Abstract
Coronaviruses have caused several zoonotic infections in the past two decades, leading to significant morbidity and mortality globally. Balanced regulation of cell death and inflammatory immune responses is essential to promote protection against coronavirus infection; however, the underlying mechanisms that control these processes remain to be resolved. Here we demonstrate that infection with the murine coronavirus mouse hepatitis virus (MHV) activated the NLRP3 inflammasome and inflammatory cell death in the form of PANoptosis. Deleting NLRP3 inflammasome components or the downstream cell death executioner gasdermin D (GSDMD) led to an initial reduction in cell death followed by a robust increase in the incidence of caspase-8– and receptor-interacting serine/threonine-protein kinase 3 (RIPK3)–mediated inflammatory cell deathafter coronavirus infection. Additionally, loss of GSDMD promoted robust NLRP3 inflammasome activation. Moreover, the amounts of some cytokines released during coronavirus infection were significantly altered in the absence of GSDMD. Altogether, our findings show that inflammatory cell death, PANoptosis, is induced by coronavirus infection and that impaired NLRP3 inflammasome function or pyroptosis can lead to negative consequences for the host. These findings may have important implications for studies of coronavirus-induced disease.
Keywords: coronavirus, mouse hepatitis virus (MHV), NLRP3, gasdermin D, caspase-8, RIPK3, inflammasome, caspase-1, pyroptosis, necroptosis, apoptosis, PANoptosis, RNA virus, plus-stranded RNA virus, infection, infectious disease, host defense
Coronaviruses are single-stranded positive sense RNA viruses with a wide range of hosts (1). Four coronavirus genera (alpha, beta, gamma, and delta) have been identified based on their genetic and serologic properties. Two members of the betacoronavirus genera, severe acute respiratory syndrome-associated coronavirus (SARS-CoV) and Middle East respiratory syndrome-associated coronavirus (MERS-CoV), have caused zoonotic events in the recent past (2). Now, a new member of the betacoronavirus genera, SARS-CoV-2, which was first isolated in Wuhan, China in 2019, has emerged and is causing a pandemic (3). SARS-CoV-2 infection leads to coronavirus disease 2019 (COVID-19), which is most often a mild to moderate respiratory illness with symptoms including fever, fatigue, and dry cough (4). However, patients can develop severe COVID-19 with acute respiratory distress syndrome and present with acute lung damage (5–7). Furthermore, cytokine storm has been suggested to be involved in the pathogenesis of severe and critical cases (8–10). Many cytokines, including proinflammatory cytokines and chemokines, have been found to be increased after SARS-CoV-2 infection (9, 11). Some of them, such as tumor necrosis factor (TNF) and IL-6, are correlated with disease development (8, 9, 12). Despite these findings, little is known about what cellular and molecular mechanisms contribute to the lung damage and determine the onset of cytokine storm in response to SARS-CoV-2 infection.
With other infectious respiratory RNA viruses such as influenza A virus (IAV) and MERS, studies have shown that aberrant programmed cell death may be a critical driver of organ failure and overt cytokine release in severe cases (13–16). Programmed cell death is a genetically encoded, actively controlled cellular process for the targeted removal of redundant, irreversibly damaged, and/or potentially deleterious cells (17). Among the programmed cell death pathways, apoptosis, pyroptosis, and necroptosis have been the best characterized and function during organismal development and infection. Apoptosis is a caspase-8/10– or -9–dependent form of cell death that is executed by caspase-3 and -7 (18, 19). Pyroptosis is executed by gasdermin family members (20), which can be activated through inflammasome activation-mediated caspase-1 cleavage of gasdermin D (GSDMD); caspase-11/4/5– or caspase-8–mediated cleavage of GSDMD; caspase-3–mediated cleavage of GSDME; or granzyme A-mediated cleavage of GSDMB (21–27). Necroptosis is characterized by RIPK3-activated MLKL oligomerization to mediate cell death (28). Numerous pathogens induce at least one of these three programmed cell death pathways during infection. Some pathogens, such as IAV and vesicular stomatitis virus (VSV), can even induce all three in the same cell population through a process termed PANoptosis (29, 30).
The concept of PANoptosis was established based on our studies that have identified cross-talk between the inflammasome/pyroptosis and apoptosis and necroptosis (29–40, 94). PANoptosis is a unique inflammatory programmed cell death regulated by the PANoptosome, which provides a molecular scaffold that allows for interactions and activation of the machinery required for inflammasome/pyroptosis (such as NLRP3, ASC, caspase-1), apoptosis (caspase-8), and necroptosis (RIPK3/RIPK1) (29, 34, 37, 38). The ability of these molecules to interact allows for intricate coregulation between cell death pathways that had previously been thought to be independent. PANoptosis has been implicated in infectious and autoinflammatory diseases, cancer, and beyond (29, 30, 32, 34, 35, 37, 40, 94). This process can trigger the release of proinflammatory cytokines and damage-associated molecular patterns, which lead to robust inflammation (29, 38). Therefore, dysregulation of these pathways can be detrimental to the host during infection.
Sporadic studies have demonstrated that betacoronavirus proteins or infection with full virus can induce apoptosis, pyroptosis, and necroptosis in specific cell types. In HEK293T cells, ORF-3a of SARS-CoV activates necroptosis (in RIPK3-reconstituted cells) (16) or caspase-1 (in inflammasome component–reconstituted cells) (16, 41), suggesting the capacity of SARS-CoV to activate pyroptosis. Furthermore, transfection assays indicate that ORF-8b (42) and ORF-3a (41) of SARS-CoV can activate the NLRP3 inflammasome in THP1 cells. In addition, the ion channel activity of the SARS-CoV envelope (E) protein has been shown to contribute to IL-1β release in vivo after SARS-CoV infection (43–45), suggesting an essential role for the E protein in regulating SARS-CoV–induced inflammasome activation. This role is further supported by work showing that transfection of the SARS-CoV E protein into Vero E6 cells reconstituted with inflammasome components leads to IL-1β release (46). Moreover, MERS and mouse hepatitis virus (MHV), another betacoronavirus, have also been shown to activate apoptosis (15, 47) and induce IL-1β release (48, 49), suggesting the activation of the inflammasome and cell death pathways. However, the mechanistic details of the cell death induced by coronaviruses and the functional consequences of this cell death have not been elucidated.
Understanding how coronaviruses activate cell death and how host factors regulate coronavirus-induced cell death and proinflammatory cytokine expression is key to identifying effective treatment strategies. In this study, we used the mouse coronavirus MHV to systematically assess coronavirus-induced cell death. MHV is the prototypical laboratory coronavirus and has been used to guide our understanding of coronavirus immune responses (50). By infecting cells lacking one or more specific programmed cell death pathway, we evaluated the cell death and inflammatory cytokine release to examine the mechanistic details of these pathways. Our results highlight the role of cell death pathways in coronavirus infection.
