Abstract
A sterilizing or functional cure for HIV is currently precluded by resting CD4+ T cells that harbor latent but replication-competent provirus. The “shock-and-kill” pharmacological ap-proach aims to reactivate provirus expression in the presence of antiretroviral therapy and target virus-expressing cells for elimination. However, no latency reversal agent (LRA) to date effectively clears viral reservoirs in humans, suggesting a need for new LRAs and LRA combinations. Here, we screened 216 compounds from the pan-African Natural Product Library and identified knipholone anthrone (KA) and its basic building block anthralin (dithranol) as novel LRAs that reverse viral latency at low micromolar concentrations in multiple cell lines. Neither agent's activity depends on protein kinase C; nor do they inhibit class I/II histone deacetylases. However, they are differentially modulated by oxidative stress and metal ions and induce distinct patterns of global gene expression from established LRAs. When applied in combination, both KA and anthralin synergize with LRAs representing multiple functional classes. Finally, KA induces both HIV RNA and protein in primary cells from HIV-infected donors. Taken together, we describe two novel LRAs that enhance the activities of multiple “shock-and-kill” agents, which in turn may inform ongoing LRA combination therapy efforts.
Keywords: drug-drug synergy, histone deacetylase (HDAC), human immunodeficiency virus (HIV), latency reversal, molecular pharmacology, natural product, protein kinase C (PKC), viral reservoirs, drug discovery, histone deacetylase inhibitor (HDAC inhibitor) (HDI)
The use of combination antiretroviral therapy (cART) has been a resounding success in terms of reducing HIV/AIDS-related morbidity and mortality as well as HIV transmission (1). As of 2018, 23.3 million people living with HIV, or 62% of the global HIV/AIDS burden, were reliably accessing cART (UNAIDS (2019) Global HIV & AIDS statistics—2019 fact sheet; https://www.unaids.org/en/resources/fact-sheet; ac-cessed September 11, 2019). However, cART does not cure HIV due to resting CD4+ T cells that persistently bear integrated and immunologically invisible provirus. As these persistent proviral reservoirs can reactivate at any time to produce infectious virus, cART must be taken for life (2–5).
One method toward developing a sterilizing or functional HIV cure involves use of latency reversal agents (LRAs) that induce HIV-1 provirus expression. HIV reactivation, coupled with immunotherapy support (6), could render infected cells “visible” to the host immune system, whereas co-administration of cART would prevent further seeding of viral reservoirs (7, 8). This approach, frequently termed “shock-and-kill,” could theoretically eliminate an individual's viral reservoir and/or reduce the viral reservoir to a point that cART-free remission is achievable, provided that sufficiently effective LRAs and immune enhancers can be identified. Numerous LRAs have been described, representing different functional classes. The majority represents protein kinase C (PKC) activators and histone deacetylase (HDAC) inhibitors, although agents that act by other mechanisms, such as BET bromodomain and DNA methyltransferase inhibition, have also been intensively studied (9, 10). However, LRAs tested to date in humans have shown limited clinical success due to extensive toxicity, poor efficacy, inconsistent viral reactivation, and/or insufficient engagement of cellular “kill” mechanisms (9, 11). New LRA-based strategies are likely to be needed to circumvent these issues.
Toward this goal, several groups report that combinations of LRAs from different functional classes can synergistically enhance latency reversal (12–14). For example, Jiang et al. (14) described that the PKC activator ingenol-3-angelate (PEP005) and the BET bromodomain inhibitor JQ1, which each alone stimulated an ∼25-fold increase in HIV transcription in vitro, could induce a 250-fold increase when applied in combination. Similar results are also reported using primary CD4+ T cells from HIV-infected donors (12–14). These observations suggest that optimized LRA combinations may promote broader latency reversal at lower concentrations, thereby maximizing virus reactivation while limiting drug toxicities and other off-target effects. Thus, discovery of LRAs that enhance the activities of existing LRA clinical candidates would support efforts to identify optimized LRA combinations.
Pure compounds isolated from natural products are a rich source of unique chemical diversity and new LRAs. For example, we previously screened a library of 257 compounds originating from marine natural products and identified four (1.6%) that reversed HIV latency in both cell line and primary cell models (15). LRA “hit” rates of 1.0% or more have also been reported by others (16, 17). Based on these observations, we hypothesized that new LRAs that can enhance the activities of existing agents could be isolated from additional natural product–based compound libraries. Here, we describe the results of a screen of the pan-African Natural Product Library (pANAPL), which contains compounds originating from African medicinal plants (18, 19). From this screen, we identified and characterized knipholone anthrone (KA), in addition to its synthetic analog anthralin (dithranol), as novel LRAs that synergize with established HIV latency reversal agents.
Results
Discovery of novel LRAs from pure natural products
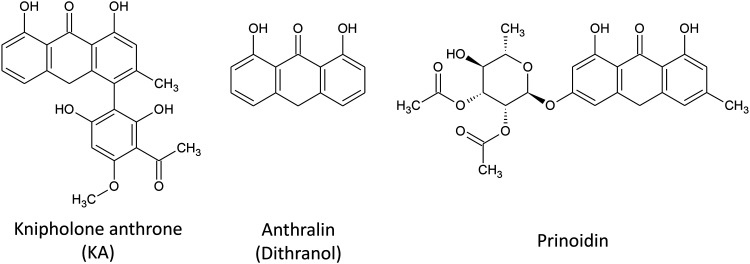
To identify new LRAs from natural product sources, we used the Jurkat-derived J-Lat 9.2 cell line, which contains a noninfectious HIV provirus where premature stop codons are engineered into env and where nef is replaced with a GFP reporter (20). Detection of GFP in these cells, as measured by flow cytometry, thus indicates HIV provirus expression. Using this assay, we screened 216 pure compounds from the pANAPL at 5 µg/ml for 24 h and identified one compound, KA (Fig. 1), which at 5 µg/ml (∼12 μm) induced GFP expression in 6.1 ± 5.2% of cells (mean ± S.D.). Based on this observation, we then screened 16 additional anthrones from pANAPL and commercially available sources at 10 μm (Fig. S1) and observed that anthralin (dithranol; Fig. 1) also induced 6.9 ± 2.4% GFP-positive cells. A third anthrone, prinoidin (Fig. 1), was also observed to induce 2.8 ± 0.7% GFP-positive cells; however, it was not explored further due to its limited availability. No other assessed anthrones induced latency reversal. KA and anthralin were therefore selected for further study.
Figure 1.
Structures of identified LRAs.
KA and anthralin reverse HIV latency in multiple in vitro cell models
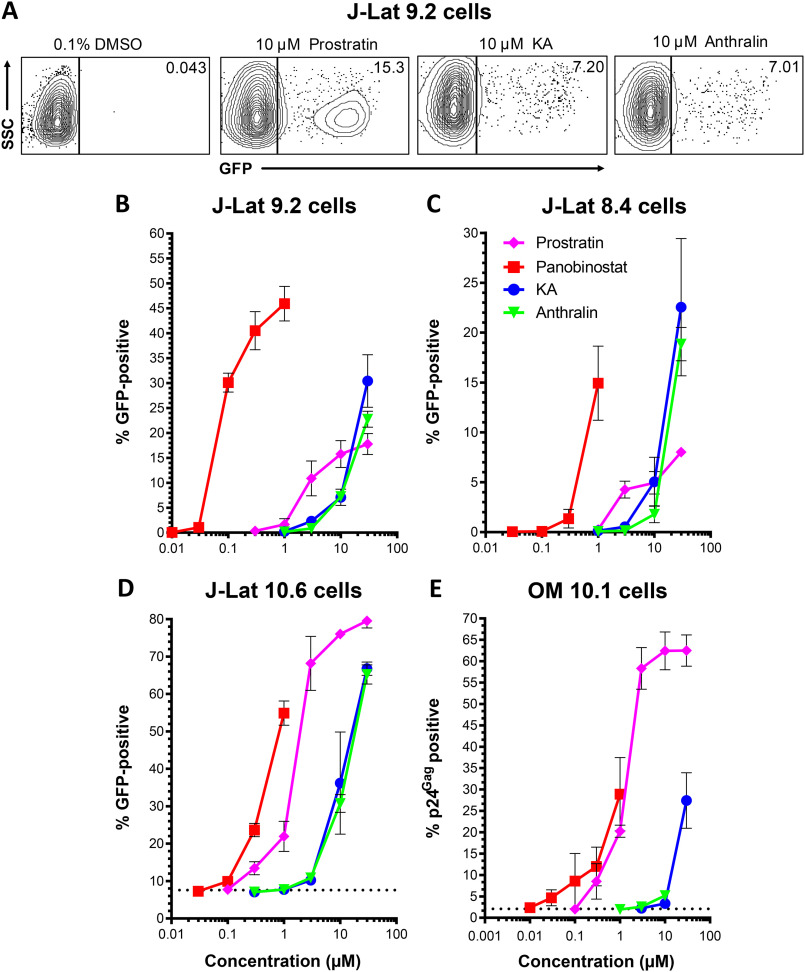
To investigate the in vitro latency reversal properties of these anthrones in detail, we next measured the dose-response profiles of KA and anthralin in live J-Lat 9.2 cells. In parallel, we also assessed the activities of control LRAs, including the PKC activator prostratin and the HDAC inhibitor panobinostat. Examples of latency reversal in J-Lat 9.2 cells, as measured by GFP expression, are shown in Fig. 2A. In these studies, treatment of J-Lat 9.2 cells with 10 μm prostratin induced 15.8 ± 2.7% GFP-positive cells, whereas stimulation with 0.3 μm panobinostat resulted in 40.5 ± 3.8% GFP-positive cells (Fig. 2B). In contrast, 10 μm KA resulted in 7.1 ± 1.6% GFP-positive cells, whereas 10 μm anthralin induced 7.2 ± 0.5% positive cells. Using the approach of Hashemi et al. (21) and normalizing to the average GFP response for 10 μm prostratin as described previously (15), the relative EC50 values for prostratin, panobinostat, KA, and anthralin were calculated to be 5.4 ± 1.4, 0.14 ± 0.02, 10.4 ± 1.0, and 12.1 ± 1.7 μm, respectively (Table 1).
Figure 2.
KA and anthralin reverse HIV latency in vitro. A, representative flow cytometry data showing latency reversal, as measured by GFP expression, in J-Lat 9.2 cells. Values indicate percentage GFP-positive cells for each condition. B–D, dose-response profiles of control LRAs panobinostat and prostratin, in addition to KA and anthralin, are shown in J-Lat 9.2 (B), J-Lat 8.4 (C), and J-Lat 10.6 (D) T cells. E, dose-response profiles of LRAs in OM10.1 promyeloid cells, as measured by cellular expression of viral p24Gag protein. Dotted lines in C and D indicate baseline levels of spontaneous latency reversal. Error bars, S.D.
Table 1.
