Abstract
The DNA glycosylase NEIL3 has been implicated in DNA repair pathways including the base excision repair and the interstrand cross-link repair pathways via its DNA glycosylase and/or AP lyase activity, which are considered canonical roles of NEIL3 in genome integrity. Compared with the other DNA glycosylases NEIL1 and NEIL2, Xenopus laevis NEIL3 C terminus has two highly conserved zinc finger motifs containing GRXF residues (designated as Zf-GRF). It has been demonstrated that the minor AP endonuclease APE2 contains only one Zf-GRF motif mediating interaction with single-strand DNA (ssDNA), whereas the major AP endonuclease APE1 does not. It appears that the two NEIL3 Zf-GRF motifs (designated as Zf-GRF repeat) are dispensable for its DNA glycosylase and AP lyase activity; however, the potential function of the NEIL3 Zf-GRF repeat in genome integrity remains unknown. Here, we demonstrate evidence that the NEIL3 Zf-GRF repeat was associated with a higher affinity for shorter ssDNA than one single Zf-GRF motif. Notably, our protein–protein interaction assays show that the NEIL3 Zf-GRF repeat but not one Zf-GRF motif interacted with APE1 but not APE2. We further reveal that APE1 endonuclease activity on ssDNA but not on dsDNA is compromised by a NEIL3 Zf-GRF repeat, whereas one Zf-GRF motif within NEIL3 is not sufficient to prevent such activity of APE1. In addition, COMET assays show that excess NEIL3 Zf-GRF repeat reduces DNA damage in oxidative stress in Xenopus egg extracts. Together, our results suggest a noncanonical role of NEIL3 in genome integrity via its distinct Zf-GRF repeat in suppressing APE1 endonuclease-mediated ssDNA breakage.
Keywords: APE1, APE2, DNA repair, genome instability, NEIL3, ssDNA, zinc finger Zf-GRF motif, DNA endonuclease, DNA damage response, DNA enzyme, genomic instability, APE1
DNA damage induced by oxidative stress is inevitable and has been implicated in the pathology of human diseases such as cancer and neurodegenerative disorders (1). To repair oxidized bases, cells have evolved several DNA glycosylases including NEIL1, NEIL2, NEIL3, NTH1, and OGG1, among others for initiating base excision repair (BER) pathway. NEIL1 and NEIL2 are bifunctional DNA glycosylases, which not only excise the damaged base to generate a AP (apurinic/apyrimidinic) site via glycosylase activity but also subsequently introduce a DNA single-strand break (SSB) at the 3′-side of the AP site via their AP lyase activity (2, 3). Whereas NEIL1 interacts with replication proteins and are involved in pre-replicative repair during S-phase (4), NEIL2 associates with RNA polymerase II and preferentially repairs oxidized bases in the transcribing genes (5). Human and mouse NEIL3 displays DNA glycosylase activity on a variety of oxidized or damaged bases such as spiroiminodihydantoin and guanidinohydantoin (6, 7). Unlike NEIL1 and NEIL2, NEIL3 can incise AP site via β-elimination with weak AP lyase activity in vitro (6, 7). Interestingly, NEIL3 preferentially recognizes and binds to base lesions in ssDNA and bubble structures, which is partially explained by its distinct structure (6, 8). NEIL3 is expressed in hematopoietic tissue specifically (9) and during embryogenesis and neurogenesis (10, 11). Furthermore, NEIL3 mRNA is also overexpressed in tumor tissues compared with normal tissue, except in testis and pancreas (11). Although NEIL3-knockout mice are viable and fertile, NEIL3 deficiency impairs the repair of hydantoin lesions in ssDNA in neural stem/progenitor cells and displays profound neuropathology including a reduced number of microglia and loss of proliferating neuronal progenitors after hypoxia-ischemia (9, 12). In addition, NEIL3 has been recently shown to unhook inter-strand cross-links (ICLs) such as psoralen-ICL and AP-ICL structures but not the cisplatin-ICL structure via its DNA glycosylase activity in the Xenopus system and mammalian cells, indicating a critical role of NEIL3 in ICL repair (13–15). Intriguingly, depending on the nature of ICL substrates and recombinant NEIL3 protein utilized in in vitro assays, the AP site after NEIL3 unhooking may or may not be subsequently cleaved by its AP lyase activity (13, 16, 17).
NEIL3 protein contains a conserved Fpg/Nei-like core glycosylase domain including an H2TH motif and a zinc finger (Fpg-Znf) motif at the N terminus, and an extended C-terminal domain harboring a Ran-binding protein-type zinc finger (RBP-Znf) motif, a nuclear localization signal, and two conserved zinc finger-GRF (Zf-GRF) motifs at the extreme C terminus (9, 18, 19). The Zf-GRF motifs contain three conserved residues glycine (Gly), arginine (Arg), and phenylalanine (Phe) and have been found in ∼100 proteins that participate in DNA/RNA metabolism (e.g. NEIL3, APE2, and Top3A) (19). As the first characterized Zf-GRF, APE2 Zf-GRF motif associates with ssDNA but not dsDNA (19). Moreover, the APE2 Zf-GRF motif consists of three β-sheets harboring Zn2+ and conserved GRF residues, which folds into crescent-shaped claw-like structures specific for ssDNA and is required for 3′-5′-end resection on damaged DNA (19). Consistent with the binding of APE2 Zf-GRF to ssDNA, a recent report also shows that NEIL3 Zf-GRF motifs interact with ssDNA (15). However, the two Zf-GRF motifs of NEIL3 are dispensable for its DNA glycosylase and potential AP lyase activities (6, 8). Therefore, the potential role of the two Zf-GRF motifs within NEIL3 in genome integrity is unknown.
As endonuclease/exonuclease/phosphatase (EEP) family member, both APE1 and APE2 are critical players in the maintenance of genome integrity (20–24). In general, APE1 displays faster AP endonuclease activity but slower exonuclease activity (25–27); however, APE2 contains faster exonuclease activity but slower AP endonuclease activity (23, 24, 28, 29). Furthermore, unlike APE1, APE2 contains a PCNA-interaction protein (PIP) box and a Zf-GRF motif in its extreme C terminus (19). APE2's Zf-GRF preferentially interacts with ssDNA but not dsDNA (19). The binding of APE2's Zf-GRF to ssDNA is essential for APE2's exonuclease activity and oxidative stress-induced ATR-Chk1 DDR pathway activation (19). Consistent with the APE2 Zf-GRF–ssDNA interaction, the Walter group (15) has recently shown that Xenopus NEIL3's Zf-GRF motifs including Zf-GRF1 (ZF1) and Zf-GRF2 (ZF2) also interact with ssDNA, respectively. APE2's Zf-GRF motif was also implicated in association with PCNA to regulate its exonuclease activity on gapped structures in in vitro biochemical assays (29). Intriguingly, it is noted that APE2 contains one Zf-GRF motif compared with APE1, and that NEIL3 displays two Zf-GRF motifs compared with NEIL1 and NEIL2.
Although NEIL3 interacts with APE1 and PCNA as well as telomere protein TRF2 to repair telomere DNA damage during S phase, it remains elusive how exactly NEIL3 regulates and/or cross-talks with APE1 (30). Here, we demonstrate evidence that the two Zf-GRF motifs within NEIL3 directly interact with APE1 and PCNA, but not APE2. We further show that NEIL3's Zf-GRF repeat (ZF1&ZF2) associates with shorter ssDNA, whereas ZF1 cannot. We also reveal unexpectedly that NEIL3's Zf-GRF repeat but not ZF1 negatively regulates APE1 AP endonuclease activity on ssDNA but not dsDNA. These findings clearly reveal previously uncharacterized features of the Zf-GRF repeat compared with a single Zf-GRF motif. Thus, we elucidate the distinct regulation of NEIL3 Zf-GRF motifs in genome integrity via the negative regulation of APE1 endonuclease activity on ssDNA to avoid unnecessary DNA breakages.
Results
NEIL3 contains distinct Zf-GRF repeat in its extreme C terminus
Compared with NEIL1 and NEIL2, the least-characterized NEIL3 displays a RBP-Znf motif and two Zf-GRF motifs (designated as ZF1 and ZF2) in its extreme C terminus (Fig. S1A). Sequence alignment of ZF1 and ZF2 across four different species (Xenopus laevis (Xl), Xenopus tropicalis (Xt), Mus musculus (Ms), and Homo sapiens (Hs)) showed highly conserved residues in each ZF motif (Figs. S1B and S2). Similar to the APE2 Zf-GRF motif (19), NEIL3 ZF1 contains the three conserved core amino acids: Gly, Arg and Phe, whereas Lys replaces Arg to produce a Zf-GKF motif in NEIL3 ZF2 (Fig. S1B). Although Lys and Arg have positive charge, their structures are much different with ionic interactions formed by Arg, which has shown to be more stable than Lys (31). In addition, this replacement has been found across all four species and indicates a possible unique function that ZF2 might add into NEIL3 Zf-GRF motifs overall.
Furthermore, both ZF1 and ZF2 within NEIL3 contain the conserved CHCC-type Zn2+ contact motif (Fig. S1B). There is a α1-helix and a proline-rich helix at the N-terminal side of the APE2 Zf-GRF motif; however, it seems that no such helix motifs were found in the two Zf-GRF motifs in NEIL3 (Fig. S1C). Essentially, the NEIL3 Zf-GRF repeat has two Zf-GRF motifs (ZF1 and ZF2), whereas APE2 contains only one Zf-GRF motif (Fig. 1A). Sequence alignment reveals the distinctiveness of NEIL3-ZF2 as the only one with lysine Lys, whereas APE2 had Arg in the Zf-GRF motif (Fig. S1C).