Results
Coronavirus infection induces PANoptosis
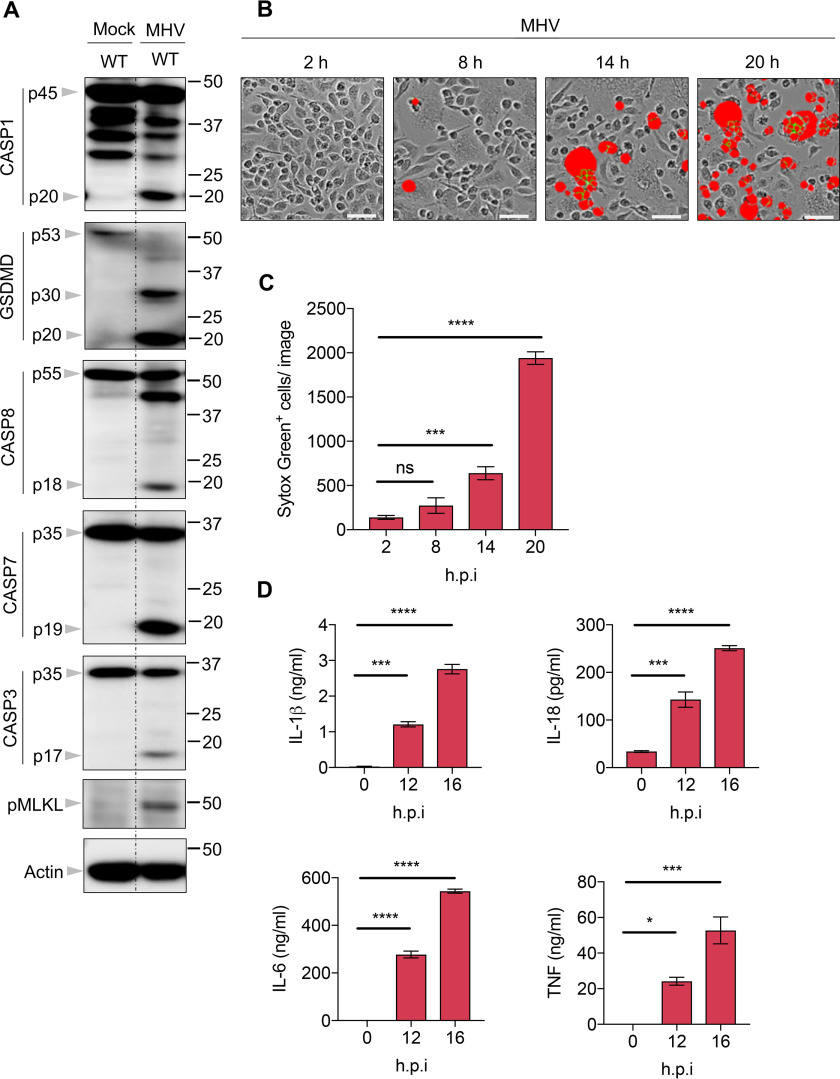
To systematically investigate the programmed cell death pathways induced by coronavirus infection, we infected bone marrow–derived macrophages (BMDMs) from mice with strain A59 of MHV and monitored the effects on cell death pathways. We found that caspase-1 was cleaved after MHV infection, indicating that inflammasome activation was occurring (Fig. 1A). Additionally, pyroptosis was induced by MHV infection, as evidenced by the presence of the p30 cleaved fragment of GSDMD (Fig. 1A). Consistent with previous reports (47, 51), MHV infection also triggered apoptosis, as indicated by the cleavage of caspase-8, -7, and -3 (Fig. 1A). Previous studies have shown that caspase-3 or -7 activation can inactivate GSDMD by processing it to produce a p20 fragment (52, 53). In line with this, a p20 band of GSDMD was observed in MHV-infected cells (Fig. 1A). Moreover, phosphorylated MLKL was detected in MHV-infected cells, suggesting that MHV could also induce necroptosis. Together, these data indicate that, similar to what we have shown previously with IAV and VSV (29, 30), MHV can induce PANoptosis.
Figure 1.
Coronavirus infection induces PANoptosis (pyroptosis, apoptosis, and necroptosis). A, immunoblot analysis of pro- (p45) and cleaved CASP1 (p20), pro- (p53), activated (p30), and inactivated (p20) GSDMD, pro- (p55) and cleaved CASP8 (p18), pro- (p35) and cleaved CASP7 (p19), pro- (p35) and cleaved CASP3 (p17), and phosphorylated MLKL (pMLKL) in BMDMs after MHV infection for 12 h. Actin was used as the internal control. B, real-time analysis of cell death in BMDMs using the IncuCyte imaging system and SYTOX Green nucleic acid staining after infection with MHV. Results at the indicated time points are shown. The red denotes the dead cells counted during the analysis. Scale bar, 50 µm. C, quantification of the cell death observed in (B). D, IL-1β, IL-18, IL-6, and TNF release from BMDMs infected with MHV at the indicated time points. Data are representative of at least three independent experiments. Data are shown as mean ± S.E. (error bars) (C and D). *p < 0.05; ***p < 0.001; and ****p < 0.0001 (one-way ANOVA); ns, not significant; hpi, hours post-infection.
To monitor the kinetics of cell death in response to MHV infection, we used the nucleic acid stain SYTOX Green and investigated cell death in real-time. It is known that the spike protein of coronaviruses can mediate viral entry into infected cells and also induce syncytia during the infection (54–56). Syncytia formation was observed following MHV infection, and the number of dead cells gradually increased over time (Fig. 1, B and C). In addition, we also evaluated cytokine release from the infected cells. In line with the cleavage of caspase-1, which indicated inflammasome assembly, the downstream cytokines IL-1β and IL-18 were detected, and their concentrations gradually increased in the supernatants of infected cells over time (Fig. 1D). Other proinflammatory cytokines like IL-6 and TNF were also detected (Fig. 1D), indicating that MHV infection can induce inflammatory responses. Taken together, these data show that coronavirus infection can initiate PANoptosis and trigger inflammatory responses.
Pyroptosis deficiency leads to increased cell death after coronavirus infection
In response to canonical inflammasome triggers, GSDMD is processed by caspase-1, resulting in the release of the N terminus of GSDMD to form pores in the cell membrane (22, 25, 57, 58). The GSDMD pore can initiate pyroptotic cell death and also facilitates the release of active inflammasome-dependent cytokines IL-1β and IL-18 (21, 59). To investigate the importance of inflammasome activation and GSDMD-mediated pyroptosis in MHV-induced cell death, we infected BMDMs deficient in caspase-1/11 or GSDMD. At 8 h post-infection, deletion of caspase-1/11 or GSDMD resulted in a decrease in cell death (Fig. 2A). Counterintuitively, deleting caspase-1/11 or GSDMD led to substantially increased cell death in response to MHV infection compared with the cell death in WT cells at 14 h post-infection (Fig. 2A). By contrast, cell death induced by IAV, another respiratory infectious RNA virus, or VSV, a single-stranded RNA virus, was not affected in BMDMs lacking caspase-1/11 or GSDMD early or later in infection (Fig. 2, B and C and Fig. S1, A and B), suggesting that losing caspase-1/11 or GSDMD specifically affects coronavirus-induced cell death. In line with this, activation of caspase-8, -7, and -3 and phosphorylation of MLKL was greatly increased after MHV infection in cells lacking caspase-1/11 or GSDMD compared with their activation in WT cells (Fig. 2D). These data suggest that without a functional inflammasome or GSDMD-mediated pyroptosis, MHV tends to induce more apoptosis and necroptosis. Of note, despite the robust cell death in caspase-1/11–deficient cells, no increase in the presence of the p30 fragment of GSDMD, which is responsible for executing pyroptosis, was detected after MHV infection (Fig. 2D). These results indicate that the pyroptotic product of GSDMD is processed by caspase-1/11 during MHV infection at the time point tested.
Figure 2.
Pyroptosis deficiency leads to increased cell death after coronavirus infection. A, real-time analysis of cell death in BMDMs using the IncuCyte imaging system and SYTOX Green nucleic acid staining after infection with MHV. Quantification (left) and images (right) of the cell death at the indicated time points are shown. The red in cell death images denotes the dead cells counted during the analysis. Scale bar, 50 µm. B and C, real-time analysis of cell death in BMDMs using the IncuCyte imaging system and SYTOX Green nucleic acid staining after infection with IAV (B) or VSV (C). D, immunoblot analysis of pro- (p45) and cleaved CASP1 (p20), pro- (p53), activated (p30), and inactivated (p20) GSDMD, pro- (p55) and cleaved CASP8 (p18), pro- (p35) and cleaved CASP7 (p19), pro- (p35) and cleaved CASP3 (p17), and pMLKL in BMDMs after MHV infection for 12 h. GAPDH was used as the internal control. Data are shown as mean ± S.E. (error bars) (A–C). *p < 0.05 and ****p < 0.0001 (one-way ANOVA). Data are representative of at least three independent experiments. ns, not significant; hpi, hours post-infection.
In addition to GSDMD, other host factors are also able to mediate pore formation leading to cell death, including MLKL, which can form pores to initiate necroptosis (28), and GSDME, which can be activated by caspase-3 to induce pyroptosis (26, 60). To investigate whether these two pore-forming proteins function similarly to GSDMD in the context of coronavirus infections, we infected Mlkl–/– or Gsdme−/− BMDMs with MHV. We found a slight decrease in MHV-induced cell death in the Mlkl−/− or Gsdme−/− BMDMs compared with WT cells (Fig. S2A). Molecularly, we found that MLKL may play a role in promoting MHV-induced inflammasome assembly, as evidenced by reduced caspase-1 activation in the Mlkl−/− cells (Fig. S2B). Whereas apoptosis markers were comparable between WT and Mlkl−/− cells, the amount of the p30 fragment of GSDMD detected was reduced in Mlkl−/− cells, which is consistent with the levels of activated caspase-1, suggesting that activation of GSDMD-mediated pyroptosis during MHV infection depends on caspase-1 (Fig. S2B). GSDME deficiency had no impact on GSDMD activation, and apoptosis and necroptosis were not impaired in GSDME-deficient cells compared with WT after MHV infection (Fig. S2C).
Together, these data suggest that inflammasome activation and GSDMD-mediated pyroptosis play a critical role in inhibiting coronavirus-induced apoptosis and necroptosis and that loss of the inflammasome/GSDMD pathway results in robust cell death.