Relative activities of LRAs in vitro. Data are presented as relative EC50 values (in µm) normalized to the average GFP response for 10 μm prostratin (i.e. the concentration required to induce 50% of the signal observed by 10 μm prostratin) (21)
| Cell line | Prostratin | Panobinostat | KA | Anthralin |
|---|---|---|---|---|
| µm | µm | µm | µm | |
| J-Lat 9.2 | 5.4 ± 1.4 | 0.14 ± 0.02 | 10.4 ± 1.0 | 12.1 ± 1.7 |
| J-Lat 8.4 | 5.0 ± 0.9 | 0.46 ± 0.12 | 7.4 ± 2.6 | 12.6 ± 1.9 |
| J-Lat 10.6 | 1.9 ± 0.9 | 0.64 ± 0.09 | 16.0 ± 5.5 | 16.2 ± 1.2 |
| OM10.1 | 2.7 ± 1.0 | 1.6 ± 0.5 | >30 | >30 |
To investigate whether latency reversal due to KA and anthralin was independent of the proviral integration site in J-Lat cells, we next assessed their dose-response profiles in the related cell lines J-Lat 8.4 and J-Lat 10.6 (Fig. 2, C and D). Whereas results were broadly consistent with those from J-Lat 9.2 cells, a few differences were observed. For example, whereas 10 μm prostratin induced 5.0 ± 1.1% GFP-positive, live J-Lat 8.4 cells, 0.3 μm panobinostat induced GFP in only 1.4 ± 0.9% of J-Lat 8.4 cells, indicating that this cell line is less responsive to this HDAC inhibitor. In contrast, KA and anthralin induced 5.0 ± 2.5 and 1.8 ± 0.8% GFP-positive live cells, respectively (Fig. 2C). When normalized to 10 μm prostratin, the relative EC50 values of prostratin, panobinostat, KA, and anthralin were 5.0 ± 0.9, 0.46 ± 0.12, 7.4 ± 2.6, and 12.6 ± 1.9 μm, respectively (Table 1). Similarly, in live J-Lat 10.6 cells, which in our hands (and as described previously (23, 24)) induced spontaneous GFP expression in 7.6 ± 0.2% of cells and as described previously (22, 23), 10 μm prostratin induced 76.0 ± 0.8% GFP-positive cells, whereas 0.3 μm panobinostat induced GFP in 23.7 ± 1.8% of cells. Comparatively, 36.2 ± 13.7 and 30.8 ± 2.4% of cells were induced by 10 μm KA and anthralin, respectively (Fig. 2D). When normalized to 10 μm prostratin, we recorded relative EC50 values of 1.9 ± 0.9, 0.64 ± 0.09, 16.0 ± 5.5, and 16.2 ± 1.2 μm for prostratin, panobinostat, KA, and anthralin (Table 1). The observation that the latency reversal properties of KA and anthralin are broadly consistent across multiple cell lines suggests that their activities are not dependent on specific proviral integration sites, at least in Jurkat-derived T cell lines.
To determine whether KA and anthralin's latency reversal was independent of cell type, we next treated OM-10.1 cells, which are derived from HL-60 promyelocyte cells and contain an integrated, replication-competent provirus. Similar to J-Lat 10.6 cells, we observed that OM10.1 cells spontaneously expressed p24Gag protein in 2.1 ± 0.2% of cells. Treatment of OM-10.1 cells with 10 μm prostratin for 24 h stimulated virus expression in 62.4 ± 4.4% of live cells, whereas 0.3 μm panobinostat induced provirus expression in only 12.0 ± 4.5% of cells. Notably, 10 μm KA elicited only 3.3 ± 0.7% p24Gag-positive live cells, whereas 10 μm anthralin induced 5.2 ± 1.2% p24Gag-positive live cells (Fig. 2E). Whereas improved responses were observed with 30 μm KA (27.4 ± 6.5% p24Gag-positive live cells), extensive toxicity precluded measurements of 30 μm anthralin. When normalized to 10 μm prostratin, the relative EC50 values for prostratin, panobinostat, KA, and anthralin were 2.7 ± 1.0, 1.6 ± 0.5, >30, and >30 μm, respectively (Table 1). Thus, KA and anthralin may more effectively reverse latency in T cell–derived lines. Taken together, these results suggest that both KA and anthralin induce provirus expression across multiple cell lines and proviral integration sites, although KA and anthralin's activities varied, depending on the cell line.
Effects of KA and anthralin on apoptosis and cell activation markers
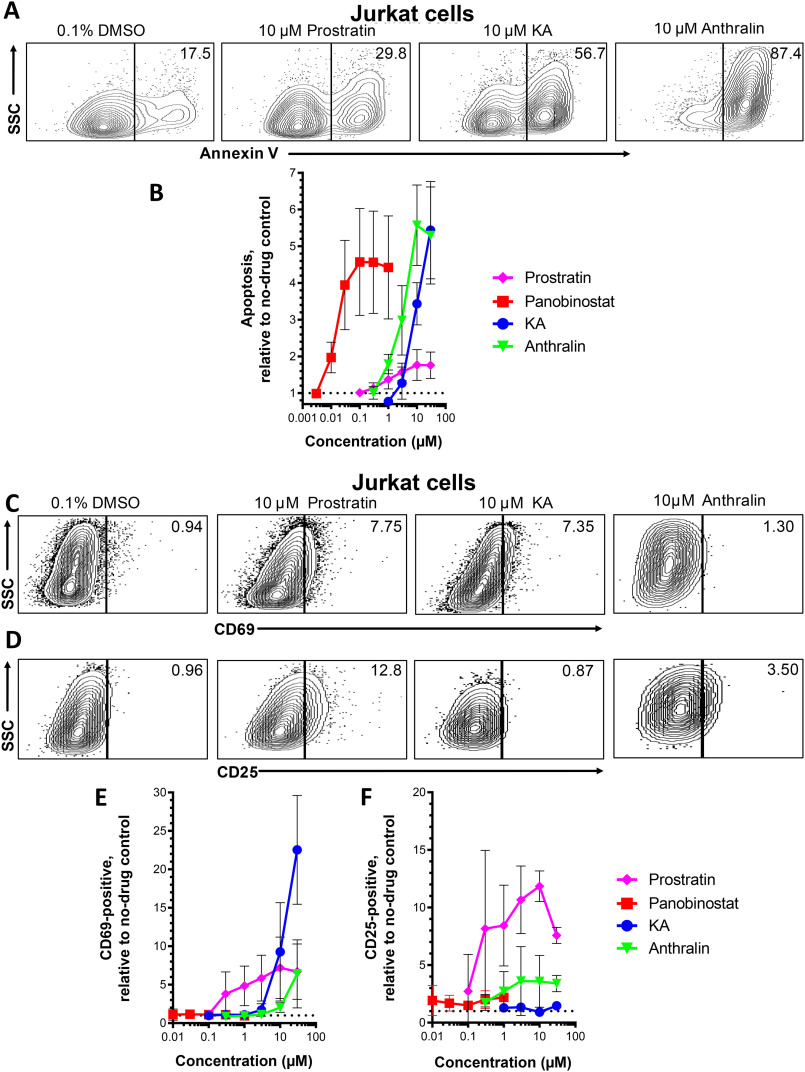
To directly assess the impact of KA and anthralin on cell viability, we next treated Jurkat cells (the parental cell line of J-Lat cells) with LRAs as described above. Following treatment, cells were then assessed for surface expression of the early apoptotic marker annexin V by flow cytometry. Fig. 3A shows representative results of apoptotic cells in the presence of LRAs, where control data are consistent with previous results (15), whereas Fig. 3B shows dose-response profiles where data were normalized to the percentage of apoptotic cells in control cell cultures treated with 0.1% DMSO. In this assay, 10 μm prostratin induced only a 1.8 ± 0.4-fold increase in apoptosis, whereas 0.3 μm panobinostat induced a 4.6 ± 1.4-fold increase, consistent with our previous results (15). Like panobinostat, both KA and anthralin also induced apoptosis: for example, 10 μm KA resulted in a 3.4 ± 0.6-fold increase in apoptotic cells, whereas 10 μm anthralin caused a 5.6 ± 1.1-fold increase (Fig. 3B). These results suggest that, like the HDAC inhibitor panobinostat, both KA and anthralin induce apoptosis in vitro at concentrations that also induce latency reversal.
Figure 3.
Effects of LRAs on in vitro cell viability and expression of cell activation markers. A, representative flow cytometry data showing Jurkat cell apoptosis, as measured by annexin V detection. Values indicate percentage of annexin-positive cells. B, dose-response profiles of panobinostat, prostratin, KA, and anthralin on apoptosis in Jurkat cells. Data are presented as -fold increase in annexin V–positive cells relative to cells treated with 0.1% DMSO (dotted line). C and D, representative flow cytometry data showing Jurkat cells stained for CD69 (C) or CD25 expression (D). Values indicate percentage of CD69- or CD25-positive cells, respectively. E and F, dose-response profiles of LRAs on CD69 (E) and CD25 (F) expression in Jurkat cells. Data are presented as -fold increases in each marker relative to cells treated with 0.1% DMSO (dotted lines). Error bars, S.D.
To assess whether KA and anthralin may also affect T cell activation, Jurkat cells were treated with LRAs for 24 h and stained for the T cell activation markers CD69 and CD25. Fig. 3 (C and D) shows representative results of CD69 and CD25 expression in live-gated Jurkat cells in the presence of LRAs, respectively, whereas Fig. 3 (E and F) shows dose-response profiles normalized to marker expression in control cells treated with 0.1% DMSO. As expected, based on previous observations (24, 25), 10 μm prostratin induced a 7.2 ± 4.0-fold increase in CD69 expression, relative to cells treated with 0.1% DMSO, whereas no detectable increase in CD69 expression was observed with any concentration of panobinostat (Fig. 3E). Notably, 10 μm KA induced a 9.3 ± 6.4-fold increase in CD69 expression, whereas 30 μm induced an up to 22.5 ± 7.1-fold increase. In contrast, 10 μm anthralin induced only a 2.0 ± 0.6-fold increase in CD69-positive cells, whereas 30 μm induced a 6.4 ± 4.4-fold increase. Also consistent with previous observations (24, 25), 10 μm prostratin induced an 11.8 ± 1.3-fold increase in CD25 expression, whereas no more than a 2.0 ± 0.8-fold increase in CD25 expression was observed with 0.3 μm panobinostat (Fig. 3F). However, neither KA nor anthralin induced CD25 expression to levels approximating those of prostratin: no increase was observed for KA at any concentration, whereas 10 μm anthralin induced only a maximal 3.6 ± 2.2-fold increase. These observations indicate that KA is a particularly potent inducer of at least a subset of T cell activation markers in vitro, whereas anthralin can also induce T cell activation markers at high concentrations.