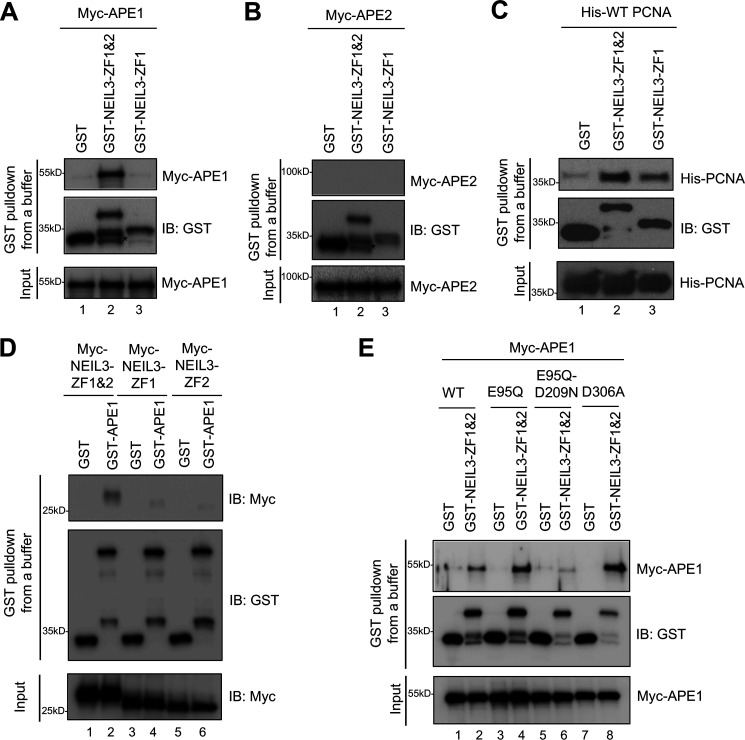
Figure 1.
NEIL3 Zf-GRF repeat interacts with APE1 and PCNA but not APE2 in vitro. A, GST pulldown assay performed between Myc-APE1 and GST-NEIL3-ZF1&2, GST-NEIL3-ZF1, or GST (4 μm) in vitro. B, interaction between Myc-APE2 and GST-NEIL3-ZF1&2, GST-NEIL3-ZF1, or GST (4 μm). C, interaction of GST-NEIL3-ZF1&2 to His-PCNA (4 μm) was detected, whereas GST-NEIL3-ZF1 had almost 50% reduction in interaction to His-PCNA. D, GST pulldown assay expanded to detect interaction between Myc-NEIL3-ZF1&2/Myc-NEIL3-ZF1/Myc-NEIL3-ZF2 and GST-APE1 (4 μm) in vitro. E, Myc-APE1 WT, E95Q, E95Q-D209N, or D306A interactions with GST-NEIL3-ZF1&2 (4 μm) for determining the possible residue within APE1 for such interaction. * indicates degradation products. IB, immunoblot.
NEIL3 Zf-GRF repeat interacts with APE1 and PCNA but not APE2
A recent study using co-immunoprecipitation assays from cell lysates expressing NEIL3-HA revealed the interaction of NEIL3 to BER proteins FEN1, APE1, and PCNA (30). Further analysis showed that the N-terminal domain of NEIL3 was not sufficient for the interaction to APE1, whereas full-length NEIL3 could (30). Although the C-terminal domain of NEIL3 seemed to be the key for such an interaction between NEIL3 and APE1, it has not been identified which part within the C-terminal domain of NEIL3 is required for APE1 interaction. To test the possibility that NEIL3-ZF1&2 interacts with APE1, we first expressed and purified GST-NEIL3-ZF1&2, GST-NEIL3-ZF1, and GST-NEIL3-ZF1&2-K553A (Fig. S3). Indeed, we found that GST-NEIL3-ZF1&2, but not GST-NEIL3-ZF1 nor GST, interacted with Myc-APE1 (Fig. 1A).
Because APE1 and APE2 share a similar EEP domain, we sought to determine whether NEIL3 Zf-GRF motifs interact with APE2. Our GST-pulldown experiment showed that neither GST-NEIL3-ZF1&2 nor GST-NEIL3-ZF1 associated with Myc-APE2, suggesting no direct interaction between NEIL3 Zf-GRF motifs and APE2 at least under the conditions tested (Fig. 1B). We recently reported that APE2 Zf-GRF associates with PCNA (29). Consistent with this, we found that GST-NEIL3-ZF1&2 but not GST associated with His-tagged PCNA in vitro (Fig. 1C). Notably, GST-NEIL3-ZF1 also interacted with His-tagged PCNA in vitro, despite a decreased interaction compared with the two Zf-GRF motifs (Fig. 1C). These observations suggest that one or both NEIL3 Zf-GRF motifs are sufficient for PCNA interaction. Future experiments are needed to determine the critical domains within PCNA and NEIL3 Zf-GRF motifs responsible for this interaction. Notably, a prior study has shown that the N-terminal domain of NEIL3 is sufficient to interact with PCNA (30). Although the exact underlying mechanism remains unknown, we speculate that NEIL3 may have two different modes of PCNA interaction (one through N-terminal domain, and another one is through Zf-GRF motifs), depending on the context of its complex with DNA and/or other repair proteins.
Furthermore, we sought to determine the domain requirement within APE1 and the Zf-GRF repeat for the NEIL3-Zf-GRF repeat–APE1 interaction. Our reciprocal experiment showed that GST-APE1, but not GST, associated with Myc-NEIL3-ZF1&2 (Fig. 1D). Notably, neither Myc-NEIL3-ZF1 nor Myc-NEIL3-ZF2 were found to interact with GST-APE1 (Fig. 1D). These observations suggest that the NEIL3 Zf-GRF repeat is required and sufficient for APE1 interaction and that one Zf-GRF within NEIL3 is insufficient for interaction to APE1 (Fig. 1, A and D).
Next, we reasoned that residues in the active site of the APE1 EEP domain may be important for interaction with the NEIL3 Zf-GRF repeat. Prior studies have shown that the human APE1 variant with D308A mutant and Xenopus APE1 variant with D306A mutant are deficient for 3′-5′-exonuclease activity but proficient in AP endonuclease activity (26, 32). Furthermore, human APE1 mutant containing E96Q-D210N and Xenopus APE1 mutant containing E95Q-D209N lack AP endonuclease activity (26, 33). To identify critical residues within APE1 for such interactions with NEIL3 Zf-GRF motifs, we generated three mutations in Myc-APE1 (i.e. E95Q, E95Q-D209N, and D306A). GST pulldown assays show that compared with WT APE1, E95Q-D209N but neither E95Q nor D306A mutant APE1 significantly reduced the binding to NEIL3-ZF1&2 (Fig. 1E). This observation suggests that the Asp-209 residue within APE1 is critical for interaction with the NEIL3 Zf-GRF repeat. Because Asp-209 in the APE1 active site is important for its endonuclease activity (26, 33), it is possible that APE1 endonuclease activity may be regulated by this distinct interaction with the NEIL3 Zf-GRF repeat.
NEIL3 Zf-GRF repeat binds to shorter ssDNA compared with NEIL3 ZF1
Our recent study has shown that the APE2 Zf-GRF motif preferentially associates with ssDNA but not dsDNA (19). Although previous study showed that the C-terminal domain of NEIL3 was responsible for ssDNA binding (30), the specific region within the C-terminal domain of NEIL3 for such ssDNA binding has not been identified. Thus, we sought to determine whether NEIL3 Zf-GRF motifs associate with ssDNA; and if so, what aspect of ssDNA binding of the two Zf-GRF motifs can distinguish from one individual Zf-GRF motif. Through biotin-ssDNA binding assays, GST-NEIL3-ZF1&2 but not GST associated with 60, 40, and 20 nt, and as short as 10 nt ssDNA (Fig. 2A). However, GST-NEIL3-ZF1 but not GST associated with 60 nt but not 40 nor 20 nt ssDNA (Fig. 2B). These observations suggest that having two Zf-GRF motifs may be beneficial for NEIL3 to recognize and remove base damage in short ssDNA sequences, such as replicating forks or telomere regions.
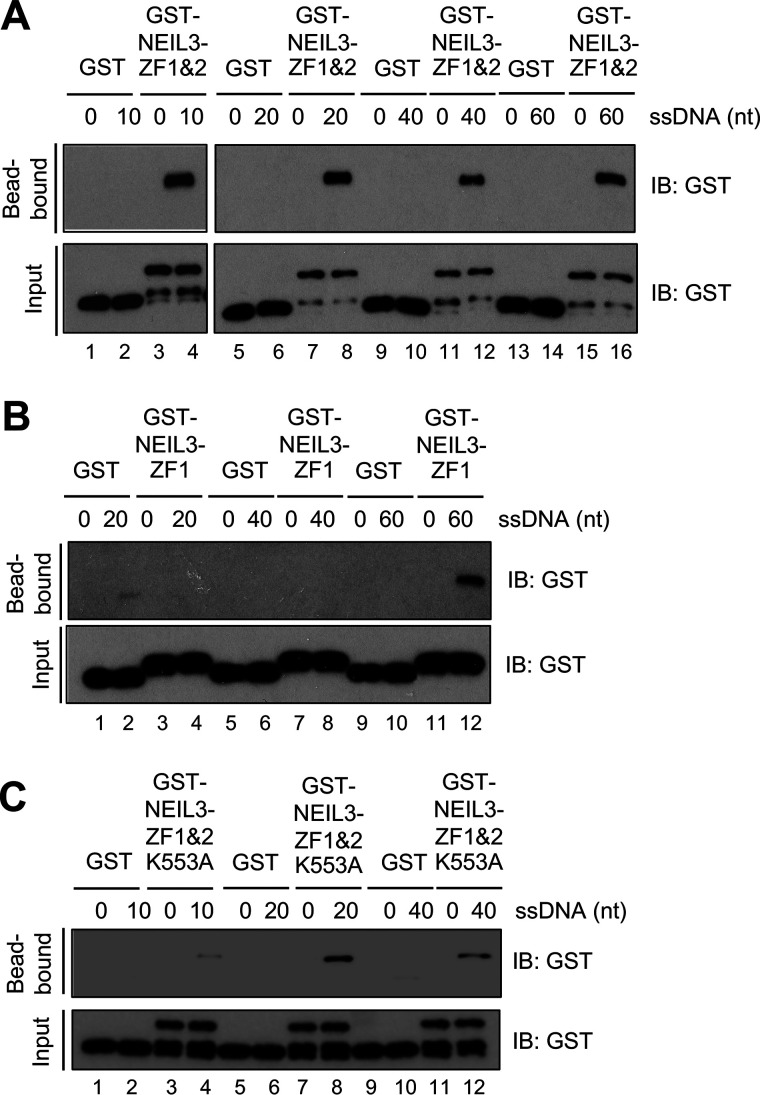
Figure 2.
NEIL3 Zf-GRF repeat binds to shorter ssDNA than NEIL3-ZF1. A, interaction between GST-NEIL3-ZF1&2 (4 μm) and biotin-labeled ssDNA structures with lengths of 10, 20, 40, or 60 nt (5 nm) via biotin-DNA binding assays. B, the interaction of GST-NEIL3-ZF1 and different biotin-labeled ssDNA structures of 20, 40, or 60 nt. C, interaction of GST-NEIL3-ZF1&2 K553A and biotin-ssDNA structure with different lengths via biotin-DNA binding assays. IB, immunoblot.