Loss of the NLRP3 inflammasome leads to increased apoptosis and necroptosis
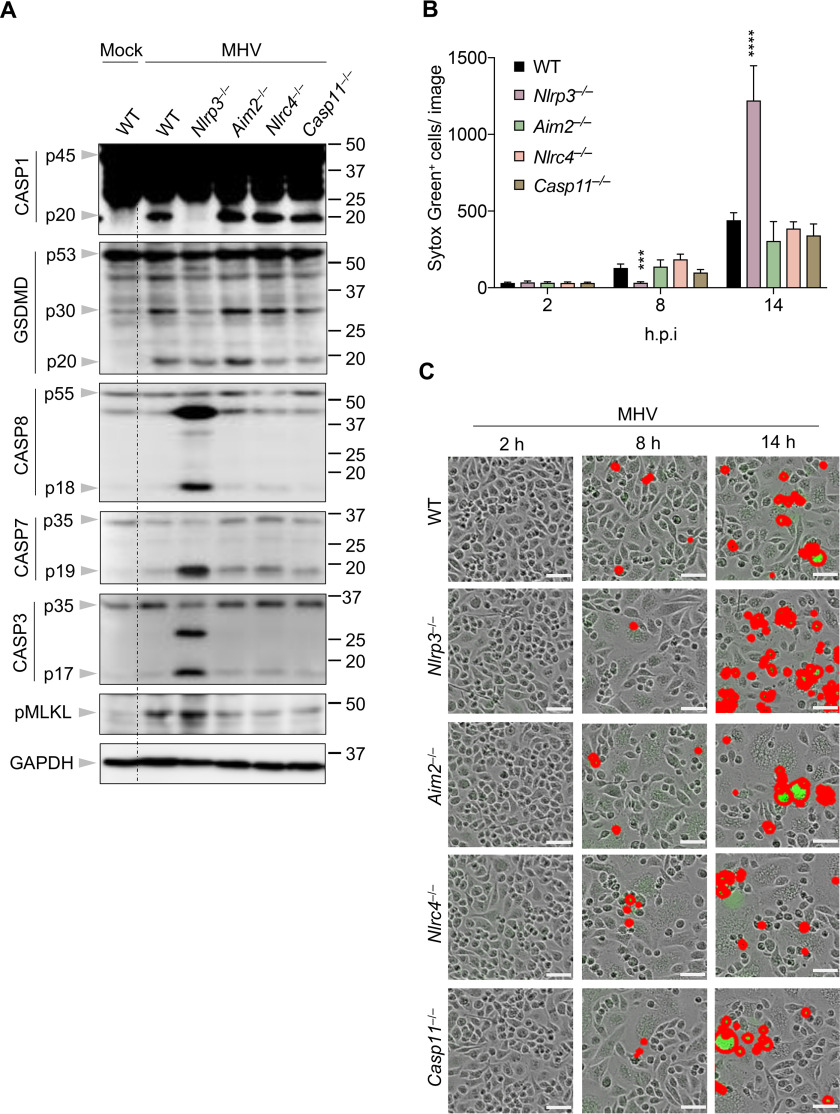
Some coronavirus proteins and certain coronaviruses are known to initiate NLRP3 inflammasome assembly (41, 42, 48, 61). However, whether they can induce the activation of other inflammasomes has not been examined. To investigate this, we infected cells deficient in NLRP3, AIM2, NLRC4, or caspase-11 with MHV. We observed that NLRP3 deficiency completely abolished caspase-1 activation following MHV infection, while deleting AIM2, NLRC4, or caspase-11 did not impair MHV-induced caspase-1 activation (Fig. 3A). Given that NLRP3 inflammasome activation requires a priming signal to up-regulate the expression of NLRP3, we checked NLRP3 protein levels following MHV infection and found that NLRP3 expression was increased after MHV infection (Fig. S3). It was previously shown that MyD88 is necessary for TNF and IL-6 release in response to MHV infection (62), suggesting that MyD88 is required to prime the inflammatory response. Consistent with this, cells lacking MyD88 showed impaired NLRP3 expression after MHV infection (Fig. S3), indicating that MyD88 is essential for the priming of the MHV-induced NLRP3 inflammasome. These findings suggest that MHV infection specifically induces the NLRP3 inflammasome. The reduced caspase-1 cleavage was further supported by impaired GSDMD cleavage to form the p30 fragment in MHV-infected Nlrp3−/− cells (Fig. 3A), suggesting pyroptosis was not executed. By contrast, AIM2–, NLRC4–, and caspase-11–deficient BMDMs showed similar pyroptotic GSDMD cleavage when compared with WT BMDMs (Fig. 3A). These data indicate that at the time point tested, MHV-induced GSDMD activation depends on the NLRP3 inflammasome.
Figure 3.
Loss of the NLRP3 inflammasome leads to increased apoptosis and necroptosis. A, immunoblot analysis of pro- (p45) and cleaved CASP1 (p20), pro- (p53), activated (p30), and inactivated (p20) GSDMD, pro- (p55) and cleaved CASP8 (p18), pro- (p35) and cleaved CASP7 (p19), pro- (p35) and cleaved CASP3 (p17), and pMLKL in BMDMs after MHV infection for 12 h. GAPDH was used as the internal control. B, real-time analysis of cell death in BMDMs using the IncuCyte imaging system and SYTOX Green nucleic acid staining after infection with MHV. Quantification of the cell death at the indicated time points is shown. C, representative cell death images from (B) are shown. The red denotes the dead cells counted during the analysis. Scale bar, 50 µm. Data are shown as mean ± S.E. (error bars) (B). Significant differences compared with WT are denoted as ***p < 0.001 and ****p < 0.0001 (one-way ANOVA). Data are representative of three independent experiments. hpi, hours post-infection.
Given that both caspase-1/11 and NLRP3 are required for GSDMD cleavage, and caspase-1/11–deficiency led to increased cell death similar to that observed in GSDMD-deficient cells after MHV infection (Fig. 2A), we investigated whether NLRP3 functions similarly to caspase-1 during MHV infection. We found that, similar to what we observed in caspase-1/11–deficient cells, the activation of caspase-8, -7, and -3 and MLKL was markedly increased in MHV-infected Nlrp3−/− cells compared with WT cells (Fig. 3A). By contrast, MHV-induced apoptosis and necroptosis activation were not increased in Aim2−/−, Nlrc4−/−, or Casp11−/− cells (Fig. 3A). In line with the cell death markers, quantitatively, more cell death was observed in MHV-infected Nlrp3−/− cells compared with WT cells at 14 h post-infection following the initial slight reduction at 8 h (Fig. 3, B and C and Fig. S2A). In the absence of AIM2, NLRC4, or caspase-11, MHV-induced cell death was not significantly affected (Fig. 3, B and C), further supporting the unique role of NLRP3 during coronavirus infection. We also noted that the increase in cell death corresponded to a decrease in MHV viral replication, because cells deficient in NLRP3, caspase-1, or GSDMD had reduced virus titers compared with that of WT cells (Fig. S4).
To confirm the role of the other inflammasome components during MHV infection, we evaluated ASC– and caspase-1–deficient cells. Consistent with the function of NLRP3, deleting caspase-1 or ASC increased cell death at 14 h post-infection (Fig. S2A). We consistently observed that there was more cell death in the WT cells at around 8 h post-infection than in cells lacking NLRP3, ASC, caspase-1, or GSDMD (Fig. 3, B and C and Fig. S2A), suggesting that early cell death during MHV infection is largely caused by inflammasome-mediated pyroptosis.
Taken together, these data imply that coronaviruses specifically activate the NLRP3 inflammasome, which supports viral replication, and that GSDMD activation during coronavirus infection depends on NLRP3 inflammasome assembly.
