Abstract
The protective effect of transthyretin (TTR) on cellular toxicity of β-amyloid (Aβ) has been previously reported. TTR is a tetrameric carrier of thyroxine in blood and cerebrospinal fluid, the pathogenic aggregation of which causes systemic amyloidosis. However, studies have documented a protective effect of TTR against cellular toxicity of pathogenic Aβ, a protein associated with Alzheimer's disease. TTR binds Aβ, alters its aggregation, and inhibits its toxicity both in vitro and in vivo. In this study, we investigate whether the amyloidogenic ability of TTR and its antiamyloid inhibitory effect are associated. Using protein aggregation and cytotoxicity assays, we found that the dissociation of the TTR tetramer, required for its amyloid pathogenesis, is also necessary to prevent cellular toxicity from Aβ oligomers. These findings suggest that the Aβ-binding site of TTR may be hidden in its tetrameric form. Aided by computational docking and peptide screening, we identified a TTR segment that is capable of altering Aβ aggregation and toxicity, mimicking TTR cellular protection. EM, immune detection analysis, and assessment of aggregation and cytotoxicity revealed that the TTR segment inhibits Aβ oligomer formation and also promotes the formation of nontoxic, nonamyloid amorphous aggregates, which are more sensitive to protease digestion. Finally, this segment also inhibits seeding of Aβ catalyzed by Aβ fibrils extracted from the brain of an Alzheimer's patient. Together, these findings suggest that mimicking the inhibitory effect of TTR with peptide-based therapeutics represents an additional avenue to explore for the treatment of Alzheimer's disease.
Keywords: amyloid, amyloid-beta (AB), Alzheimer's disease, neuroprotection, neuroscience, cytotoxicity, peptide, transthyretin
The physiological importance of transthyretin in Alzheimer's disease was first reported by Schwarzman et al. in 1994 (1). One of the hallmarks of Alzheimer's disease (AD) is the formation of brain plaques composed of β-amyloid peptide (Aβ). A 42-residue-long β-amyloid peptide (Aβ42) is the predominant variant in neuritic plaques of AD patients, with higher amyloidogenicity and cellular toxicity in vitro (2, 3). Many studies have shown that transthyretin (TTR) binds to Aβ, alters its aggregation, and inhibits its toxicity both in vitro and in vivo (4–8). In vitro, TTR co-aggregates with Aβ oligomers into large nontoxic assemblies, thereby inhibiting cellular toxicity (9, 10). In vivo, TTR sequesters Aβ and facilitates its clearance in the brain (1). More recently, the Buxbaum laboratory (6) showed that overexpression of WT human TTR suppressed disease progression in the APP23 transgenic AD mouse model. They also showed that silencing the endogenous TTR gene in AD transgenic mice accelerated Aβ42 deposition (5).
What makes the pair of TTR and Aβ42 particularly interesting is the amyloid nature of the two elements. In health, TTR functions as a transporter of retinol and thyroxine in blood, cerebrospinal fluid, and the eye and is secreted by the liver, choroid plexus, and retinal epithelium, respectively. However, dissociation of tetrameric TTR leads to amyloid fibril formation and systemic TTR amyloid deposition in patients of transthyretin amyloidosis (11–13). Whether the amyloidogenicity of TTR is linked to its interaction to Aβ42 is still under debate. Previous work showed that different aggregation propensities of TTR result in distinct interaction capabilities: fewer amyloidogenic variants had increased affinity for Aβ (7). However, this did not impact on the levels of inhibition of Aβ aggregation, and cytotoxicity protection was not assessed. Here we attempt to fill the experimental gap by studying the protective effect of TTR variants at different aggregated states over Aβ42 cellular toxicity. In addition, we show the identification and characterization of a segment of TTR that binds Aβ42 triggering the formation of nonamyloid amorphous aggregates that are more sensitive to protease digestion, therefore mimicking TTR inhibitory effect over Aβ42.
Results
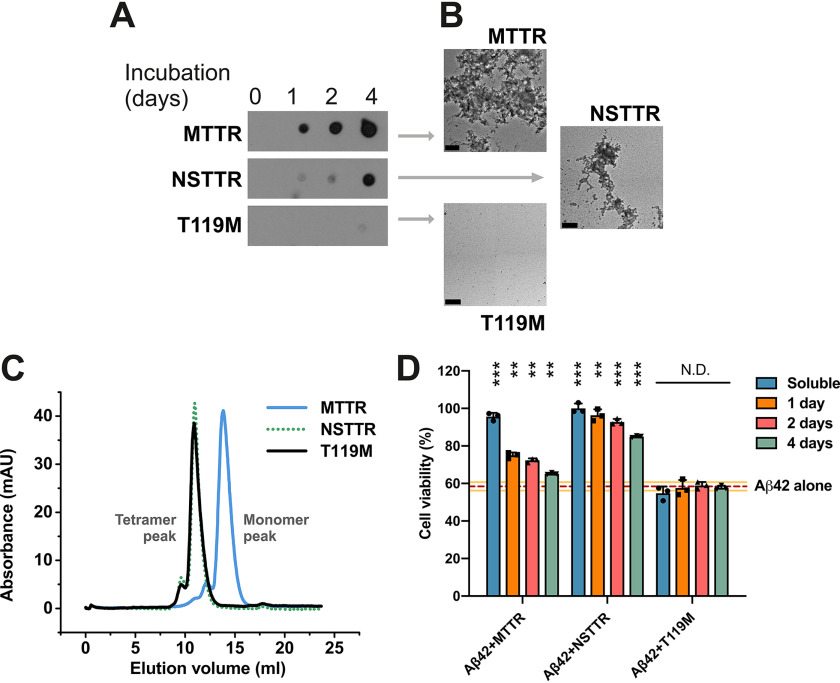
We first evaluated the protection against Aβ42 cytotoxicity by nine TTR variants with distinct aggregation propensities in several aggregated states (Table 1). The amyloidogenic behavior of three representative TTR variants is shown in Fig. 1 (A and B). For this assay, recombinant transthyretin was incubated at 37 °C, pH 4.3; protein aggregation was followed by immunodot blot of the insoluble fractions collected by centrifugation at several time points (Fig. 1A); and EM was performed after 4 days of incubation (Fig. 1B). We found that M-TTR, a monomeric-engineered form of transthyretin that is soluble at physiological pH (14), shows notable aggregation after 1 day of incubation at pH 4.3 (Fig. 1, A–C). NSTTR is an artificially mutated variant of TTR carrying the double mutation N99R/S100R that, although remaining tetrameric in solution (Fig. 1C), shows a slower aggregation pattern than M-TTR (Fig. 1, A and B). T119M is a very stable mutant that did not show any sign of aggregation in 4 days (Fig. 1, A–C). Consistently, T119M results in a significant delay of the onset of hereditary neuropathic TTR amyloidosis in patients who carry both ttr-V30M and ttr-T119M genes (15). We then incubated Aβ42 with the samples obtained from Fig. 1A for 16 h and evaluated its cytotoxicity by following MTT reduction. Three observations were made.
Table 1.
List of TTR variants evaluated in this study
| TTR variant | Mutation | Oligomeric state in solution |
|---|---|---|
| M-TTR | F87M/L110M | Monomer |
| NSTTR | N99R/S100R | Tetramer |
| T119M | T119M | Tetramer |
| WT | None | Tetramer |
| A108R/L110R | A108R/L110R | Tetramer + dimer + monomer |
| M13R/L17R | M13R/L17R | Dimer + monomer |
| S112I | S112I | Dimer |
| L55P | L55P | Tetramer |
| L55P/T119M | L55P/T119M | Tetramer |
Figure 1.