In addition, we failed to express and purify GST-NEIL3-ZF2 recombinant protein after many trials. To test the role of NEIL3-ZF2 for ssDNA binding, we decided to mutate the unique residue in NEIL3-ZF2 from Lys to Ala, which is a nonpolar amino acid (i.e. GST-NEIL3-ZF1&2-K553A). Amazingly, the binding of GST-NEIL3-ZF1&2-K553A was observed clearly at 20- and 40-nt ssDNA but reduced significantly in a 10-nt ssDNA (Fig. 2C). This observation indicates that the conserved Lys-553 residue within NEIL3-ZF2 is important for the Zf-GRF repeat binding to short ssDNA (10 nt). In conclusion, our findings reveal that two NEIL3 Zf-GRF motifs associate with ssDNA, and that two Zf-GRF motifs but not one individual Zf-GRF motif are needed to bind to shorter ssDNA (10 nt). Thus, our findings identified binding to shorter ssDNA as the second feature of the Zf-GRF repeat compared with individual Zf-GRF motif.
NEIL3 Zf-GRF repeat interacts with different ssDNA structures
Next, we sought to determine the sequence and nature of ssDNA for interacting with NEIL3 Zf-GRF motifs. We utilized EMSA with different 5′-FAM–labeled ssDNA with or without the AP site to quantify the binding ability of NEIL3 Zf-GRF motifs. Consistent with findings from our biotin DNA-binding assays in Fig. 2A, GST-NEIL3-ZF1&2 formed a complex with 39-nt ssDNA starting as low as 3 μm (Fig. 3A). Notably, GST-NEIL3-ZF1 did not form a complex with 39-nt ssDNA even under a concentration of 15 μm (Fig. 3B). Furthermore, the complex formation of GST-NEIL3-ZF1&2-K553A with 39-nt ssDNA was significantly reduced even under a concentration of 15 μm, suggesting that Lys-553 is important for ssDNA interaction (Fig. 3C). We then tested the binding of NEIL3 Zf-GRF motifs to ssDNA, which contains an AP site. Interestingly, the presence of an AP site in the 39-nt ssDNA did not increase the interactions with GST-NEIL3-ZF1&2, GST-NEIL3-ZF1, and GST-NEIL3-ZF1&2-K553A (Fig. 3, D–F). This observation suggests that NEIL3 Zf-GRF motifs do not preferentially associate with AP site on ssDNA.
Figure 3.
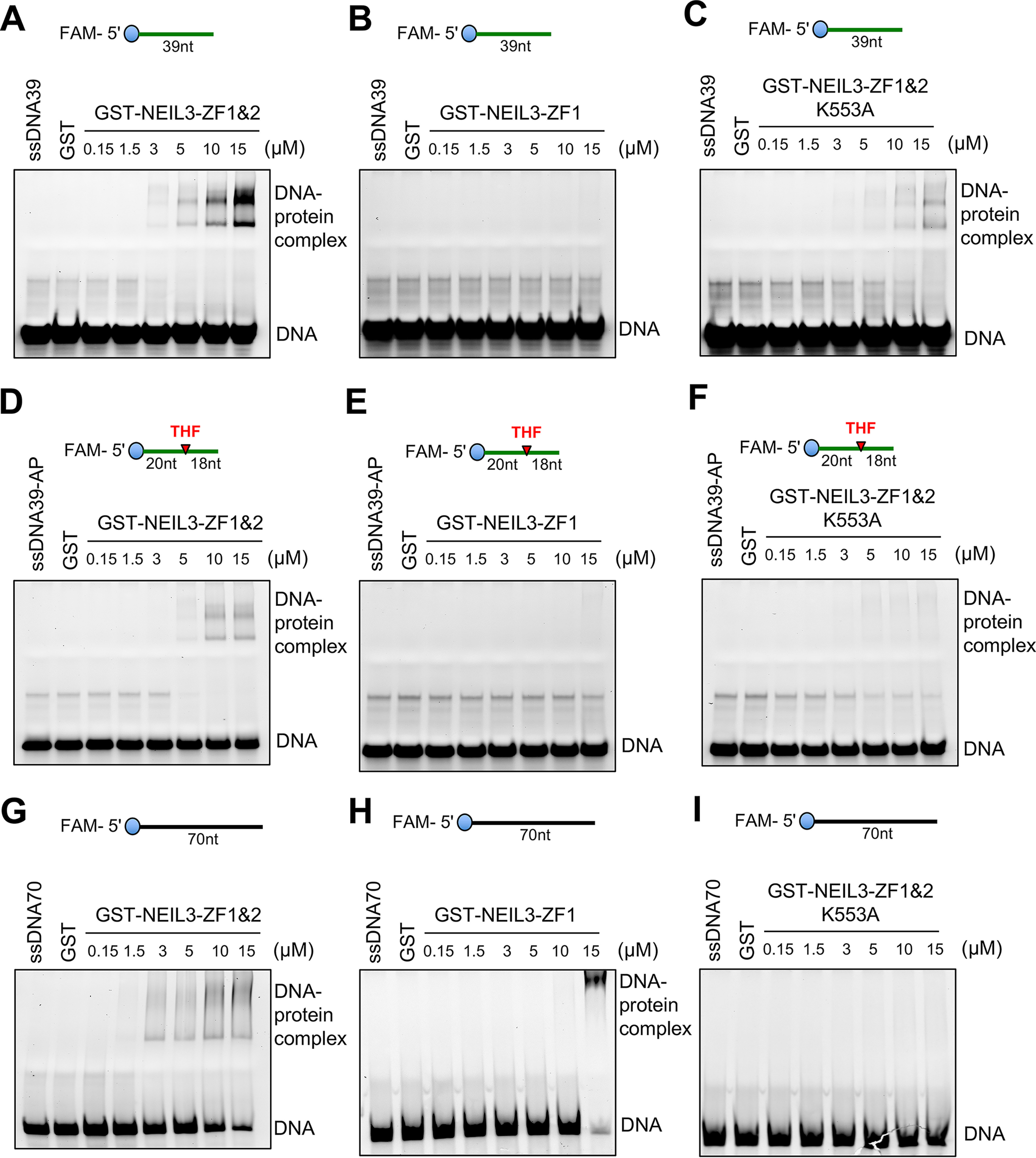
NEIL3 Zf-GRF motifs interact with different ssDNA structures. A–C, EMSA show the interaction between ssDNA39 and GST-NEIL3-ZF1&2, GST-NEIL3-ZF1, or GST-NEIL3-ZF1&2 K553A at different doses as indicated at room temperature for 60 min. D-F, EMSA reveal interaction between ssDNA39-AP and increased concentrations of GST-NEIL3-ZF1&2 or GST-NEIL3-ZF1 or GST-NEIL3-ZF1&2 K553A in vitro. G-I, interaction between ssDNA70 and GST-NEIL3-ZF1&2 or GST-NEIL3-ZF1 but not with GST-NEIL3-ZF1&2 K553A via EMSA.
To determine whether the interaction between NEIL3 Zf-GRF motifs and ssDNA is sequence-dependent, we also tested another longer ssDNA (70 nt) with a different DNA sequence compared with the 39-nt ssDNA. Similar to the binding to 39-nt ssDNA, GST-NEIL3-ZF1&2 associated with 70-nt ssDNA at a concentration as low as 3 μm in EMSA (Fig. 3G); however, the GST-NEIL3-ZF1&2-K553A mutant showed very weak to almost no binding to 70-nt ssDNA even at 15 μm (Fig. 3I). Notably, GST-NEIL3-ZF1 formed a complex with 70-nt ssDNA at around 15 μm, whereas such an interaction in shorter 39-nt ssDNA was absent (Fig. 3, B and H). This observation is consistent with the finding that GST-NEIL3-ZF1 binds to 60-nt ssDNA but not shorter ssDNA in our biotin-DNA binding assays (Fig. 2B).
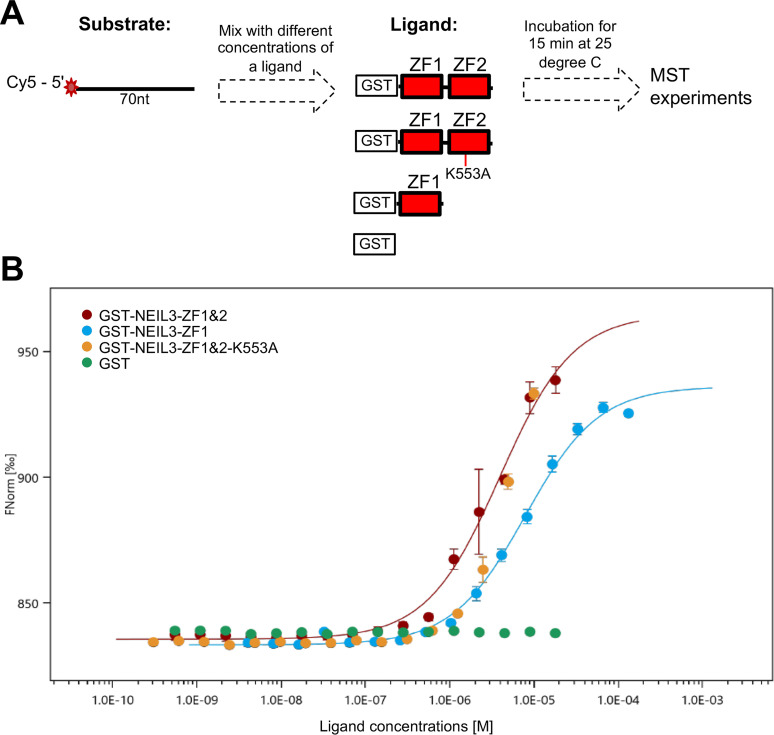
Furthermore, we quantified the binding affinity of NEIL3-ZF1&2, NEIL3-ZF1, and NEIL3-ZF1&2-K553A to ssDNA using microscale thermophoresis (MST) assays with Cy5-labeled 70-nt ssDNA as substrate (i.e. Cy5-ssDNA70, Fig. 4A). The substrate was mixed with different concentrations of four ligands for 15 min at 25 °C, respectively (Fig. 4A). Notably, we found that GST-NEIL3-ZF1&2 and GST-NEIL3-ZF1, but not GST, associated with Cy5-ssDNA70 (Fig. 4B). The Kd value for GST-NEIL3-ZF1&2 and GST-NEIL3-ZF1 was 3.9 ± 0.55 and 7.9 ± 0.68 μm, respectively. The MST observations suggest higher affinity of two Zf-GRF motifs than one individual Zf-GRF to 70-nt ssDNA. Consistent with our EMSA result (Fig. 3I), there were not enough binding points accumulated to generate binding curves for NEIL3-ZF1&2-K553A, suggesting compromised binding of NEIL3-ZF1&2-K553A to 70-nt ssDNA (Fig. 4B).