Increased cell death in the absence of NLRP3/pyroptosis depends on caspase-8 and RIPK3
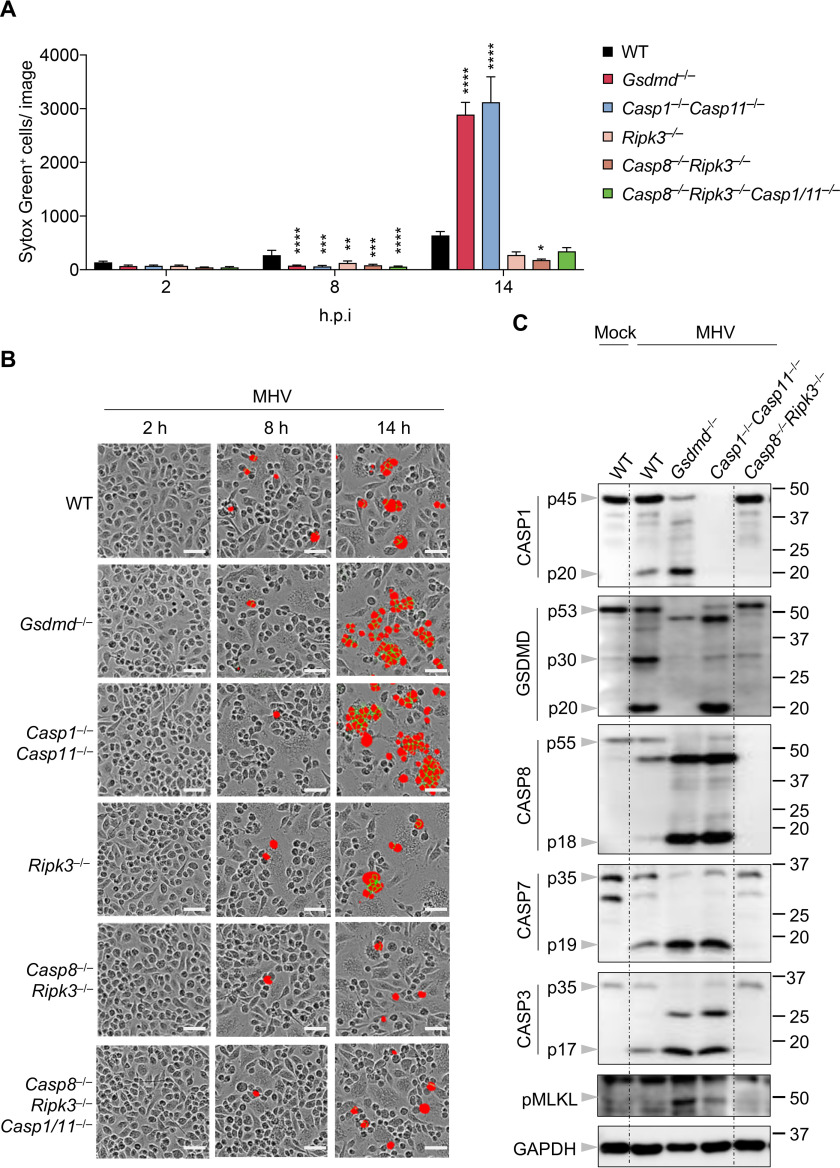
We have previously shown that IAV- and VSV-induced PANoptosis activation completely (IAV) or heavily (VSV) depend on caspase-8 and RIPK3 (29, 30). Consistent with these previous results, we again observed IAV- and VSV-induced cell death were abolished or dramatically reduced, respectively, in Casp8−/−Ripk3−/− cells (Fig. S1, A and B). We then investigated whether Casp8−/−Ripk3−/− cells have a similar phenotype following MHV infection. While Casp1−/−Casp11−/− and Gsdmd−/− cells showed robust cell death at 14 h after MHV infection, RIPK3 deficiency slightly decreased MHV-induced cell death, and cell death in Casp8−/−Ripk3−/− cells was much less than that in WT cells (Fig. 4, A and B). Furthermore, deleting caspase-8 and RIPK3 can block the increase in cell death caused by the loss of caspase-1/11 in response to MHV infection (Fig. 4, A and B). These observations suggest that MHV-induced cell death largely depends on caspase-8 and RIPK3. Additionally, we also observed consistently increased activation of caspase-8, -7, and -3 and MLKL in cells deficient in caspase-1/11 or GSDMD, whereas there was little activation of the inflammasome (caspase-1), pyroptosis (GSDMD), apoptosis (caspase-8, -7, and -3), or necroptosis (pMLKL) detected in Casp8−/−Ripk3−/− cells (Fig. 4C), further supporting the critical role of caspase-8 and RIPK3 in MHV-induced cell death. In sum, these data suggest that coronavirus-induced cell death predominantly signals through caspase-8 and RIPK3 together, whereas GSDMD activation can inhibit caspase-8– and RIPK3–mediated cell death.
Figure 4.
Increased cell death in the absence of NLRP3/pyroptosis depends on caspase-8 and RIPK3. A, real-time analysis of cell death in BMDMs using the IncuCyte imaging system and SYTOX Green nucleic acid staining after infection with MHV. Quantification of the cell death at the indicated time points is shown. B, representative cell death images from (A) are shown. The red denotes the dead cells counted during the analysis. Scale bar, 50 µm. C, immunoblot analysis of pro- (p45) and cleaved CASP1 (p20), pro- (p53), activated (p30), and inactivated (p20) GSDMD, pro- (p55) and cleaved CASP8 (p18), pro- (p35) and cleaved CASP7 (p19), pro- (p35) and cleaved CASP3 (p17), and pMLKL in BMDMs after MHV infection for 12 h. GAPDH was used as the internal control. Data are shown as mean ± S.E. (error bars) (A). Significant differences compared with WT are denoted as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (one-way ANOVA). Data are representative of at least three independent experiments. hpi, hours post-infection.
GSDMD deficiency affects cytokine release in response to MHV infection
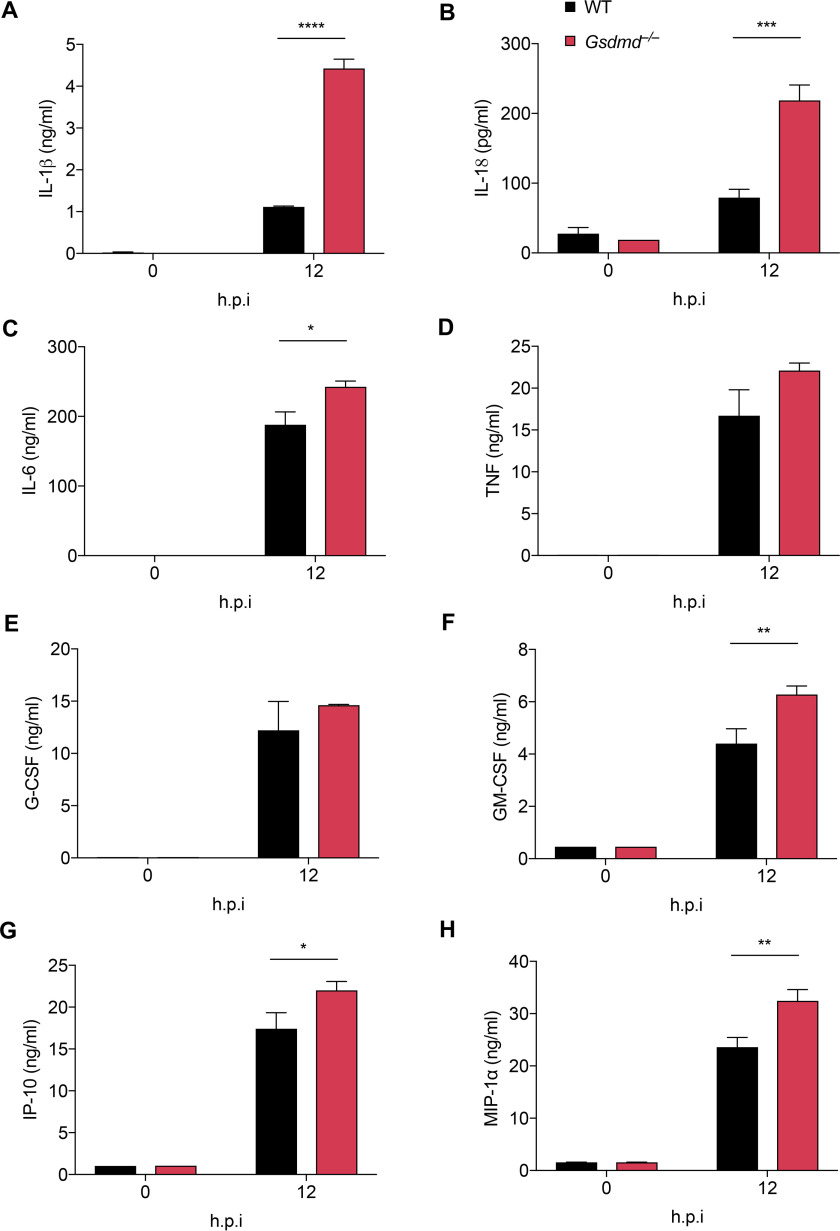
Given that the loss of GSDMD had remarkable effects on MHV-induced inflammatory cell death, we investigated whether this loss would also affect MHV-induced cytokine release. Consistent with the remarkably increased caspase-1 activation in Gsdmd−/− cells, IL-1β and IL-18 levels were significantly higher in Gsdmd−/− cells than in WT cells following MHV infection (Fig. 5, A and B), indicating that blocking GSDMD alone cannot suppress inflammasome-dependent cytokine release and instead promotes this release during coronavirus infection.