The inhibition of Aβ42 cytotoxicity by TTR depends on the aggregation propensity of each TTR variant. A, aggregation assay of three TTR variants followed by immunodot blot of insoluble fractions collected prior to incubation or after 1, 2, and 4 days of incubation at 37° C, as labeled. All samples were treated equally and onto the same membrane. Cropping was applied for ethetical reasons. B, electron micrographs of aggregated TTR variants after 4 days of incubation. Scale bar, 200 nm. C, size-exclusion chromatography of soluble TTR variants at pH 7.4. D, cytotoxicity assay of Aβ42 in the presence of TTR variants at different stages of aggregation, followed by MTT reduction. A 5-fold molar excess of soluble TTR (50 μm, considering monomeric concentration) and aggregated samples collected after 1, 2 and 4 days of incubation were added to soluble 10 μm Aβ42 and incubated overnight. The samples were added to HeLa cells, and MTT reduction was measured after 24 h. Buffer-treated cells were considered 100% viability and used for normalization (n = 3). All replicates are shown. Error bars, S.D. **, p ≤ 0.005; ***, p ≤ 0.0005. N.D., significance not detected. These results suggest that the dissociation of the tetrameric TTR structure precedes Aβ42 cytotoxicity protection, whereas aggregated TTR does not exert any effect.
The first observation is that once aggregated, TTR does not prevent Aβ42 toxicity, for any of the variants (Fig. 1D). In addition, the variant NSTTR, which aggregates at a slow pace, resulted to be cytoprotective longer than M-TTR (Fig. 1D). The third observation is that T119M, which does not dissociate and aggregate, does not prevent toxicity either (Fig. 1D). We observed the same phenomena when the other six TTR variants were evaluated (Fig. S1). Although variants S112I, M13R/L17R, or A108R/L110R are mainly monomeric or dimeric in solution, the cytotoxic protection at day 0 is not greater than NSTTR or L55P that are mainly tetrameric in solution (Fig. 1C and Fig. S1B). This finding suggests that the capacity to protect from Aβ42 toxicity does not correlate with the initial oligomeric state of soluble variants (Fig. 1C and Fig. S1B) but rather with the dissociation state at the moment of the co-incubation with Aβ42 (Fig. 1, A and B, and Fig. S1C). A control experiment shows that the TTR variants used in this assay were not cytotoxic in the absence of Aβ42, with the exception of M-TTR, which resulted in a 20% reduction of cell viability (Fig. S1D). Overall, our results indicate that dissociation of TTR is required for the inhibition of Aβ cytotoxicity. In addition, the data suggest that a more stable dissociated TTR exerts more Aβ42 inhibition than a more amyloidogenic variant. These findings led us to wonder whether a segment of TTR that is only exposed when dissociated but not in the aggregate might be similarly capable of altering Aβ toxicity.
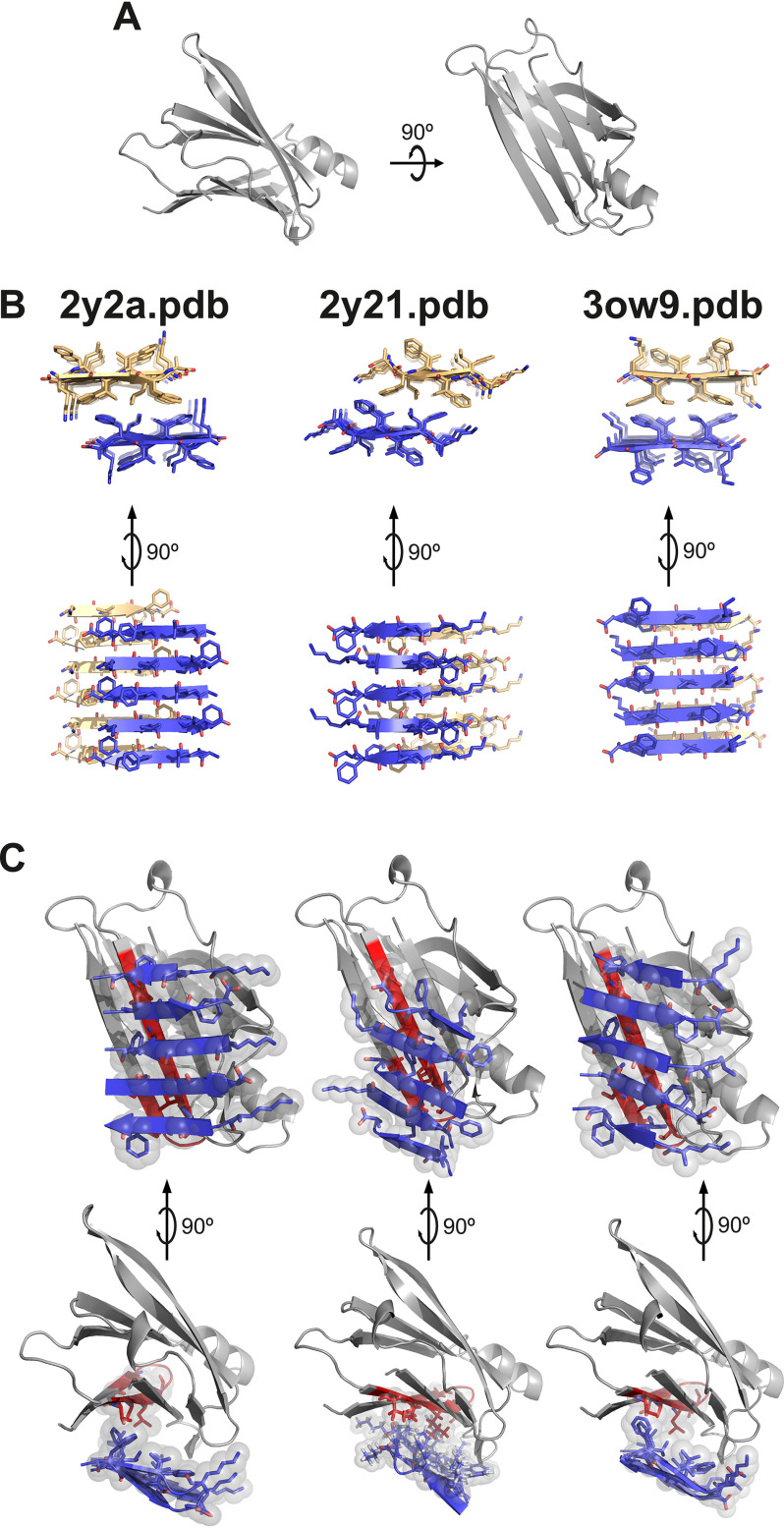
Computational modeling of the interaction of a fibrillar segment of Aβ42 and the TTR monomer shows a tight packing of the thyroxine-binding pocket and the fibrillar structure (Fig. 2). Others have shown that the amyloidogenic segment KLVFFA is protected when TTR is bound to Aβ (16). In previous studies, the Eisenberg laboratory determined the structures of three fibrillar polymorphs of KLVFFA by X-ray microcrystallography (17). We used these three polymorphs, as well as the monomeric form of WT TTR (PDB code 4TLT) (18) to perform protein–protein computational docking (Fig. 2). For every fibrillar form, the segment TTR(105–117) was the longest interacting TTR segment. The segment TTR(105–117) is only exposed in the monomeric or dimeric form of TTR. We wondered whether this segment might mimic TTR protective effects when isolated.
Figure 2.