Figure 4.
NEIL3 Zf-GRF repeat has higher binding affinity to Cy5-labeled 70-nt ssDNA than NEIL3-ZF1 using MST assays. A, a diagram showed the overall steps and conditions used for MST experiments including the structure of substrate (target) and structure of each of four ligands. B, binding curves constructed for NEIL3-ZF1&2 (n = 4), NEIL3-ZF1 (n = 3), and NEIL3-ZF1&2 K553A (n = 3) with GST (n = 3) as control using 50 nm Cy5-labeled ssDNA in Buffer A. Kd value for GST-NEIL3-ZF1&2 and GST-NEIL3-ZF1 was 3.9 ± 0.55 and 7.9 ± 0.68 μm, respectively. Fnorm is normalized fluorescence measured by F1 as fluorescence after thermodiffusion (at 4-5 s range) divided by F0 as initial fluorescence.
APE1 endonuclease activity on ssDNA is compromised by NEIL3 Zf-GRF repeat
Our data have revealed that NEIL3-ZF1&2 associate with APE1 and ssDNA (Figs. 1–4). To determine the biological significance of the interaction between NEIL3 Zf-GRF motifs and APE1, we aimed to test whether APE1 endonuclease activity is affected by NEIL3 Zf-GRF motifs as NEIL3 Zf-GRF motifs are dispensable for NEIL3 DNA glycosylase activity.
First, we tested the potential effects of NEIL3 Zf-GRF motifs on APE1 endonuclease activity targeting the AP site on dsDNA. We found that increasing doses of NEIL3-ZF1&2 had almost no effect on the cleavage of 5′-FAM–labeled dsDNA39-AP by GST-APE1 (Fig. 5A). Furthermore, as APE1 displays very robust AP endonuclease activity targeting the AP site on dsDNA, we sought to determine whether APE1 endonuclease activity on dsDNA at low doses is affected by NEIL3 Zf-GRF motifs. We tested APE1 endonuclease assays on dsDNA with a serial dilution of recombinant GST-APE1 and found that the addition of NEIL3-ZF1&2 had almost no effect on APE1 endonuclease activity on dsDNA (Fig. 5B). Although NEIL3 Zf-GRF motifs interact with APE1, we confirmed that APE1 endonuclease activity on dsDNA is not affected by NEIL3 Zf-GRF motifs at least under the conditions we tested.
Figure 5.
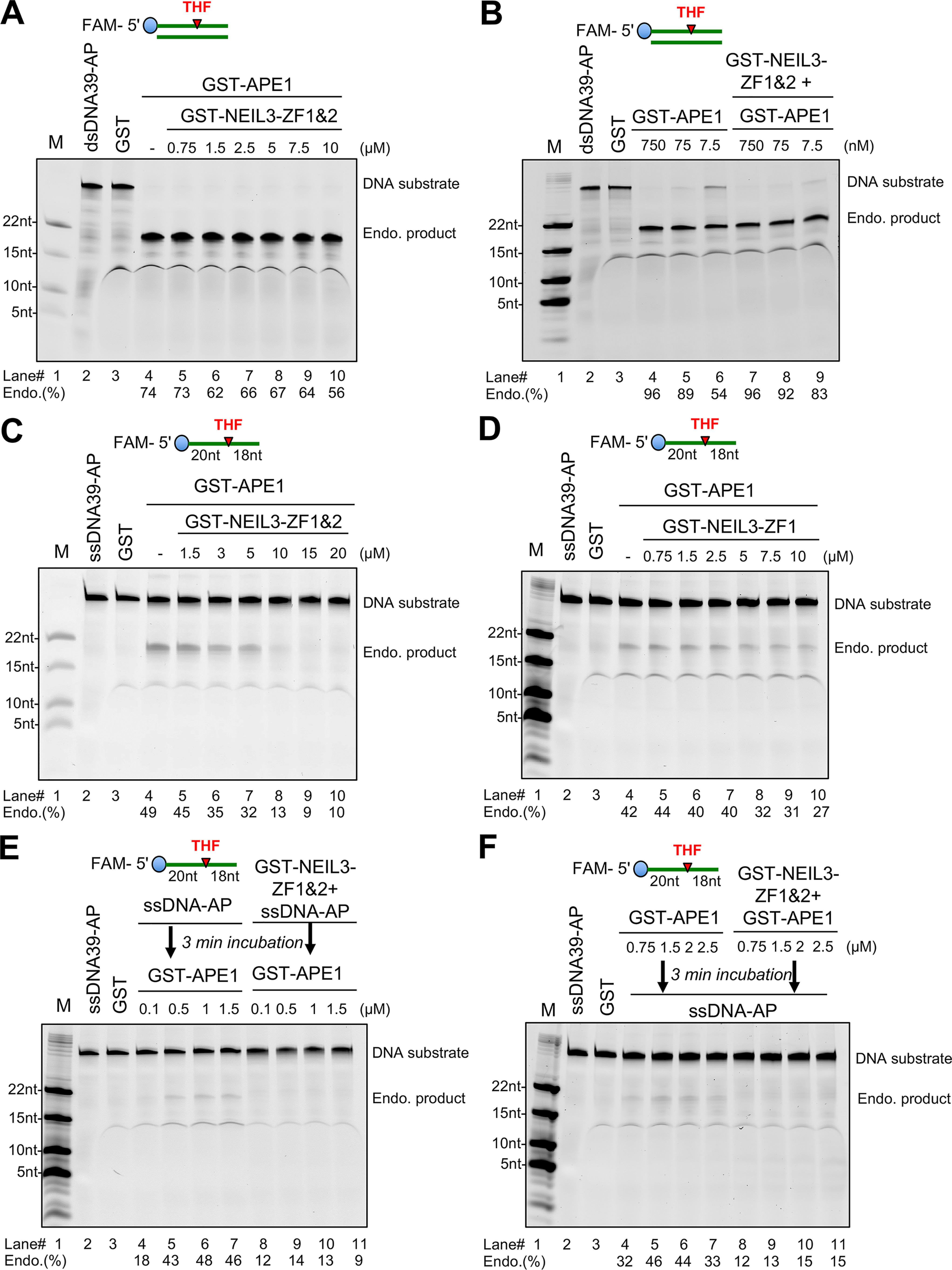
NEIL3 Zf-GRF repeat compromises APE1 endonuclease activity on ssDNA but not dsDNA. A, GST-APE1 endonuclease activity detected with or without the presence of different doses of GST-NEIL3-ZF1&2 using FAM-dsDNA39-AP as substrate. B, effect of GST-NEIL3-ZF1&2 on GST-APE1 endo activity targeting FAM-dsDNA39-AP using different concentrations of GST-APE1. C, endonuclease assay using 5′-FAM–ssDNA39-AP in the presence of GST-APE1 with or without GST-NEIL3-ZF1&2 at different doses. D, a similar assay repeated using different doses of GST-NEIL3-ZF1 instead for detecting any effects on APE1 endonuclease activity. E, 5′-FAM–ssDNA39-AP with or without GST-NEIL3-ZF1&2 incubated at 37 °C for 3 min before the addition of GST-APE1 at various doses as indicated, then incubated at 37 °C for 60 min for the shielding effect of NEIL3-ZF1&2 on ssDNA-AP. F, different doses of GST-APE1 incubated with or without GST-NEIL3-ZF1&2 at 37 °C for 3 min before the addition of FAM-ssDNA39-AP for another 60-min incubation for the inhibition effect through direct interaction between two proteins. Endo.(%) was calculated by the percentage of intensity of catalyzed products divided by the intensity of catalyzed products and uncatalyzed substrates. Intensity of bands was measured with ImageJ.
Second, we sought to determine the role of the NEIL3 Zf-GRF repeat on APE1 endonuclease activity on ssDNA. Notably, the endonuclease activity of GST-APE1 targeting 5′-FAM–labeled ssDNA with an AP mimicking site was compromised by the addition of GST-NEIL3-ZF1&2 in a dose-dependent manner (Fig. 5C). Interestingly, the addition of GST-NEIL3-ZF1 had almost no noticeable effect on the APE1 endonuclease activity targeting the AP site on ssDNA (Fig. 5D). The deficiency of NEIL3-ZF1 in APE1 endonuclease activity on ssDNA may be due to its failed interaction with APE1 and/or reduced interaction with ssDNA (Figs. 1, A and D, 2B, and 3, B and E).
To further explore how APE1 endonuclease activity targeting the AP site on ssDNA is compromised by NEIL3-ZF1&2, we tested two different experimental scenarios: a shielding effect and an inhibition effect. First, for the shielding effect experiment, we hypothesize that the negative effect of NEIL3-ZF1&2 is due to its direct interaction to ssDNA-AP to prevent APE1 from recognizing and binding to the AP site. We incubated NEIL3-ZF1&2 with ssDNA-AP before the addition of GST-APE1 for endonuclease assays and found that once bound with GST-NEIL3-ZF1&2, the 39-nt ssDNA with an AP site was not cleaved by the GST-APE1 (Fig. 5E). This observation supports the shielding effect of NEIL3 Zf-GRF motifs for APE1 endonuclease activity on ssDNA. Second, for the inhibition effect experiment, we hypothesize that NEIL3 Zf-GRF motifs may inhibit APE1 endonuclease activity directly because NEIL3-ZF1&2 interacts with APE1 in vitro. Thus, we incubated GST-APE1 with GST-NEIL3-ZF1&2 before the addition of ssDNA-AP substrate, and found that APE1 endonuclease activity targeting the AP site on ssDNA was compromised by the preincubation of NEIL3-ZF1&2 as well (Fig. 5F). This observation suggests that NEIL3 Zf-GRF motifs inhibit APE1 endonuclease activity targeting the AP site on ssDNA through direct interaction. This result is also supported by the observation that NEIL3 Zf-GRF motifs interact with APE1 via its active site including the Asp-209 residue shown in Fig. 1E. Overall, the shielding effect and inhibition effect are not mutually exclusive to each other for the regulation of APE1 endonuclease activity on ssDNA by NEIL3 Zf-GRF motifs.