KA and anthralin do not function as PKC activators or HDAC inhibitors
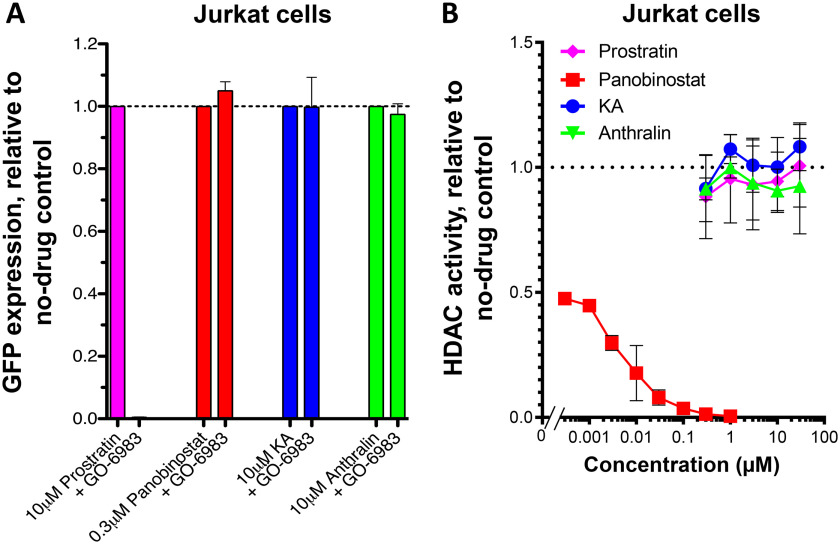
To date, numerous LRAs have been reported to act through two major cellular pathways: PKC activation and HDAC inhibition (9, 10). To determine whether KA and anthralin function as PKC activators, we asked whether their activities in live J-Lat 9.2 cells were antagonized by the pan-PKC inhibitor GÖ-6983 (26). Following a 24-h treatment with LRAs, additional co-treatment with 1 μm GÖ-6983 resulted in complete suppression of GFP expression induced by a 10 μm concentration of the PKC activator prostratin (0.4% GFP-positive cells relative to cells treated with only prostratin), but not by a 0.3 μm concentration of the HDAC inhibitor panobinostat (Fig. 4A). Furthermore, no loss of GFP expression was observed when 1 μm GÖ-6983 was added to J-Lat cultures treated with either 10 μm KA or anthralin (in both cases, >95% of cells treated without GÖ-6983 maintained GFP). This indicates that KA and anthralin do not function as PKC activators in vitro.
Figure 4.
KA and anthralin are not PKC activators or HDAC inhibitors. A, effects of panobinostat, prostratin, KA, and anthralin on latency reversal in J-Lat 9.2 cells in the presence of pan-PKC inhibitor GÖ-6983. B, effects of LRAs on cellular HDAC activity, as measured by HDAC-Glo assay. Error bars, S.D.
To assess whether KA and anthralin function as HDAC inhibitors, we used a commercially available HDAC-Glo I/II assay kit, which quantifies class I and II HDAC activity in live Jurkat cells via a cell-permeable, acetylated, luminogenic peptide substrate. With this approach, HDAC inhibitors should antagonize cellular deacetylation of the fluorogenic substrate and reduce luminescence readout. As expected, panobinostat was a potent cellular HDAC inhibitor in this assay: 0.1 μm inhibited 96.4 ± 1.2% of luminescence, whereas 0.3 nm inhibited 52.5 ± 1.2% (Fig. 4B). In contrast, up to 30 μm prostratin did not affect cellular HDAC activity. Similarly, neither KA nor anthralin had any activity at up to 30 μm, indicating that these LRAs also do not function as HDAC inhibitors in vitro.
Latency reversal by KA and anthralin are regulated by reactive oxygen species and/or metal ions
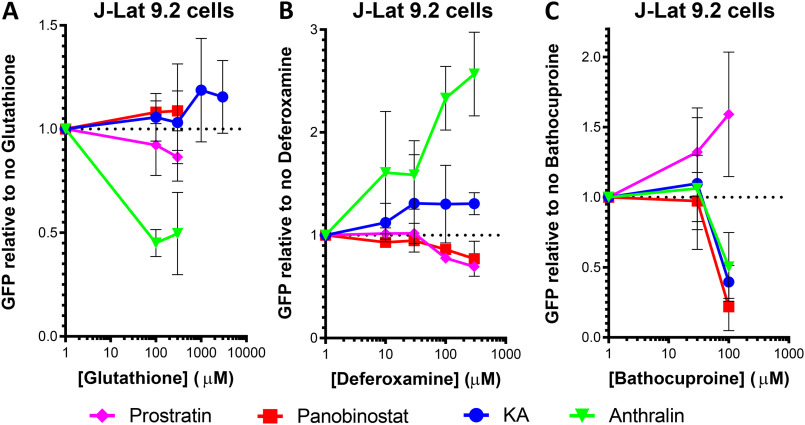
Both KA and anthralin are reported to promote oxidative stress in cells (27–29). KA is also reported to chelate metal ions (27). If one or more of these properties are required by KA or anthralin for latency reversal, then blocking these pathways should antagonize these LRAs. To test this hypothesis, we first treated J-Lat 9.2 cells with 10 μm prostratin, 0.1 μm panobinostat, 10 μm KA, or 10 μm anthralin for 24 h in the presence or absence of modulators of oxidative stress or free metal ions (Fig. 5). Modulators included GSH (a scavenger of reactive oxygen species), deferoxamine (an iron chelator), and bathocuproine (a Cu(I) chelator).
Figure 5.
Effects of LRAs on HIV latency reversal in J-Lat 9.2 cells in the presence of the anti-oxidant GSH (A), iron chelator deferoxamine (B), and copper chelator bathocuproine (C). Error bars, S.D.
In the presence of increasing concentrations of GSH, we observed that anthralin-induced GFP, but not GFP induced by KA, prostratin, or panobinostat, was inhibited (Fig. 5A). Whereas complete inhibition of anthralin's activity was not observed, treatment of J-Lat 9.2 cells with 100 μm GSH reduced anthralin-dependent latency reversal to 45.0 ± 6.5% of cells treated with anthralin in the absence of GSH. In contrast, no inhibition of KA was observed with GSH concentrations as high at 3 mm. GSH in the absence of LRAs had no effect on GFP expression (data not shown). These observations suggest that the latency reversal properties of anthralin, but not KA or control LRAs, are dependent on cellular oxidative stress.
In contrast, treatment of J-Lat 9.2 cells with increasing concentrations of deferoxamine resulted in enhanced latency reversal when co-incubated with KA, where 300 μm boosted the activity of 10 μm KA to 130.7 ± 10.6% of cells expressing GFP in the absence of deferoxamine (Fig. 5B). In the presence of 10 μm anthralin, 300 μm deferoxamine caused a 2.6 ± 0.4-fold increase in GFP expression relative to cells treated with anthralin alone. In contrast, both prostratin and panobinostat were slightly inhibited by 300 μm deferoxamine (69.5 ± 3.9 and 77.1 ± 17.1% GFP-positive cells, respectively). These results suggest that the activities of both KA and anthralin are either inhibited by iron ions or otherwise stimulated by deferoxamine in vitro.
Finally, treatment of cells with 100 μm bathocuproine inhibited GFP expression induced by 10 μm KA, where only 39.6 ± 11.8% of GFP-positive cells were observed relative to KA-treated cells without bathocuproine. This suggests that KA's activity depends on the presence of copper ions. GFP expression induced by anthralin and panobinostat was also affected by 100 μm bathocuproine (50.2 ± 24.6 and 21.9 ± 17.1% of GFP expression, respectively; Fig. 5C), suggesting that their activity is also dependent on copper. In contrast, prostratin's activity was enhanced in the presence of bathocuproine, where, for example, 100 μm boosted GFP expression to 159.1 ± 44.4% of cells treated with prostratin alone. Taken together, these results suggest that the latency reversal properties of anthralin are dependent on both oxidative stress and metal ions, whereas KA's properties are dependent only on metal ions.
KA and anthralin induce distinct gene expression profiles in vitro
To identify potential mechanisms by which KA and anthralin may induce HIV transcription in vitro, we next treated three independent preparations of J-Lat 10.6 cells for 24 h with 0.1% DMSO vehicle control, prostratin (10 μm), panobinostat (0.1 μm), or KA or anthralin (10 μm). Cells were then harvested for RNA extraction, and global transcriptional profiles were assessed by RNA-Seq. High-quality sequence data were ob-tained for all samples with the potential exception of cells treated with anthralin, which contained ∼21.5% of total transcriptome reads observed in other samples, which was suggestive of cellular toxicity. From these data, we identified a total of 8707 unique differentially expressed genes (false discovery rate (FDR) < 5%) affected by at least one treatment when compared with DMSO-treated samples. Anthralin and panobinostat had the largest effects on overall transcription with overlap of over 2000 genes (Fig. 6A).
Figure 6.
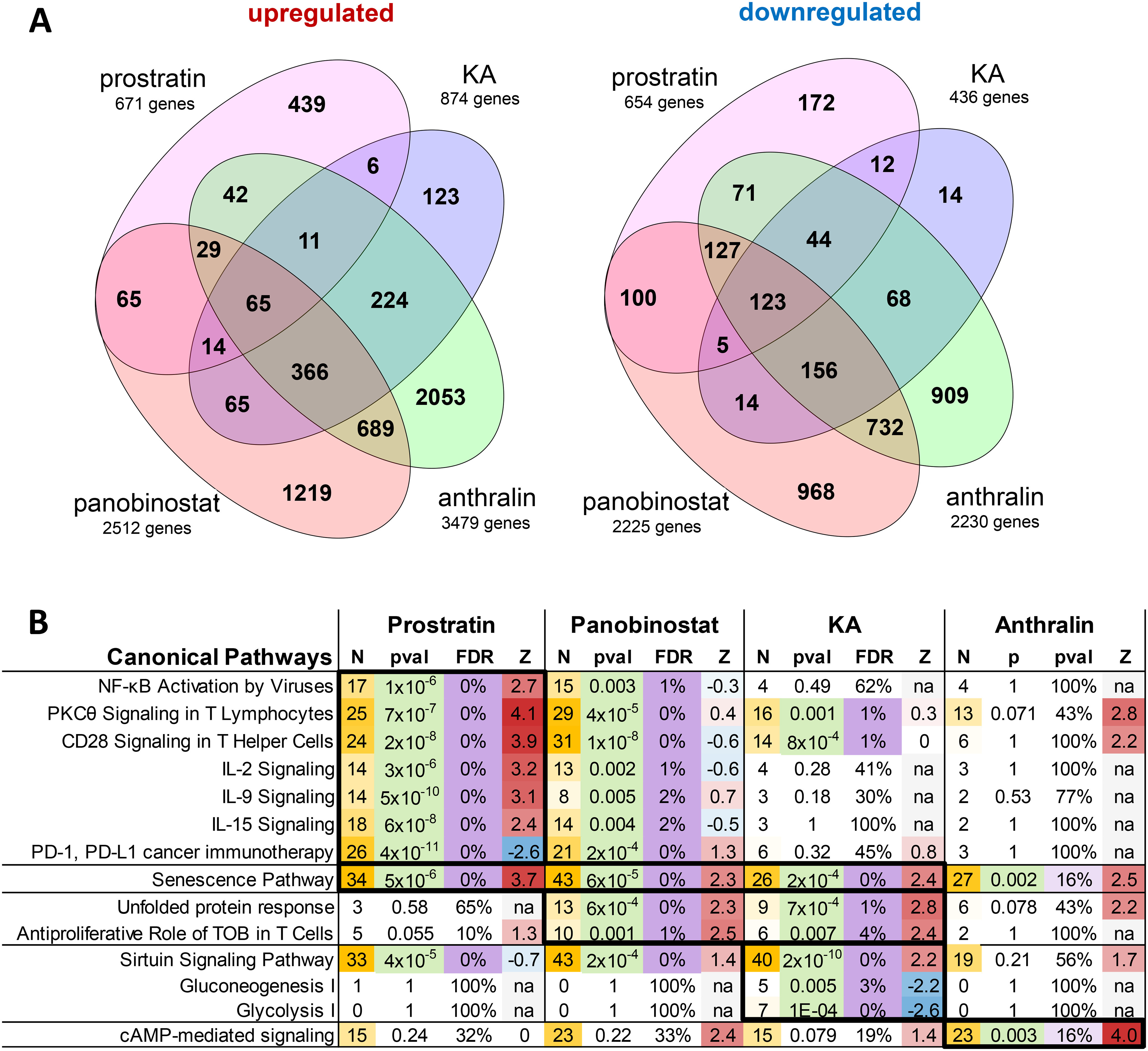
Effects of LRAs on in vitro global gene expression as measured by RNA-Seq. A, Venn diagrams showing number of significantly up-regulated or down-regulated genes (FDR < 5%) in J-Lat 10.6 cells treated with prostratin, panobinostat, KA, or anthralin, when compared with cells treated with 0.1% DMSO. B, Ingenuity Pathway Analysis results of genes identified in A that passed FDR < 5% and |Z-score| > 2 thresholds. For each pathway, data listed include Z-scores (Z) for predicted pathway state (where positive and negative values indicate activation or inhibition by treatment, respectively), number of affected genes (N), p value (pval), and FDR. p values < 0.05, FDR < 5% (or <20% for anthralin) thresholds are highlighted. Number of genes and Z-scores are highlighted as scales.
