was measured using the “measure” tool within the region of interest manager. Lipid droplets and vesicles were excluded from the analysis based on their location (vacuole versus cytoplasm), brightness (lipid droplets were significantly brighter), and size (vesicles were 5–10-fold smaller) relative to autophagosomes.
PhosphoMAPK analysis
MAP kinase activation was measured using quantitative immunoblotting as previously described (83). Briefly, BY4741 Kss1-9xMyc cells were grown to A600 ∼ 1 in SCD medium and transferred to SCD(-nitrogen) or SCD(-potassium) medium or treated with 3 μm α factor pheromone (positive control). Aliquots were collected at the indicated time points, mixed with 5% (w/v) TCA, and collected by centrifugation at 4,000 × g for 2 min. Cell pellets were resuspended in TCA buffer and protein extracts were normalized with resuspension buffer and sample buffer. Proteins were resolved at 150 V for 1.5 h at 25 °C with 10% SDS-PAGE gels containing 50 μm Phos-tag (Wako Chemicals) and 100 μm Zn(NO3)2. Proteins were then transferred to PVDF membranes (Millipore, IPVH00010) in Phos-tag transfer buffer at 20 V for 20 h at 4 °C. Blots were probed with Myc antibodies (2276, Cell Signaling Technology, 1:1,000) or glucose 6-phosphate dehydrogenase (8866, Cell Signaling Technology, 1:1,000) antibodies and quantified as detailed above.
RNA-Seq
The cells were grown to A600 ∼ 1 in SCD medium, then transferred to SCD(-potassium) or SCD(-nitrogen) media by centrifugation and resuspension and shaken at 30 °C for 1 h. For each condition, 8 × 107 cells were collected by centrifugation at 11,000 × g for 30 s at 4 °C. The supernatant was aspirated and cell pellets were frozen in liquid nitrogen and stored at −80 °C. Total RNA was extracted using the RNeasy Mini kit (Qiagen, 74104) with on-column removal of DNA using the RNase-Free DNase Set (Qiagen, 79254). The cDNA library was prepared with the KAPA mRNA HyperPrep Kit (Roche, 08098115702, KAPA Code KK8580), barcoded with NEBNext Multiplex Oligos (Illumina, E7710S), and sequenced with Illumina NextSeq 500 for 75-bp single-end reads.
Quality checks of raw sequence data were performed using the FastQC algorithm (91). Genome indices for yeast were downloaded from Ensembl Genome Browser and sequence alignment was performed using the STAR algorithm (92). Transcripts were quantified with the SALMON algorithm (93). Data were filtered to remove genes with <10 counts across all conditions and analyzed with DESeq2 package in R, while controlling for batch effect (94, 95). All reported log2 fold-change values and adjusted p-values for genes were directly obtained from the DESeq2 model. GO term enrichment analysis was performed with the GO Term Finder (version 0.86) from the Saccharomyces Genome Database.
Data availability
All data described herein are presented within this article and in the supporting information. All gene expression data reported in this article can be accessed through the NCBI GEO database under accession number GSE151898.
Supplementary Material
Acknowledgments
We thank Paul Cullen for valuable feedback on the manuscript, Lee Bardwell for helpful comments, Patrick Brennwald for help with interpretation of TEM images, Amy Pomeroy for help with cell viability analysis, and Sara Kimiko Suzuki and Shu Zhang for assistance with strain construction and immunoblotting.
This article contains supporting information.
Author contributions—N. R. and H. G. D. conceptualization; N. R. data curation; N. R. formal analysis; N. R. validation; N. R., I. K., S. L., P. D., K. K. W., V. J. M., and H. G. D. investigation; N. R., I. K., P. D., K. K. W., and V. J. M. methodology; N. R. and H. G. D. writing-original draft; N. R., S. L., K. K. W., V. J. M., and H. G. D. writing-review and editing; S. L., K. K. W., and V. J. M. visualization; H. G. D. resources; H. G. D. supervision; H. G. D. funding acquisition.