Overexpression of NEIL3 Zf-GRF repeat reduces the generation of DNA damage in oxidative stress in Xenopus egg extracts
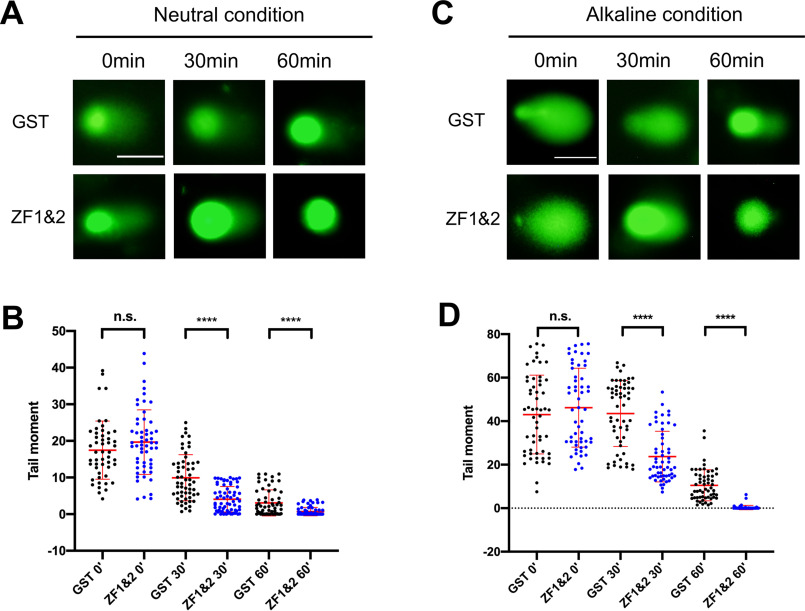
Based on the above observations, we hypothesize that the NEIL3 Zf-GRF repeat may help to prevent the generation of DNA damage such as replication-derived DSBs in oxidative stress. To further support the biological significance of our biochemical characterization, we performed COMET assays to test whether the excess addition of GST-NEIL3-ZF1&2 has any effect on oxidative stress-derived DNA damage in Xenopus egg extracts. As shown in Fig. 6, A and B, the Tail moment in COMET assays under neutral conditions after the excess addition of GST-NEIL3-ZF1&2 was significantly reduced compared with the addition of GST after 30 and 60 min of hydrogen peroxide treatment. This observation suggests that the impaired APE1 endonuclease activity on ssDNA by the excess NEIL3 Zf-GRF repeat may lead to reduction of the generation of replication-derived DSBs. Similarly, we observed the Tail moment in COMET assays under alkaline conditions after the excess addition of GST-NEIL3-ZF1&2 was also significantly reduced compared with the addition of GST after 30 and 60 min of hydrogen peroxide treatment (Fig. 6, C and D). This observation suggests that the generation of AP sites, SSBs, and DSBs in oxidative stress is reduced with overexpression of the NEIL3 Zf-GRF repeat. Taken together, this line of evidence from COMET assays under neutral and alkaline conditions supports the biological significance of the NEIL3 Zf-GRF repeat in the maintenance of ssDNA stability via distinct regulation of APE1. Future studies are needed to further delineate more in vivo function of the NEIL3 Zf-GRF repeat in genome integrity using a mammalian cell line system.
Figure 6.
Overexpression of the NEIL3 Zf-GRF repeat reduces the generation of DNA damage in oxidative stress in Xenopus egg extracts. A and C, GST-NEIL3-ZF1&2 (designated as ZF1&2) or GST (100 ng/µg) were added to Xenopus egg extracts supplemented with sperm chromatin DNA and hydrogen peroxide. After different incubation times (i.e. 0, 30, or 60 min), COMET assays under neutral condition (A) or alkaline condition (C) were performed. Representative images were shown. Scale bar in A and C represents 1 μm. B and D, Tail moment of images from COMET assays was quantified with the COMET Assay IV software (B for A and D for C, respectively) and statistical analysis was performed with GraphPad Prism8 (unpaired t test). n.s. indicates no significance (p > 0.05). ****, indicates p < 0.0001. n = 50, 56, 57, 67, 60, and 71, respectively, in B. n = 56, 55, 56, 57, 56, and 61, respectively, in D.
Discussion
Regulation of APE1 endonuclease activity on ssDNA by NEIL3 Zf-GRF repeat
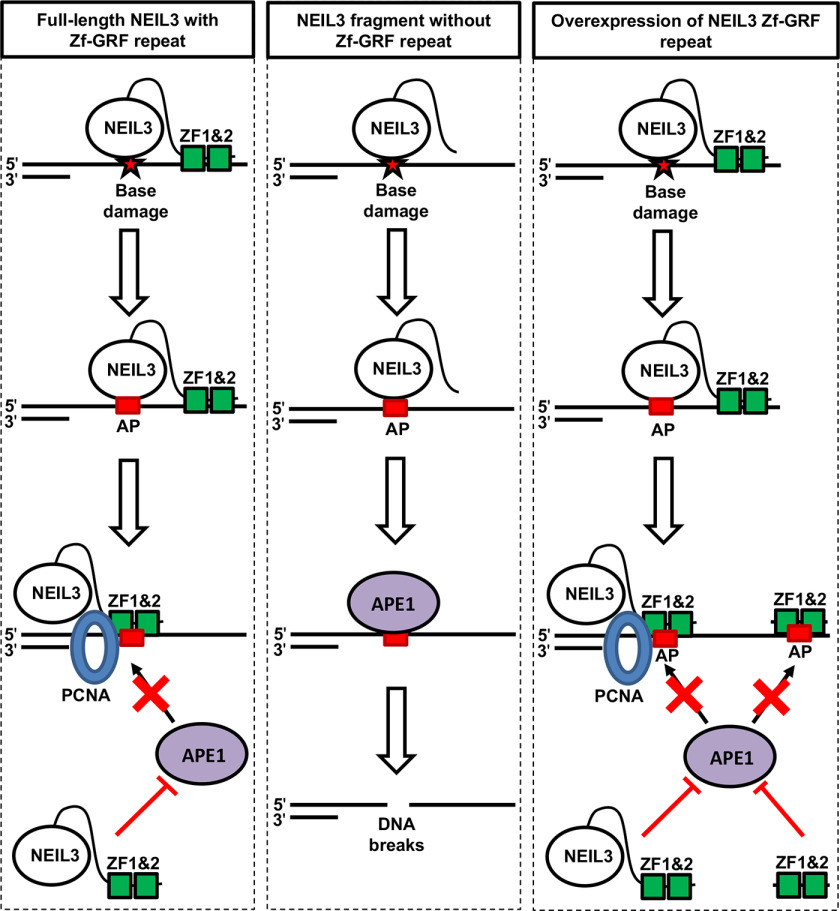
In this study, we have provided evidence showing (i) that NEIL3 Zf-GRF motifs interact with APE1 and PCNA but not APE2 (Fig. 1); (ii) that NEIL3 Zf-GRF repeat associates with shorter ssDNA and has higher affinity to ssDNA compared with one single Zf-GRF motif (Figs. 2–4); and (iii) that APE1 endonuclease activity on ssDNA but not dsDNA is compromised by NEIL3 Zf-GRF repeat (Fig. 5). These findings suggest a previously uncharacterized negative regulation of APE1 endonuclease on ssDNA by NEIL3 Zf-GRF repeat. We propose a working model of how the NEIL3 Zf-GRF repeat regulates APE1 endonuclease activity on ssDNA to maintain ssDNA stability (Fig. 7). Base damage is recognized and removed by NEIL3 to generate an AP site on the ssDNA region at the replication fork or telomeres. On one hand, with the presence of the NEIL3 Zf-GRF repeat, the AP site on ssDNA may be shielded from APE1 endonuclease activity by two Zf-GRF motifs, or the NEIL3 Zf-GRF repeat may interact with APE1 directly and compromise its endonuclease activity. On the other hand, without NEIL3 Zf-GRF repeat, the AP site on ssDNA is recognized and cleaved by APE1 to generate SSBs, leading to genome instability. In addition, excess addition of the GST-NEIL3 Zf-GRF repeat but not GST may prevent the APE1-mediated cleavage of AP site in ssDNA, which are generated by NEIL3 and/or other DNA glycosylases, via the two possible mechanisms (i.e. shielding effect and inhibition effect).
Figure 7.
A working model for the regulation of APE1 endonuclease activity on ssDNA by NEIL3-ZF1&2. Base damage is recognized and removed by NEIL3. Left panel (full-length NEIL3 with Zf-GRF repeat): base damage in ssDNA is converted into the AP site via NEIL3's DNA glycosylase activity. AP site may be shielded from APE1 endonuclease activity (shielding effect). Alternatively, NEIL3 Zf-GRF motifs may interact with APE1 and inhibit its AP endonuclease activity (inhibition effect). Middle panel (NEIL3 fragment without Zf-GRF repeat): base damage in ssDNA is converted into the AP site via DNA glycosylase activity of a fragment of NEIL3 that lacks the Zf-GRF repeat. APE1 recognizes the AP site and cleaves it into SSB, leading to genome instability. Right panel (overexpression of NEIL3 Zf-GRF repeat): AP site is generated by endogenous NEIL3 or other DNA glycosylases. APE1-mediated cleavage of the AP site is impaired by excess addition of the NEIL3 Zf-GRF repeat. These different possible mechanisms of action may explain the significance of the NEIL3 Zf-GRF repeat in the maintenance of ssDNA stability from APE1-mediated ssDNA breakage and DNA shortening. See text for details.
Our findings suggest a distinct mechanism of the regulation of APE1's endonuclease activity on ssDNA by NEIL3 Zf-GRF repeats. APE1 cleaves the AP site on both ssDNA and dsDNA, although its AP endonuclease was rather weak on ssDNA. Nevertheless, APE1 definitely could generate SSBs from ssDNA. Thus, it is extremely significant for cells to prevent SSB formation in ssDNA regions of the genome. Furthermore, APE1 and NEIL3 interact with each other and are functional at telomeres and at replication forks during S phase when long ssDNA is generated (30). The novel negative regulation of APE1 function on ssDNA by NEIL3 Zf-GRF motifs is not dependent on NEIL3's catalytic function as a glycosylase or AP lyase. Our findings are of significance because more breakages would be generated on ssDNA by APE1 with the absence of NEIL3 Zf-GRF motifs, leading to ssDNA instability.