Figure 5.
GSDMD deficiency leads to increased cytokine release after MHV infection. IL-1β (A), IL-18 (B), IL-6 (C), TNF (D), G-CSF (E), GM-CSF (F), IP-10 (G), and MIP-1α (H) release from BMDMs infected with MHV at the indicated time points. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (one-way ANOVA). Data are representative of three independent experiments. Data are shown as mean ± S.E. (error bars) (A–H). hpi, hours post-infection
It has been recently suggested that cytokine storm contributes to disease development during SARS-CoV-2 infection (10, 63), but little is known about how these cytokines become dysregulated in patients with severe or critical disease. Several cytokines including IL-6, TNF, G-CSF, GM-CSF, IP-10, and MIP-1α have been found to be increased following SARS-CoV-2 infection and correlate with COVID-19 development (8, 9, 12). We therefore examined whether deleting GSDMD would affect the release of these proinflammatory cytokines or chemokines during MHV infection. We found that the levels of IL-6, TNF, G-CSF, GM-CSF, IP-10, and MIP-1α were greatly increased in the supernatant of MHV-infected cells at 12 h post-infection (Fig. 5, C–H). Remarkably, loss of GSDMD significantly increased the release of IL-6, GM-CSF, IP-10, and MIP-1α in response to MHV infection (Fig. 5, C–H). These results suggest that impairing GSDMD function could affect both inflammasome-related and nonrelated cytokine release, showing that GSDMD has a significant impact on coronavirus-induced cytokine release.
Discussion
In this study, we systematically investigated cell death and inflammasome activation and found that coronavirus infection activated PANoptosis and specifically induced NLRP3 inflammasome assembly. Deleting components of the NLRP3 inflammasome, i.e. NLRP3, ASC, or caspase-1, resulted in a substantial increase in apoptosis and necroptosis beyond 8 h of infection. The increased cell death was likely a result of the loss of GSDMD activation, because cells lacking GSDMD showed a similar phenotype. Our data suggest that GSDMD inhibits coronavirus-induced cell death. The underlying mechanism by which GSDMD mediates the inhibition is unknown, but this phenotype is uncommon. In the case of the classical inflammasome triggers, deleting inflammasome components or GSDMD causes a delay and a reduction in cell death compared with the cell death in WT cells (22, 64, 65). Additionally, in the case of two other RNA viruses we tested, IAV and VSV, which are both known to activate PANoptosis, neither showed increased cell death in caspase-1/11– or GSDMD–deficient cells after infection. Within the gasdermin family, GSDME has also been shown to be critical in mediating pyroptosis in macrophages (52). However, deleting GSDME had little effect on the induction of coronavirus-induced PANoptosis. Similar results were observed with MLKL-deficient cells. These data indicate that GSDMD plays a unique role in addition to mediating pyroptosis during coronavirus infection. It was noteworthy that we observed a clear transition in cell death over the time course of MHV infection; NLRP3–, caspase-1/11–, or GSDMD–deficient cells showed much less cell death compared with WT cells at around 8 h post-infection, but then exhibited greatly elevated cell death at later times. These data suggest that early during infection, the coronavirus induced the activation of the NLRP3 inflammasome and GSDMD. The activated GSDMD then controlled the induction of other forms of coronavirus-induced cell death to prevent dysregulated cell death from occurring, and this dysregulated cell death was detrimental to viral replication.
The substantially increased IL-1β and IL-18 release in GSDMD-deficient cells showed that blocking GSDMD during coronavirus infection is unlikely to inhibit the release of these cytokines and may actually increase the release. Because MHV induced PANoptosis, it is likely that without GSDMD pores, IL-1β and IL-18 were released through the MLKL or GSDME pore or both. It will be interesting to investigate the IL-1β and IL-18 release in cells lacking all three of these pore-forming molecules after coronavirus infection.
Our study focuses on MHV, the prototypical laboratory coronavirus. MHV does not infect humans and has key differences from the human coronaviruses, including the host cell receptor used for invasion (66, 67); this difference between the murine virus and SARS-CoV-2 prevents SARS-CoV-2 from being studied in WT mouse models. However, MHV and other mammalian coronaviruses have been important in guiding our understanding of coronavirus immune responses, particularly in the context of emerging infections where immune responses are poorly understood (50). For example, shortly after the SARS-CoV outbreak began, clinicians observed an inflammatory storm in patients that was similar to what had been seen previously with MHV (68), and now a similar phenomenon seems to occur with SARS-CoV-2 (10, 63, 69–71). The extensive history of studies with MHV has served as a key starting point for investigations of human coronaviruses, including the initial identification of the spike protein as a potential vaccine target (72). Therefore, it is important for future studies to extend our findings in MHV to the pathogenic human coronaviruses.
Severe forms of disease caused by the human coronaviruses, including SARS, MERS, and COVID-19, have been characterized by lung damage (5, 7, 73, 74) and cytokine storm (10, 63, 69–71). Additionally, the mortality rate of all three diseases is extremely high in people over the age of 65 compared with younger populations (75–77). It is possible that the elderly tend to have increased incidence of underlying medical conditions that contribute to this death rate. However, based on the observation that loss of NLRP3 resulted in increased cell death in the context of MHV, it will be important to extend these findings to human coronaviruses and determine whether compromised NLRP3 inflammasome function in the elderly could be a contributing factor. Older mice have impaired NLRP3 inflammasome activity in response to respiratory infectious agents, including S. pneumoniae and IAV (78, 79). Macrophages and dendritic cells isolated from these mice show attenuated expression of NLRP3, ASC, and caspase-1 compared with the expression in young mice, which could partially explain the decreased NLRP3 inflammasome activation in older mice. Additionally, older people have reduced NLRP3 expression compared with younger individuals in response to IAV infection (80). Given that loss of a functional NLRP3 inflammasome resulted in increased cell death and that loss of the downstream executioner GSDMD led to increased proinflammatory cytokine and chemokine release after MHV infection, it will be important to explore whether impaired NLRP3 inflammasome activation contributes to the high mortality rate of COVID-19 in the elderly population.
Overall, our study identified that coronavirus infection induced NLRP3 inflammasome assembly and activated PANoptosis. Furthermore, the inhibition of the NLRP3 inflammasome/pyroptosis may be detrimental to the host and result in increased cell death and inflammatory cytokine release in the context of coronaviruses. It is critical for future studies to extend our findings with MHV to human coronaviruses, particularly SARS-CoV-2, to understand the basic biological processes that occur during the infection and inform which treatments should proceed for clinical evaluation. These findings here improve our understanding of coronavirus pathogenesis as it relates to cell death and cytokine release and may provide some constructive insight to inform studies of COVID-19.
Experimental procedures
Mice
Nlrp3−/− (81), Casp11−/− (82), Gsdmd−/− (83), Gsdme−/− (84), Mlkl−/− (85), Asc−/− (86), Nlrc4−/− (87), Aim2−/− (88), Ripk3−/− (89), Casp8−/−Ripk3−/− (90), Casp8−/−Ripk3−/−Casp1/11−/− (35), and MyD88−/− (91) mice have been described previously. All mice were bred at the Animal Resource Center at St. Jude Children's Research Hospital and were backcrossed to the C57BL/6 background. Animal study protocols were approved by St. Jude Children's Research Hospital committee on the care and use of animals.
Bone marrow–derived macrophages
Primary BMDMs were cultured for 6 days in Iscove's modified Dulbecco's medium (Thermo Fisher Scientific, 12440-053) supplemented with 10% FBS (Biowest, S1620), 30% L929-conditioned medium, 1% nonessential amino acids (Thermo Fisher Scientific, 11140-050), and 1% penicillin and streptomycin (Thermo Fisher Scientific, 15070-063). Then BMDMs were seeded into 12-well plates at a density of 1 million cells/well and incubated overnight before use.
Murine hepatitis virus culture
The murine hepatitis virus (A59 strain from Dr. Channappanavar) (95) was propagated in 17Cl-1 cells as previously described (92). Virus titer was measured by plaque assay in 17Cl-1 cells.
Influenza A virus culture
The influenza A virus (A/Puerto Rico/8/34, H1N1 (PR8)) was rescued by reverse genetics as previously described (93). Virus stocks were prepared by inoculation of the rescued virus into the allantoic cavity of 9- to 11-day-old embryonated chicken eggs. The virus was titrated by plaque assay in Madin-Darby canine kidney cells.
Vesicular stomatitis virus culture
The Indiana strain of vesicular stomatitis virus was propagated in Vero cells, and virus titer was measured by plaque assay in Vero cells.