Funding and additional information—Research reported in the publication was supported by the National Institutes of Health Grant R35GM118105 (to H. G. D.). P. D. was supported by an individual summer undergraduate research fellowship from the Office for Undergraduate Research at UNC Chapel Hill. The Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, is supported in part by P30 CA016086 National Cancer Institute Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PI
- phosphatidylinositol
- ATG
- autophagy-related gene
- SCD
- synthetic complete medium with dextrose
- YNB
- yeast nitrogen base
- TEM
- transmission electron microscopy
- MAPK
- mitogen-activated protein kinase
- DEG
- differentially expressed gene
- TORC
- target of rapamycin complex
- GABARAP
- GABA type A receptor-associated protein
- GATE-16
- golgi-associated ATPase Eenhancer of 16 kDa
- UVRAG
- UV radiation resistance associated
- GO
- Gene Ontology
- VPS
- vacuolar protein sorting.
References
- 1. Jona G., Choder M., and Gileadi O. (2000) Glucose starvation induces a drastic reduction in the rates of both transcription and degradation of mRNA in yeast. Biochim. Biophys. Acta 1491, 37–48 [DOI] [PubMed] [Google Scholar]
- 2. Lillie S. H., and Pringle J. R. (1980) Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143, 1384–1394 10.1128/JB.143.3.1384-1394.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martínez-Pastor M. T., and Estruch F. (1996) Sudden depletion of carbon source blocks translation, but not transcription, in the yeast Saccharomyces cerevisiae. FEBS Lett. 390, 319–322 10.1016/0014-5793(96)00683-7 [DOI] [PubMed] [Google Scholar]
- 4. Nilsson A., Påhlman I. L., Jovall P. A., Blomberg A., Larsson C., and Gustafsson L. (2001) The catabolic capacity of Saccharomyces cerevisiae is preserved to a higher extent during carbon compared to nitrogen starvation. Yeast 18, 1371–1381 10.1002/yea.786 [DOI] [PubMed] [Google Scholar]
- 5. Dikic I. (2017) Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 10.1146/annurev-biochem-061516-044908 [DOI] [PubMed] [Google Scholar]
- 6. Takeshige K., Baba M., Tsuboi S., Noda T., and Ohsumi Y. (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitchener J. S., Shelburne J. D., Bradford W. D., and Hawkins H. K. (1976) Cellular autophagocytosis induced by deprivation of serum and amino acids in HeLa cells. Am. J. Pathol. 83, 485–492 [PMC free article] [PubMed] [Google Scholar]
- 8. Schmelzle T., and Hall M. N. (2000) TOR, a central controller of cell growth. Cell 103, 253–262 10.1016/S0092-8674(00)00117-3 [DOI] [PubMed] [Google Scholar]
- 9. De Virgilio C., and Loewith R. (2006) Cell growth control: little eukaryotes make big contributions. Oncogene 25, 6392–6415 10.1038/sj.onc.1209884 [DOI] [PubMed] [Google Scholar]
- 10. Cebollero E., and Reggiori F. (2009) Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793, 1413–1421 10.1016/j.bbamcr.2009.01.008 [DOI] [PubMed] [Google Scholar]
- 11. Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., and Emr S. D. (1993) Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- 12. Stack J. H., Herman P. K., Schu P. V., and Emr S. D. (1993) A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 12, 2195–2204 10.1002/j.1460-2075.1993.tb05867.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Backer J. M. (2016) The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem. J. 473, 2251–2271 10.1042/BCJ20160170 [DOI] [PubMed] [Google Scholar]
- 14. Herman P. K., Stack J. H., and Emr S. D. (1991) A genetic and structural analysis of the yeast Vps15 protein kinase: evidence for a direct role of Vps15p in vacuolar protein delivery. EMBO J. 10, 4049–4060 10.1002/j.1460-2075.1991.tb04981.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stack J. H., DeWald D. B., Takegawa K., and Emr S. D. (1995) Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, 321–334 10.1083/jcb.129.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kametaka S., Okano T., Ohsumi M., and Ohsumi Y. (1998) Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273, 22284–22291 10.1074/jbc.273.35.22284 [DOI] [PubMed] [Google Scholar]