Protein–protein docking of TTR monomer with three KLVFFA polymorphs. Protein–protein docking was performed using monomeric TTR and three KLVFFA polymorphs identified previously (17): PDB codes 2y2a, 2y21, and 3ow9. A, monomeric TTR obtained from chain A of the model of PDB code 4TLT (18) is shown in gray as a secondary structure. On the left is shown a view down the hydrophobic pocket. On the right is a lateral view. B, fibrillar structures of KLVFFA from PDB codes 2y2a (left panel), 2y21 (middle panel), and 3ow9 (right panel). Only one sheet of each KLVFFA polymer was included in the docking, shown in blue. The resulting docking models, shown in C, suggest that TTR may interact with Aβ42 through residues 105–117. Monomeric TTR is shown in gray with the segment TTR(105–117) in red. Top row, lateral view of interface. Bottom row, view down the binding interface. Residues involved in the interaction between monomeric TTR and KLVFFA are shown as sticks. Spheres represent the van der Waals radii of the side chain atoms of the tightly packed binding interface.
The peptide TTR(105–117) was found to be highly amyloidogenic. We first analyzed the amyloidogenicity of TTR sequence by ZipperDB, which measures the propensity of every six-residue segment to form amyloid fibrils (19, 20) (Fig. S2A). TTR predictions show that the region that contains this segment is highly prone to form amyloid structures. In fact, we found the segment TTR(105–117) and the shorter peptide TTR(106–117) to be amyloidogenic in solution (Fig. S2B), which may explain the lack of inhibitory effect in previous studies (21). In contrast to the amorphous aggregates generated by recombinant full-length TTR, these peptides form amyloid-like fibrils (Fig. S2B). We then explored several sequence modifications to increase solubility, by eliminating the first tyrosine or adding a charged tag to the N-terminal end. The sequences and names of all the analyzed peptides are listed in Table 2. We found that TTR(105–117) amyloidogenicity was fully hindered by the addition of a polyarginine tag (Fig. S2B).
Table 2.
List of peptides evaluated in this study
| Peptide name | Sequence |
|---|---|
| Controls | |
| K5 | KKKKK |
| R5 | RRRRR |
| S12 | TIAALLSPYSYS |
| S13 | YTIAALLSPYSYS |
| Optimizations | |
| K14 | YTIAALLSPYSYSK |
| K16 | YTIAALLSPYSYSKKK |
| K18 | YTIAALLSPYSYSKKKKK |
| R14 | YTIAALLSPYSYSR |
| R16 | YTIAALLSPYSYSRRR |
| K17 | TIAALLSPYSYSKKKKK |
| R17 | TIAALLSPYSYSRRRRR |
| TTR-S | YTIAALLSPYSYSRRRRR |
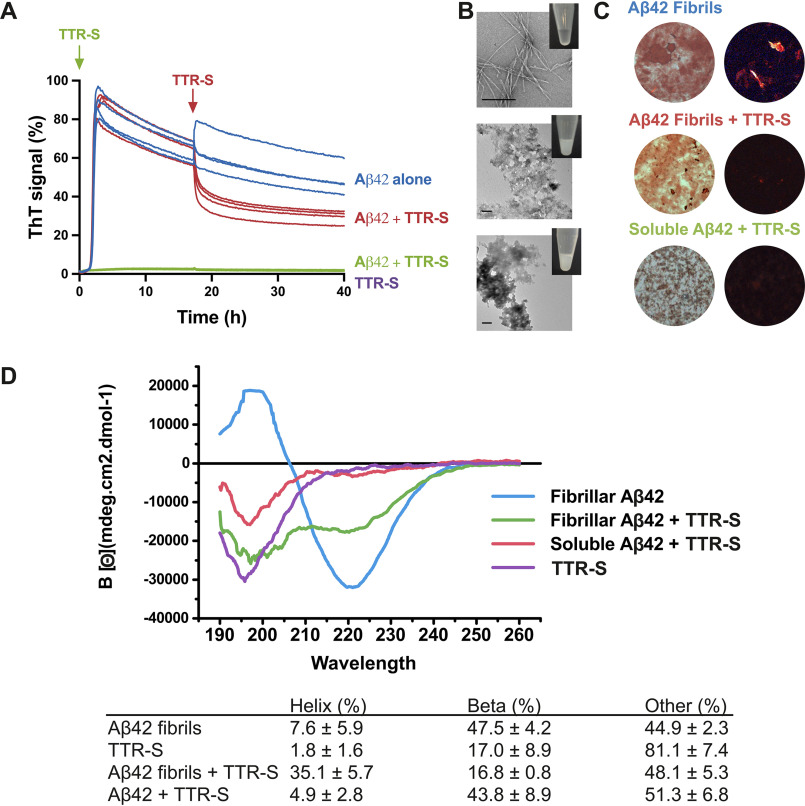
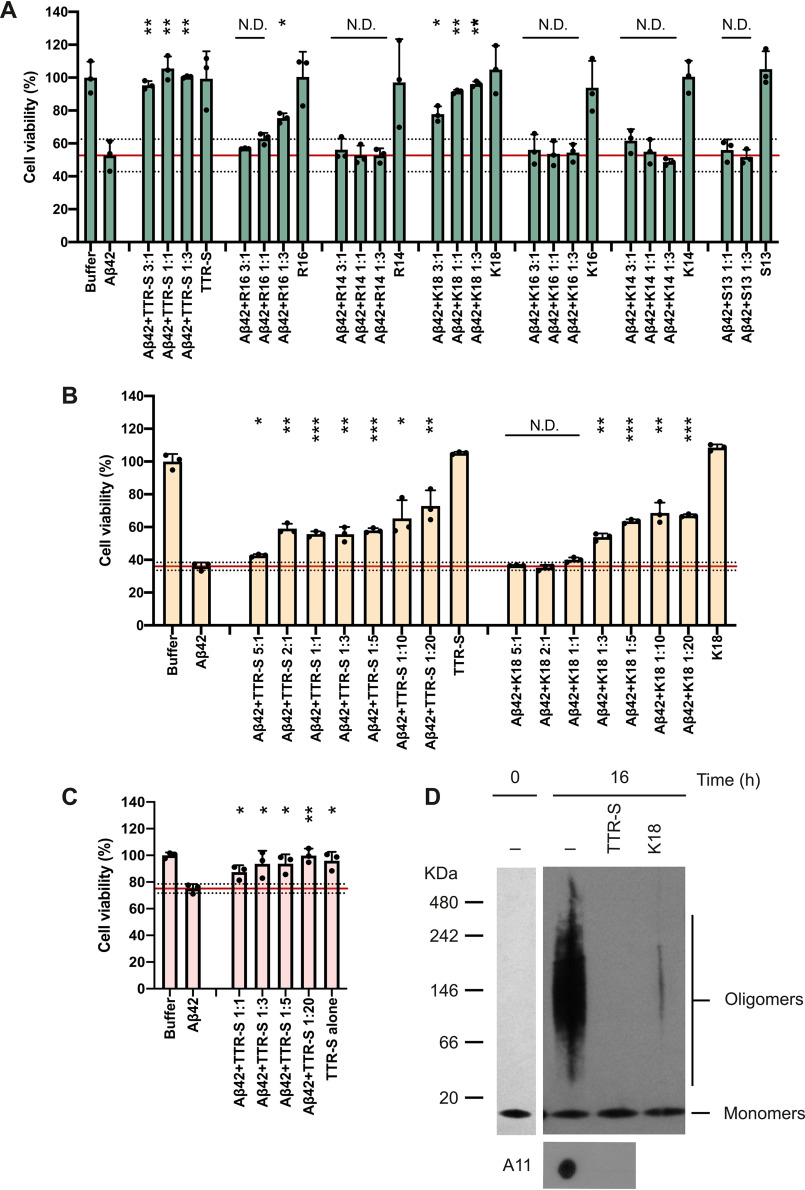
The soluble tagged derivatives of the segment TTR(105–117) inhibited Aβ42 fibril formation (Fig. 3 and Figs. S3 and S4). We first evaluated the inhibitory effect of TTR-derived peptides over Aβ42 fibril formation in thioflavin T (ThT) assays (Fig. 3A and Fig. S3). Of all the peptides that we analyzed, the best Aβ42 inhibitory effect was obtained with TTR-S (sequence YTIAALLSPYSYSRRRRR), which contains a four-arginine tag followed by TTR(105–117) (Fig. S3). After an incubation of 2 days, we analyzed the samples by EM and found that the addition of TTR-S promoted the formation of amorphous aggregates (Fig. 3B) that were not birefringent when stained with Congo red (Fig. 3C). Remarkably, we found that TTR-S also promoted the formation of amorphous species when incubated with preformed Aβ42 fibrils (Fig. 3B). These aggregates were also found to be thioflavin T–negative (Fig. 3A), and nonbirefringent when stained with Congo red (Fig. 3C). The structural characterization of Aβ42 amorphous aggregates by CD showed that the addition of TTR-S to preformed fibrils resulted in a significant structural shift from β to helical secondary motifs (Fig. 3D). Additionally, we found that this structural modification results in cytotoxicity protection (Fig. 4). HeLa, PC12, and SH-5YSY cells were subjected to Aβ42 in the absence and presence of TTR-S at different molar ratios, and cell metabolic activity was followed by MTT reduction (Fig. 4, A–D). We found that the addition of TTR-S results in a significant reduction of Aβ42 cellular toxicity. This protective effect may be explained by the inhibition of the formation of A11-positive oligomers as a result of the incubation with TTR-S (Fig. 4D). It is worth noting that the larger aggregates did not resolve in the SDS-PAGE gel.