The four sets of genes affected by each individual treatment were then analyzed for enrichment of canonical pathways using Ingenuity Pathway Analysis (Fig. 6B). Consistent with the established role of prostratin as a PKC agonist, we identified distinct up-regulation of genes involved in PKCθ signaling (Z-score = 4.1; see “Experimental procedures”) and NF-κB activation by viruses (Z-score = 2.7; Fig. 6B). Additionally, we found activation of CD28 signaling and cytokine signaling and production, whereas PD-1/PD-L1 pathways were inhibited, consistent with T cell activation (Fig. 6B). In contrast, cells treated with panobinostat activated cellular senescence, unfolded protein response, and the antiproliferative role of TOB (transducer of ErbB2) in T cell signaling, suggestive of reduced cellular activation and mild to moderate cytotoxicity. Also, consistent with induction of T cell quiescence in cells treated with panobinostat, genes affected by prostratin were frequently induced in the opposite direction of T cell activation and cytokine signaling (Fig. 6B).
Cells treated with KA also had several affected gene pathways that were also observed in panobinostat-treated cells. However, KA-treated cells also had particularly strong and unique activation of the sirtuin signaling pathway (Z-score = 2.2). This result suggests that KA might disproportionately support latency reversal via sirtuin-mediated enhancement of Tat deacetylation and priming to initiate new cycles of viral transcription (30). Additional pathways unique to KA treatment included inhibition of gluconeogenesis and glycolysis (Z-scores = −2.2 and −2.6, respectively; Fig. 6B).
Finally, although cells treated with anthralin resulted in the largest number of significantly affected genes (n = 5709; Fig. 6A), no signaling pathways were identified at FDR < 5% significance. However, among the most strongly affected pathways unique to anthralin treatment was cAMP-mediated signaling, which exhibited borderline significance (p = 0.003; FDR = 0.16; Z-score = 4.0), which is suggestive of anthralin uniquely acting in part by driving cAMP or protein kinase A–mediated viral transcription (31, 32). Taken together, these results suggest that KA and anthralin affect gene expression and induction of pathways distinct from those of control LRAs and further suggest that they induce latency reversal by distinct mechanisms.
KA and anthralin synergize with multiple LRAs
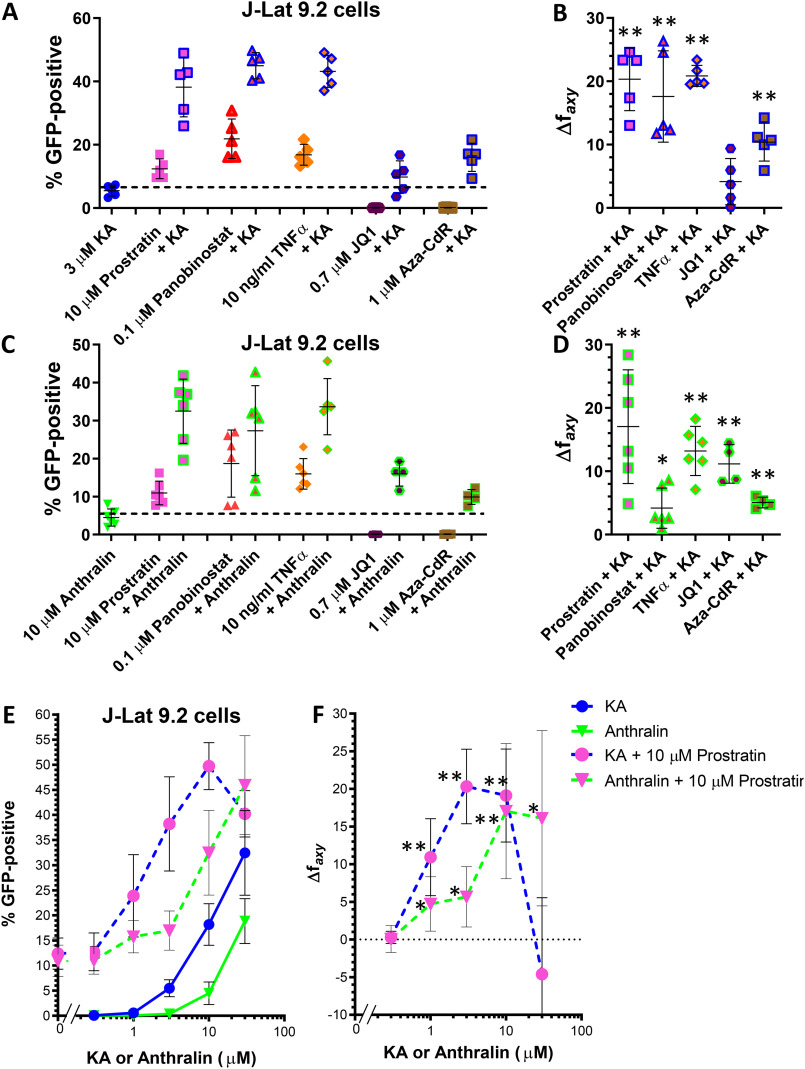
When applied in combination, LRAs with similar mechanisms of action tend to exhibit additive effects, whereas LRAs representing different functional classes frequently result in synergistic (i.e. greater than additive) effects (12–15). To examine whether KA and anthralin can enhance the activities of established LRAs, we treated J-Lat 9.2 cells with KA or anthralin in combination with control LRAs at concentrations that induce submaximal GFP expression. We first assessed whether 3 μm KA and 10 μm anthralin could synergize with control LRAs tested at a single concentration. Control LRAs included prostratin (10 μm), panobinostat (0.1 μm), TNFα (10 ng/ml), the BET bromodomain inhibitor JQ1 (0.7 μm) (12–14), and the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (Aza-CdR; 1 μm) (33). For each combination, synergism was assessed using the Bliss independence model (12, 14), where a calculated Δfaxy value > 0 indicates evidence of synergistic effects (see “Experimental procedures”). In this study, we conservatively defined evidence of synergism as a Δfaxy value > 1.
In all cases, KA enhanced the activities of control LRAs (Fig. 7, A and B). For example, when administered alone, 3 μm KA induced GFP in 5.5 ± 1.7% of live cells, whereas 10 μm prostratin induced GFP in 12.4 ± 3.1% of live J-Lat 9.2 cells. However, when cells were co-incubated with 10 μm prostratin and 3 μm KA, we observed 38.2 ± 9.4% GFP-positive cells (Fig. 7A). This represented a 2.1-fold increase over what would be expected by additive effects (∼17.9%) and significant evidence of synergism as measured by the Bliss independence model (Δfaxy = 20.3 ± 5.0; p = 7.8 × 10−4; Fig. 7B). KA also synergized with 0.1 μm panobinostat, which induced 21.9 ± 6.2% GFP expression on its own but 45.0 ± 4.1% GFP expression in combination with KA (Δfaxy of 17.6 ± 7.2; p = 0.0055; Fig. 7, A and B). Similarly, whereas 10 ng/ml TNFα induced 16.8 ± 3.3% GFP-positive cells on its own, the addition of KA resulted in 43.2 ± 5.1% GFP-positive cells (Δfaxy of 20.8 ± 1.7; p = 9.7 × 10−6). When 3 μm KA was combined with a 0.7 μm concentration of the BET bromodomain inhibitor JQ1 (which exhibited no LRA activity on its own in J-Lat cells), we observed 9.8 ± 5.1% GFP-positive cells and a borderline significant Δfaxy of 4.2 ± 3.7 (p = 0.064). Finally, whereas 1 μm Aza-CdR also exhibited no activity on its own in live J-Lat cells, the addition of KA increased GFP-positive cells to 16.0 ± 4.5% GFP-positive cells and a Δfaxy of 10.4 ± 3.0 (p = 0.0015). These results indicate that 3 μm KA significantly synergizes with four of five control LRAs at these concentrations in vitro.
Figure 7.
KA and anthralin synergize with LRAs from multiple functional classes in vitro. A, effects of 3 μm KA on latency reversal in J-Lat 9.2 cells in the presence of 10 μm prostratin, 0.1 μm panobinostat, 10 ng/ml TNFα, 0.7 μm JQ1, and 1 μm Aza-Cdr. B, extent of synergism in J-Lat 9.2 cells treated with KA plus control LRAs in A, as measured by the Bliss independence model. C and D, effects of 10 μm anthralin on latency reversal (C) and synergism (D) in J-Lat 9.2 cells in the presence of control LRAs. Data are arrayed as described in A and B. E and F, effects of KA and anthralin on latency reversal (E) and synergism (F) in the presence of 10 μm prostratin. In E, “0” on the x axis indicates the activity of 10 μm prostratin in the absence of KA or anthralin. Data shown for 10 μm prostratin plus 3 μm KA or 10 μm anthralin are the same data shown in A–D. *, p < 0.05; **, p < 0.01 between the observed and predicted responses assuming strictly additive effects (i.e. Bliss independence model-based synergism) from at least four independent experiments. Error bars, S.D.
Similar results were observed when control LRAs were co-incubated with anthralin (Fig. 7, C and D). For example, when administered alone, 10 μm anthralin induced 4.5 ± 2.3% GFP expression in live J-Lat 9.2 cells, whereas 10 μm prostratin induced 11.0 ± 3.1% GFP-positive cells in paired experiments. However, when cells were co-incubated with 10 μm prostratin plus anthralin, we observed 32.5 ± 8.4% live GFP-positive cells (Fig. 7C). This represented a 1.8-fold increase over the expected additive effects (∼16.5%) and a significant Δfaxy of 17.0 ± 9.0(p = 0.0056; Fig. 7D). In addition, 10 μm anthralin also enhanced the activity of 0.1 μm panobinostat and 10 ng/ml TNFα (Δfaxy = 4.2 ± 3.2 and 13.2 ± 3.9; p = 0.023 and 4.0 × 10−4, respectively; Fig. 7, C and D). Co-administration of 10 μm anthralin with 0.7 μm JQ1 also resulted in a Δfaxy value of 11.1 ± 3.0 (p = 0.0053), whereas anthralin plus 1 μm Aza-CdR resulted in a Δfaxy value of 5.1 ± 0.8; p = 0.0013). Thus, 10 μm anthralin significantly synergized with all control LRAs at these concentrations in vitro. In contrast, no obvious enhancement of GFP expression was observed when KA was combined with anthralin (data not shown), although the poor viability of cells treated with both agents made interpretation of these data difficult.