- 17. Kihara A., Noda T., Ishihara N., and Ohsumi Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan H. X., Russell R. C., and Guan K. L. (2013) Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 9, 1983–1995 10.4161/auto.26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., and Ohsumi Y. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 20. Abeliovich H., Dunn W. A. Jr, Kim J., and Klionsky D. J. (2000) Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151, 1025–1034 10.1083/jcb.151.5.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujita N., Itoh T., Omori H., Fukuda M., Noda T., and Yoshimori T. (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 10.1091/mbc.e07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie Z., Nair U., and Klionsky D. J. (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 10.1091/mbc.e07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaufmann A., Beier V., Franquelim H. G., and Wollert T. (2014) Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell 156, 469–481 10.1016/j.cell.2013.12.022 [DOI] [PubMed] [Google Scholar]
- 24. Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., and Elazar Z. (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee Y. K., and Lee J. A. (2016) Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 49, 424–430 10.5483/bmbrep.2016.49.8.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rubinsztein D. C., Shpilka T., and Elazar Z. (2012) Mechanisms of autophagosome biogenesis. Curr. Biol. 22, R29–R34 10.1016/j.cub.2011.11.034 [DOI] [PubMed] [Google Scholar]
- 27. Merchan S., Bernal D., Serrano R., and Yenush L. (2004) Response of the Saccharomyces cerevisiae Mpk1 mitogen-activated protein kinase pathway to increases in internal turgor pressure caused by loss of Ppz protein phosphatases. Eukaryot. Cell 3, 100–107 10.1128/ec.3.1.100-107.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Page M. J., and Di Cera E. (2006) Role of Na+ and K+ in enzyme function. Physiol. Rev. 86, 1049–1092 10.1152/physrev.00008.2006 [DOI] [PubMed] [Google Scholar]
- 29. Lubin M., and Ennis H. L. (1964) On the role of intracellular potassium in protein synthesis. Biochim. Biophys. Acta 80, 614–631 10.1016/0926-6550(64)90306-8 [DOI] [PubMed] [Google Scholar]
- 30. Mancias J. D., Pontano Vaites L., Nissim S., Biancur D. E., Kim A. J., Wang X., Liu Y., Goessling W., Kimmelman A. C., and Harper J. W. (2015) Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. Elife 4, 10.7554/eLife.10308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Medina D. L., and Ballabio A. (2015) Lysosomal calcium regulates autophagy. Autophagy 11, 970–971 10.1080/15548627.2015.1047130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Medina D. L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., Settembre C., Wang W., Gao Q., Xu H., Sandri M., et al. (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kawamata T., Horie T., Matsunami M., Sasaki M., and Ohsumi Y. (2017) Zinc starvation induces autophagy in yeast. J. Biol. Chem. 292, 8520–8530 10.1074/jbc.M116.762948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pottier M., Masclaux-Daubresse C., Yoshimoto K., and Thomine S. (2014) Autophagy as a possible mechanism for micronutrient remobilization from leaves to seeds. Front. Plant Sci. 5, 11 10.3389/fpls.2014.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Horie T., Kawamata T., Matsunami M., and Ohsumi Y. (2017) Recycling of iron via autophagy is critical for the transition from glycolytic to respiratory growth. J. Biol. Chem. 292, 8533–8543 10.1074/jbc.M116.762963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sutter B. M., Wu X., Laxman S., and Tu B. P. (2013) Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 154, 403–415 10.1016/j.cell.2013.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosado C. J., Mijaljica D., Hatzinisiriou I., Prescott M., and Devenish R. J. (2008) Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 4, 205–213 10.4161/auto.5331 [DOI] [PubMed] [Google Scholar]