Figure 3.
Effect of TTR-S on both soluble and fibrillar Aβ42. A, ThT fluorescence was measured from samples containing 10 μm Aβ42 in the presence and absence of a 3-fold molar excess of TTR-S. TTR-S was added before incubation (green) or after 18 h of incubation (red). TTR-S alone (purple) and buffer (not shown) were used as negative controls (n = 4). All replicates are shown. For a complete assessment of Aβ42 inhibition by TTR-derived peptides, see Fig. S4. B, electron micrographs of Aβ42 fibrils formed in the absence of inhibitor (top panel), Aβ42 amorphous aggregates formed when TTR-S was added after 18 h of incubation (middle panel), and Aβ42 amorphous aggregates formed when TTR-S was added prior to incubation (bottom). Scale bar, 400 nm. Insets, pictures of test tubes containing the same samples shown by EM but 10 times more concentrated. These pictures were taken after 30 min of incubation and reveal obvious precipitation upon addition of TTR-S. C, Congo red staining of samples shown in A under bright field (left panels) and polarized light (right panels). D, CD traces of Aβ42 fibrils, Aβ42 fibrils after addition of TTR-S, and soluble Aβ42 after addition of TTR-S. The table below shows the calculated percentage of various structural conformations: helix, β-stranded, and others (turns and unstructured). These analyses indicate that the TTR-derived peptide TTR-S inhibits Aβ42 aggregation and promotes the formation of nonamyloid amorphous aggregates.
Figure 4.
TTR-S inhibition of Aβ42 cytotoxicity. A–C, cell viability assay followed by MTT reduction in which HeLa (A), PC12 (B), and SH-5YSY (C) cells were treated with 10 μm Aβ42 in the absence and the presence of increasing concentrations of peptides. Molar ratios of Aβ42 and peptides are labeled. As a negative control, cytotoxicity of peptides alone was also measured. Buffer-treated cells were considered 100% viability and used for normalization. Cell toxicity measured in the absence of peptides is marked with a red continuous line (mean) and dotted lines (S.D.; n = 3). All replicates are shown. Error bars, S.D. *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005. N.D., significance not detected. D, nondenaturing electrophoresis followed by immunoblot of Aβ42 before and after 16 h of incubation at 37 °C without and with TTR-S. Two gels were run to analyze Aβ42 before and after incubation and are shown in separate insets. The same samples were analyzed by dot blot with the oligomer specific A11 antibody (bottom panel). These results suggest that the inhibition of Aβ42 cytotoxicity by TTR-S results from the elimination of toxic oligomeric species.
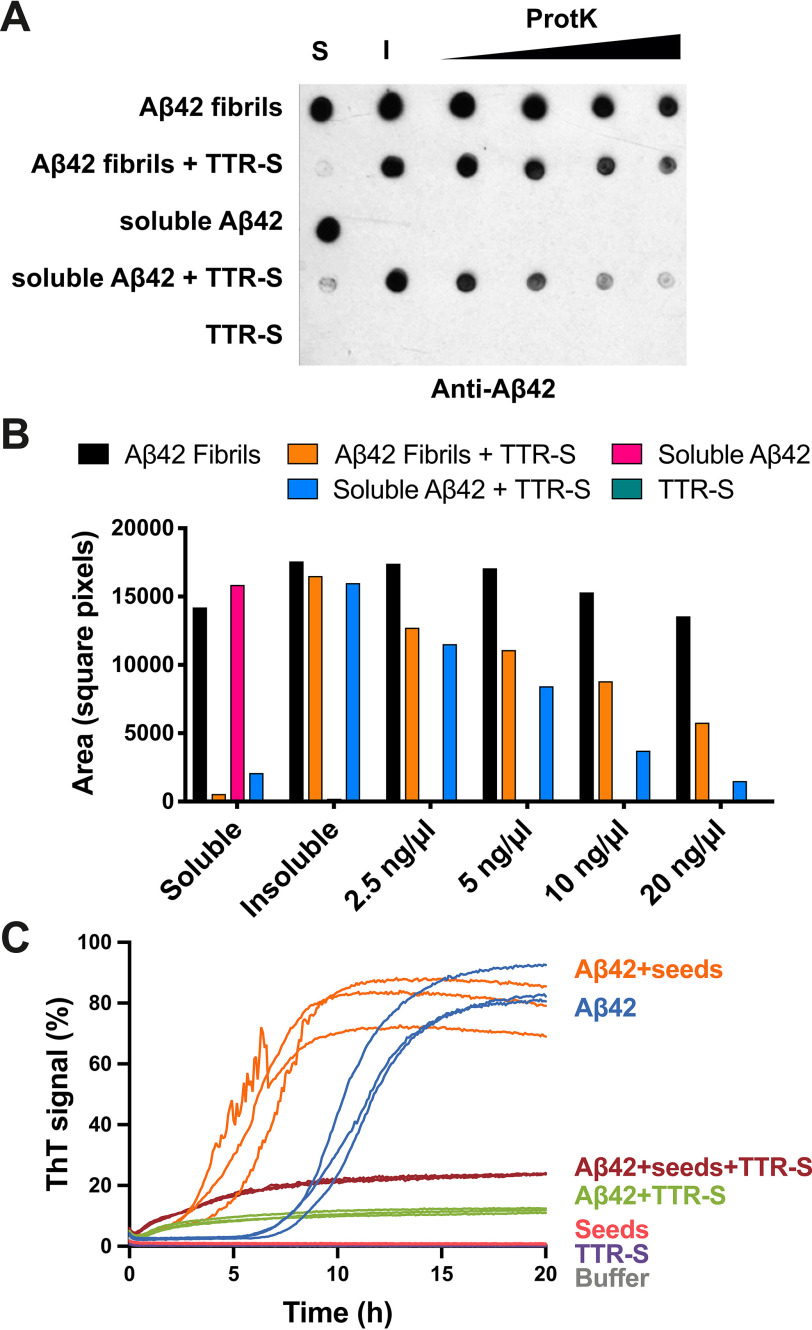
Next we found that the binding of TTR-S to soluble and fibrillar Aβ42 alters their protease resistance. Others have shown that TTR increases Aβ42 clearance in vivo (1). We reasoned that the large assemblies that form upon binding to TTR might be more sensitive to proteolytic activity than amyloid fibrils, thereby facilitating clearance. This hypothesis would also explain the protective effect found in vivo (6). We explored this hypothesis by analyzing proteolytic sensitivity of TTR-S–derived amorphous aggregates (Fig. 5). We incubated soluble and fibrillar Aβ42 with TTR-S overnight and collected the insoluble fractions by centrifugation. The immunodot blot of insoluble fractions showed that amorphous aggregates that result from the incubation of TTR-S with both soluble and fibrillar Aβ42 were easily digested by proteinase K after 1 h (Fig. 5, A and B). In contrast, Aβ42 fibrils showed a significantly higher resistance to proteinase K digestion. We included soluble Aβ42 in the assay as a control of proteolytic activity.