We next asked what concentrations of KA and anthralin were required to achieve synergy with a control LRA. In this experiment, J-Lat 9.2 cells were treated with 10 μm prostratin in the presence of multiple concentrations of KA or anthralin (Fig. 7, E and F). We observed additionally synergistic provirus expression when 10 μm prostratin was co-incubated with 1 and 10 μm KA (Fig. 7E), with calculated Δfaxy values of 10.9 ± 5.1 and 19.1 ± 6.2, respectively (p = 0.0088 and 0.0023, respectively; Fig. 7F), in addition to the previously described synergy with 3 μm KA (i.e. Fig. 7A). Prostratin also synergized with 1, 3, and 30 μm anthralin (Fig. 7E), where Δfaxy values were calculated as 4.7 ± 3.6, 5.7 ± 4.0, and 16.1 ± 11.6, respectively (p = 0.024, 0.018, and 0.019, respectively; Fig. 7F), in addition to the previously described synergy with 10 μm anthralin (Fig. 7C). In summary, these results indicate that both KA and anthralin synergize with control LRAs representing multiple functional classes and at concentrations as low as 1 μm.
KA but not anthralin induces HIV-1 expression ex vivo
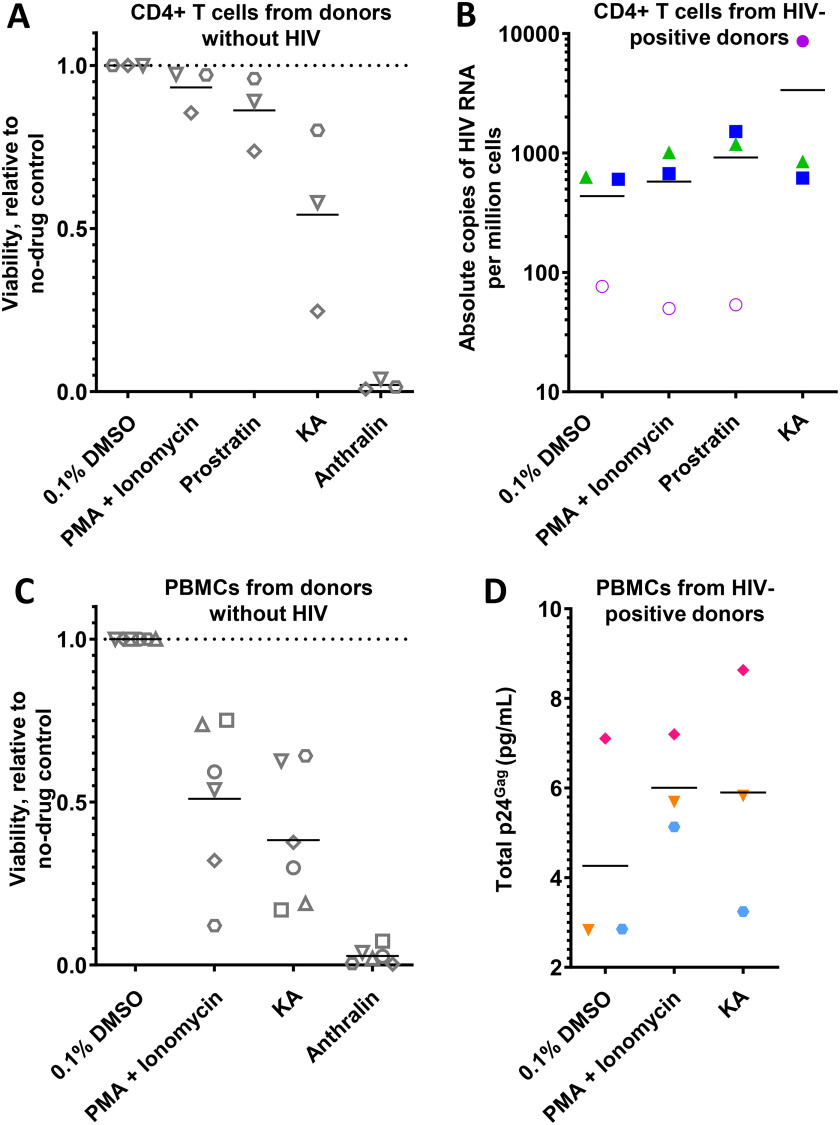
We next sought to investigate whether KA and/or anthralin can reactivate HIV proviruses in primary cells directly isolated from HIV-infected, cART-suppressed individuals. However, we first assessed the extent to which LRAs affected viability of isolated CD4+ cells obtained from three uninfected donors, as determined using ViaCount cell viability dye (34). LRAs assessed in this assay included 10 μm prostratin, KA, or anthralin, whereas 100 ng/ml phorbol myristate acetate (PMA) + 0.1 µg/ml ionomycin was applied as a positive control. For each condition, 106 cells were cultured in 1 ml of medium in the presence of test agents and 100 units/ml IL-2 for 24 h before assessment of cell viability. Consistent with in vitro observations (Fig. 3, A and B), the effects of LRAs on cell viability after 24-h incubation extended to primary CD4+ cells (Fig. 8A). For example, CD4+ cells treated with PMA + ionomycin resulted in 93.3 ± 6.8% viability relative to cells treated with 0.1% DMSO, whereas 10 μm prostratin resulted in 86.2 ± 11.4% viability. Cells treated with 10 μm KA resulted in 54.2 ± 27.9% viability, indicating detectable but moderate cytotoxicity. In contrast, almost complete loss of cell viability was observed with CD4+ cells treated with 10 (2.0 ± 1.6% viability; Fig. 8A) or 3 μm anthralin (data not shown).
Figure 8.
Effects of LRAs on primary cells. A, effects of LRA treatments (100 ng/ml PMA + 1 μm ionomycin, or 10 μm prostratin, KA, or anthralin) on viability of CD4+ T cells from three uninfected donors after 24 h, as measured by ViaCount viability stain. B, effects of LRAs on total HIV RNA copies per million CD4+ T cells isolated from three cART-suppressed, HIV-infected donors. Empty circles indicate the limit of detection, where no viral RNA was observed. C, effects of LRA treatments on viability of PBMCs from six uninfected donors after 24 h, as measured by ViaCount viability stain. D, effects of LRAs on p24Gag production in total cell lysate and cell culture supernatants of PBMCs from three additional cART-suppressed, HIV-infected donors. For each panel, shapes and/or colors denote individual donors.
We next assessed whether PMA + ionomycin, prostratin, KA, or anthralin at the same concentrations and treatment durations described above could induce viral RNA expression, as measured by quantitative PCR (qPCR), from 106 isolated CD4+ T cells obtained from three HIV-infected donors (cultured in 1 ml of medium; Fig. 8B). qPCR results were normalized to co-amplified 18S housekeeping gene copy number and respective copy number standards to determine absolute copies of HIV per million CD4+ cells (Table S1). Following treatment, PMA + ionomycin increased viral RNA expression in two of three donors compared with baseline expression in cells treated with 0.1% DMSO control (average across three donors of 576 ± 486 viral RNA copies per million CD4+ cells treated with PMA + ionomycin versus 435 ± 311 copies/million cells in DMSO-treated cells; Table S1). Similar results were also observed in CD4+ cells treated with prostratin (average across three donors of 916 ± 765 copies/million cells). Notably, increased viral RNA expression was also observed in two of three donors treated with KA (average 3365 ± 4563 copies/million cells; Fig. 8B and Table S1). These results suggest that KA induces total viral RNA expression in primary CD4+ T cells on par with established LRAs. In contrast, CD4+ cells treated with anthralin resulted in extensive cell death, as observed by microscopy, and viral RNA was not detected (data not shown).To validate whether KA enhances HIV production ex vivo, we also assessed its effects on peripheral blood mononuclear cells (PBMCs). When 3 × 105 PBMCs from six uninfected donors were cultured at 1.5 × 106 cells/ml and assessed for viability following 24-h incubation with PMA + ionomycin, we observed 51.0 ± 24.7% viability relative to PBMCs treated with 0.1% DMSO (Fig. 8C). However, cells treated with 10 μm KA resulted in 38.3 ± 20.8% viability, indicating elevated but comparable toxicity when compared with control PMA + ionomycin in whole PBMC cultures. In contrast, nearly complete loss of cell viability was again observed with PBMCs treated with 10 μm (2.8 ± 2.6% viability; Fig. 8C) or 3 μm anthralin (data not shown).
We next treated 3 × 105 PBMCs from three additional HIV-infected donors (separate from those in Fig. 8B) with PMA + ionomycin or KA at 1.5 × 106 cell/ml for 24 h, as described above, and quantified combined lysate and supernatant p24Gag protein using a commercially available ELISA kit (Fig. 8D) (15). Data were then normalized to internal kit standards to calculate total p24Gag protein. Per donor, PMA + ionomycin induced an average 1.6 ± 0.5-fold increase in p24Gag relative to PBMCs treated with 0.1% DMSO, with two of three donors exhibiting at least 1.8-fold increases. Across all donors, this resulted in a combined average of 6.0 ± 1.1 pg/ml p24Gag protein in PMA + ionomycin–treated PBMCs versus 5.0 ± 2.5 pg/ml in DMSO-treated PBMCs (Fig. 8D). In contrast, KA induced an average 1.5 ± 0.5-fold increase per donor, with 1 donor inducing a 2.1-fold increase in supernatant p24Gag, and an average of 5.9 ± 2.7 pg/ml detected in PBMCs across all donors. These results further support KA as having ex vivo efficacy. Conversely, extensive cell death, as visualized microscopically, and no p24Gag protein were detected from PBMCs treated with 10 or 3 μm anthralin (data not shown), further supporting poor ex vivo activity of this LRA. Taken together, these results suggest that KA, but not anthralin, enhances the production of both viral RNA and protein from HIV-positive CD4+ T cells and PBMCs, respectively.
Discussion
New LRAs and LRA combinations are needed to improve existing “shock-and-kill” HIV therapeutic approaches (9, 35). Here, we screened 216 compounds from pANAPL and 18 chemical analogues to identify KA, anthralin, and prinoidin as novel LRAs, which continues to suggest that chemical libraries of pure compounds from natural products can be rich sources of LRAs (15–17). Although both KA and anthralin, due to their toxicities, are unlikely to be used as LRAs in humans, their distinct activities in vitro make them useful probes to further understand the cellular mechanisms of HIV latency and latency reversal. This in turn can aid in the development of future LRAs with reduced toxicities, in addition to LRA combinations with improved efficacies and fewer side effects in clinical studies.
KA is a phenylanthraquinone that was originally isolated from Ethiopian Kniphofia foliosa and initially reported to display antiprotozoal activity in vitro (36, 37). KA is also reported to possess both pro- and antioxidant activities, depending on the reaction partners and culture or solvent conditions (27, 37). Since its initial discovery, several phenylanthraquinones have been isolated from plants of South Africa, Botswana, Lesotho, Germany, Australia, and Japan (38). In contrast, anthralin (dithranol) is a KA-like molecule and a licensed topical therapy for psoriasis, dermatitis, and eczema, where mechanisms of action include the induction of excessive oxidative stress in targeted cells (28, 29) in addition to induction of antiproliferative and proapoptotic signaling pathways (39). Whereas KA was recently reported by us to inhibit HIV-1 replication in PBMCs infected in vitro, with an EC50 of 4.3 μm (40), to our knowledge, this is the first report of anthrones affecting HIV latency.