- 38. Rangarajan N., Gordy C. L., Askew L., Bevill S. M., Elston T. C., Errede B., Hurst J. H., Kelley J. B., Sheetz J. B., Suzuki S. K., Valentin N. H., Young E., and Dohlman H. G. (2019) Systematic analysis of F-box proteins reveals a new branch of the yeast mating pathway. J. Biol. Chem. 294, 14717–14731 10.1074/jbc.RA119.010063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lang M. J., Martinez-Marquez J. Y., Prosser D. C., Ganser L. R., Buelto D., Wendland B., and Duncan M. C. (2014) Glucose starvation inhibits autophagy via vacuolar hydrolysis and induces plasma membrane internalization by down-regulating recycling. J. Biol. Chem. 289, 16736–16747 10.1074/jbc.M113.525782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adachi A., Koizumi M., and Ohsumi Y. (2017) Autophagy induction under carbon starvation conditions is negatively regulated by carbon catabolite repression. J. Biol. Chem. 292, 19905–19918 10.1074/jbc.M117.817510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Primo C., Ferri-Blázquez A., Loewith R., and Yenush L. (2017) Reciprocal regulation of target of rapamycin complex 1 and potassium accumulation. J. Biol. Chem. 292, 563–574 10.1074/jbc.M116.746982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barreto L., Canadell D., Valverde-Saubí D., Casamayor A., and Ariño J. (2012) The short-term response of yeast to potassium starvation. Environ. Microbiol. 14, 3026–3042 10.1111/j.1462-2920.2012.02887.x [DOI] [PubMed] [Google Scholar]
- 43. Brewster J. L., de Valoir T., Dwyer N. D., Winter E., and Gustin M. C. (1993) An osmosensing signal transduction pathway in yeast. Science 259, 1760–1763 10.1126/science.7681220 [DOI] [PubMed] [Google Scholar]
- 44. English J. G., Shellhammer J. P., Malahe M., McCarter P. C., Elston T. C., and Dohlman H. G. (2015) MAPK feedback encodes a switch and timer for tunable stress adaptation in yeast. Sci. Signal. 8, ra5 10.1126/scisignal.2005774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yenush L., Merchan S., Holmes J., and Serrano R. (2005) pH-responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell Biol. 25, 8683–8692 10.1128/MCB.25.19.8683-8692.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haro R., and Rodríguez-Navarro A. (2002) Molecular analysis of the mechanism of potassium uptake through the TRK1 transporter of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1564, 114–122 10.1016/S0005-2736(02)00408-X [DOI] [PubMed] [Google Scholar]
- 47. Lauff D. B., and Santa-Maria G. E. (2010) Potassium deprivation is sufficient to induce a cell death program in Saccharomyces cerevisiae. FEMS Yeast Res. 10, 497–507 10.1111/j.1567-1364.2010.00628.x [DOI] [PubMed] [Google Scholar]
- 48. Deere D., Shen J., Vesey G., Bell P., Bissinger P., and Veal D. (1998) Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 14, 147–160 [DOI] [PubMed] [Google Scholar]
- 49. Du Toit A., Hofmeyr J. S., Gniadek T. J., and Loos B. (2018) Measuring autophagosome flux. Autophagy 14, 1060–1071 10.1080/15548627.2018.1469590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Backues S. K., Chen D., Ruan J., Xie Z., and Klionsky D. J. (2014) Estimating the size and number of autophagic bodies by electron microscopy. Autophagy 10, 155–164 10.4161/auto.26856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sugawara K., Suzuki N. N., Fujioka Y., Mizushima N., Ohsumi Y., and Inagaki F. (2004) The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9, 611–618 10.1111/j.1356-9597.2004.00750.x [DOI] [PubMed] [Google Scholar]
- 52. Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C. M., Adams P. D., Adeli K., Adhihetty P. J., Adler S. G., Agam G., Agarwal R., Aghi M. K., et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheong H., and Klionsky D. J. (2008) Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 451, 1–26 10.1016/S0076-6879(08)03201-1 [DOI] [PubMed] [Google Scholar]