Distinct features of Zf-GRF motifs
Zf-GRF (EMBL-EBI Family PF06839) is a less characterized zinc finger motif that contains conserved GRXF (X represents any amino acid) and has been implicated in ∼100 DNA/RNA metabolism (19). Accumulating evidence shows distinctive features of Zf-GRF motifs in genome integrity.
First, Zf-GRF motif preferentially interacts with ssDNA. APE2 Zf-GRF motif has been found in association with ssDNA but not dsDNA, and this interaction is essential for its exonuclease activity in genome integrity (19). Consistent with APE2 Zf-GRF binding with ssDNA, each of the two NEIL3 Zf-GRF motifs was also reported to associate with ssDNA (15). Our observations suggest that the NEIL3 Zf-GRF repeat can associate with shorter ssDNA, whereas individual NEIL3 ZF1 requires at least 60-nt ssDNA for binding (Figs. 2 and 3). Because the sequence of the 39-nt ssDNA and 70-nt ssDNA is different, it seems that the ssDNA interaction by Zf-GRF motif is sequence-independent. Notably, the presence of an AP-mimicking THF site did not increase binding of the NEIL3 Zf-GRF motif to ssDNA (Fig. 3), suggesting that Zf-GRF motifs do not recognize and bind with AP site specifically. Taken together, the NEIL3 Zf-GRF motif interaction with ssDNA is length-dependent but sequence/damage-independent.
Second, Zf-GRF motifs associate with PCNA. Prior studies have shown that APE2 interacts with PCNA via APE2's PIP box (24, 34). It has been revealed recently that APE2 Zf-GRF also interacts with PCNA in vitro and such interaction is important for APE2's role in SSB repair and signaling (29). Therefore, two modes of APE2–PCNA interaction have been proposed through APE2's PIP box and Zf-GRF motif, despite the exact mechanisms of the transition and dynamics between the two modes remain unknown (29, 35). We found in this study that both Zf-GRF repeat and ZF1 in NEIL3 associated with PCNA, although NEIL3–ZF1 interaction with PCNA was mildly reduced (Fig. 1C). Therefore, PCNA interaction is the second feature of Zf-GRF motifs at least in APE2 and NEIL3 so far.
Third, Zf-GRF repeat but not individual Zf-GRF motif interacts with APE1. Our evidence from this study shows that the Zf-GRF repeat within NEIL3 associates with APE1 but not APE2 (Fig. 1). Intriguingly, NEIL3-ZF1 itself is not sufficient for APE1 interaction. Although it remains unknown what residues within the NEIL3 Zf-GRF repeat are essential for the APE1 interaction, the Asp-209 residue within APE1 is important for interaction with the NEIL3 Zf-GRF repeat, suggesting a potential effect of NEIL Zf-GRF repeat on APE1 catalytic function. Consistent with this prediction, APE1 endonuclease activity on ssDNA was compromised by the NEIL3 Zf-GRF repeat but not NEIL3-ZF1 (Fig. 5). In addition, Top3A also contains two Zf-GRF motifs in its C terminus (19). However, it seems that the two Zf-GRF motifs within Top3A are about 30-40 amino acids apart from each other. Future experiments are needed to test whether the two Zf-GRF motifs within Top3A interact with APE1, and if so, whether Top3A regulates APE1 endonuclease activity.
What is the potential role of the NEIL3 Zf-GRF repeat in NEIL3's glycosylase and AP lyase activity? It has been shown that mouse NEIL3 fragment that lacks the Zf-GRF repeat is sufficient to remove damaged or oxidized bases in in vitro biochemical assays, suggesting that the Zf-GRF repeat is dispensable for NEIL3's DNA glycosylate activity (6). Although defined oligos containing AP-ICL can be processed sequentially by NEIL3's DNA glycosylate and AP lyase activity in vitro biochemical reconstitution systems, the AP site after NEIL3 unhooking on defined plasmids containing psoralen- or AP-ICL were not cleaved by its AP lyase activity in Xenopus egg extracts (13). The catalytic fragment of human NEIL3 was shown to incise damaged DNA via its lyase activity (7). Recent studies reveal that the dA-AP ICL in slayed DNA structures can be processed by the catalytic fragment of mouse NEIL3 through a two-step mechanism: first is unhooking by its DNA glycosylase activity to generate AP site, and second is cleavage of the 3′-side of the resulting AP site via AP lyase activity (16, 17). These observations suggest that the NEIL3 Zf-GRF repeat may be dispensable for NEIL3 AP lyase activity. Overall, prior studies suggest that the NEIL3 Zf-GRF repeat is not required for its DNA glycosylase and AP lyase activities. However, we cannot exclude the possibility that the NEIL3 Zf-ZGF repeat may interact with its catalytic domain in an intra- and/or inter-molecular fashion to regulate its glycosylase/AP lyase activity. Alternatively, NEIL3 Zf-GRF repeat may be important to recruit NEIL3 to different DNA damage sites for its glycosylase/AP lyase activity. Future studies are needed to test these different scenarios directly.
ssDNA stability in DNA repair and DNA damage response pathways
There are several scenarios to generate ssDNA in genome. Uncoupling of helicase and DNA polymerase at stalled DNA replication forks can generate ssDNA during replication stress (36). Transcription and R-loops also generate ssDNA, which is the complementary strand of RNA-DNA hybrids (37). Furthermore, HR-mediated DSB end resection will generate 3′-ssDNA overhangs (38). After generation, RPA is recruited to ssDNA to protect from nuclease degradation and to coordinate DNA damage response and DNA repair pathways (39, 40). Despite the RPA protection, ssDNAs are still vulnerable to generate chemical modifications on their bases such as oxidized bases from oxidative stress (41, 42). DNA glycosylases such as NEIL3 and UNG2 are able to recognize and remove damaged bases in ssDNA to generate AP sites (39).
Although the AP site on dsDNA can be cleaved by APE1 into a SSB structure and repaired by APE1- and APE2-dependent SSB repair pathways (26, 29, 43), the AP site on ssDNA is cleaved by APE1, leading to ssDNA instability and chromosome breakages. Therefore, it is significant to determine how APE1 endonuclease activity is regulated on ssDNA. Our findings from this study suggest a negative regulation of APE1 endonuclease activity on ssDNA by a NEIL3 Zf-GRF repeat to maintain ssDNA stability. Our observations provide two possible mechanisms: shield effect and inhibition effect (Figs. 5 and 7). Shield effect requires NEIL3 Zf-GRF repeat binding to ssDNA to prevent APE1 from accessing to AP site on ssDNA, whereas inhibition effect requires NEIL3 Zf-GRF repeat to directly interact with APE1 first to compromise its endonuclease activity. Despite these two mechanisms are not mutually exclusive to each other, future structural analysis is needed to test these different scenarios.
Physiological relevance of the regulation of APE1 endonuclease on ssDNA by NEIL3
NEIL3 has been implicated in BER and inter-strand cross-link repair (13, 30), whereas APE1 has been shown to be critical for DNA repair, transcriptional regulation, and DDR pathways (26, 44). Abnormal expression of APE1 and NEIL3 has been implicated in cancer development (11, 21, 45, 46). In a query of 83,622 samples from 80,293 cancer patients in 269 studies using Cbioportal analysis on April 16, 2020, a total of 336 mutation events (265 missense, 67 truncating, 3 nonstart, and 1 fusion) were found in NEIL3 (Fig. S4). In particular, missense mutations or truncations were revealed on 23 amino acids within the 92-amino acid region of NEIL3-ZF1&2 (i.e. amino acids 505-596), which accounts for ∼25% of amino acids in the NEIL3 Zf-GRF repeat. We speculate these mutation events may impair NEIL3 binding to ssDNA and/or its regulation on APE1 endonuclease, leading to DNA ssDNA breakages or mutagenesis. Future studies are needed to test this hypothesis directly. Thus, our research may provide a novel avenue to therapeutic treatment for cancer patients with abnormal expression or mutants of APE1 and NEIL3.
Together, we elucidate the distinct features of NEIL3 Zf-GRF repeat and reveal the previously uncharacterized regulation of APE1 endonuclease activity on ssDNA by NEIL3. Our findings may shed new light on the novel regulatory mechanism by which NEIL3 interacts and regulates APE1 through its noncatalytic function in genome integrity.
Experimental procedures
Preparation of recombinant plasmids and recombinant proteins
Recombinant plasmid pGEX4T1-NEIL3-ZF1&2 was prepared by PCR coding sequences for two NEIL3 ZF-GRF from X. laevis NEIL3 cDNA (GenBank BC072255.1) with a pair of primers (FP#1 and RP#1) (Table S1) and inserting into pGEX4T1 vector at BamHI and XhoI sites. A similar procedure was used to prepare pGEX4T1-NEIL3-ZF1 with a different pair of primers (FP#1 and RP #2) (Table S1). To generate pGEX4T1-NEIL3-ZF1&2-K553A, two single-primer PCR were set up with methylated parental plasmid pGEX4T1-NEIL3-ZF1&2 as template and either FP#2 or RP#3 (Table S1). The two PCR were combined and denatured at 95 °C and cooled for annealing, followed by DpnI treatment to digest methylated, nonmutated parental DNA strands at 37 °C overnight. Then DH5 α cells were used to amplify the recombinant plasmid. Sequencing was used to validate the right mutation within the plasmid as designed. To generate recombinant plasmids pCS2+MT-NEIL3-ZF1&2, pCS2+MT-NEIL3-ZF1, and pCS2+MT-NEIL3-ZF2, subcloning PCR product with pGEX4T1-NEIL3-ZF1&2 as template and with different pairs of primers (FP#3 and RP#1, FP#3 and RP #2, and FP#4 and RP#1, respectively) (Table S1) into the pCS2+MT vector at EcoRI and XhoI sites were used.
Recombinant plasmids pGEX4T1-APE1, pCS2+MT-APE1, pCS2+MT-APE1-D306A, and pCS2+MT-APE2 were described previously (19, 26, 47). Recombinant plasmid pCS2+MT-APE1-E95Q was generated on pCS2+MT-APE1 template using designed primers (FP#5 and RP#5) with the site-directed mutagenesis kit (Table S1). Recombinant plasmid pCS2+MT-APE1-E95Q-D209N was prepared on pCS2+MT-APE1-E95Q template with another pair of primers (FP#6 and RP#6) (Table S1). The mutant plasmids were validated by DNA sequencing.