We observed that both KA and anthralin reversed HIV latency across multiple cell line models with dose dependence, indicating that their activities were independent of the viral integration site, at least in lymphoid-derived cell lines. When assessed in vitro, both KA and anthralin also induced cellular apoptosis at levels approximating those of the control HDAC inhibitor panobinostat. Both KA and anthralin, like prostratin, also induced expression of T cell activation markers CD69 and CD25, although KA induced exceptionally strong (i.e. up to 22-fold) up-regulation of CD69 but not CD25, whereas anthralin induced weak CD69 and CD25 expression. Thus, despite the observed toxicity and proactivation properties of KA and anthralin, their distinct profiles, when compared with prostratin and panobinostat, suggested that their effects on T cells may differ from these control PKC activator and HDAC inhibitor functional classes of LRAs. In support of this, our results using both a pharmacological pan-inhibitor of PKC signaling and a cell-based HDAC activity assay indicate that KA and anthralin are neither PKC activators nor HDAC inhibitors.
Based on the known cellular mechanisms of KA and anthralin (27–29), we initially hypothesized that both would reverse HIV latency by promoting “oxidative stress” or enhanced redox traffic, namely the formation of redox-reactive radicals derived from oxygen and from the anthrones themselves. The effects of reactive oxygen species on HIV latency reversal have been described extensively (41), and agents that intensify and uncouple cellular redox processes have been reported as LRAs or inducers of latency reversal in vitro and ex vivo (16, 42). Redox-reactive species like superoxide induce T cell activation but also promote cell death, whereas factors that mitigate redox-reactive species production tend to favor the maintenance of HIV latency (41). In support of this model, a scavenger of oxidizing species, GSH, inhibited in vitro latency reversal induced by anthralin, consistent with the known reactivity of anthralin in promoting oxidative stress. Contrary to our initial hypothesis, however, latency reversal by KA was not inhibited by GSH, indicating that the latency reversal properties of KA cannot solely rest on the anthrone moiety it has in common with anthralin, but that one or more distinct mechanisms of action appear to operate.
The latency reversal effects of both KA and anthralin, in addition to panobinostat, were further inhibited by the copper-sequestering agent bathocuproine. Anthrones are reported to show selectivity toward copper ions and are more easily oxidized in the presence of copper (43). This corroborates that the oxidation reaction could be responsible for driving latency reversal (27, 40). In line with this, latency reversal by KA and anthralin were enhanced by the iron chelator deferoxamine, indicating that KA and anthralin maintain latency reversal following iron sequestration and that iron inhibits them. Superoxide also leads to oxidative dimerization of anthrone derivatives, and physiologically, superoxide is reduced by copper- or iron-containing enzymes (44). Taken together, latency reversal by KA and anthralin appears to require a specific oxidation pathway that is connected to superoxide metabolism and promotes cellular activation, and this pathway is tightly modulated by particular metal ion species.
Despite years of research on both KA and anthralin (27–29, 38), direct molecular targets of these compounds are not yet elucidated. Whereas the exact molecular targets of KA and anthralin in the context of HIV latency reversal also require further investigation, analysis of in vitro global gene expression following KA or anthralin treatment indicates distinct expression profiles when compared with control LRAs prostratin or panobinostat. Most notably, KA treatment robustly induced expression of genes related to sirtuin signaling pathways, whereas responses to anthralin treatment were suggestive of up-regulation of cAMP-mediated signaling. To our knowledge, this is the first report linking these agents to these signaling pathways. These leads warrant further validation in cellular models, although studies would likely require derivatives of anthralin and perhaps KA with reduced cytotoxicities.
We further showed that both KA and anthralin synergize with the activities of LRAs representing PKC activators, HDAC inhibitors, cytokines, BET bromodomain inhibitors, and DNA methyltransferase inhibitors. In most cases, this synergism was statistically significant as measured using the Bliss independence model. These results suggest that future compounds with improved preclinical profiles but similar mechanisms of action to KA or anthralin may be capable of enhancing the activities of existing LRAs which currently exhibit suboptimal efficacies in clinical studies (9, 11).
The ability of KA to promote latency reversal was further confirmed ex vivo, as it enhanced production of both viral RNA from CD4+ T cells and p24Gag protein from PBMCs from HIV-infected donors. The moderate toxicity observed with KA also raises the possibility of its use at lower concentrations, where toxicity is less likely, as part of future LRA combination therapies with synergistic effects on latency reversal. The extent to which combination LRA therapies that include KA can be optimized in primary cells also requires further study. In contrast to KA, we were unable to demonstrate ex vivo latency reversal by anthralin, where severe oxidative stress is likely driving cell toxicity even at relatively low concentrations. In contrast to our results, a previous study identified 5-hydroxynaphthalene-1,4-dione as a novel LRA in latently infected, Bcl-2–transduced primary CD4+ cells that also acted through enhanced oxidative stress (16). In this study, 5-hydroxynaphthalene-1,4-dione exhibited an EC50 of 0.5 μm and 50% cytotoxic concentration of 7.7 μm in primary cells, indicating an improved therapeutic range relative to anthralin. Like anthralin, this compound also induced weak expression of T cell activation markers, and proviral effects could be mitigated by co-treatment with antioxidant agents. Thus, further support of this latency reversal mechanism in primary cells and in LRA combinations may be better modeled using anthralin analogues with higher therapeutic indices.
Several aspects of our results warrant further study. Most notably, the limited efficacies and toxicities of KA and especially anthralin in vitro and ex vivo are likely to limit the potential of these agents as future clinical candidates. However, their ability to synergize with numerous LRAs representing different functional classes suggests that chemical derivatives that function like KA or anthralin, but harbor improved preclinical parameters, could eventually contribute to future LRA combination strategies that maximize LRA efficacy while minimizing off-target toxicities in humans.
Experimental procedures
Cells and reagents
Jurkat T cells (clone E6-1) were obtained from the American Tissue Culture Collection (ATCC). J-Lat T cells (clones 8.4, 9.2, and 10.6) and OM-10.1 cells were obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID, National Institutes of Health (contributed by Drs. Eric Verdin and Salvatore Butera, respectively) (20, 45). Cells were cultured in R10+ medium (RPMI 1640 with HEPES and l-glutamine, 10% fetal bovine serum (FBS), 100 units/ml of penicillin, and 100 µg/ml streptomycin (Sigma)).
PBMCs were collected from three uninfected donors in addition to three HIV-infected donors on stably suppressive combination antiretroviral therapy for at least 3 years. Study protocols were approved by the institutional review boards of Simon Fraser University and the University of British Columbia–Providence Health Care Research Institute (REB: H15-03077 (approved March 8, 2016) and H16-02474 (approved November 15, 2016)) and abide by the Declaration of Helsinki principles. CD4+ T cells were also obtained from an additional three HIV-infected donors on stably suppressive combination antiretroviral therapy (<50 copies/ml of plasma viral load) for 3 years. These study participants were recruited in accordance with the human subject research guidelines of the United States Department of Health and Human Services under the supervision of the Wistar and Philadelphia FIGHT institutional review boards. Written informed consent was obtained from all study participants.
KA was obtained from pANAPL chemical stocks and as described previously (40, 46, 47). Anthralin, prostratin, panobinostat, Aza-CdR, GÖ-6983, deferoxamine, GSH, and bathocuproine were commercially obtained from Sigma. (+)-JQ1 was obtained from Cayman Chemical Co. (Ann Arbor, MI). Annexin V-APC and HIV p24Gag antibody KC57-RD1 were purchased from BioLegend (San Diego, CA). CD25-FITC and CD69-phycoerythrin were purchased from BD Biosciences (Mississauga, Ontario, Canada).
In vitro latency reversal assays
J-Lat and OM-10.1 cells were resuspended in fresh R10+ to a concentration of 106 cells/ml, and 2 × 105 cells were aliquoted into 96-well plates alongside test agents at defined concentrations or 0.1% DMSO vehicle control and incubated for 24 h. For each experiment, all conditions were performed in duplicate. Following incubation, for experiments using OM-10.1 cells and select experiments with J-Lat cells, p24Gag viral antigen was detected by staining cells with anti-p24Gag antibody and using the Cytofix/Cytoperm fixation/permeabilization kit (BD Biosciences) according to the manufacturer's instructions prior to flow cytometric analysis. 5000 live cells (as estimated from cells displaying the characteristic forward- and side-scatter parameters of cells treated with 0.1% DMSO) (19) from each cell culture were collected for detection of GFP and/or p24Gag expression by flow cytometry (Guava EasyCyte 8HT, EMD Millipore).
Detection of cell viability and T cell activation markers
To detect in vitro cell viability directly, Jurkat cells were treated with test agents at defined concentrations or 0.1% DMSO vehicle control in duplicate for 24 h and stained with annexin V-APC according to the manufacturer's instructions (BioLegend). To detect markers of cell activation, Jurkat T cells were stained with CD25-FITC or CD69-phycoerythrin antibodies using the Cytofix/Cytoperm fixation/permeabilization kit (BD Biosciences) according to the manufacturer's instructions. Flow cytometry was then performed as described above. Viability in uninfected PBMCs was measured in the presence of test agents for 24 h using Guava ViaCount reagent (Millipore) and flow cytometry as described previously (34).
In vitro HDAC activity
Cellular HDAC activity was measured in the presence of test agents using the HDAC-Glo I/II assay kit (Promega) according to the manufacturer's instructions. Briefly, Jurkat cells were resuspended in phenol red and FBS-free RPMI 1640 at 3.0 × 105 cells/ml, and 10-μl cell cultures were aliquoted into white 384-well plates in the presence of test agents or 0.1% DMSO diluted in 10 μl of HDAC-Glo Buffer. Following incubation at 37 °C for 90 min, 20 μl of HDAC-Glo I/II Reagent plus 1% Triton X-100 was added to each well, mixed, and incubated at room temperature for 30 min. Luminescence was detected using an Infinity M200 multimode plate reader (Tecan Life Sciences). Wells containing only medium were processed in parallel to control for signal background. For each experiment, four replicates of each condition were performed.
RNA-Seq and data analysis
RNA-Seq and data analysis were performed as described previously (48). RNA was extracted from cells using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) with on-column DNase treatment (Qiagen RNase-free DNase set). 100 ng of DNase-treated total RNA was then used for library preparation using the Quant-Seq 3′ mRNA-Seq Library Preparation Kit (Lexogen, Vienna, Austria). Library quantity was determined by qPCR (KAPA Biosystems, Inc., Wilmington, MA), and library size was determined using the Agilent TapeStation and DNA High Sensitivity D5000 ScreenTape (Agilent, Santa Clara, CA). Libraries were pooled in equimolar amounts and denatured, and high-output, single-read, 75-base pair sequencing was performed using a NextSeq 500 (Illumina, San Diego, CA).
RNA-Seq data were aligned against the human genome (version hg19) using STAR (49). Raw read counts were estimated using RSEM version 1.2.12 software (50) with Ensemble transcriptome information version GRCh37.13. Raw counts were normalized and tested by DESeq2 (51) to estimate significance of differential expression, where genes that passed the FDR < 5% threshold were considered significant. Gene set enrichment analysis of significant genes was performed using Ingenuity Pathway Analysis software (Qiagen) using the “canonical pathways” category. Nominal p values were adjusted for multiple testing using the Benjamini–Hochberg procedure to estimate the FDR (52). Pathways enriched at FDR < 5% and with a predicted activation |Z-score| > 2 in at least one treatment were reported. Predicted activation Z-score was calculated by Ingenuity Pathway Analysis software based on the direction of gene expression changes and known effect on pathway activity.