Figure 5.
Evaluation of the inhibitory effect of TTR-S by protease digestion and amyloid seeding. A, anti-Aβ42 immunodot blot of Aβ42 aggregates after proteinase K digestion. Soluble Aβ42 and preformed fibrils were incubated with TTR-S. Half of the sample was subjected to centrifugation, and soluble (S) and insoluble (I) fractions were collected. Increasing concentrations of proteinase K were added to the other half of the sample and incubated at 37° C for 1 h. The samples were analyzed by dot blot using E610 specific anti-Aβ42 antibody. As negative control, we included a sample with TTR-S alone. B, the signal was quantified using ImageJ and plotted as shown. This assay shows that Aβ42 amorphous aggregates generated after the addition of TTR-S are degraded by proteinase K more readily than Aβ42 fibrils. C, amyloid seeding assay followed by ThT signal. Soluble Aβ42 was incubated with AD ex vivo seeds in the presence or absence of TTR-S. Samples with only TTR-S, with only AD seeds, or with buffer were included as negative control. Note that the addition of TTR-S results in inhibition of Aβ42 seeding (n = 3). All replicates are shown.
Finally, we evaluated TTR-S ability to hinder amyloid seeding caused by fibrils extracted from the brain of an AD patient (Fig. 5C). The extraction of Aβ fibrils was performed by several cycles of homogenization and ultracentrifugation as described by Tycko (22). Amyloid seeding was measured by thioflavin T fluorescence. As expected, we found that the addition of sonicated patient-derived fibrils to soluble Aβ42 resulted in the acceleration of fibril formation. In contrast, incubation with TTR-S resulted in inhibition of Aβ42 fibril formation even in the presence of patient-derived Aβ seeds.
Discussion
Our results indicate that the dissociation of the TTR tetramer is required to prevent cytotoxicity from Aβ oligomers. These results are consistent with the model proposed by Buxbaum and co-workers (16), in which dissociated TTR monomers efficiently bind Aβ oligomers, which are thought to be responsible for cellular Aβ-associated toxicity (23). They also found that tetrameric TTR binds to soluble monomeric Aβ, thereby suppressing aggregation in vitro (16). Others have also found an association between genetic stabilization of transthyretin and decreased risk of cerebrovascular disease (24). Consistently, an additional study found that the stabilization of the tetrameric form of TTR promotes Aβ clearance in a mouse model of Alzheimer's disease (25). In our study, we did not find any effect of tetrameric TTR on Aβ42 cytotoxicity. For instance, the tetrameric nonamyloidogenic T119M variant did not inhibit Aβ42 cytotoxicity (Fig. 1). Consistently, the nonamyloidogenic variant L55P/T119M did not inhibit cytotoxicity, whereas the amyloidogenic L55P variant did (Fig. S1). Taken together, other studies and our results suggest that tetrameric TTR may be protective in vivo perhaps not by recruiting toxic oligomers, but by sequestering and promoting clearance of Aβ monomers. In contrast, dissociated TTR seems to inhibit Aβ cytotoxicity by binding to toxic oligomeric species triggering the formation of large nontoxic assemblies.
Using computational tools, we generated a TTR-derived peptide that inhibits Aβ42 fibril formation and cellular toxicity (Fig. 3). This TTR-derived peptide, called TTR-S, contains the highly amyloidogenic segment TTR(105–117) followed by a polyarginine tag (Fig. S3). Our results are consistent with previous NMR studies showing that the interaction of TTR residues Lys15, Leu17, Ile107, Ala108, Ala109, Leu110, Ala120, and Val121 with Aβ contributes to the inhibition of Aβ fibril formation in vitro (16). In addition, others have shown that the segment TTR(106–117), one residue shorter, was capable of binding Aβ when immobilized on a membrane (8). However, the same peptide was not effective inhibiting Aβ aggregation in solution (21), unless the peptide was made cyclic, therefore not amyloidogenic (26), probably because of the high amyloidogeneicity of this segment (Fig. S3). In our study, we solubilized TTR(105–117) by adding a charged tag, which confers higher solubility and also hinders self-aggregation (Fig. S3). The addition of a polyarginine tag was sufficient to convert a highly amyloidogenic peptide into an inhibitor of Aβ42 cellular toxicity even at substoichiometric concentrations (Fig. 4A).
TTR-S inhibits Aβ42 oligomer formation while promoting the formation of nontoxic, nonamyloid, amorphous aggregates. Previous studies of the inhibition of Aβ cytotoxicity by TTR have revealed that TTR co-aggregates with Aβ oligomers into large nontoxic assemblies, thereby inhibiting cellular toxicity in vitro (9). TTR-S mimics this effect and promotes the formation of large aggregates (Fig. 3B) that do not bind thioflavin T (Fig. 3A), do not show birefringence upon staining with Congo red (Fig. 3C), and display a non–β-secondary structure when subjected to CD (Fig. 3D). These amorphous species share some resemblance with the large unstructured aggregates found after the addition of EGCG to intrinsically disordered Aβ and other amyloid proteins (27). Similar to EGCG-derived Aβ aggregates, TTR-S–derived aggregates display a unique secondary structure that results in reduced thioflavin T fluorescence and a lack of birefringence upon binding to Congo red (Fig. 3, A and C).
Aβ42 amorphous aggregates generated upon binding to TTR-S are more sensitive to protease digestion (Figs. 4 and 5). As discussed above, TTR inhibits Aβ oligomer toxicity by co-aggregation into large nontoxic assemblies (9). However, those studies did not assess protease sensitivity of Aβ species. We observe that TTR-S–derived aggregates, from both soluble and fibrillar Aβ, are more sensitive to proteinase K than preformed Aβ fibrils. We speculate that TTR may promote Aβ clearance in vivo by a similar mechanism; this is, the formation of large assemblies that are more prone to digestion and clearance.
Finally, we evaluated the inhibitory effect of TTR-S on Aβ amyloid seeding. As shown previously, we found that the addition of ex vivo fibril extracts to soluble Aβ accelerates fibril formation (28, 29). In contrast, we found that TTR-S inhibits amyloid seeding catalyzed by Aβ extracted from the brain tissue of an AD patient (Fig. 5C).
In summary, our results suggest that the dissociation of the TTR tetramer into monomers is required to prevent cytotoxicity from Aβ oligomers. In addition, we found that a segment derived from TTR, TTR-S, exerts antiamyloid activity, thereby inhibiting Aβ42 cytotoxicity and amyloid seeding. We also found that TTR-S binds to soluble and fibrillar species, causing a structural rearrangement that leads to an increase of protease sensitivity. Finally, we found that TTR-S inhibits amyloid seeding catalyzed by Aβ fibrils extracted from the brain of an AD patient. Our results encourage further evaluation of the inhibitory effect of TTR-derived peptides by additional experimentation. Additional experiments may include independent cell death assays to confirm our MTT-based cytotoxicity assays and evaluation of the species that result from the inhibition of seeded polymerization by ex vivo fibrils using techniques such as EM imaging, CD, and protease digestion analysis. The present study represents an expansion of the current knowledge on the mechanism of protection of TTR over Aβ42 cellular toxicity and opens a potential therapeutic avenue.