Recombinant Myc-tagged proteins were expressed in the TnT SP6 Quick Coupled Transcription/Translation system at 30 °C for 90 min using the respective recombinant plasmid subcloned in pCS2+MT vector. Recombinant GST or GST-tagged proteins were expressed in DE3/BL21 Escherichia coli cells after isopropyl 1-thio-β-d-galactopyranoside induction with the respective recombinant plasmid subcloned in pGEX4T1 vector and purified using the vendor's procedures. Furthermore, His-PCNA has been described previously (26).
Preparation of 5′-labeled ssDNA/dsDNA structures
Different sizes of Biotin-ssDNA: 10, 20, 40, and 60 nt were generated with biotin labeled to the 5'-side as described by the vendor with sequences indicated below (Table S2).
The 5′-FAM–labeled 39-nt ssDNA with the AP site-mimicking THF (designed as ssDNA39-AP), the 5′-FAM–labeled 39-nt ssDNA without THF site (designed as ssDNA39), the 5′-FAM–labeled 70-nt sDNA (designed as ssDNA70), and the 5′-Cy5–labeled 70-nt sDNA (designed as Cy5-ssDNA70) was synthesized by vendor (FP#7, FP#8, FP#9, and FP#10), respectively (Table S1). In addition, the 5′-FAM–labeled 39-bp dsDNA containing the AP site mimicking THF in the middle of the one strand (designed as dsDNA39-AP) was generated by combining forward and reverse primers (FP#7 and RP#7) (Table S1). The mixture was incubated at 95-100 °C for 5 min with mixing every minute during incubation. The mixture was slowly cooled down naturally at room temperature for 30-45 min.
Biotin-DNA binding assays
Different biotin-labeled DNA structures (5 nm) were coupled to streptavidin Dynabeads using the approach previously described (29). The beads were mixed with 4 μm GST or GST-tagged recombinant proteins including GST-NEIL3-ZF1&2 and GST-NEIL3-ZF1 at room temperature for 60 min. The beads were washed by Buffer A (80 mm NaCl, 20 mm β-glycerophosphate, 2.5 mm EGTA, 0.01% Nonidet P-40, and 10 mm HEPES-KOH, pH 7.5). The input and bead-bound fractions were examined via immunoblotting analysis using anti-GST antibodies.
GST pulldown assays
GSH beads was incubated with 4 μm GST or GST-tagged recombinant proteins in Buffer B (100 mm NaCl, 5 mm MgCl2, 10% glycerol (v/v), 0.01% Nonidet P-40, 20 mm Tris-HCl, pH 8.0) for 1 h binding at room temperature. Then, WT His-PCNA (4 μm) or 10 μl of TnT SP6 reactions containing various Myc-tagged recombinant proteins were added and the mixtures were incubated overnight at 4 °C with rotation. After incubation, the samples were washed by Buffer C (100 mm NaCl, 5 mm MgCl2, 10% glycerol (v/v), 0.1% Nonidet P-40, 20 mm Tris-HCl, pH 8.0) twice to remove nonspecific binding proteins. The input and bead-bound fractions were examined via immunoblotting analysis.
Electrophoretic mobility shift assays
Different doses of GST-NEIL3 ZF1&2/GST-NEIL3-ZF1/GST-NEIL3-ZF1&2 K553A were incubated with 500 nm of different 5′-FAM–labeled ssDNA structures in Buffer D (10 mm Tris, pH 8.0, 50 mm NaCl, 0.2 mm tris(2-carboxyethyl)phosphine, and 5% glycerol) at room temperature for 60 min with mixing every 20 min. Then, 5× Hi-Density TBE Sample Buffer was added to each sample before running. Samples were resolved on 5% TBE gel at 150 V for 2-3 h in cold 0.5× TBE running buffer. Then, the gel was visualized using BIO-RAD ChemiDoc MP Imaging System.
MST assays
The Cy5-ssDNA70 (50 nm) was used as substrate and was checked via Pretest function in MST Monolith NT.115 to determine fluorescence intensity, adsorption on capillaries, variations, and sample aggregation at 25 °C. Once verified, the Binding Affinity option in the MST instrument was selected with a maximum dose of GST-NEIL3-ZF1&2 used as base line for other proteins: GST, GST-NEIL3-ZF1, and GST-NEIL3-ZF1&2 K553A. 36 μm NEIL3-ZF1&2 was used for each trial, for a total of four trials. Series dilutions were generated following the direction indicated in the MST instrument. Buffer A was used for both substrate and ligand dilutions. The results obtained were analyzed via MO Affinity Analysis version 2.3 software as a combination of trials for all ligands tested.
In vitro endonuclease assays
The 5′-FAM dsDNA39-AP or ssDNA39-AP structures (500 nm) were treated with different concentrations of purified GST-NEIL3-ZF1&2 or GST-NEIL3-ZF1 and 0.75 μm GST-APE1 in Buffer E (20 mm KCl, 10 mm MgCl2, 2 mm DTT, 50 mm HEPES, pH 7.5) with GST only as control at 37 °C for 60 min. Then, the reactions were quenched with equal volumes of 2× TBE-urea Sample Buffer, denatured at 95 °C for 5 min. After a quick 10-s spin, samples were resolved on a 18% TBE-urea gel in 1× TBE running buffer at 20,000 watts for 2 h. Gels were viewed using the BIO-RAD ChemiDoc MP Imaging System. ImageJ was used to measure intensity of uncleaved substrates and cleaved products. Different dilutions of GST-APE1, as indicated, were incubated with or without 10 μm GST-NEIL3-ZF1&2 together with the 5′-FAM–labeled dsDNA39-AP structure in Buffer E (Fig. 5, A and B). All samples were incubated at 37 °C for 60 min and resolved on TBE-urea gels.
For the shielding effect experiment (Fig. 5E), the 5′-FAM–labeled ssDNA39-AP (500 nm) was incubated with or without 10 μm GST-NEIL3-ZF1&2 for 3 min at 37 °C. Different concentrations of purified GST-APE1 were added together with Buffer E into each sample. For the inhibition effect experiment (Fig. 5F), different concentrations of GST-APE1 were incubated with or without 10 μm GST-NEIL3-ZF1&2 for 3 min at 37 °C. Then, the 5′-FAM–labeled ssDNA39-AP (500 nm) was added into each sample together with Buffer E. All samples were incubated at 37 °C for 60 min prior to the addition of 2× TBE-urea sample buffer. Later steps were repeated similarly as indicated above.
Experimental procedure for COMET assays with Xenopus egg extracts and sperm chromatin
The care and use of X. laevis were approved by University of North Carolina Charlotte's Institutional Animal Care and Use Committee (IACUC). The preparation of Xenopus low-speed supernatant egg extracts and sperm chromatin was described previously (48–50). Recombinant GST or GST-NEIL3-ZF1&2 (100 ng/µg) were incubated in Xenopus egg extracts for 10 min before the addition of sperm chromatin and hydrogen peroxide (100 mm), followed by different incubation times (i.e. 0, 30, and 60 min) at room temperature. OxiSelect COMET Assay Kit (Cell BioLabs Inc.) was utilized to perform COMET assays in neutral conditions (pH ∼ 7.0) and alkaline condition (pH > 13) using a similar procedure as described recently (43). COMET Images were collected using DP Controller software (Olympus Corporation, Japan) and examined with the COMET Assay IV Lite software (Instem, United Kingdom). GraphPad Prism8 was used for statistical analysis.
Data availability
All data described in the manuscript are present in the main text, figures, and the supporting figures and tables.
Supplementary Material
Acknowledgments
We thank Dr. Irina Nesmelova for the assistance in MST analysis and Dr. Didier Dréau for the assistance in COMET imaging experiments.
This article contains supporting information.
Author contributions—A. H. and S. Y. conceptualization; A. H. formal analysis; A. H. and Y. L. investigation; A. H. and Y. L. methodology; A. H. writing-original draft; Y. L. and S. Y. writing-review and editing; S. Y. resources; S. Y. supervision; S. Y. funding acquisition; S. Y. project administration.
Funding and additional information—The Yan lab was supported, in part, by National Institutes of Health NCI Grant R01CA225637, NIGMS Grant R15GM114713, and National Institutes of Health Grant S10OD026970 (to S. Y.). A. H. was supported by a National Science Foundation REU Grant 1359271. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- BER
- base excision repair
- AP
- apurinic/apyrimidinic
- EEP
- endonuclease/exonuclease/phosphatase
- EMSA
- electrophoretic mobility shift assays
- ICL
- inter-strand cross-link
- MST
- microscale thermophoresis
- GST
- glutathione S-transferase
- RBP-Znf
- Ran-binding protein-type zinc finger
- ZF1
- Zf-GRF1;
- ZF2
- Zf-GRF2
- PCNA
- proliferating cell nuclear antigen
- PIP
- PCNA-interaction protein
- ssDNA
- single-strand DNA
- dsDNA
- double-strand DNA
- nt
- nucleotide(s)
- TBE
- Tris borate-EDTA
- DSB
- double-strand break
- SSB
- single-strand break.