- 54. Noda T., and Klionsky D. J. (2008) The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 451, 33–42 10.1016/S0076-6879(08)03203-5 [DOI] [PubMed] [Google Scholar]
- 55. Kanki T., Kang D., and Klionsky D. J. (2009) Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy 5, 1186–1189 10.4161/auto.5.8.9854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eberhart T., and Kovacs W. J. (2018) Pexophagy in yeast and mammals: an update on mysteries. Histochem. Cell Biol. 150, 473–488 10.1007/s00418-018-1724-3 [DOI] [PubMed] [Google Scholar]
- 57. Wang X., Wang P., Zhang Z., Farré J. C., Li X., Wang R., Xia Z., Subramani S., and Ma C. (2020) The autophagic degradation of cytosolic pools of peroxisomal proteins by a new selective pathway. Autophagy 16, 154–166 10.1080/15548627.2019.1603546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klein B., Wörndl K., Lütz-Meindl U., and Kerschbaum H. H. (2011) Perturbation of intracellular K(+) homeostasis with valinomycin promotes cell death by mitochondrial swelling and autophagic processes. Apoptosis 16, 1101–1117 10.1007/s10495-011-0642-9 [DOI] [PubMed] [Google Scholar]
- 59. Kanki T., and Klionsky D. J. (2008) Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32386–32393 10.1074/jbc.M802403200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kanki T., and Klionsky D. J. (2009) Atg32 is a tag for mitochondria degradation in yeast. Autophagy 5, 1201–1202 10.4161/auto.5.8.9747 [DOI] [PubMed] [Google Scholar]
- 61. Okamoto K., Kondo-Okamoto N., and Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- 62. Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., Yonezawa K., and Ohsumi Y. (2010) Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell Biol. 30, 1049–1058 10.1128/MCB.01344-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gimeno C. J., Ljungdahl P. O., Styles C. A., and Fink G. R. (1992) Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68, 1077–1090 10.1016/0092-8674(92)90079-R [DOI] [PubMed] [Google Scholar]
- 64. Breitkreutz A., Boucher L., and Tyers M. (2001) MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11, 1266–1271 10.1016/S0960-9822(01)00370-0 [DOI] [PubMed] [Google Scholar]
- 65. Roberts C. J., Nelson B., Marton M. J., Stoughton R., Meyer M. R., Bennett H. A., He Y. D., Dai H., Walker W. L., Hughes T. R., Tyers M., Boone C., and Friend S. H. (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873–880 10.1126/science.287.5454.873 [DOI] [PubMed] [Google Scholar]
- 66. Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., Fink G. R., and Young R. A. (2003) Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113, 395–404 10.1016/S0092-8674(03)00301-5 [DOI] [PubMed] [Google Scholar]
- 67. Paliwal S., Iglesias P. A., Campbell K., Hilioti Z., Groisman A., and Levchenko A. (2007) MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 446, 46–51 10.1038/nature05561 [DOI] [PubMed] [Google Scholar]
- 68. Liu H., Styles C. A., and Fink G. R. (1993) Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262, 1741–1744 10.1126/science.8259520 [DOI] [PubMed] [Google Scholar]
- 69. Kusari A. B., Molina D. M., Sabbagh W. Jr, Lau C. S., and Bardwell L. (2004) A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 164, 267–277 10.1083/jcb.200310021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ma J., Jin R., Jia X., Dobry C. J., Wang L., Reggiori F., Zhu J., and Kumar A. (2007) An interrelationship between autophagy and filamentous growth in budding yeast. Genetics 177, 205–214 10.1534/genetics.107.076596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brückner S., Kern S., Birke R., Saugar I., Ulrich H. D., and Mösch H. U. (2011) The TEA transcription factor Tec1 links TOR and MAPK pathways to coordinate yeast development. Genetics 189, 479–494 10.1534/genetics.111.133629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nagiec M. J., McCarter P. C., Kelley J. B., Dixit G., Elston T. C., and Dohlman H. G. (2015) Signal inhibition by a dynamically regulated pool of monophosphorylated MAPK. Mol. Biol. Cell 26, 3359–3371 10.1091/mbc.E15-01-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. You Y. J., Kim J., Cobb M., and Avery L. (2006) Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 3, 237–245 10.1016/j.cmet.2006.