Experimental procedures
Patients and tissue material
Post-mortem brain tissue from the occipital lobe of an 83-year-old female patient of Alzheimer's disease was obtained from Dr. Vinters at the Pathology Department of the University of California, Los Angeles. The patient was previously evaluated for the presence of amyloid plaques by immunohistochemistry and pathologically diagnosed for Alzheimer's disease. The University of California, Los Angeles Office of the Human Research Protection Program granted exemption from internal review board review because the specimen was anonymized.
Recombinant protein purification
TTR mutants were cloned and purified as previously described (18). Briefly, exponentially growing Escherichia coli RosettaTM(DE3)pLysS competent cells (Millipore) were treated with 1 mm of isopropyl β-d-thiogalactopyranoside for 3 h. Mutant TTR were purified by nickel-affinity chromatography using HisTrap columns (GE Healthcare), followed by gel-filtration chromatography using a HiLoad 16/60 Superdex 75 column (GE Healthcare) running on an AKTA FPLC system. β-Amyloid peptide (Aβ42) was overexpressed through E. coli recombinant expression system and was purified as reported previously (30). The fusion construct for Aβ42 expression contains an N-terminal His tag, followed by 19 repeats of Asn-Ala-Asn-Pro, TEV protease site, and the human Aβ42 sequence. Briefly, the fusion construct was expressed into inclusion bodies in E. coli BL21(DE3) cells. 8 m urea was used to solubilize the inclusion bodies. Fusion proteins were purified through HisTrap HP columns, followed by reversed-phase HPLC. After TEV cleavage, Aβ42 peptide was purified from the cleavage solution by reversed-phase HPLC followed by lyophilization. To disrupt preformed aggregation, lyophilized Aβ42 was resuspended in 100% hexafluoroisopropanol, which was finally removed by evaporation.
Size-exclusion chromatography with multiangle light scatter and refractometer detection
Components of 0.5-µg protein samples were separated by a silica-based size-exclusion column (Toso Biosep G3000SWXL; 5 μm beads, 7.8 × 300 mm with guard column). The elution buffer contained 0.025 m NaH2PO4, 0.1 m Na2SO4, 1 mm NaN3, which was weighed on an analytical scale, titrated together to pH 6.5 with NaOH, and diluted to final volume in a 2-liter volumetric flask. The flow rate was 0.4 ml/min. Samples were 0.1-μm filtered (Millipore UltraFree MC centrifugal filters). Each injection was 40–50 µl. The peaks were detected by UV absorbance at 280 nm (Waters 2487), light scatter, and index of refraction (Wyatt Technologies MiniDAWN and OPTILAB DSP). Molecular weights were calculated with Astra software, from the light scatter and refractometer signals, transferring the dn/dc and normalization parameters determined from the monomer peak of BSA.
TTR aggregation assay
TTR aggregation assays were performed as previously described (31). Briefly, 1 mg/ml TTR sample in 10 mm sodium acetate, pH 4.3, 100 mm KCl, and 10 mm EDTA was incubated at 37° C during a maximum of 7 days. Transmission EM micrographs were taken after 7 days of incubation. Insoluble fractions were sampled and used to follow TTR aggregation by immunodot blot.
Transmission EM
Transmission EM was performed to visualize TTR mutant aggregation and the fibrillation of Aβ42 in the presence of TTR mutants or TTR-derived inhibitors. 5 μl of solution was spotted onto freshly glow-discharged carbon-coated EM grids (Ted Pella, Redding, CA, USA). The grids were rinsed three times with 5 μl of distilled water after 3 min of incubation, followed by staining with 2% uranyl acetate for 2 min. A T12 Quick CryoEM electron microscope at an accelerating voltage of 120 kV was used to examine the specimens. The images were recorded digitally by a Gatan 2kX2k CCD camera.
Cell lines
HeLa, PC-12 Adh, and SH-SY5Y (catalog nos. CRL-2, CRL-1721.1, and CRL-2266, respectively; ATCC) cell lines were used for measuring the toxicity of Aβ42. HeLa cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, PC-12 cells were cultured in ATCC-formulated F-12K medium (ATCC; catalog no. 30-2004) with 2.5% fetal bovine serum and 15% horse serum.
MTT-based cell assay
We performed MTT-based cell viability assay to assess the cytotoxicity of Aβ42 with or without the addition of TTR mutants or TTR-derived peptide inhibitors. A CellTiter 96 aqueous nonradioactive cell proliferation assay kit (MTT) (catalog no. G4100, Promega, Madison, WI, USA) was used. Prior to toxicity test, HeLa, PC-12, and SH-5YSY cells were plated at 10,000, 15,000, and 10,000 cells/well, respectively, in 96-well plates (catalog no. 3596, Costar, Washington, D.C., USA). The cells were cultured in 96-well plates for 20 h at 37 °C in 5% CO2. For Aβ42 and inhibitors samples preparation, purified Aβ42 was dissolved in PBS at the final concentration of 10 μm, followed by the addition of TTR mutants or TTR-derived inhibitors at indicated concentrations. The mixtures were filtered with a 0.2-μm filter and further incubated for 16 h at 37 °C without shaking for fiber formation. To start the MTT assay, 10 μl of preincubated mixture was added to each well containing 90 μl medium. After 24 h of incubation at 37 °C in 5% CO2, 15 μl of dye solution (catalog no. G4102, Promega) was added into each well. After incubation for 4 h at 37 °C, 100 μl of solubilization solution/stop mix (catalog no. G4101, Promega) was added to each well. After 12 h of incubation at room temperature, the absorbance was measured at 570 nm with background absorbance recorded at 700 nm. Three replicates were measured for each of the samples. The MTT cell viability assay measured the percentage of survival cell upon the treatment of the mixture of Aβ42 and inhibitors. The cell viability (%) after treatment with Aβ42 with and without TTR-derived peptide inhibitors was calculated by normalizing the cell survival rate using the PBS buffer–treated cells as 100% viability and 2% SDS-treated cells as 0% viability.
Computational docking of fibrillar KLVFFA and monomeric TTR
Protein–protein docking between monomeric TTR and Aβ42 fibrillar segments was performed using the ClusPro server (32). Monomeric TTR was generated by removing chain B of the asymmetric unit of PDB code 4TLT (18). Three fibrillar polymorphs of the Aβ42 segment KLVFFA were analyzed: PDB codes 2Y2A, 2Y21, and 3OW9 (17). To increase binding surface, only one sheet of each polymorph was included in the modeling.
Thioflavin T fibrillation assay
Purified Aβ42 was dissolved in 10 mm NaOH at the concentration of 300 μm. Aβ42 was diluted into PBS buffer at the final concentration of 30 μm and was mixed with 30 μm ThT and different concentrations of TTR mutants or TTR-derived peptide inhibitors. The reaction mixture was split into four replicates and placed in a 394-well plate (black with flat optic bottom). The ThT fluorescence signal was measured every 5 min using the Varioskan plate reader (Thermo Scientific, Inc.) or FLUOstar Omega plate reader (BMG Labtech) with excitation and emission wavelengths of 444 and 484 nm, respectively, at 37 °C.