Measures of HIV-1 latency reversal and viability in primary CD4+ T cells
CD4+ T cells were isolated from the PBMCs of three HIV-infected antiretroviral therapy–suppressed individuals using the EasySepTM Human CD4+ T Cell Enrichment Kit (STEMCELL) and allowed to recover in RPMI plus 20% FBS at 37 °C for 24 h. For each donor, 106 CD4+ T cells were then cultured in 1 ml of RPMI plus 20% FBS plus test agent and 100 units/ml IL-2 (Sigma–Aldrich) at defined concentrations for an additional 24 h. Total RNA was then extracted using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) with on-column DNase treatment. Cell-associated total elongated HIV-1 RNA was then quantified with a qPCR TaqMan assay using long terminal repeat–specific and control PCR primers and conditions described previously (53). Nucleic acid input from 5 μl of isolated total RNA was normalized for cell number by comparing the 18S housekeeping gene copy number with co-amplified copy number standards. Viral RNA per million cells was then determined by comparing results with co-amplified HIV copy number standards. Viability of CD4+ T cells from three donors without HIV was measured using the culture conditions described above and ViaCount dye (Millipore) according to the manufacturer's instructions.
Measures of HIV-1 latency reversal and viability in PBMCs
3 × 105 PBMCs were incubated in 200 μl of R10+ plus for 24 h at 37 °C before the addition of test agents at defined concentrations or 0.1% DMSO in duplicate and further incubation for an additional 24 h. Total cell lysates and supernatants were then measured for detection of p24Gag protein by ELISA (Xpress Bio, Frederick, MD) according to the manufacturer's instructions. Results were normalized to kit p24Gag protein standards to calculate total p24Gag per sample (pg/ml). Viability of PBMCs from six donors without HIV was measured using the culture conditions described above and ViaCount dye (Millipore) according to the manufacturer's instructions.
Cellular data analysis
Flow cytometry data were analyzed using FlowJo version 10.5.3 software (FlowJo LLC, Ashland, OR). For studies using flow cytometry, background GFP signals in J-Lat cells treated with 0.1% DMSO were set to an average of 0.05–0.15% positive cells, whereas background GFP signals in CEM-GXR cells treated with 0.1% DMSO were set to an average of 1.0%. For flow cytometry experiments measuring CD25/CD69 and p24Gag, background fluorescence signals were set to an average of 1.0%. Synergism from LRA combinations was calculated using the Bliss independence model as described previously (12, 15). Here, synergy was defined by the equation,
| (Eq. 1) |
where faxy,P represents the predicted fractional response due to drugs x + y assuming strictly additive effects, given the observed fractional responses of drug x (fax) and drug y (fay). The experimentally observed fractional response due to drugs x + y (faxy,O) was then compared with the predicted fractional response.
| (Eq. 2) |
For a given drug combination, a Δfaxy > 0 indicates a fractional response greater than what is expected for additive effects. The statistical significance of this difference (i.e. faxy,O versus faxy,P) was determined using Student's paired (for cell lines) or unpaired (for PBMCs) t test, where a two-sided p value of 0.05 was considered significant.
All data are reported as mean ± S.D. from at least three independent experiments. For in vitro drug combination/synergy studies, all data are reported as mean ± S.D. from at least four independent experiments.
Data availability
All data are contained within the article and supporting material with the exception of RNA-Seq data, which were generated at the Wistar Institute and are available from the corresponding author upon request (I.T.; itietjen@wistar.org).
Supplementary Material
Acknowledgments
We are indebted to the study participants for provision of primary cell samples.
This article contains supporting information.
Author contributions—K. R., C. S., L. B. G., T. K., A. V. K., and I. T. formal analysis; K. R., C. S., L. B. G., J. R.-O., S. R., T. K., N. N. K., A. S., R. F., S. W., M. H., K. M., A. V. K., K. A.-M., and I. T. investigation; K. R., L. B. G., J. R.-O., S. R., T. K., K. M., A. V. K., M. A.-M., and I. T. methodology; K. R., P. I., and I. T. writing-original draft; L. B. G. and J. R.-O. validation; L. B. G., J. R.-O., Z. L. B., M. A. B., A. V. K., M. A.-M., K. A.-M., L. J. M., and I. T. writing-review and editing; P. I., Z. L. B., M. A. B., A. V. K., K. A.-M., and I. T. conceptualization; P. I., Z. L. B., M. A. B., A. V. K., M. A.-M., K. A.-M., L. J. M., and I. T. supervision; P. I., Z. L. B., M. A. B., A. V. K., K. A.-M., and I. T. project administration; Z. L. B., M. A. B., M. A.-M., K. A.-M., L. J. M., and I. T. funding acquisition; K. M. data curation; A. V. K. software.
Funding and additional information—This work was supported by Canadian Institutes for Health Research Grants CIHR PJT-153057 (to I. T., M. A. B., and Z. L. B.) and CIHR PJT-159625 (to Z. L. B.), the Canadian Foundation for AIDS Research (CANFAR) (to I. T., M. A. B., and Z. L. B.), New Frontiers in Research Fund – Explorations Grant NRFRE-2018-01386 (to I. T.), the German Federal Ministry for Education and Research (BmBF), and the German Academic Exchange Service (DAAD) through the PhytoSustain/Trisustain project (to R. F., S. W., P. I., and K. A.-M.). This work was also supported through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE) DELTAS Africa Initiative Grant DEL-15-006 (to K. R., K. A.-M., and I. T.). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)'s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from Wellcome Trust Grant 107752/Z/15/Z and the UK government. M. A.-M. is supported by National Institutes of Health Grants R01 DK123733, R01 AG062383, R01 NS117458, R21 AI143385, R21 AI129636, and R21 NS106970 and Penn Center for AIDS Research Grant P30 AI 045008. This work was also supported by the following grants to Luis J. Montaner: Beyond Antiretroviral Treatment (BEAT)-HIV Delaney Collaboratory Grant UM1AI126620, co-funded by NIAID, NIMH, NINDS, and NIDA, National Institutes of Health. This work was also supported by Merck, Inc; the Philadelphia Field Initiating Group for HIV-1 Trials (Philadelphia FIGHT); the CLAWS Foundation; the Philadelphia Foundation (Robert I. Jacobs Fund); Ken Nimblett and the Summerhill Trust; Penn Center for AIDS Research Grant P30 AI 045008; and the Herbert Kean, M.D., Family Professorship. K. R. was a recipient of a Canadian Queen Elizabeth II Diamond Jubilee Scholarship, a partnership between the Rideau Hall Foundation, Community Foundations of Canada, and Universities Canada, in addition to a SANTHE Ph.D. fellowship. C. S. and N. N. K. were supported by CIHR Frederick Banting and Charles Best M.Sc. Awards, and N. N. K. now holds a Vanier Doctoral award. M. A. B. holds a Tier 2 Canada Research Chair in viral pathogenesis and immunity. Z. L. B. is supported by a Scholar Award from the Michael Smith Foundation for Health Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- cART
- combination antiretroviral therapy
- Aza-CdR
- 5-aza-2′-deoxycytidine
- FBS
- fetal bovine serum
- FDR
- false discovery rate
- HDAC
- histone deacetylase
- KA
- knipholone anthrone
- LRA
- latency reversal agent
- pANAPL
- pan-African Natural Product Library
- PBMC
- peripheral blood mononuclear cells
- PKC
- protein kinase C
- PMA
- phorbol myristate acetate
- qPCR
- quantitative PCR.
References
- 1. Ghosn J., Taiwo B., Seedat S., Autran B., and Katlama C. (2018) HIV. Lancet 392, 685–697 10.1016/S0140-6736(18)31311-4 [DOI] [PubMed] [Google Scholar]
- 2. Finzi D., Hermankova M., Pierson T., Carruth L. M., Buck C., Chaisson R. E., Quinn T. C., Chadwick K., Margolick J., Brookmeyer R., Gallant J., Markowitz M., Ho D. D., Richman D. D., and Siliciano R. F. (1997) Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 10.1126/science.278.5341.1295 [DOI] [PubMed] [Google Scholar]
- 3. Siliciano J., Kajdas J., Finzi D., Quinn T. C., Chadwick K., Margolick J. B., Kovacs C., Gange S. J., and Siliciano R. F. (2003) Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728 10.1038/nm880 [DOI] [PubMed] [Google Scholar]
- 4. Strain M. C., Günthard H. F., Havlir D. V., Ignacio C. C., Smith D. M., Leigh-Brown A. J., Macaranas T. R., Lam R. Y., Daly O. A., Fischer M., Opravil M., Levine H., Bacheler L., Spina C. A., Richman D. D., et al. (2003) Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. U.S.A. 100, 4819–4824 10.1073/pnas.0736332100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wong J. K., Hezareh M., Günthard H. F., Havlir D. V., Ignacio C. C., Spina C. A., and Richman D. D. (1997) Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 10.1126/science.278.5341.1291 [DOI] [PubMed] [Google Scholar]
- 6. Sadowski I., and Hashemi F. B. (2019) Strategies to eradicate HIV from infected patients: elimination of latent provirus reservoirs. Cell. Mol. Life Sci. 76, 3583–3600 10.1007/s00018-019-03156-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deeks S. G. (2012) HIV: shock and kill. Nature 487, 439–440 10.1038/487439a [DOI] [PubMed] [Google Scholar]
- 8. Bullen C. K., Laird G. M., Durand C. M., Siliciano J. D., and Siliciano R. F. (2014) Novel ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429 10.1038/nm.3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abner E., and Jordan A. (2019) HIV “shock and kill” therapy: in need of revision. Antiviral Res. 166, 19–34 10.1016/j.antiviral.2019.03.008 [DOI] [PubMed] [Google Scholar]