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Oh D. Y., Talukdar S., Bae E. J., Imamura T., Morinaga H., Fan W., Li P., Lu W. J., Watkins S. M., and Olefsky J. M. (2010) GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698 10.1016/j.cell.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Aránguiz-Urroz P., Canales J., Copaja M., Troncoso R., Vicencio J. M., Carrillo C., Lara H., Lavandero S., and Díaz-Araya G. (2011) β(2)-adrenergic receptor regulates cardiac fibroblast autophagy and collagen degradation. Biochim. Biophys. Acta 1812, 23–31 10.1016/j.bbadis.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 76. Lizaso A., Tan K. T., and Lee Y. H. (2013) β-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 9, 1228–1243 10.4161/auto.24893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang L., Lu K., Hao H., Li X., Wang J., Wang K., Wang J., Yan Z., Zhang S., Du Y., and Liu H. (2013) Decreased autophagy in rat heart induced by anti-β1-adrenergic receptor autoantibodies contributes to the decline in mitochondrial membrane potential. PLoS ONE 8, e81296 10.1371/journal.pone.0081296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wauson E. M., Zaganjor E., Lee A. Y., Guerra M. L., Ghosh A. B., Bookout A. L., Chambers C. P., Jivan A., McGlynn K., Hutchison M. R., Deberardinis R. J., and Cobb M. H. (2012) The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol. Cell 47, 851–862 10.1016/j.molcel.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., and Dohlman H. G. (2006) Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein α subunit at the endosome. Cell 126, 191–203 10.1016/j.cell.2006.04.045 [DOI] [PubMed] [Google Scholar]
- 80. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 81. Guan J., Stromhaug P. E., George M. D., Habibzadegah-Tari P., Bevan A., Dunn W. A. Jr, and Klionsky D. J. (2001) Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell 12, 3821–3838 10.1091/mbc.12.12.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sikorski R. S., and Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shellhammer J. P., Pomeroy A. E., Li Y., Dujmusic L., Elston T. C., Hao N., and Dohlman H. G. (2019) Quantitative analysis of the yeast pheromone pathway. Yeast 36, 495–518 10.1002/yea.3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- 85. Shintani T., and Klionsky D. J. (2004) Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tanaka K., Kitamura E., and Tanaka T. U. (2010) Live-cell analysis of kinetochore-microtubule interaction in budding yeast. Methods 51, 206–213 10.1016/j.ymeth.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vermulst M., Denney A. S., Lang M. J., Hung C. W., Moore S., Moseley M. A., Thompson J. W., Madden V., Gauer J., Wolfe K. J., Summers D. W., Schleit J., Sutphin G. L., Haroon S., Holczbauer A., et al. (2015) Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat. Commun. 6, 8065 10.1038/ncomms9065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wright R. (2000) Transmission electron microscopy of yeast. Microsc. Res. Tech. 51, 496–510 [DOI] [PubMed] [Google Scholar]
- 90. Reynolds E. S. (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Andrews S. (2010) FastQC: A quality control tool for high throughput sequence data [Google Scholar]
- 92. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Patro R., Duggal G., Love M. I., Irizarry R. A., and Kingsford C. (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Core Team R. (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- 95. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described herein are presented within this article and in the supporting information. All gene expression data reported in this article can be accessed through the NCBI GEO database under accession number GSE151898.