Western and immunodot blot
The aggregation of His-tagged TTR mutants was followed by immunodot blot analysis as described by Saelices et al. (18) using SuperSignal® West HisProbeTM kit following the manufacturer's instructions (Life Technologies). Briefly, 100 μl of samples was spun at 13,000 rpm for 30 min, and the pellet was resuspended in the same volume of fresh buffer and spun again. The final pellet was resuspended in 6 m guanidine chloride and dotted onto nitrocellulose membranes (0.2 μm, Bio-Rad). We used a concentration of the HisProbe antibody of 1:10,000. The aggregation of Aβ42 was followed by Western and immunodot blotting analysis. For the Western blots, 7.5 μl of 6 μm samples were separated by SDS-PAGE (NuPAGE 4–12% Bis-Tris gel, Life Technologies) or native gels (NativePAGE 4–16% Bis-Tris gel, Life Technologies) and transferred onto a nitrocellulose membrane (iBlot® 2 NC mini stacks, Life Technologies) by iBlot® 2 membrane transfer system (Life Technologies). For the immunodot blot analysis, 2–15-µl of samples were dotted onto a nitrocellulose membrane (Bio-Rad). We used concentrations of 6E10 antibody of 1:2000 (Biolegend®) (33), OC antibody of 1:25,000, and A11 antibody of 1:500 (Millipore and Life Technologies, respectively) (34). The horseradish peroxidase–conjugated anti-mouse IgG antibody (Sigma–Aldrich) was used as secondary antibody at a concentration of 1:2000, and anti-rabbit IgG antibody (Thermo Scientific) was used to detect OC and A11 at a concentration of 1:1000. The membrane was finally developed using the SuperSignal® West Pico chemiluminescent substrate kit (Life Technologies), following the manufacturer's instructions.
CD spectroscopy
Secondary structures of Aβ42 samples were analyzed by CD spectroscopy. The samples (200 µl) were placed into a 1-mm-path length quartz cell (Hëllma Analytics). A Jasco J-810 UV-visible spectropolarimeter was employed. The spectra were obtained in a wavelength range of 190–250 nm, with a time response of 2 s, a scan speed of 50 nm/min, and a step resolution of 0.1 nm. Each spectrum was the average of five accumulations. All of the samples were assayed at a concentration of 50 µg/ml in 15 mm phosphate buffer (pH 7.4). The spectra were recorded at 25° C. The results are expressed as the mean residue molar ellipticity,
where θ is the ellipticity in degrees, l is the optical path in cm, C is the concentration in mg/ml, M is the molecular mass, and n is in the number of residues in of the sample molecule. The ellipticity of the samples of Aβ after addition of TTR-S was normalized by using the ellipticity of TTR-S as a blank. The percentage of the various structural conformations was calculated by using the software package CDPro, which includes three programs to determine the secondary structure fractions: CONTIN, SELCON, and CDSSTR (35). The results are displayed as the mean values and standard deviations of the three methods.
Proteinase K assay
Soluble Aβ42 was prepared in PBS and filtered (0.22 nm). Aβ42 fibrils were obtained after 1–2 weeks of incubation in PBS at room temperature. Three-fold molar excess of TTR-S was added to soluble or fibrillar Aβ42 and incubated for 16 h. Soluble and insoluble fractions were extracted by centrifugation of half of the sample prior to incubation. PK digestion was performed in a reaction volume of 100 μl containing 10 μm of Aβ42 samples and varying concentrations of PK (from 0.2 to 50 μg/ml) in 0.1 m Tris-HCl buffer, pH 7.5, for 1 h at 37 °C. The reactions were quenched with 1 mm phenylmethylsulfonyl fluoride, and the samples were analyzed dot blot using E610 specific anti-Aβ42 antibody. As negative control, we included a sample with TTR-S alone. The signal was quantified using ImageJ.
Statistical analysis
Statistical analysis of cell assays was performed with Prism 7 for Mac (GraphPad Software) using an unpaired t test. All samples were included in the analysis. All quantitative experiments are presented as independent replicates or as means ± S.D. of at least three independent experiments.
Data availability
The data sets and images used and/or analyzed during the current study are available from the corresponding author on reasonable request. All remaining data are contained within the article.
Supplementary Material
Acknowledgments
We thank Prof. David Eisenberg and Prof. Cong Liu for significant support. We also thank Dr. Harry V. Vinters for supplying the AD brain tissue and the patient who generously donated it.
Author contributions—Q. C. and L. S. data curation; Q. C. and L. S. validation; Q. C., D. H. A., W. Y. L., J. C., and L. S. investigation; L. S. conceptualization; L. S. formal analysis; L. S. supervision; L. S. funding acquisition; L. S. visualization; L. S. methodology; L. S. writing-original draft; L. S. project administration; L. S. writing-review and editing.
Funding and additional information—This work was supported by Amyloidosis Foundation Grants 20160759 and 20170827 (to L. S.) and the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007–2013 under Research Executive Agency Grant Agreement 298559 (to L. S.)
Conflict of interest—L. S. is a consultant for ADRx, Inc.
- AD
- Alzheimer's disease
- TTR
- transthyretin
- Aβ
- amyloid-β peptide
- Aβ42
- amyloid-β peptide 1–42
- ThT
- thioflavin T
- M-TTR
- monomeric TTR variant
- NSTTR
- N98R/S99R TTR variant
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- TTR-S
- TTR segment modified with a polyarginine tag
- PDB
- Protein Data Bank.
References
- 1. Schwarzman A. L., Gregori L., Vitek M. P., Lyubski S., Strittmatter W. J., Enghilde J. J., Bhasin R., Silverman J., Weisgraber K. H., Coyle P. K. (1994) Transthyretin sequesters amyloid β protein and prevents amyloid formation. Proc. Natl. Acad. Sci. U.S.A. 91, 8368–8372 10.1073/pnas.91.18.8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roher A. E., Lowenson J. D., Clarke S., Woods A. S., Cotter R. J., Gowing E., and Ball M. J. (1993) β-Amyloid-(1–42) is a major component of cerebrovascular amyloid deposits: implications for the pathology of Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 90, 10836–10840 10.1073/pnas.90.22.10836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burdick D., Soreghan B., Kwon M., Kosmoski J., Knauer M., Henschen A., Yates J., Cotman C., and Glabe C. (1992) Assembly and aggregation properties of synthetic Alzheimer's A4/β amyloid peptide analogs. J. Biol. Chem. 267, 546–554 [PubMed] [Google Scholar]
- 4. Stein T. D., Anders N. J., DeCarli C., Chan S. L., Mattson M. P., and Johnson J. A. (2004) Neutralization of transthyretin reverses the neuroprotective effects of secreted amyloid precursor protein (APP) in APPSW mice resulting in tau phosphorylation and loss of hippocampal neurons: support for the amyloid hypothesis. J. Neurosci. 24, 7707–7717 10.1523/JNEUROSCI.2211-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi S. H., Leight S. N., Lee V. M., Li T., Wong P. C., Johnson J. A., Saraiva M. J., and Sisodia S. S. (2007) Accelerated Aβ deposition in APPswe/PS1deltaE9 mice with hemizygous deletions of TTR (transthyretin). J. Neurosci. 27, 7006–7010 10.1523/JNEUROSCI.1919-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buxbaum J. N., Ye Z., Reixach N., Friske L., Levy C., Das P., Golde T., Masliah E., Roberts A. R., and Bartfai T. (2008) Transthyretin protects Alzheimer's mice from the behavioral and biochemical effects of Aβ toxicity. Proc. Natl. Acad. Sci. U.S.A. 105, 2681–2686 10.1073/pnas.0712197105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Costa R., Gonçalves A., Saraiva M. J., and Cardoso I. (2008) Transthyretin binding to A-β peptide: impact on A-β fibrillogenesis and toxicity. FEBS Lett. 582, 936–942 10.1016/j.febslet.2008.02.034 [DOI] [PubMed] [Google Scholar]