- 10. Hashemi P., and Sadowski I. (2019) Diversity of small molecule HIV-1 latency reversing agents identified in low- and high-throughput small molecule screens. Med. Res. Rev. 40, 881–908 10.1002/med.21638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zerbato J. M., Purves H. V., Lewin S. R., and Rasmussen T. A. (2019) Between a shock and a hard place: challenges and developments in HIV latency reversal. Curr. Opin. Virol. 38, 1–9 10.1016/j.coviro.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laird G. M., Bullen C. K., Rosenbloom D. I. S., Martin A. R., Hill A. L., Durand C. M., Siliciano J. D., and Siliciano R. F. (2015) Ex vivo analysis identifies effective HIV-1 latency–reversing drug combinations. J. Clin. Invest. 125, 1901–1912 10.1172/JCI80142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Darcis G., Kula A., Bouchat S., Fujinaga K., Corazza F., Ait-Ammar A., Delacourt N., Melard A., Kabeya K., Vanhulle C., Van Driessche B., Gatot J. S., Cherrier T., Pianowski L. F., Gama L., et al. (2015) An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+JQ1 and ingenol-B+JQ1 to potently reactivate viral gene expression. PLoS Pathog. 11, e1005063 10.1371/journal.ppat.1005063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang G., Mendes E. A., Kaiser P., Wong D. P., Tang Y., Cai I., Fenton A., Melcher G. P., Hildreth J. E., Thompson G. R., Wong J. K., and Dandekar S. (2015) Synergistic reactivation of latent HIV expression by ingenol-3-Angelate, PEP005, targeted NF-κB signaling in combination with JQ1 induced p-TEFb activation. PLoS Pathog. 11, e1005066 10.1371/journal.ppat.1005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richard K., Williams D. E., de Silva E. D., Brockman M. A., Brumme Z. L., Andersen R. J., and Tietjen I. (2018) Identification of novel HIV-1 latency-reversing agents from a library of marine natural products. Viruses 10, 348 10.3390/v10070348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang H.-C., Xing S., Shan L., O'Connell K., Dinoso J., Shen A., Zhou Y., Shrum C. K., Han Y., Liu J. O., Zhang H., Margolick J. B., and Siliciano R. F. (2009) Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 119, 3473–3485 10.1172/JCI39199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doyon G., Sobolewski M. D., Huber K., McMahon D., Mellors J. W., and Sluis-Cremer N. (2014) Discovery of a small molecule agonist of phosphatidylinositol 3-kinase p110 that reactivates latent HIV-1. PLoS ONE. 9, e84964 10.1371/journal.pone.0084964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ntie-Kang F., Amoa Onguéné P., Fotso G. W., Andrae-Marobela K., Bezabih M., Ndom J. C., Ngadjui B. T., Ogundaini A. O., Abegaz B. M., and Meva'a L. M. (2014) Virtualizing the p-ANAPL library: a step towards drug discovery from African medicinal plants. PLoS ONE 9, e90655 10.1371/journal.pone.0090655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tietjen I., Ntie-Kang F., Mwimanzi P., Onguéné P. A., Scull M. A., Idowu T. O., Ogundaini A. O., Meva'a L. M., Abegaz B. M., Rice C. M., Andrae-Marobela K., Brockman M. A., Brumme Z. L., and Fedida D. (2015) Screening of the pan-African Natural Product Library identifies ixoratannin A-2 and boldine as novel HIV-1 inhibitors. PLoS ONE 10, e0121099 10.1371/journal.pone.0121099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jordan A., Bisgrove D., and Verdin E. (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22, 1868–1877 10.1093/emboj/cdg188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashemi P., Barreto K., Bernhard W., Lomness A., Honson N., Pfeifer T. A., Harrigan P. R., and Sadowski I. (2018) Compounds producing an effective combinatorial regimen for disruption of HIV-1 latency. EMBO Mol. Med. 10, 160–174 10.15252/emmm.201708193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cummins N. W., Sainski-Nguyen A. M., Natesampillai S., Aboulnasr F., Kaufmann S., and Badley A. D. (2017) Maintenance of the HIV reservoir is antagonized by selective BCL2 inhibition. J. Virol. 91, e00012–17 10.1128/JVI.00012-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williams S. A., Chen L. F., Kwon H., Fenard D., Bisgrove D., Verdin E., and Greene W. C. (2004) Prostratin antagonizes HIV latency by activating NF-κB. J. Biol. Chem. 279, 42008–42017 10.1074/jbc.M402124200 [DOI] [PubMed] [Google Scholar]
- 24. Korin Y. D., Brooks D. G., Brown S., Korotzer A., and Zack J. A. (2002) Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 76, 8118–8123 10.1128/jvi.76.16.8118-8123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gustafson K. R., Cardellina J. H. 2nd, McMahon J. B., Gulakowski R. J., Ishitoya J., Szallasi Z., Lewin N. E., Blumberg P. M., Weislow O. S., Beutler J. A., Buckheit R. W. Jr., Cragg G. M., Cox P. A., Bader J. P., and Boyd M. R. (1992) A nonpromoting phorbol from the samoan medicinal plant Homalanthus nutans inhibits cell killing by HIV-1. J. Med. Chem 35, 1978–1986 10.1021/jm00089a006 [DOI] [PubMed] [Google Scholar]
- 26. Tietjen I., Ngwenya B. N., Fotso G., Williams D. E., Simonambango S., Ngadjui B. T., Andersen R. J., Brockman M. A., Brumme Z. L., and Andrae-Marobela K. (2018) The Croton megalobotrys Müll Arg. traditional medicine in HIV/AIDS management: documentation of patient use, in vitro activation of latent HIV-1 provirus, and isolation of active phorbol esters. J. Ethnopharmacol. 211, 267–277 10.1016/j.jep.2017.09.038 [DOI] [PubMed] [Google Scholar]
- 27. Habtemariam S., and Dagne E. (2009) Prooxidant action of knipholone anthrone: copper dependent reactive oxygen species generation and DNA damage. Food Chem. Toxicol. 47, 1490–1494 10.1016/j.fct.2009.03.032 [DOI] [PubMed] [Google Scholar]
- 28. Müller K. (1996) Antipsoriatic anthrones: aspects of oxygen radical formation, challenges and prospects. Gen. Pharmacol. 27, 1325–1335 10.1016/S0306-3623(96)00075-4 [DOI] [PubMed] [Google Scholar]
- 29. Kemény L., Ruzicka T., and Braun-Falco O. (1990) Dithranol: a review of the mechanism of action in the treatment of psoriasis vulgaris. Skin Pharmacol. 3, 1–20 10.1159/000210836 [DOI] [PubMed] [Google Scholar]
- 30. Pinzone M. R., Cacopardo B., Condorelli F., Di Rosa M., and Nunnari G. (2013) Sirtuin-1 and HIV-1: an overview. Curr. Drug Targets 14, 648–652 10.2174/1389450111314060005 [DOI] [PubMed] [Google Scholar]
- 31. Rabbi M. F., Al-Harthi L., and Roebuck K. A. (1997) TNFα cooperates with the protein kinase A pathway to synergistically increase HIV-1 LTR transcription via downstream TRE-like cAMP response elements. Virology 237, 422–429 10.1006/viro.1997.8798 [DOI] [PubMed] [Google Scholar]
- 32. Banerjee A., Li L., Pirrone V., Krebs F. C., Wigdahl B., and Nonnemacher M. R. (2017) cAMP signaling enhances HIV-1 long terminal repeat (LTR)-directed transcription and viral replication in bone marrow progenitor cells. Clin. Med. Insights Pathol. 10, 1179555717694535 10.1177/1179555717694535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kauder S. E., Bosque A., Lindqvist A., Planelles V., and Verdin E. (2009) Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5, e1000495 10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mwimanzi P., Tietjen I., Miller S. C., Shahid A., Cobarrubias K., Kinloch N. N., Baraki B., Richard J., Finzi A., Fedida D., Brumme Z. L., and Brockman M. A. (2016) Novel acylguanidine-based inhibitor of HIV-1. J. Virol. 90, 9495–9508 10.1128/JVI.01107-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rasmussen T. A., and Lewin S. R. (2016) Shocking HIV out of hiding: where are we with clinical trials of latency reversing agents? Curr. Opin. HIV AIDS 11, 394–401 10.1097/COH.0000000000000279 [DOI] [PubMed] [Google Scholar]
- 36. Bringmann G., Menche D., Bezabih M., Abegaz B. M., and Kaminsky R. (1999) Antiplasmodial activity of knipholone and related natural phenylanthraquinones. Planta Med. 65, 757–758 10.1055/s-2006-960859 [DOI] [PubMed] [Google Scholar]
- 37. Habtemariam S. (2007) Antioxidant activity of Knipholone anthrone. Food Chem. 102, 1042–1047 10.1016/j.foodchem.2006.06.040 [DOI] [Google Scholar]
- 38. Bringmann G., Mutanyatta-Comar J., Knauer M., and Abegaz B. M. (2008) Knipholone and related 4-phenylanthraquinones: structurally, pharmacologically, and biosynthetically remarkable natural products. Nat. Prod. Rep. 25, 696–718 10.1039/b803784c [DOI] [PubMed] [Google Scholar]
- 39. Ronpirin C., and Tencomnao T. (2012) Effects of the antipsoriatic drug dithranol on E2A and caspase-9 gene expression in vitro. Genet. Mol. Res. 11, 412–420 10.4238/2012.February.17.3 [DOI] [PubMed] [Google Scholar]
- 40. Feilcke R., Arnouk G., Raphane B., Richard K., Tietjen I., Andrae-Marobela K., Erdmann F., Schipper S., Becker K., Arnold N., Frolov A., Reiling N., Imming P., and Fobofou S. A. T. (2019) Biological activity and stability analyses of knipholone anthrone, a phenyl anthraquinone derivative isolated from Kniphofia foliosa Hochst. J. Pharm. Biomed. Anal. 174, 277–285 10.1016/j.jpba.2019.05.065 [DOI] [PubMed] [Google Scholar]
- 41. Benhar M., Shytaj I. L., Stamler J. S., and Savarino A. (2016) Dual targeting of the thioredoxin and glutathione systems in cancer and HIV. J. Clin. Invest. 126, 1630–1639 10.1172/JCI85339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Savarino A., Mai A., Norelli S., El Daker S., Valente S., Rotili D., Altucci L., Palamara A. T., and Garaci E. (2009) “Shock and kill” effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology 6, 52 10.1186/1742-4690-6-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mittal S. K., Kaur K., Kumar S. K. A., Kumar S., and Kumar A. (2013) Anthrone derivatives as voltammetric sensors for applications in metal ion detection. Sensor Lett. 11, 223–236 10.1166/sl.2013.2715 [DOI] [Google Scholar]
- 44. D'Ischia M., and Prota G. (1985) Generation and role of superoxide ion in the autoxidation of 1,8-dihydroxy-9-anthrone. Gazzetta Chimica Italiana 115, 511–514 [Google Scholar]
- 45. Butera S. T., Perez V. L., Wu B. Y., Nabel G. J., and Folks T. M. (1991) Oscillation of the human immunodeficiency virus surface receptor is regulated by the state of viral activation in a CD4+ cell model of chronic infection. J. Virol. 65, 4645–4653 10.1128/JVI.65.9.4645-4653.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dagne E., and Steglich W. (1984) Knipholone: a unique anthraquinone derivative from Kniphofia foliosa. Phytochemistry 23, 1729–1731 10.1016/S0031-9422(00)83479-2 [DOI] [Google Scholar]
- 47. Dagne E., and Yenesew A. (1993) Knipholone anthrone from Kniphofia foliosa. Phytochemistry 34, 1440–1441 10.1016/0031-9422(91)80048-6 [DOI] [Google Scholar]
- 48. Giron L. B., Tanes C. E., Schleimann M. H., Engen P. A., Mattei L. M., Anzurez A., Damra M., Zhang H., Bittinger K., Bushman F., Kossenkov A., Denton P. W., Tateno H., Keshavarzian A., Landay A. L., et al. (2020) Sialylation and fucosylation modulate inflammasome-activating eIF2 signaling and microbial translocation during HIV infection. Mucosal Immunol. 13, 753–766 10.1038/s41385-020-0279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Benjamini Y., Drai D., Elmer G., Kafkafi N., and Golani I. (2001) Controlling the false discovery rate in behavior genetics research. Behavioural Brain Res. 125, 279–284 10.1016/S0166-4328(01)00297-2 [DOI] [PubMed] [Google Scholar]
- 53. Abdel-Mohsen M., Chavez L., Tandon R., Chew G. M., Deng X., Danesh A., Keating S., Lanteri M., Samuels M. L., Hoh R., Sacha J. B., Norris P. J., Niki T., Shikuma C. M., Hirashima M., et al. (2016) Human galectin-9 is a potent mediator of HIV transcription and reactivation. PLoS Pathog. 12, e1005677 10.1371/journal.ppat.1005677 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting material with the exception of RNA-Seq data, which were generated at the Wistar Institute and are available from the corresponding author upon request (I.T.; itietjen@wistar.org).