Cell stimulation/infection
For MHV infection, BMDMs seeded in 12-well plates were infected at a multiplicity of infection (MOI) of 0.1 in DMEM plain media (Sigma, D6171). For IAV and VSV infection, cells were infected at an MOI of 20 or 1, respectively. After 2 h of incubation with each virus, cells were supplemented with 10% FBS and then incubated for the indicated time.
Immunoblot analysis
For caspase protein analysis, BMDMs were lysed with the supernatant using 50 µl of caspase lysis buffer (1× protease inhibitors, 1× phosphatase inhibitors, 10% Nonidet P-40, and 25 mm DTT), followed by the addition of 125 µl 4× SDS loading buffer. For analysis of all other proteins, the supernatants were removed, and cells were washed once with PBS. Then BMDMs were lysed with radioimmune precipitation buffer. Electrophoresis was utilized to separate proteins in 10%–12% polyacrylamide gels, after which proteins were transferred onto PVDF membranes. The membranes were blocked with 5% skim milk for 1 h at room temperature. All the primary antibodies were incubated overnight at 4 °C, and secondary antibodies with HRP were incubated for 1 h at room temperature. Images were developed using a GE Amersham Biosciences Imager 600.
The antibodies used were as follows: anti-caspase-1 (AdipoGen, AG-20B-0042, 1:2000), anti-caspase-3 (Cell Signaling Technology (CST), 9662, 1:1000), anti-cleaved caspase-3 (CST, 9661, 1:1000), anti-caspase-7 (CST, 9492, 1:1000), anti-cleaved caspase-7 (CST, 9491, 1:1000), anti-caspase-8 (CST, 4927, 1:1000), anti-cleaved caspase-8 (CST, 8592, 1:1000), anti-pMLKL (CST, 37333, 1:1000), anti-GSDMD (Abcam, ab209845, 1:1000), anti-NLRP3 (Adipogen, AG-20B-0014, 1:2000), anti-β-actin (Proteintech, 66009-1-IG, 1:5000), anti-GAPDH (CST, 5174, 1:1000), and HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, anti-rabbit (111-035-047), 1:5000; anti-mouse (315-035-047), 1:5000).
Real-time cell death analysis
Real-time cell death analysis was conducted as previously described (32). In brief, BMDMs were seeded in 24-well plates (0.5 × 106 cells/well) and infected with the indicated virus. After 2 h of incubation, 100 nm SYTOX Green (Thermo Fisher Scientific, S7020) was added to the cells together with FBS. Images were analyzed using IncuCyte S3 software.
Growth kinetics of MHV
BMDMs were seeded in 24-well plates (0.5 × 106 cells/well) and infected with MHV at an MOI of 0.1. After 2 h of incubation, the virus was removed, and the cells were washed twice with PBS. Then fresh media was added into each well, and supernatants were collected at the indicated time points. Viral titers were determined by plaque assay using 17Cl-1 cells.
Cytokine analysis
Cytokines were analyzed by multiplex ELISA (Millipore, MCYTOMAG-70K) or IL-18 ELISA (Invitrogen, BMS618-3) according to the manufacturer's instructions.
Statistical analysis
GraphPad Prism 7.0 software was used for data analysis. Data are presented as mean ± S.E. The one-way analysis of variance (ANOVA) with Dunnett's multiple comparisons test was used to determine the statistical significance. p values less than 0.05 were considered statistically significant where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Data availability
All data generated for this study are included within this article and in the supporting information.
Supplementary Material
Acknowledgments
We thank all the members of the Kanneganti laboratory for comments and suggestions. We thank A. Burton (St. Jude Children's Research Hospital) for technical support. We also thank R. Tweedell, PhD, for scientific editing and writing support. We thank R. Channappanavar (University of Tennessee Health Sciences Center) for providing MHV and 17Cl-1 cells and M.A. Whitt (University of Tennessee Health Science Center) for providing VSV. We thank the Wellcome Trust Sanger Institute Mouse Genetics Project and its funders for providing the mutant mouse line Gsdmetm1a(KOMP)Wtsi, and INFRAFRONTIER/EMMA. Funding information may be found at the Sanger Mouse Resource Portal, and associated primary phenotypic information may be found through the International Mouse Phenotyping Consortium.
This article contains supporting information.
Author contributions—M. Z., R. K., and T.-D. K. conceptualization; M. Z., R. K. S. M., and R. K. formal analysis; M. Z., E. P. W., R. K. S. M., B. B., and A. B. investigation; M. Z., R. K., R. W., R. C., and C. B. J. methodology; M. Z. and T.-D. K. writing-original draft; M. Z., E. P. W., R. K. S. M., R. K., B. B., A. B., R. W., R. C., C. B. J., and T.-D. K. writing-review and editing; T.-D. K. funding acquisition.
Funding and additional information—Work from our laboratory is supported by the United States National Institutes of Health Grants AI101935, AI124346, AR056296, and CA253095 (to T.-D. K.) and by the American Lebanese Syrian Associated Charities (to T.-D. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MHV
- mouse hepatitis virus
- GSDM
- gasdermin
- RIPK
- receptor-interacting serine/threonine-protein kinase
- NLRP
- nucleotide-binding oligomerization domain-like receptor pyrin domain
- CoV
- coronavirus
- SARS
- severe acute respiratory syndrome
- MERS
- Middle East respiratory syndrome
- COVID-19
- coronavirus disease 2019
- TNF
- tumor necrosis factor
- IAV
- influenza A virus
- MLKL
- mixed lineage kinase domain-like
- pMLKL
- phosphorylated MLKL
- VSV
- vesicular stomatitis virus
- BMDM
- bone marrow–derived macrophages
- AIM
- absent in melanoma
- NLRC
- nucleotide-binding oligomerization domain-like receptor caspase activation and recruitment domain
- MyD
- myeloid differentiation primary response
- ASC
- apoptosis-associated speck-like protein containing a caspase activation and recruitment domain
- G-CSF
- granulocyte colony-stimulating factor
- GM-CSF
- granulocyte/macrophage CSF
- MIP
- macrophage inflammatory protein
- IP
- IFN-γ–induced protein
- MOI
- multiplicity of infection
- CASP
- caspase
- ANOVA
- analysis of variance.
References
- 1. MacLachlan N. J., and Dubovi E. J. (2017) Chapter 24 Coronaviridae in Fenner's Veterinary Virology, 5E, 435–461, Academic Press, Boston: 10.1016/B978-0-12-800946-8.00024-6 [DOI] [Google Scholar]
- 2. Chan J. F. W., Lau S. K. P., To K. K. W., Cheng V. C. C., Woo P. C. Y., and Yuen K.-Y. (2015) Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 28, 465–522 10.1128/CMR.00102-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jaimes J. A., André N. M., Chappie J. S., Millet J. K., and Whittaker G. R. (2020) Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 10.1016/j.jmb.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou M., Zhang X., and Qu J. (2020) Coronavirus disease 2019 (COVID-19): a clinical update. Front. Med. 14, 126–135 10.1007/s11684-020-0767-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fox S. E., Akmatbekov A., Harbert J. L., Li G., Brown J. Q., and Heide R. S. V. (2020) Pulmonary and cardiac pathology in Covid-19: the first autopsy series from New Orleans. 8, 681–686 10.1016/S2213-2600(20)30243-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo W., Yu H., Gou J., Li X., Sun Y., Li J., and Liu L. (2020) Clinical pathology of critical patient with novel coronavirus pneumonia (COVID-19) Preprints [Google Scholar]
- 7. Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., et al. (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 8, 420–422 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., et al. (2020) Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. medRxiv 10.1101/2020.04.19.20068015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395, 497–506 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moore J. B., and June C. H. (2020) Cytokine release syndrome in severe COVID-19. Science. 368, 473–474 10.1126/science.abb8925 [DOI] [PubMed] [Google Scholar]
- 11. Blanco-Melo D., Nilsson-Payant B. E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T. X., Oishi K., Panis M., Sachs D., Wang T. T., Schwartz R. E., Lim J. K., Albrecht R. A., and tenOever B. R. (2020) Imbalanced host response to Sars-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Y., Shen C., Li J., Yuan J., Yang M., Wang F., Li G., Li Y., Xing L., Peng L., Wei J., Cao M., Zheng H., Wu W., Zou R., et al. (2020) Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. medRxiv 10.1101/2020.03.02.20029975 [DOI] [Google Scholar]
- 13. Fujikura D., and Miyazaki T. (2018) Programmed cell death in the pathogenesis of influenza. Int. J. Mol. Sci. 19, 2065 10.3390/ijms19072065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herold S., Becker C., Ridge K. M., and Budinger G. R. S. (2015) Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J. 45, 1463–1478 10.1183/09031936.00186214 [DOI] [PubMed] [Google Scholar]