- 8. Du J., Cho P. Y., Yang D. T., and Murphy R. M. (2012) Identification of β-amyloid–binding sites on transthyretin. Protein Eng. Des. Sel. 25, 337–345 10.1093/protein/gzs026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cascella R., Conti S., Mannini B., Li X., Buxbaum J. N., Tiribilli B., Chiti F., and Cecchi C. (2013) Transthyretin suppresses the toxicity of oligomers formed by misfolded proteins in vitro. Biochim. Biophys. Acta 1832, 2302–2314 10.1016/j.bbadis.2013.09.011 [DOI] [PubMed] [Google Scholar]
- 10. Garai K., Posey A. E., Li X., Buxbaum J. N., and Pappu R. V. (2018) Inhibition of amyloid β fibril formation by monomeric human transthyretin. Protein Sci. 27, 1252–1261 10.1002/pro.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Westermark P., Sletten K., Johansson B., and Cornwell G. G. (1990) Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc. Natl. Acad. Sci. U.S.A. 87, 2843–2845 10.1073/pnas.87.7.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colon W., and Kelly J. W. (1992) Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry 31, 8654–8660 10.1021/bi00151a036 [DOI] [PubMed] [Google Scholar]
- 13. Saelices L., Chung K., Lee J. H., Cohn W., Whitelegge J. P., Benson M. D., and Eisenberg D. S. (2018) Amyloid seeding of transthyretin by ex vivo cardiac fibrils and its inhibition. Proc. Natl. Acad. Sci. U.S.A. 115, E6741–E6750 10.1073/pnas.1805131115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang X., Smith C. S., Petrassi H. M., Hammarström P., White J. T., Sacchettini J. C., and Kelly J. W. (2001) An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry 40, 11442–11452 10.1021/bi011194d [DOI] [PubMed] [Google Scholar]
- 15. Coelho T., Carvalho M., Saraiva M. J., Alves I., Almeida M. R., and Costa P. P. (1993) A strikingly benign evolution of FAP in an individual found to be a compound heterozygote for two TTR mutations: TTR MET 30 and TTR MET 119. J. Rheumatol. 20, 179 [Google Scholar]
- 16. Li X., Zhang X., Ladiwala A. R., Du D., Yadav J. K., Tessier P. M., Wright P. E., Kelly J. W., and Buxbaum J. N. (2013) Mechanisms of transthyretin inhibition of β-amyloid aggregation in vitro. J. Neurosci. 33, 19423–19433 10.1523/JNEUROSCI.2561-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Colletier J. P., Laganowsky A., Landau M., Zhao M., Soriaga A. B., Goldschmidt L., Flot D., Cascio D., Sawaya M. R., and Eisenberg D. (2011) Molecular basis for amyloid-β polymorphism. Proc. Natl. Acad. Sci. U.S.A. 108, 16938–16943 10.1073/pnas.1112600108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saelices L., Johnson L. M., Liang W. Y., Sawaya M. R., Cascio D., Ruchala P., Whitelegge J., Jiang L., Riek R., and Eisenberg D. S. (2015) Uncovering the mechanism of aggregation of human transthyretin. J. Biol. Chem. 290, 28932–28943 10.1074/jbc.M115.659912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldschmidt L., Teng P. K., Riek R., and Eisenberg D. (2010) Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A. 107, 3487–3492 10.1073/pnas.0915166107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson M. J., Sievers S. A., Karanicolas J., Ivanova M. I., Baker D., and Eisenberg D. (2006) The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 4074–4078 10.1073/pnas.0511295103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho P. Y., Joshi G., Johnson J. A., and Murphy R. M. (2014) Transthyretin-derived peptides as β-amyloid inhibitors. ACS Chem. Neurosci. 5, 542–551 10.1021/cn500014u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tycko R. (2014) Physical and structural basis for polymorphism in amyloid fibrils. Protein Sci. 23, 1528–1539 10.1002/pro.2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisenberg D., and Jucker M. (2012) The amyloid state of proteins in human diseases. Cell 148, 1188–1203 10.1016/j.cell.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hornstrup L. S., Frikke-Schmidt R., Nordestgaard B. G., and Tybjærg-Hansen A. (2013) Genetic stabilization of transthyretin, cerebrovascular disease, and life expectancy. Arterioscler. Thromb. Vasc. Biol. 33, 1441–1447 10.1161/ATVBAHA.113.301273 [DOI] [PubMed] [Google Scholar]
- 25. Ribeiro C. A., Oliveira S. M., Guido L. F., Magalhães A., Valencia G., Arsequell G., Saraiva M. J., and Cardoso I. (2014) Transthyretin stabilization by iododiflunisal promotes amyloid-β peptide clearance, decreases its deposition, and ameliorates cognitive deficits in an Alzheimer's disease mouse model. J. Alzheimers Dis. 39, 357–370 10.3233/JAD-131355 [DOI] [PubMed] [Google Scholar]
- 26. Cho P. Y., Joshi G., Boersma M. D., Johnson J. A., and Murphy R. M. (2015) A cyclic peptide mimic of the β-amyloid binding domain on transthyretin. ACS Chem. Neurosci. 6, 778–789 10.1021/cn500272a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palhano F. L., Lee J., Grimster N. P., and Kelly J. W. (2013) Toward the molecular mechanism(s) by which EGCG treatment remodels mature amyloid fibrils. J. Am. Chem. Soc. 135, 7503–7510 10.1021/ja3115696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jarrett J. T., Berger E. P., and Lansbury P. T. J. (1993) The carboxy terminus of the β amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer's disease. Biochemistry 32, 4693–4697 10.1021/bi00069a001 [DOI] [PubMed] [Google Scholar]
- 29. Paravastu A. K., Leapman R. D., Yau W. M., and Tycko R. (2008) Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 18349–18354 10.1073/pnas.0806270105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finder V. H., Vodopivec I., Nitsch R. M., and Glockshuber R. (2010) The recombinant amyloid-β peptide Aβ1–42 aggregates faster and is more neurotoxic than synthetic Aβ1–42. J. Mol. Biol. 396, 9–18 10.1016/j.jmb.2009.12.016 [DOI] [PubMed] [Google Scholar]
- 31. Saelices L., Nguyen B. A., Chung K., Wang Y., Ortega A., Lee J. H., Coelho T., Bijzet J., Benson M. D., and Eisenberg D. S. (2019) A pair of peptides inhibits seeding of the hormone transporter transthyretin into amyloid fibrils. J. Biol. Chem. 294, 6130–6141 10.1074/jbc.RA118.005257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozakov D., Hall D. R., Xia B., Porter K. A., Padhorny D., Yueh C., Beglov D., and Vajda S. (2017) The ClusPro web server for protein–protein docking. Nat. Protoc. 12, 255–278 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pirttilä T., Kim K. S., Mehta P. D., Frey H., and Wisniewski H. M. (1994) Soluble amyloid β-protein in the cerebrospinal fluid from patients with Alzheimer's disease, vascular dementia and controls. J. Neurol. Sci. 127, 90–95 10.1016/0022-510X(94)90140-6 [DOI] [PubMed] [Google Scholar]
- 34. Sarsoza F., Saing T., Kayed R., Dahlin R., Dick M., Broadwater-Hollifield C., Mobley S., Lott I., Doran E., Gillen D., Anderson-Bergman C., Cribbs D. H., Glabe C., and Head E. (2009) A fibril-specific, conformation-dependent antibody recognizes a subset of Aβ plaques in Alzheimer disease, Down syndrome and Tg2576 transgenic mouse brain. Acta Neuropathol. 118, 505–517 10.1007/s00401-009-0530-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 10.1006/abio.2000.4880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets and images used and/or analyzed during the current study are available from the corresponding author on reasonable request. All remaining data are contained within the article.