References
- 1. Yan S., Sorrell M., and Berman Z. (2014) Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol. Life Sci. 71, 3951–3967 10.1007/s00018-014-1666-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hazra T. K., Izumi T., Boldogh I., Imhoff B., Kow Y. W., Jaruga P., Dizdaroglu M., and Mitra S. (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 3523–3528 10.1073/pnas.062053799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hazra T. K., Kow Y. W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S. K., Mitra S., and Izumi T. (2002) Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 277, 30417–30420 10.1074/jbc.C200355200 [DOI] [PubMed] [Google Scholar]
- 4. Hegde M. L., Hegde P. M., Bellot L. J., Mandal S. M., Hazra T. K., Li G. M., Boldogh I., Tomkinson A. E., and Mitra S. (2013) Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E3090–3099 10.1073/pnas.1304231110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banerjee D., Mandal S. M., Das A., Hegde M. L., Das S., Bhakat K. K., Boldogh I., Sarkar P. S., Mitra S., and Hazra T. K. (2011) Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 286, 6006–6016 10.1074/jbc.M110.198796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu M., Bandaru V., Bond J. P., Jaruga P., Zhao X., Christov P. P., Burrows C. J., Rizzo C. J., Dizdaroglu M., and Wallace S. S. (2010) The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 4925–4930 10.1073/pnas.0908307107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krokeide S. Z., Laerdahl J. K., Salah M., Luna L., Cederkvist F. H., Fleming A. M., Burrows C. J., Dalhus B., and Bjørås M. (2013) Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst.) 12, 1159–1164 10.1016/j.dnarep.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu M., Imamura K., Averill A. M., Wallace S. S., and Doublié S. (2013) Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure 21, 247–256 10.1016/j.str.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torisu K., Tsuchimoto D., Ohnishi Y., and Nakabeppu Y. (2005) Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J. Biochem. 138, 763–772 10.1093/jb/mvi168 [DOI] [PubMed] [Google Scholar]
- 10. Regnell C. E., Hildrestrand G. A., Sejersted Y., Medin T., Moldestad O., Rolseth V., Krokeide S. Z., Suganthan R., Luna L., Bjørås M., and Bergersen L. H. (2012) Hippocampal adult neurogenesis is maintained by Neil3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Rep. 2, 503–510 10.1016/j.celrep.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 11. Hildrestrand G. A., Neurauter C. G., Diep D. B., Castellanos C. G., Krauss S., Bjørås M., and Luna L. (2009) Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 10, 45 10.1186/1471-2202-10-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sejersted Y., Hildrestrand G. A., Kunke D., Rolseth V., Krokeide S. Z., Neurauter C. G., Suganthan R., Atneosen-Åsegg M., Fleming A. M., Saugstad O. D., Burrows C. J., Luna L., and Bjørås M. (2011) Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc. Natl. Acad. Sci. U.S.A. 108, 18802–18807 10.1073/pnas.1106880108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Semlow D. R., Zhang J., Budzowska M., Drohat A. C., and Walter J. C. (2016) Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167, 498–511e414 10.1016/j.cell.2016.09.008 10.1016/j.cell.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li N., Wang J., Wallace S. S., Chen J., Zhou J., and D'Andrea A. D. (2020) Cooperation of the NEIL3 and Fanconi anemia/BRCA pathways in interstrand crosslink repair. Nucleic Acids Res. 48, 3014–3028 10.1093/nar/gkaa038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu R. A., Semlow D. R., Kamimae-Lanning A. N., Kochenova O. V., Chistol G., Hodskinson M. R., Amunugama R., Sparks J. L., Wang M., Deng L., Mimoso C. A., Low E., Patel K. J., and Walter J. C. (2019) TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 567, 267–272 10.1038/s41586-019-1002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Z., Nejad M. I., Varela J. G., Price N. E., Wang Y., and Gates K. S. (2017) A role for the base excision repair enzyme NEIL3 in replication-dependent repair of interstrand DNA cross-links derived from psoralen and abasic sites. DNA Repair (Amst.) 52, 1–11 10.1016/j.dnarep.2017.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imani Nejad M., Housh K., Rodriguez A. A., Haldar T., Kathe S., Wallace S. S., Eichman B. F., and Gates K. S. (2020) Unhooking of an interstrand cross-link at DNA fork structures by the DNA glycosylase NEIL3. DNA Repair (Amst.) 86, 102752 10.1016/j.dnarep.2019.102752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takao M., Oohata Y., Kitadokoro K., Kobayashi K., Iwai S., Yasui A., Yonei S., and Zhang Q. M. (2009) Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells 14, 261–270 10.1111/j.1365-2443.2008.01271.x [DOI] [PubMed] [Google Scholar]
- 19. Wallace B. D., Berman Z., Mueller G. A., Lin Y., Chang T., Andres S. N., Wojtaszek J. L., DeRose E. F., Appel C. D., London R. E., Yan S., and Williams R. S. (2017) APE2 Zf-GRF facilitates 3'-5' resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 114, 304–309 10.1073/pnas.1610011114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whitaker A. M., and Freudenthal B. D. (2018) APE1: a skilled nucleic acid surgeon. DNA Repair (Amst.) 71, 93–100 10.1016/j.dnarep.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M., and Wilson D. M. 3rd (2014) Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal. 20, 678–707 10.1089/ars.2013.5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tell G., Quadrifoglio F., Tiribelli C., and Kelley M. R. (2009) The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid. Redox Signal. 11, 601–620 10.1089/ars.2008.2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hadi M. Z., Ginalski K., Nguyen L. H., and Wilson D. M. 3rd (2002) Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol. 316, 853–866 10.1006/jmbi.2001.5382 [DOI] [PubMed] [Google Scholar]
- 24. Burkovics P., Hajdú I., Szukacsov V., Unk I., and Haracska L. (2009) Role of PCNA-dependent stimulation of 3'-phosphodiesterase and 3'-5' exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res. 37, 4247–4255 10.1093/nar/gkp357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuznetsova A. A., Fedorova O. S., and Kuznetsov N. A. (2018) Kinetic features of 3'-5' exonuclease activity of human AP-endonuclease APE1. Molecules 23, 2101 10.3390/molecules23092101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin Y., Raj J., Li J., Ha A., Hossain M. A., Richardson C., Mukherjee P., and Yan S. (2020) APE1 senses DNA single-strand breaks for repair and signaling. Nucleic Acids Res. 48, 1925–1940 10.1093/nar/gkz1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Whitaker A. M., Flynn T. S., and Freudenthal B. D. (2018) Molecular snapshots of APE1 proofreading mismatches and removing DNA damage. Nat. Commun. 9, 399 10.1038/s41467-017-02175-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., and Nakabeppu Y. (2001) Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 29, 2349–2360 10.1093/nar/29.11.2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin Y., Bai L., Cupello S., Hossain M. A., Deem B., McLeod M., Raj J., and Yan S. (2018) APE2 promotes DNA damage response pathway from a single-strand break. Nucleic Acids Res. 46, 2479–2494 10.1093/nar/gky020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou J., Chan J., Lambele M., Yusufzai T., Stumpff J., Opresko P. L., Thali M., and Wallace S. S. (2017) NEIL3 repairs telomere damage during S phase to secure chromosome segregation at mitosis. Cell Rep. 20, 2044–2056 10.1016/j.celrep.2017.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sokalingam S., Raghunathan G., Soundrarajan N., and Lee S. G. (2012) A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PLoS ONE 7, e40410 10.1371/journal.pone.0040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gelin A., Redrejo-Rodriguez M., Laval J., Fedorova O. S., Saparbaev M., and Ishchenko A. A. (2010) Genetic and biochemical characterization of human AP endonuclease 1 mutants deficient in nucleotide incision repair activity. PLoS ONE 5, e12241 10.1371/journal.pone.0012241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McNeill D. R., and Wilson D. M. 3rd (2007) A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol. Cancer Res. 5, 61–70 10.1158/1541-7786.MCR-06-0329 [DOI] [PubMed] [Google Scholar]
- 34. Unk I., Haracska L., Gomes X. V., Burgers P. M., Prakash L., and Prakash S. (2002) Stimulation of 3' → 5' exonuclease and 3'-phosphodiesterase activities of yeast apn2 by proliferating cell nuclear antigen. Mol. Cell Biol. 22, 6480–6486 10.1128/mcb.22.18.6480-6486.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hossain M. A., Lin Y., and Yan S. (2018) Single-strand break end resection in genome integrity: mechanism and regulation by APE2. Int. J. Mol. Sci. 19, 2389 10.3390/ijms19082389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Byun T. S., Pacek M., Yee M. C., Walter J. C., and Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aguilera A., and García-Muse T. (2012) R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 38. Daley J. M., Niu H., Miller A. S., and Sung P. (2015) Biochemical mechanism of DSB end resection and its regulation. DNA Repair (Amst.) 32, 66–74 10.1016/j.dnarep.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anindya R. (2020) Single-stranded DNA damage: protecting the single-stranded DNA from chemical attack. DNA Repair (Amst.) 87, 102804 10.1016/j.dnarep.2020.102804 [DOI] [PubMed] [Google Scholar]
- 40. Maréchal A., and Zou L. (2015) RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 10.1038/cr.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Degtyareva N. P., Saini N., Sterling J. F., Placentra V. C., Klimczak L. J., Gordenin D. A., and Doetsch P. W. (2019) Mutational signatures of redox stress in yeast single-strand DNA and of aging in human mitochondrial DNA share a common feature. PLoS Biol. 17, e3000263 10.1371/journal.pbio.3000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roberts S. A., Sterling J., Thompson C., Harris S., Mav D., Shah R., Klimczak L. J., Kryukov G. V., Malc E., Mieczkowski P. A., Resnick M. A., and Gordenin D. A. (2012) Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46, 424–435 10.1016/j.molcel.2012.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cupello S., Lin Y., and Yan S. (2019) Distinct roles of XRCC1 in genome integrity in Xenopus egg extracts. Biochem. J. 476, 3791–3804 10.1042/BCJ20190798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dyrkheeva N. S., Lebedeva N. A., and Lavrik O. I. (2016) AP Endonuclease 1 as a key enzyme in repair of apurinic/apyrimidinic sites. Biochemistry 81, 951–967 10.1134/S0006297916090042 [DOI] [PubMed] [Google Scholar]
- 45. Shinmura K., Kato H., Kawanishi Y., Igarashi H., Goto M., Tao H., Inoue Y., Nakamura S., Misawa K., Mineta H., and Sugimura H. (2016) Abnormal expressions of DNA glycosylase genes NEIL1, NEIL2, and NEIL3 are associated with somatic mutation loads in human cancer. Oxid. Med. Cell Longev. 2016, 1546392 10.1155/2016/1546392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jensen K. A., Shi X., and Yan S. (2020) Genomic alterations and abnormal expression of APE2 in multiple cancers. Sci. Rep. 10, 3758 10.1038/s41598-020-60656-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Willis J., Patel Y., Lentz B. L., and Yan S. (2013) APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 110, 10592–10597 10.1073/pnas.1301445110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cupello S., Richardson C., and Yan S. (2016) Cell-free Xenopus egg extracts for studying DNA damage response pathways. Int. J. Dev. Biol. 60, 229–236 10.1387/ijdb.160113sy [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Willis J., DeStephanis D., Patel Y., Gowda V., and Yan S. (2012) Study of the DNA damage checkpoint using Xenopus egg extracts. J. Vis. Exp. e4449 10.3791/4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lebofsky R., Takahashi T., and Walter J. C. (2009) DNA replication in nucleus-free Xenopus egg extracts. Methods Mol. Biol 521, 229–252 10.1007/978-1-60327-815-7_13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are present in the main text, figures, and the supporting figures and tables.