- 15. Yeung M.-L., Yao Y., Jia L., Chan J. F. W., Chan K.-H., Cheung K.-F., Chen H., Poon V. K. M., Tsang A. K. L., To K. K. W., Yiu M.-K., Teng J. L. L., Chu H., Zhou J., Zhang Q., et al. (2016) MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 1, 16004 10.1038/nmicrobiol.2016.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yue Y., Nabar N. R., Shi C.-S., Kamenyeva O., Xiao X., Hwang I.-Y., Wang M., and Kehrl J. H. (2018) SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 9, 1–15 10.1038/s41419-018-0917-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galluzzi L., Vitale I., Aaronson S. A., Abrams J. M., Adam D., Agostinis P., Alnemri E. S., Altucci L., Amelio I., Andrews D. W., Annicchiarico-Petruzzelli M., Antonov A. V., Arama E., Baehrecke E. H., Barlev N. A., et al. (2018) Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elmore S. (2007) Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kesavardhana S., Malireddi R. K. S., and Kanneganti T.-D. (2020) Caspases in cell death, inflammation, and pyroptosis. Annu. Rev. Immunol. 38, 567–595 10.1146/annurev-immunol-073119-095439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi J., Gao W., and Shao F. (2017) Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 42, 245–254 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 21. He W. T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z. H., Zhong C. Q., and Han J. (2015) Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., et al. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526, 666–671 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- 23. Orning P., Weng D., Starheim K., Ratner D., Best Z., Lee B., Brooks A., Xia S., Wu H., Kelliher M. A., Berger S. B., Gough P. J., Bertin J., Proulx M. M., Goguen J. D., et al. (2018) Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069 10.1126/science.aau2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sarhan J., Liu B. C., Muendlein H. I., Li P., Nilson R., Tang A. Y., Rongvaux A., Bunnell S. C., Shao F., Green D. R., and Poltorak A. (2018) Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. U. S. A. 115, E10888–E10897 10.1073/pnas.1809548115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 26. Wang Y., Gao W., Shi X., Ding J., Liu W., He H., Wang K., and Shao F. (2017) Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 547, 99–103 10.1038/nature22393 [DOI] [PubMed] [Google Scholar]
- 27. Zhou Z., He H., Wang K., Shi X., Wang Y., Su Y., Wang Y., Li D., Liu W., Zhang Y., Shen L., Han W., Shen L., Ding J., and Shao F. (2020) Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 368, eaaz7548 10.1126/science.aaz7548 [DOI] [PubMed] [Google Scholar]
- 28. Galluzzi L., Kepp O., and Kroemer G. (2014) MLKL regulates necrotic plasma membrane permeabilization. Cell Research 24, 139–140 10.1038/cr.2014.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Christgen S., Zheng M., Kesavardhana S., Karki R., Malireddi R. K. S., Banoth B., Place D. E., Briard B., Sharma B. R., Tuladhar S., Samir P., Burton A., and Kanneganti T.-D. (2020) Identification of the PANoptosome: a molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 237 10.3389/fcimb.2020.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuriakose T., Man S. M., Malireddi R. K., Karki R., Kesavardhana S., Place D. E., Neale G., Vogel P., and Kanneganti T. D. (2016) ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 10.1126/sciimmunol.aag2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., and Núñez G. (2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics 7, 2350–2363 10.1074/mcp.M800132-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malireddi R. K. S., Gurung P., Mavuluri J., Dasari T. K., Klco J. M., Chi H., and Kanneganti T.-D. (2018) TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. J. Exp. Med. 215, 1023–1034 10.1084/jem.20171922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malireddi R. K. S., Kesavardhana S., and Kanneganti T.-D. (2019) ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis). Front. Cell. Infect. Microbiol. 9, 10.3389/fcimb.2019.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malireddi R. K. S., Gurung P., Kesavardhana S., Samir P., Burton A., Mummareddy H., Vogel P., Pelletier S., Burgula S., and Kanneganti T.-D. (2020) Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity–independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J. Exp. Med. 217, 10.1084/jem.20191644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gurung P., Burton A., and Kanneganti T.-D. (2016) NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1β-mediated osteomyelitis. Proc. Natl. Acad. Sci. U. S. A. 113, 4452–4457 10.1073/pnas.1601636113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gurung P., Anand P. K., Malireddi R. K., Vande Walle L., Van Opdenbosch N., Dillon C. P., Weinlich R., Green D. R., Lamkanfi M., and Kanneganti T. D. (2014) FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol. 192, 1835–1846 10.4049/jimmunol.1302839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng M., Karki R., Vogel P., and Kanneganti T.-D. (2020) Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell 181, 674–687 10.1016/j.cell.2020.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samir P., Malireddi R. K. S., and Kanneganti T.-D. (2020) The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 238 10.3389/fcimb.2020.00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malireddi R. K. S., Ippagunta S., Lamkanfi M., and Kanneganti T.-D. (2010) Cutting edge: proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes. J. Immunol. 185, 3127–3130 10.4049/jimmunol.1001512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lukens J. R., Gurung P., Vogel P., Johnson G. R., Carter R. A., McGoldrick D. J., Bandi S. R., Calabrese C. R., Walle L. V., Lamkanfi M., and Kanneganti T.-D. (2014) Dietary modulation of the microbiome affects autoinflammatory disease. Nature 516, 246–249 10.1038/nature13788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siu K.-L., Yuen K.-S., Castaño-Rodriguez C., Ye Z.-W., Yeung M.-L., Fung S.-Y., Yuan S., Chan C.-P., Yuen K.-Y., Enjuanes L., and Jin D.-Y. (2019) Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 33, 8865–8877 10.1096/fj.201802418R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi C.-S., Nabar N. R., Huang N.-N., and Kehrl J. H. (2019) SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 5, 101 10.1038/s41420-019-0181-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DeDiego M. L., Nieto-Torres J. L., Jimenez-Guardeño J. M., Regla-Nava J. A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., and Enjuanes L. (2014) Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 194, 124–137 10.1016/j.virusres.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nieto-Torres J. L., DeDiego M. L., Verdiá-Báguena C., Jimenez-Guardeño J. M., Regla-Nava J. A., Fernandez-Delgado R., Castaño-Rodriguez C., Alcaraz A., Torres J., Aguilella V. M., and Enjuanes L. (2014) Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathogens 10, e1004077 10.1371/journal.ppat.1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nieto-Torres J. L., Verdiá-Báguena C., Castaño-Rodriguez C., Aguilella V. M., and Enjuanes L. (2015) Relevance of viroporin ion channel activity on viral replication and pathogenesis. Viruses 7, 3552–3573 10.3390/v7072786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nieto-Torres J. L., Verdiá-Báguena C., Jimenez-Guardeño J. M., Regla-Nava J. A., Castaño-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V. M., and Enjuanes L. (2015) Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 485, 330–339 10.1016/j.virol.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. An S., Chen C. J., Yu X., Leibowitz J. L., and Makino S. (1999) Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 73, 7853–7859 10.1128/JVI.73.9.7853-7859.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo S., Yang C., Diao B., Huang X., Jin M., Chen L., Yan W., Ning Q., Zheng L., Wu Y., and Chen Y. (2015) The NLRP3 inflammasome and IL-1β accelerate immunologically mediated pathology in experimental viral fulminant hepatitis. PLoS Pathog. 11, e1005155 10.1371/journal.ppat.1005155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang Y., Li J., Teng Y., Sun H., Tian G., He L., Li P., Chen Y., Guo Y., Li J., Zhao G., Zhou Y., and Sun S. (2019) Complement receptor C5aR1 inhibition reduces pyroptosis in hDPP4-transgenic mice infected with MERS-CoV. Viruses 11, 39 10.3390/v11010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sariol A., and Perlman S. (2020) Lessons for COVID-19 immunity from other coronavirus infections. Immunity 53, 248–263 10.1016/j.immuni.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belyavsky M., Belyavskaya E., Levy G. A., and Leibowitz J. L. (1998) Coronavirus MHV-3-induced apoptosis in macrophages. Virology 250, 41–49 10.1006/viro.1998.9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen K. W., Demarco B., Heilig R., Shkarina K., Boettcher A., Farady C. J., Pelczar P., and Broz P. (2019) Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. EMBO J. 38, 10.15252/embj.2019101638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taabazuing C. Y., Okondo M. C., and Bachovchin D. A. (2017) Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 24, 507–514e4 10.1016/j.chembiol.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Belouzard S., Millet J. K., Licitra B. N., and Whittaker G. R. (2012) Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4, 1011–1033 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lavi E., Wang Q., Weiss S. R., and Gonatas N. K. (1996) Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology 221, 325–334 10.1006/viro.1996.0382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., et al. (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., Sun H., Wang D. C., and Shao F. (2016) Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 10.1038/nature18590 [DOI] [PubMed] [Google Scholar]
- 58. Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V. G., Wu H., and Lieberman J. (2016) Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 10.1038/nature18629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Evavold C. L., Ruan J., Tan Y., Xia S., Wu H., and Kagan J. C. (2018) The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity 48, 35–44.e6 10.1016/j.immuni.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rogers C., Fernandes-Alnemri T., Mayes L., Alnemri D., Cingolani G., and Alnemri E. S. (2017) Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat. Commun. 8, 14128 10.1038/ncomms14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen I.-Y., Moriyama M., Chang M.-F., and Ichinohe T. (2019) Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol. 10, 50 10.3389/fmicb.2019.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou H., Zhao J., and Perlman S. (2010) Autocrine interferon priming in macrophages but not dendritic cells results in enhanced cytokine and chemokine production after coronavirus infection. mBio 1, e00219–10 10.1128/mBio.00219-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mehta P., McAuley D. F., Brown M., Sanchez E., Tattersall R. S., and Manson J. J. (2020) COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 395, 1033–1034 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pierini R., Juruj C., Perret M., Jones C. L., Mangeot P., Weiss D. S., and Henry T. (2012) AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 19, 1709–1721 10.1038/cdd.2012.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tsuchiya K., Nakajima S., Hosojima S., Thi Nguyen D., Hattori T., Manh Le T., Hori O., Mahib M. R., Yamaguchi Y., Miura M., Kinoshita T., Kushiyama H., Sakurai M., Shiroishi T., and Suda T. (2019) Caspase-1 initiates apoptosis in the absence of gasdermin D. Nat. Commun. 10, 2091 10.1038/s41467-019-09753-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., and Pöhlmann S. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Williams R. K., Jiang G. S., and Holmes K. V. (1991) Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 88, 5533–5536 10.1073/pnas.88.13.5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perlman S., and Dandekar A. A. (2005) Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5, 917–927 10.1038/nri1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huang K.-J., Su I.-J., Theron M., Wu Y.-C., Lai S.-K., Liu C.-C., and Lei H.-Y. (2005) An interferon-γ-related cytokine storm in SARS patients. J. Med. Virol. 75, 185–194 10.1002/jmv.20255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., Trilling M., Lu M., Dittmer U., and Yang D. (2020) Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J.Med. Virol. 92, 491–494 10.1002/jmv.25709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Prompetchara E., Ketloy C., and Palaga T. (2020) Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 38, 1–9 10.12932/AP-200220-0772 [DOI] [PubMed] [Google Scholar]
- 72. Daniel C., and Talbot P. J. (1990) Protection from lethal coronavirus infection by affinity-purified spike glycoprotein of murine hepatitis virus, strain A59. Virology 174, 87–94 10.1016/0042-6822(90)90057-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hocke A. C., Becher A., Knepper J., Peter A., Holland G., Tönnies M., Bauer T. T., Schneider P., Neudecker J., Muth D., Wendtner C. M., Rückert J. C., Drosten C., Gruber A. D., Laue M., et al. (2013) Emerging human Middle East respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am. J. Respir. Crit. Care Med. 188, 882–886 10.1164/rccm.201305-0954LE [DOI] [PubMed] [Google Scholar]
- 74. Nicholls J., Dong X.-P., Jiang G., and Peiris M. (2003) SARS: clinical virology and pathogenesis. Respirology. 8, S6–S8 10.1046/j.1440-1843.2003.00517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ioannidis J. P. A., Axfors C., and Contopoulos-Ioannidis D. G. (2020) Population-level COVID-19 mortality risk for non-elderly individuals overall and for non-elderly individuals without underlying diseases in pandemic epicenters. Environ Res 188, 109890 10.1016/j.envres.2020.109890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. World Health Organization (2003) WHO issues consensus document on the epidemiology of SARS. Wkly. Epidemiol. Rec. 78, 373–375 [PubMed] [Google Scholar]
- 77. World Health Organization (2019) WHO Eastern Mediterranean Regional Office, MERS-CoV, MERS situation update, September 2019. Epidemic and Pandemic-Prone Diseases (online) Accessed May 11, 2020 [Google Scholar]
- 78. Cho S. J., Rooney K., Choi A. M. K., and Stout-Delgado H. W. (2018) NLRP3 inflammasome activation in aged macrophages is diminished during Streptococcus pneumoniae infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 314, L372–L387 10.1152/ajplung.00393.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stout-Delgado H. W., Vaughan S. E., Shirali A. C., Jaramillo R. J., and Harrod K. S. (2012) Impaired NLRP3 inflammasome function in elderly mice during influenza infection is rescued by treatment with nigericin. J. Immunol. 188, 2815–2824 10.4049/jimmunol.1103051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Piasecka B., Duffy D., Urrutia A., Quach H., Patin E., Posseme C., Bergstedt J., Charbit B., Rouilly V., MacPherson C. R., Hasan M., Albaud B., Gentien D., Fellay J., Albert M. L., et al. and Milieu Intérieur Consortium (2018) Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges. Proc. Natl. Acad. Sci. U. S. A. 115, E488–E497 10.1073/pnas.1714765115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kanneganti T. D., Body-Malapel M., Amer A., Park J. H., Whitfield J., Franchi L., Taraporewala Z. F., Miller D., Patton J. T., Inohara N., and Núñez G. (2006) Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281, 36560–36568 10.1074/jbc.M607594200 [DOI] [PubMed] [Google Scholar]
- 82. Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W. P., Roose-Girma M., and Dixit V. M. (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 10.1038/nature10558 [DOI] [PubMed] [Google Scholar]
- 83. Karki R., Lee E., Place D., Samir P., Mavuluri J., Sharma B. R., Balakrishnan A., Malireddi R. K. S., Geiger R., Zhu Q., Neale G., and Kanneganti T.-D. (2018) IRF8 Regulates Transcription of Naips for NLRC4 Inflammasome Activation. Cell 173, 920–933.e13 10.1016/j.cell.2018.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., et al. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Murphy J. M., Czabotar P. E., Hildebrand J. M., Lucet I. S., Zhang J.-G., Alvarez-Diaz S., Lewis R., Lalaoui N., Metcalf D., Webb A. I., Young S. N., Varghese L. N., Tannahill G. M., Hatchell E. C., Majewski I. J., et al. (2013) The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 10.1016/j.immuni.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 86. Ozören N., Masumoto J., Franchi L., Kanneganti T.-D., Body-Malapel M., Ertürk I., Jagirdar R., Zhu L., Inohara N., Bertin J., Coyle A., Grant E. P., and Núñez G. (2006) Distinct roles of TLR2 and the adaptor ASC in IL-1β/IL-18 secretion in response to Listeria monocytogenes. J. Immunol. 176, 4337–4342 10.4049/jimmunol.176.7.4337 [DOI] [PubMed] [Google Scholar]
- 87. Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., and Dixit V. M. (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- 88. Jones J. W., Kayagaki N., Broz P., Henry T., Newton K., O'Rourke K., Chan S., Dong J., Qu Y., Roose-Girma M., Dixit V. M., and Monack D. M. (2010) Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. U. S. A. 107, 9771–9776 10.1073/pnas.1003738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Newton K., Sun X., and Dixit V. M. (2004) Kinase RIP3 is dispensable for normal NF-κ Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell Biol. 24, 1464–1469 10.1128/MCB.24.4.1464-1469.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Oberst A., Dillon C. P., Weinlich R., McCormick L. L., Fitzgerald P., Pop C., Hakem R., Salvesen G. S., and Green D. R. (2011) Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471, 363–367 10.1038/nature09852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., and Akira S. (1998) Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9, 143–150 10.1016/S1074-7613(00)80596-8 [DOI] [PubMed] [Google Scholar]
- 92. Schickli J. H., Zelus B. D., Wentworth D. E., Sawicki S. G., and Holmes K. V. (1997) The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 71, 9499–9507 10.1128/JVI.71.12.9499-9507.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hoffmann E., Neumann G., Kawaoka Y., Hobom G., and Webster R. G. (2000) A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97, 6108–6113 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Karki R., Sharma B. R., Lee E., Banoth B., Malireddi R. K. S., Samir P., Tuladhar S., Mummareddy H., Burton A. R., Vogel P., and Kanneganti T.-D. (2020) Interferon regulatory factor 1 regulates PANoptosis to prevent colorectal cancer. JCI Insight. 5, e136720 10.1172/jci.insight.136720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Grunewald M. E., Fehr A. R., Athmer J., and Perlman S. (2018) The coronavirus nucleocapsid protein is ADP-ribosylated. Virology 517, 62–68 10.1016/j.virol.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated for this study are included within this article and in the supporting information.